Abstract
The spatial distribution of the target (t-)SNARE proteins (syntaxin and SNAP-25) on the plasma membrane has been extensively characterized. However, the protein conformations and interactions of the two t-SNAREs in situ remain poorly defined. By using super-resolution optical techniques and fluorescence lifetime imaging microscopy, we observed that within the t-SNARE clusters syntaxin and SNAP-25 molecules interact, forming two distinct conformations of the t-SNARE binary intermediate. These are spatially segregated on the plasma membrane with each cluster exhibiting predominantly one of the two conformations, representing the two- and three-helical forms previously observed in vitro. We sought to explain why these two t-SNARE intermediate conformations exist in spatially distinct clusters on the plasma membrane. By disrupting plasma membrane lipid order, we found that all of the t-SNARE clusters now adopted a single conformational state corresponding to the three helical t-SNARE intermediates. Together, our results define spatially distinct t-SNARE intermediate states on the plasma membrane and how the conformation adopted can be patterned by the underlying lipid environment.
Keywords: Exocytosis, Fluorescence Resonance Energy Transfer (FRET), Lipid Raft, Membrane Proteins, Microscopic Imaging
Introduction
The soluble NSF attachment protein receptor (SNARE)4 proteins form a highly conserved family with a central role in the fusion of lipid membranes in both intracellular trafficking and exocytosis. They are well characterized at the atomic structure, domain organization, interaction stoichiometry, and gross membrane localization levels (1–6). In exocytosis, their function relies on the pairing of t-SNAREs (syntaxin and SNAP-25) on the plasma membrane with a vesicle SNARE (synaptobrevin). The t-SNAREs have been observed to colocalize in clusters on the plasma membrane and have been proposed to define the sites of exocytosis (4, 7, 8). Morphological characterization of these t-SNARE clusters has mirrored the advancements in optical microscopy from the diffraction-limited identification of 200–400-nm t-SNARE clusters (7–9) to the observation of 50-nm assemblies using STED microscopy (10). In addition to observing t-SNARE membrane organization, several studies have sought to uncover the mechanisms driving t-SNARE cluster formation. Lipid order has been shown to play a role in t-SNARE cluster organization using sheets of plasma membrane (7). SNAP-25, which has a palmitoylated cysteine-rich linker region essential for membrane targeting (3), may be directed to ordered cholesterol-rich regions in the plasma membrane (11); however, in vitro, the individual t-SNAREs were shown to absolutely prefer disordered lipid domains (12). These findings led to the hypothesis that plasma membrane t-SNARE protein clusters may be maintained by membrane rafts (13) or other cholesterol-dependent domains (7) by either sequestration or exclusion. Recently, however, syntaxin self-association has been proposed as a major factor in membrane protein cluster organization (10, 14), implying that SNARE proteins can self-organize into clusters by virtue of their inherent properties. From these studies, the ability of the t-SNAREs to interact in vitro and to form clusters at the plasma membrane is not in doubt, but the protein-protein interactions within these clusters remains elusive.
EXPERIMENTAL PROCEDURES
Cell culture reagents were supplied by Invitrogen. Anti-SNAP-25 (SMI81) was obtained from Sternberger Monoclonals (Lutherville, MD). Anti-syntaxin1a (HPC-1) was supplied by Sigma. The rabbit anti-syntaxin1a was a generous gift from Dr. Bazbek Davletov (Laboratory of Molecular Biology, Cambridge, UK). Anti-mouse IgG and anti-rabbit IgG conjugated to Alexafluor-647 or Alexafluor-488, propidium iodide, and calcein AM were obtained from Invitrogen. Protease inhibitor tablets were from Roche Applied Science. All other reagents were obtained from Sigma.
Cell Culture
Neuroblastoma 2a cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 10 mm l-glutamine, 50 units of penicillin, 50 μg/ml streptomycin and maintained at 37 °C in 5% (v/v) CO2, 95% (v/v) air. Pheochromocytoma (PC-12) cells were grown in RPMI 1640 medium supplemented with 10% horse serum, 5% fetal bovine serum, 10 mm Glutamax (Invitrogen), 50 μg/ml gentamicin and maintained at 37 °C in 7.5% (v/v) CO2, 92.5% (v/v) air. Transfections were performed using ExGen 500 (Fermentas) or Lipofectamine 2000 (Invitrogen). To deplete cholesterol, cells were incubated with 10 mm methyl-β-cyclodextrin (MβCD) in serum-free medium for 15–30 min at 37 °C. To assess cell viability following cholesterol depletion, both propidium iodide and calcein AM (Invitrogen) were added to a final concentration of 3 μm, incubated for 5 min, and then imaged immediately. Live cells were maintained on a heated stage (37 °C) in a chamber containing 5% (v/v) CO2, 95% (v/v) air during imaging.
Vectors and Transfection
The vector pEGFP-C2 was obtained from Clontech. An EYFP-SNAP-25 fusion was generated by ligation of SNAP-25-(1–206) into BamHI/EcoRI sites of pEGFP-C, followed by the replacement of enhanced green fluorescent protein with EYFP. SNAP-25-EGFP in pEGFP-N1 was a gift from M. Linder. The plasmid pmCerulean-syntaxin1a-(1–288), was described previously (15). Plasmids encoding PACherry-SNAP-25-(1–206) and PACherry-syntaxin1a-(1–288) were generated from the plasmids above by replacement of DNA encoding EYFP or mCerulean with PACherry using AgeI/BsrGI. All cells were cultured on glass coverslips, and transfections were performed using ExGen500 (Fermentas).
Cellular Labeling Procedures
To label cholesterol, neuroblastoma 2a cells were rinsed in phosphate-buffered saline and fixed with 4% (w/v) paraformaldehyde on ice for 30 min. Following phosphate-buffered saline washes, cells were incubated with filipin (50 μg/ml) with 10% (v/v) fetal bovine serum for 2 h. Filipin fluorescence was excited using a titanium sapphire two-photon laser at 780 nm. The same settings were employed for all samples so that their relative staining intensities could be compared. To quantify lipid order, we used laurdan (6-dodecanoyl-2-dimethylaminonaphthalene; Invitrogen) as described previously (16), except that 780-nm two-photon excitation was used.
Confocal Laser Scanning Microscopy and Image Analysis
All images were acquired on a Zeiss Axiovert 100 M confocal microscope fitted with an LSM510 scanning head. Images were sampled at the Nyquist sampling frequency with the photomultiplier tube detector gain and amplifier offset adjusted so that voxel intensities were spread over the full dynamic range. All three-dimensional image data were deconvolved using Huygens software (Scientific Volume Imaging, Hilversum, The Netherlands), using a theoretical or measured point spread function, prior to further analysis.
Photoactivatable Localization Microscopy (PALM)
PALM data (supplemental Fig. 1) were acquired from fixed cells transfected with constructs expressing either SNAP-25 or syntaxin fused to PACherry. Cycles of brief activation at 405 nm, followed by rapid imaging in total internal reflection fluorescence microscopy mode at 561 nm were performed using an Olympus IX-81 microscope equipped with the Olympus Cell-R acquisition software and an ImageEM EM-CCD 512 × 512 camera (Hamamatsu UK). All PALM imaging used an Olympus 150X UApo 1.45NA oil lens with a resulting pixel size of 106 nm. Activation and bleaching steps in each cycle were optimized to ensure a sparse distribution of single molecules was activated and photo-destroyed during each cycle. PALM data analysis was performed using a Matlab routine written by Dr. Samuel Hess (Maine). Spatial distributions of localized single molecules were analyzed using Ripley's analyses using custom-written algorithms in Matlab (Mathworks, Inc., Cambridge, UK).
Preparation of Plasma Membrane Sheets and Immunostaining for STED Microscopy
For the STED experiments, cells were plated onto polylysine-coated glass coverslips and grown between 36 and 40 h. The plated cells were then incubated with or without 10 mm MβCD (Sigma) in Dulbecco's modified Eagle's medium (Invitrogen) at 37 °C for 15 min. After treatment, the cells were sonicated as described previously (17) using a 100-ms ultrasound pulse in ice-cold KGlu sonication buffer (120 mm potassium glutamate, 20 mm potassium acetate, 10 mm EGTA, and 20 mm HEPES, pH 7.2) and fixed in 4% paraformaldehyde in phosphate-buffered saline for 1 h. Paraformaldehyde was quenched using 50 mm NH4Cl in phosphate-buffered saline for 20 min and immunostained for endogenous syntaxin1a and synaptobrevin-2 using mouse monoclonal HPC-1 (Sigma) and rabbit polyclonal antiserum (Synaptic Systems). Primary antibodies were detected using goat anti-rabbit Cy3 (Dianova) and Atto647N-labeled sheep anti-mouse (provided by Dept. of Nanobiophotonics, Max-Planck Institute for Biophysical Chemistry, Göttingen, Germany) essentially as described previously (7). The samples were embedded in Mowiol (Merck) before imaging.
STED Microscopy
The samples were imaged using a TCS-STED (Leica Microsystems) super-resolution fluorescence microscope equipped with a 1.4 NA ×100 objective. Multichannel confocal images of immunostained plasma membrane sheets were then obtained simultaneously (at 100-Hz scan speed) for Cy3 (synaptobrevin 2) and Atto647N (syntaxin1a) followed by imaging Atto647N applying STED resolution (at 10-Hz scan speed). Pixel size was 21.6 nm. STED excitation was performed with a 635-nm diode laser, and depletion was achieved via a MaiTai tunable laser (Spectra-Physics) at 750 nm. Emission was collected at 645–720 nm for the Atto647N and at 551–602 nm for Cy3. Confocal imaging was performed using photomultiplier tubes, and an avalanche photodiode was used for STED image acquisition. The system resolution limit was ∼70–90 nm, measured by analysis of 20 nm crimson-fluorescent beads (Invitrogen). Metamorph (Universal Imaging) was used for the alignment of confocal and STED images. The Cy3- and STED-resolved channels were aligned using the alignment of the Atto647N STED image to the Atto647N confocal image as reference.
Fluorescence Lifetime Imaging Microscopy and Analysis
Time-correlated single photon counting (TCSPC) measurements were made under 800–820-nm two-photon excitation that efficiently excited cerulean, without any detectable direct excitation or emission from EYFP, using a fast photomultiplier tube (H7422; Hamamatsu Photonics UK) coupled directly to the rear port of the Axiovert microscope (supplemental Figs. 3 and 4). Full frame TCSPC recordings were acquired between 30 and 60 s, with mean photon counts between 105 and 106 counts/s. Images were recorded at 256 × 256 pixels from a 1024 × 1024 image scan with 256 time bins over a 12-ns period. Off-line FLIM data analysis used pixel-based fitting software (SPCImage, Becker & Hickl). The optimization of the fit parameters was performed by using the Levenberg-Marquardt algorithm, minimizing the weighted χ2 quantity. As controls for nonspecific FRET, or FRET between green fluorescent proteins that may form dimers spontaneously when overexpressed in cells, we determined the fluorescence lifetimes of cerulean-Syx-(1–288) alone, cerulean alone, or cerulean-Syx-(1–288) cotransfected with EYFP (15). No FRET was detected in any of these experiments.
RESULTS
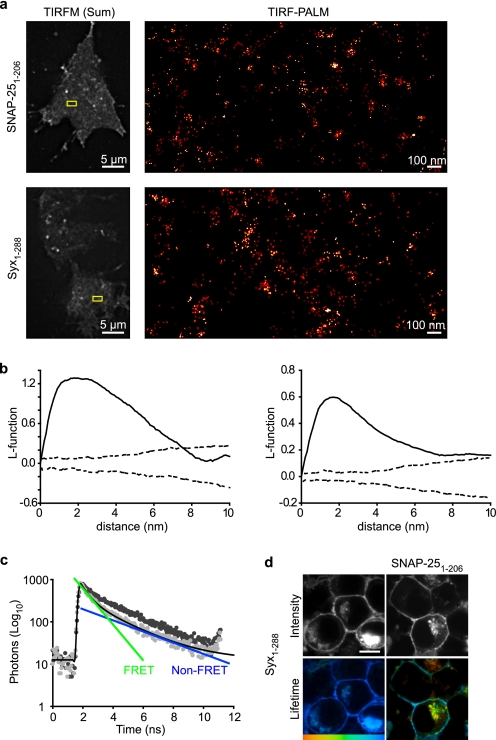
We first examined the localization of single molecules of SNAP-25 (Fig. 1a, upper panels) in PC12 cells. Using PALM (18, 19) (supplemental Fig. 1 and supplemental movie 1), allowing nanoscale localization of proteins, we found single molecules of SNAP-25 are not distributed homogeneously across the plasma membrane (Fig. 1a). Similarly, syntaxin1a molecules are concentrated into ensembles interspersed with largely single molecules (Fig. 1a, lower panels). Statistical analysis of the spatial distributions of these molecules was performed. Hopkin's statistics compares the nearest neighbor distances from a random subset of data events to the nearest neighbor distances from points placed at random in the data to be tested (20, 21). We thus compared the XY coordinates of single t-SNARE molecules to synthetic data exhibiting complete spatial randomness, finding that single SNARE molecules are subject to higher order organization into clusters (data not shown). Furthermore, Ripley's K and L function analyses (Fig. 1b) (22) of these datasets revealed that the molecular spatial organization of SNAP-25 and syntaxin1a is nonrandom. Together, these data demonstrate higher order organization of the t-SNAREs in the living plasma membrane at the molecular level. PALM cannot provide an exact measure of the number of molecules in a region as it can never be known that every molecule has been counted (18); however, it can provide a lower limit for the number of molecules per unit area. Our data provided an estimate of ∼30–40 syntaxin molecules in an area covered by a 50-nm diameter spot (over the dense ensembles). This complements a recent study (10), using another super-resolution imaging technique, STED, in combination with molecular modeling that predicted 70 molecules per 50-nm cluster. PALM allows the further localization of single molecules within and between these macromolecular assemblies. To test whether the distributions of these two proteins overlap, we quantified the covariance of SNAP-25 and syntaxin1a, finding a high degree of overlap (>86%; supplemental Fig. 2).
FIGURE 1.
Syntaxin and SNAP-25 molecules interact in clusters on the plasma membrane. a, TIRF-PALM localization map showing single molecules of SNAP-25 and syntaxin (Syx-(1–288)) on the plasma membrane. The boxed area highlighted in the summed total internal reflection fluorescence microscopy image (left panels) is zoomed in the TIRF-PALM-rendered image (right panels), where molecular clustering is clearly visible. b, L-function is a commonly used transformation of Ripley's K function and transforms spatially random data into a straight line. Representative L-function plots are shown for syntaxin (right) and SNAP-25 (left). Each representative dataset, consisting of XY coordinates describing the precise localization of individual t-SNARE molecules, was repeatedly randomized, and the L-function was re-derived. Confidence envelopes showing the maximum and minimum L-function values from 1000 randomizations are shown (dashed line). This approach thus compares the L-function between experimental data and an identical number and density of random points. Values above the confidence envelopes indicate nonrandom spatial organization. c, TCSPC-FLIM analysis confirmed that a syntaxin fluorescent donor (cerulean-syntaxin) followed a single fluorescence decay (dark gray). In the presence of a proximal acceptor (EYFP-SNAP-25), this decay was described by a bi-exponential decay (light gray). The time constants of the bi-exponential functions correspond to a short FRET lifetime (green) and a longer fluorescence lifetime identical to the non-FRET control (blue). Both the FRET and non-FRET components are present in every region of the cell in varying proportions. d, TCSPC-FLIM of a cell containing cerulean-syntaxin alone (left panels) or in the presence of EYFP-SNAP-25 (right panels) showing intensity (gray scale) and lifetime on a color scale from 1250 to 2250 ps. The lifetime of syntaxin is shortened in the presence of SNAP-25 indicating an interaction. All scale bars are 5 μm unless otherwise stated.
Super-resolution optical approaches lack sufficient resolution to provide insight into protein interactions at the molecular level. Therefore, we quantified the interaction between SNAP-25 and syntaxin using TCSPC-FLIM (for explanation of FLIM analysis and presentation see supplemental Fig. 3), an imaging spectroscopy approach we used previously to measure SNARE protein interactions in living cells (15, 23, 24). FLIM has the advantage that it delivers data unaffected by donor or acceptor concentration, the emission light path, sample scatter, or donor/acceptor ratio (25). The donor, mCerulean-Syx-(1–288), in the presence of unfused EYFP, had a single fluorescence lifetime at the plasma membrane (shown in Fig. 1, c and d) in agreement with our previous findings in neuroblastoma 2a cells (15, 23). FLIM reports protein interactions as a specific quenching of the donor fluorescence lifetime in the presence of a proximal acceptor. To exclude any possibility that high concentrations of donor and acceptor molecules, in clusters at the cell surface, could lead to donor fluorescence lifetime quenching due merely to density, we analyzed cells expressing mCerulean-syntaxin and EYFP-syntaxin. These fluorescent proteins coexisted at the plasma membrane as expected (10), and no donor fluorescence lifetime quenching could be detected (supplemental Fig. 4). However, in the presence of the FRET acceptor EYFP-SNAP-25-(1–206), a change in the donor, mCerulean-syntaxin, fluorescence lifetime was reported, indicating a specific protein-protein interaction (i.e. a second fluorescence lifetime component appeared; Fig. 1b). TCSPC-FLIM uniquely delivers information describing both interaction (lifetime) and the proportion of donor molecules participating in energy transfer in each pixel of the image. This allows the influence of proximity and number of interacting partners to be dissected. A fuller description of this type of analysis is included in supplemental Fig. 3; in addition, a SNARE-specific FLIM review (26) and a general text (27) on time-correlated single photon counting provides a complete background to this technique.
Spatial Segregation of t-SNARE Heterodimer Conformations
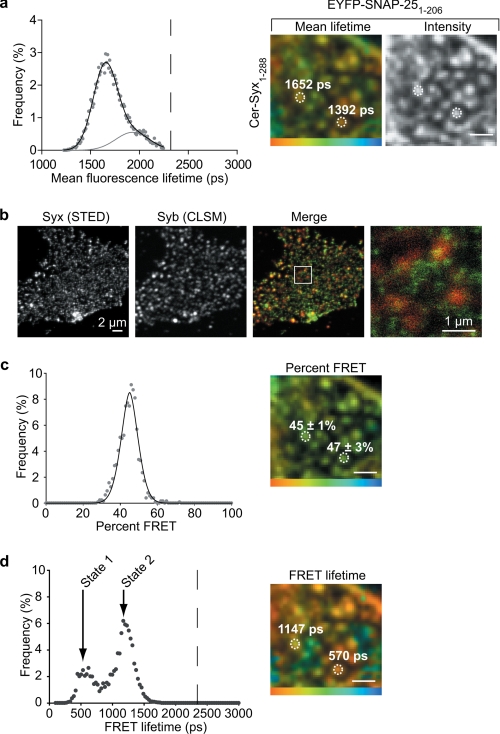
Further analysis at the cell surface measured an interaction between syntaxin and SNAP-25 within the clusters on the plasma membrane (Fig. 2, a, c, and d). Importantly, the weighted mean fluorescence lifetime data (containing both interacting and noninteracting values and their respective amplitudes; supplemental Fig. 3) were bimodal (Fig. 2a, left panel) due to two spatially segregated types of SNARE cluster containing different FRET “efficiencies” (Fig. 2a, right panels). The t-SNARE dimer is thought to serve as an acceptor for the vesicular SNARE, synaptobrevin. It was therefore theoretically possible that the two spatially segregated states we observed could be due to the presence, or not, of synaptobrevin. To address this, we employed super-resolution STED imaging (previously used to resolve single SNARE clusters (10)) to ascertain the level of colocalization between synaptobrevin and single t-SNARE clusters on the cell surface (Fig. 2b). This showed that the large majority of single syntaxin clusters do not have coincident synaptobrevin. In addition, we found only 9 ± 5% overlap (Mander's overlap coefficient; mean ± S.E., n = 8 independent experiments), in immunostained cells using deconvolution microscopy (supplemental Fig. 2). This low level of overlap between the vesicle SNARE and the t-SNAREs is insufficient to account for the observed cluster heterogeneity across the cell surface.
FIGURE 2.
Distinct t-SNARE heterodimer conformations are spatially segregated on the plasma membrane. a, histogram of the amplitude weighted mean lifetime of the time constants of both FRET and non-FRET components measured from the plasma membrane of a cell expressing cerulean-Syx and EYFP-SNAP-25 is best fit by the sum of two Gaussian distributions (left panel). The dashed line corresponds to the mean non-FRET value. These two fluorescence lifetimes are spatially segregated on the plasma membrane, indicating heterogeneous interaction states (right panel). Color scale, 1250–2250 ps. b, STED and standard confocal laser scanning microscopy demonstrate minimal coincidence between syntaxin and synaptobrevin at the plasma membrane, insufficient to account for the variable t-SNARE interaction status observed. The highlighted region in the merge panel is shown enlarged (right panel). c, heterogeneity between clusters is not due to differences in the proportion of interacting molecules. The amplitude of the FRET lifetime component (“FRET percent,” corresponding to the proportion of interacting molecules) is uni-modal as shown in the histogram (left panel) with clusters exhibiting only a small amount of variance around this mean (right panel). Color scale, 0–100%. d, two spatially segregated populations of t-SNARE heterodimer conformation are the cause of the observed heterogeneity. The FRET lifetime is bi-modal (termed state 1 and state 2) as shown in the histogram (left panel); a result of the spatial segregation of FRET lifetimes into different clusters (right panel). Color scale, 0–2000 ps. All scale bars 1 μm unless otherwise stated.
Differences in FRET “efficiency” can be a result of either a change in the number of interacting molecules or differences in the inter-molecular proximity at specific cellular locations. However, FLIM can distinguish between these two possibilities (supplemental Fig. 3), allowing control for any interference from unlabeled SNAREs, which would ordinarily go undetected. First, we quantified the proportion of interacting molecules within every diffraction-limited cluster (containing 30–70 SNARE molecules each) in an image. This is described by a single distribution (Fig. 2c, left panel) and is further emphasized by an image showing spatial uniformity of this measurement (Fig. 2c, right panel). Therefore, differences in the number of interacting molecules between clusters cannot account for the observed spatial variations. Next, we analyzed the fluorescence lifetime component derived from only the proportion of interacting molecules within clusters. This revealed two distinct populations, for simplicity here termed state 1 and state 2 (Fig. 2d, left panel). Importantly, not only do these two states possess different molecular conformations, they are also spatially segregated on the plasma membrane into distinct clusters where one of the two forms predominates (Fig. 2d, right panel).
Distinct Heterodimer Conformations Are Due to Differential Engagement of SNAP-25
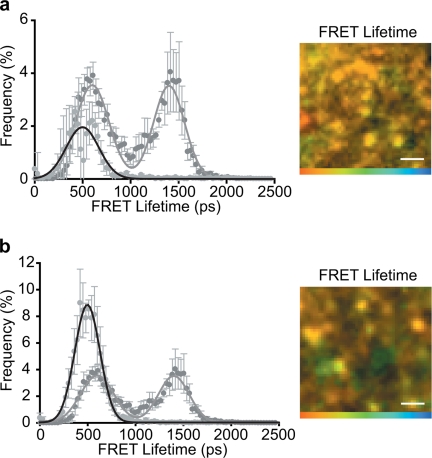
To gain structural insights into the two observed t-SNARE heterodimer states, we took advantage of the thorough biochemical and structural understanding of SNARE proteins. Recent work suggested that the t-SNAREs can exist in different conformational forms in vitro (28), where one form corresponded to a parallel three-helical bundle, whereas a second major form was shown to have the second SNAP-25 helix dissociated. We therefore tested whether these forms could account for the two states we observe in intact cells. We employed another mutant of SNAP-25 to mimic the cleavage product of botulinum neurotoxin serotype E (BoNT/E); this specific cleavage of SNAP-25 (removing the 25 C-terminal amino acids) is known to disrupt t-SNARE interactions (4) and abolish exocytosis (29). This mutant maintained micro-patterns morphologically indistinguishable from full-length SNAP-25; however, state 2 (the three helical bundle) was reduced to undetectable levels (Fig. 3a). To further understand this, a mutant SNAP-25, where the entire second SNARE helix was deleted (SNAP-25-(1–121)), still formed clusters at the plasma membrane (Fig. 3b). Importantly, however, the heterodimer conformation was now uniform across the membrane, with the complete loss of state 2 (Fig. 3b). These findings indicate that the first SNARE helix of SNAP-25 is sufficient to initiate the interaction with syntaxin1a but that state 2 relies on the second SNARE helix of SNAP-25 to be present in the heterodimer complex. A previously reported substitution in SNAP-25 (G43D), a mutation in this first helix, was proposed to prevent the formation of a four helical bundle containing two SNARE helices from syntaxin and two N-terminal SNARE helices of SNAP-25 (30). We re-examined the effect of this mutation on the interaction between full-length SNAP-25 with syntaxin in vitro; in all assays, we found little or no significant effects (supplemental Fig. 5). We conclude that the heterodimer states we observe represent two different conformations of the t-SNARE heterodimer resulting from a differential engagement of the second SNARE helix of SNAP-25.
FIGURE 3.
Two t-SNARE conformations are a result of differential SNAP-25 engagement. a, truncation of the C terminus of SNAP-25 to mimic BoNT/E cleavage (SNAP-25-(1–181)) abolished the state 2 conformation as shown in the histogram of FRET lifetimes (left panel) resulting in a largely uniform spatial distribution of FRET lifetimes (right panel) with a lifetime corresponding to the state 1 conformation. b, truncation of the entire C-terminal helix of SNAP-25 (SNAP-25-(1–120)) also abolished the state 2 conformation (left panel) resulting in a more uniform spatial distribution of FRET lifetimes (right panel). All color scales: 0–2000 ps. All scale bars 1 μm unless otherwise stated. Graphs show means ± S.E., n = at least five independent experiments.
Underlying Lipid Order Affects t-SNARE Heterodimer Conformation
How are the two t-SNARE conformations spatially segregated? It has been suggested that these clusters may in some way be affected by the surrounding lipids (7, 13). However, this was at the gross topological or biochemical level and could not examine the interaction status of the proteins in situ. Therefore, we addressed the role of lipid order in plasma membrane cluster organization at the molecular level, reasoning that lipid order may play a role in segregating the two conformational states we have defined.
We examined the effect of sublethal (supplemental Fig. 6) cholesterol depletion on the lipid order of the plasma membrane, using laurdan, which reports changes in the packing of surrounding lipids as a spectral shift (31). Generalized polarization scale (GP) values can be calculated, with higher GP values corresponding to more “ordered” lipid environments (16). This revealed two different membrane populations in living neuroendocrine cells, with distinct lipid orders; the first peak (centered at −0.25) corresponds to intracellular membranes and the second peak (centered at 0) to the plasma membrane (Fig. 4a). Thus, the plasma membrane, which is normally enriched in cholesterol, typically at levels high enough to induce the liquid ordered phase (30–40 mol % (32)), is more ordered than intracellular membranes, in agreement with previous studies of diverse cell types (16, 33–35). Treatment of the same cells with 10 mm MβCD resulted in a depletion of 21 ± 4% of plasma membrane cholesterol (t test, p < 0.001, n = 12; see supplemental Fig. 6) leading to the loss of the lipid order in the plasma membrane (Fig. 4a, middle and right panels), while maintaining cell viability. It has been reported extensively that varying cholesterol concentration causes abrupt nonlinear discontinuities in membrane phase properties at 37 °C (36). For this reason, we chose as high a sublethal concentration of MβCD as possible to permit subsequent analyses. This loss of lipid order resulted in no change in the topological arrangement of single clusters as reported by super-resolution STED imaging (Fig. 4b) or the colocalization between the t-SNAREs on the plasma membrane (supplemental Fig. 2).
FIGURE 4.
Lipid disordering patterns the conformational states of the t-SNAREs. a, laurdan was used to report lipid packing in cell membranes. The GP values were calculated on a pixel-by-pixel basis and are shown on a color scale before and after treatment with 10 mm MβCD (left and center panels). Color scale, −0.5 (blue) to +0.5 (red). The histogram of GP values is plotted (right panel), showing two separate populations (gray) corresponding to the plasma membrane (high GP; ordered lipid domains) and intracellular membranes (low GP; disordered domains). After cholesterol depletion with 10 mm MβCD (black), the lipid order on the plasma membrane was completely disrupted. b, STED microscopy of endogenous syntaxin on the plasma membrane before and after disruption of lipid order with 10 mm MβCD. Endogenous syntaxin clusters remain intact after loss of lipid order in the plasma membrane. Shown for comparison are standard confocal laser scanning microscopy images of the same sample. The highlighted regions in the STED images are shown enlarged (right panels). c, TCSPC-FLIM analyses of donor mCerulean-syntaxin in the presence of proximal EYFP-SNAP-25, in control (untreated, gray), and MβCD-treated (black) cells (left panel). Upon cholesterol depletion, the state 1 population is reduced, but the SNARE clusters remain intact (right panel). Color scale, 0–2000 ps. Scale bar, 1 μm. Graphs show means ± S.E. of donor fluorescence FRET lifetime data, n = at least five independent experiments. d, model describing the two molecular conformations of the t-SNAREs (syntaxin, red; SNAP-25 N- and C-terminal helices, light and dark green, respectively) observed on the plasma membrane. These states are spatially segregated in clusters, with the propensity of each form patterned by lipid order. BoNT/E resulted in state 1 predominating which is unable to support ternary SNARE complex formation and fusion.
As there was no topological change in the t-SNARE distribution in the disordered plasma membrane, we next examined the effect of lipid order on the two heterodimer conformational states. Combining FLIM datasets confirmed that the FRET data from cells containing mCerulean-syntaxin and EYFP-SNAP-25 followed a bimodal distribution (reporting two conformational states) as before. After lipid order disruption, state 1 (SNAP-25 second helix not fully associated) was significantly reduced (special sum of squares F-test; p < 0.001, n = 5). The small right shift likely reflects a previous averaging of the data because of the partial overlap of the two peaks, with state 2 now better resolved by the specific decrease in state 1 conformation (the inverse effect can be seen in the minor left shift of the data after BoNT/E treatment). Approximately 30% of the interacting molecules adopted the state 1 conformation (Fig. 4c), although after disruption of lipid order, it was reduced to 17%. In contrast, the state 2 population (involving both SNARE helices of SNAP-25) increased in frequency from 70 to 83%. As a control, treatment of mCerulean syntaxin-expressing cells (in the absence of a FRET acceptor) with 10 mm MβCD did not affect the donor fluorescence lifetime data (data not shown). Plasma membrane lipid order thus does not affect t-SNARE cluster morphology at the gross microscopic level; however, the conformational composition of each cluster is patterned by the underlying lipid environment.
DISCUSSION
Our data demonstrate that two distinct t-SNARE heterodimer conformations coexist at the plasma membrane of living neuroendocrine cells, segregated spatially and functionally into macromolecular clusters where one of two conformations predominates. This segregation is organized at least in part by lipid order and results in some clusters (state 1) where the C terminus of SNAP-25 is not engaged with syntaxin. As this protein complex cannot act as an acceptor for synaptobrevin, nor can it support exocytosis, these findings provide a model where proteins can be functionally and spatially patterned on the plasma membrane by the underlying lipid environment, at the level of their protein interactions. In contrast, state 2 can support exocytosis, and these states are spatially segregated. Furthermore, MβCD treatment results in an increased proportion of t-SNAREs adopting the functionally competent state 2 conformation.
We propose that cells could possibly regulate the position of release sites and the magnitude of the exocytotic response by modulating the lipid environment within which t-SNAREs reside. These two conformational t-SNARE heterodimer states may be further stabilized by accessory factors, including munc18-1, as has been observed in vitro (28). Further work will be required to determine whether accessory proteins preferentially associate with specific t-SNARE conformational states and their resulting influence.
How do the clusters of interacting t-SNAREs relate to previous findings on the morphological distribution of plasma membrane SNAREs? Syntaxin1 was shown previously to form clusters 50 nm in size, modeled to contain ∼70 syntaxin molecules (10). The mechanism for this association was in part attributed to a homotypic interaction utilizing the transmembrane spanning region of syntaxin (37, 38). A proposed model from this work predicted a high degree of molecular crowding of syntaxin in the 50-nm dense clusters, hypothesized to be deleterious for interactions with other protein species. Importantly, however, no evidence for or against this sequestration of syntaxin from t-SNARE interactions has been reported in the literature. As all of the syntaxin clusters we observed reported interactions with SNAP-25, two possibilities exist. First, the presence of syntaxin in clusters does not preclude interaction with other proteins, including SNAP-25. Second, only the syntaxin on the periphery of the cluster is able to interact with SNAP-25. The core of the cluster would thus act as a reserve of syntaxin ready to replenish t-SNARE intermediates consumed in the fusion process. Our findings show a similar number of SNAP-25 molecules (∼40–50) and syntaxin molecules (∼30–40) in dense ensembles. This could support the first hypothesis; however, PALM approaches can only provide a lower estimate for the number of molecules in a region, and so this technique cannot exclude either possibility. Recent work examining t-SNARE interactions at the plasma membrane also reported differential t-SNARE intermediate interactions (39), providing further support to our conclusions but no information to distinguish between these two models. As the spatial resolution of the TCSPC-FLIM approach is insufficient to distinguish between t-SNARE interactions throughout the cluster or restricted to the periphery, this question will require the development of single molecule FRET approaches to provide a definitive answer.
Recent work demonstrated that the C-terminal transmembrane domain of syntaxin forms a continuous α-helical structure with the SNARE motif (40). These data suggest that the transmembrane domain of syntaxin may not be long enough if structured to span the plasma membrane bilayer. Therefore, changes in the thickness of the bilayer, due to cholesterol intercalation, may be transduced to alter the conformation of syntaxin depending upon the lipid microenvironment. Furthermore, SNAP-25 is associated with the inner leaflet of the plasma membrane through palmitoylation; it is likely that SNAP-25 targeting, function, and conformation is modulated by cholesterol through differential insertion of palmitate groups into the membrane (41). In addition to cholesterol, phosphatidylinositol 4,5-bisphosphate has been reported to influence both the spatial and functional properties of syntaxin at the plasma membrane (42, 43). In conclusion, it is now becoming clear that the lipids of the plasma membrane play an active role in determining SNARE protein behavior in living cells (44).
Supplementary Material
Acknowledgments
pEGFP-N1-SNAP-25 was a gift from Maurine Linder (Washington University, St. Louis). A plasmid encoding photoactivatable mCherry was a gift from Vladislav V. Verkhusha (Yeshiva University). PALM data analysis was made possible using a Matlab routine generously gifted by Samuel Hess (Maine).
This work was supported by Wellcome Trust grants (to R. R. D. and L. H. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3, Movie 1, and an additional reference.
- SNARE
- soluble NSF attachment protein receptor
- t-SNARE
- target SNARE
- FRET
- fluorescence resonance energy transfer
- EYFP
- enhanced yellow fluorescent protein
- MβCD
- methyl-β-cyclodextrin
- PALM
- photoactivatable localization microscopy
- STED
- stimulated emission depletion
- FLIM
- fluorescence lifetime imaging microscopy
- TCSPC
- time correlated single photon counting
- GP
- generalized polarization
- BoNT/E
- botulinum neurotoxin serotype E.
REFERENCES
- 1.Sutton R. B., Fasshauer D., Jahn R., Brunger A. T. (1998) Nature 395, 347–353 [DOI] [PubMed] [Google Scholar]
- 2.Söllner T., Whiteheart S. W., Brunner M., Erdjument-Bromage H., Geromanos S., Tempst P., Rothman J. E. (1993) Nature 362, 318–324 [DOI] [PubMed] [Google Scholar]
- 3.Gonzalo S., Greentree W. K., Linder M. E. (1999) J. Biol. Chem. 274, 21313–21318 [DOI] [PubMed] [Google Scholar]
- 4.Rickman C., Meunier F. A., Binz T., Davletov B. (2004) J. Biol. Chem. 279, 644–651 [DOI] [PubMed] [Google Scholar]
- 5.Fasshauer D., Margittai M. (2004) J. Biol. Chem. 279, 7613–7621 [DOI] [PubMed] [Google Scholar]
- 6.Fasshauer D., Sutton R. B., Brunger A. T., Jahn R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 15781–15786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lang T., Bruns D., Wenzel D., Riedel D., Holroyd P., Thiele C., Jahn R. (2001) EMBO J. 20, 2202–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lang T., Margittai M., Holzler H., Jahn R. (2002) J. Cell Biol. 158, 751–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohara-Imaizumi M., Nakamichi Y., Nishiwaki C., Nagamatsu S. (2002) J. Biol. Chem. 277, 50805–50811 [DOI] [PubMed] [Google Scholar]
- 10.Sieber J. J., Willig K. I., Kutzner C., Gerding-Reimers C., Harke B., Donnert G., Rammner B., Eggeling C., Hell S. W., Grubmüller H., Lang T. (2007) Science 317, 1072–1076 [DOI] [PubMed] [Google Scholar]
- 11.Melkonian K. A., Ostermeyer A. G., Chen J. Z., Roth M. G., Brown D. A. (1999) J. Biol. Chem. 274, 3910–3917 [DOI] [PubMed] [Google Scholar]
- 12.Bacia K., Schuette C. G., Kahya N., Jahn R., Schwille P. (2004) J. Biol. Chem. 279, 37951–37955 [DOI] [PubMed] [Google Scholar]
- 13.Chamberlain L. H., Burgoyne R. D., Gould G. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5619–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laage R., Rohde J., Brosig B., Langosch D. (2000) J. Biol. Chem. 275, 17481–17487 [DOI] [PubMed] [Google Scholar]
- 15.Rickman C., Medine C. N., Bergmann A., Duncan R. R. (2007) J. Biol. Chem. 282, 12097–12103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaus K., Gratton E., Kable E. P., Jones A. S., Gelissen I., Kritharides L., Jessup W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15554–15559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avery J., Ellis D. J., Lang T., Holroyd P., Riedel D., Henderson R. M., Edwardson J. M., Jahn R. (2000) J. Cell Biol. 148, 317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Betzig E., Patterson G. H., Sougrat R., Lindwasser O. W., Olenych S., Bonifacino J. S., Davidson M. W., Lippincott-Schwartz J., Hess H. F. (2006) Science 313, 1642–1645 [DOI] [PubMed] [Google Scholar]
- 19.Hess S. T., Girirajan T. P., Mason M. D. (2006) Biophys. J. 91, 4258–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Numahara T., Nakagawa T., Takaiwa T. (1992) J. Dermatol. Sci. 4, 202–207 [DOI] [PubMed] [Google Scholar]
- 21.Hopkins B., Skellam J. G. (1954) Ann. Bot. 18, 213–226 [Google Scholar]
- 22.Ripley B. D. (1977) J. R. Stat. Soc. 39, 172–212 [Google Scholar]
- 23.Medine C. N., Rickman C., Chamberlain L. H., Duncan R. R. (2007) J. Cell Sci. 120, 4407–4415 [DOI] [PubMed] [Google Scholar]
- 24.Valkonen M., Kalkman E. R., Saloheimo M., Penttilä M., Read N. D., Duncan R. R. (2007) J. Biol. Chem. 282, 22775–22785 [DOI] [PubMed] [Google Scholar]
- 25.Lakowicz J. R. (1999) Principles of Fluorescence Spectroscopy, 2nd Ed., Plenum Publishing Corp., New York [Google Scholar]
- 26.Altenbach K., Duncan R. R., Valkonen M. (2010) Fungal Biol. Rev., in press [Google Scholar]
- 27.Becker W. (2005) Advanced Time-correlated Single Photon Counting Techniques, Springer-Verlag, Berlin [Google Scholar]
- 28.Weninger K., Bowen M. E., Choi U. B., Chu S., Brunger A. T. (2008) Structure 16, 308–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Binz T., Blasi J., Yamasaki S., Baumeister A., Link E., Südhof T. C., Jahn R., Niemann H. (1994) J. Biol. Chem. 269, 1617–1620 [PubMed] [Google Scholar]
- 30.An S. J., Almers W. (2004) Science 306, 1042–1046 [DOI] [PubMed] [Google Scholar]
- 31.Parasassi T., Gratton E., Yu W. M., Wilson P., Levi M. (1997) Biophys. J. 72, 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMullen T. P., Lewis R. N., McElhaney R. N. (2004) Curr. Opin. Colloid Interface Sci. 8, 459–468 [Google Scholar]
- 33.Vieira O. V., Gaus K., Verkade P., Fullekrug J., Vaz W. L., Simons K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18556–18561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fitzner D., Schneider A., Kippert A., Möbius W., Willig K. I., Hell S. W., Bunt G., Gaus K., Simons M. (2006) EMBO J. 25, 5037–5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stetzkowski-Marden F., Gaus K., Recouvreur M., Cartaud A., Cartaud J. (2006) J. Lipid Res. 47, 2121–2133 [DOI] [PubMed] [Google Scholar]
- 36.Parasassi T., Di Stefano M., Loiero M., Ravagnan G., Gratton E. (1994) Biophys. J. 66, 120–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kroch A. E., Fleming K. G. (2006) J. Mol. Biol. 357, 184–194 [DOI] [PubMed] [Google Scholar]
- 38.Hofmann M. W., Peplowska K., Rohde J., Poschner B. C., Ungermann C., Langosch D. (2006) J. Mol. Biol. 364, 1048–1060 [DOI] [PubMed] [Google Scholar]
- 39.Halemani N. D., Bethani I., Rizzoli S. O., Lang T. (2010) Traffic [DOI] [PubMed] [Google Scholar]
- 40.Stein A., Weber G., Wahl M. C., Jahn R. (2009) Nature 460, 525–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salaün C., Gould G. W., Chamberlain L. H. (2005) J. Biol. Chem. 280, 19449–19453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murray D. H., Tamm L. K. (2009) Biochemistry 48, 4617–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lam A. D., Tryoen-Toth P., Tsai B., Vitale N., Stuenkel E. L. (2008) Mol. Biol. Cell 19, 485–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lang T., Halemani N. D., Rammner B. (2008) Prog. Lipid Res. 47, 461–469 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.