Abstract
Human concentrative nucleoside transporter-3 (hCNT3) is a sodium-coupled nucleoside transporter that exhibits high affinity and broad substrate selectivity, making it the most suitable candidate for mediating the uptake and cytotoxic action of most nucleoside-derived drugs. The drug of this class most commonly used in the treatment of chronic lymphocytic leukemia (CLL) is the pro-apoptotic nucleoside analog fludarabine (Flu), which enters CLL cells primarily through human equilibrative nucleoside transporters (hENTs). Although CLL cells lack hCNT3 activity, they do express this transporter protein, which is located mostly in the cytosol. The aim of our study was to identify agents and mechanisms capable of promoting hCNT3 trafficking to the plasma membrane. Here, we report that all-trans-retinoic acid (ATRA), currently used in the treatment of acute promyelocytic leukemia (APL), increases hCNT3-related activity through a mechanism that involves trafficking of pre-existing hCNT3 proteins to the plasma membrane. This effect is mediated by the autocrine action of transforming growth factor (TGF)-β1, which is transcriptionally activated by ATRA in a p38-dependent manner. TGF-β1 acts through activation of ERK1/2 and the small GTPase RhoA to promote plasma membrane trafficking of the hCNT3 protein.
Keywords: Drug Resistance/Cancer Therapy/Chemotherapy; Drug Resistance/Transport; Membrane/Proteins; Membrane/Trafficking; Nucleoside/Nucleotide/Transport; Transport; Intracellular Trafficking; ATRA, hCNT3, Traffic, ERK 1/2, RhoA, p38
Introduction
Natural nucleosides and nucleoside derivatives used in anticancer therapies are generally hydrophilic and are thus taken up by cells via nucleoside-specific membrane transporters (NT).3 These transporters belong to the solute carrier families SLC28 and SLC29. The SLC28 genes encode the concentrative NT (CNT) proteins and comprise three members: CNT1, CNT2, and CNT3. These proteins mediate high affinity sodium-dependent translocation of nucleosides into cells and are mostly expressed in epithelial and immune system cells (1). The SLC29 genes encode the four members of the equilibrative NT (ENT) family, which are found in almost all studied cell types and facilitate uptake of nucleosides and nucleoside-derived drugs. The ENT transporters exhibit broad selectivity but have lower affinity compared with CNT proteins (2).
Chronic lymphocytic leukemia (CLL) is the most common leukemia in adults in western countries. CLL results from the progressive accumulation of slowly proliferating CD5+ B lymphocytes (3) because of abnormal programmed cell death. Increasing evidence indicates that the accumulation of CLL cells in vivo depends on essential survival factors that delay spontaneous and drug-induced-apoptosis (4, 5). Although at present there is no curative treatment for CLL patients, nucleoside analogs, particularly fludarabine (Flu), have substantially improved clinical outcomes during the past 20 years (6). Flu is a purine nucleoside analog (NA) prodrug that is converted to the nucleoside 9-b-d-arabinosyl-2-fluoroadenine (F-ara-A), which enters cells and accumulates mainly as the 5′-triphosphate, F-ara-ATP. F-ara-ATP has multiple mechanisms of action, most of which are predominantly DNA-directed; thus, F-ara-ATP induces apoptosis (7).
CLL cells express hENT1, hENT2, hCNT2, and hCNT3 mRNA, although most of the biological activity responsible for Flu uptake is associated with ENT-type transporters (8, 9). Interestingly, Flu-resistant populations of CLL have been reported to express high levels of cytosolic hCNT3 protein, without detectable hCNT3-related plasma membrane transport activity (10, 11). This lack of hCNT3 activity, despite high levels of hCNT3 protein, is probably due to localization of hCNT3 protein primarily in intracellular compartments of CLL cells.
All-trans-retinoic acid (ATRA) is a natural vitamin A derivative that is used in the treatment of acute promyelocytic leukemia (APL), an acute myeloid leukemia (AML) characterized by the morphology of blast cells (12). APL cells contain a t(15;17) translocation, which fuses part of the promyelocytic leukemia (PML) gene to the retinoic acid receptor (RAR) α gene to encode PML-RAR (13). ATRA induces differentiation of abnormal promyelocytes into mature granulocytes in APL in vitro and in vivo (14), presumably by degradation of the PML-RAR fusion protein (15). Through this mechanism, ATRA treatment is capable of achieving complete remission in 80–90% of newly diagnosed and first-relapsing cases of APL. Currently, combined treatment with ATRA and chemotherapy is the standard therapy for patients with APL.
In addition to the use of ATRA in APL, ATRA-induced differentiation of NB4 cells, a cell line containing the APL disease-specific t(15;17) chromosomal translocation, has been shown to result in biphasic changes in guanosine transport (16). The authors showed that ATRA induces transient increases in a CNT-type activity that is not yet well characterized but is presumably associated with a polymorphic variant of known CNTs (17). Moreover, ATRA treatment has been shown to increase ara-C transport and ara-C-induced apoptosis (18, 19).
The current study is based on the apparent ability of ATRA to regulate nucleoside transporter function in APL cells and the observation that hCNT3 protein is accumulated, but not functional, in CLL cells. The goal of the study was to investigate the effects of ATRA on the expression, localization, and activity of hCNT3.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Colchicine, cycloheximide, cytochalasin B, Y27632, ATRA, TGF-β1, and wortmannin were purchased from Sigma-Aldrich. PD98059, SB203580, and SP600125 were from Calbiochem (San Diego, CA). Uridine ([5,6-3H], 39 Ci/mmol), and d-[1-14C]mannitol were purchased from Amersham Biosciences (Little Chalfont, UK). Cytidine ([5-3H(N)], 22.2 Ci/mmol) and guanosine ([8-3H(N)], 15 Ci/mmol) were from Moravek Biochemicals (Brea, CA). Uridine, cytidine, and guanosine were obtained from Sigma-Aldrich. The monoclonal antibody against the α1-subunit of Na+/K+-ATPase was from The Developmental Studies Hybridoma Bank; anti-human TGF-β1 was from R&D Systems (Berlin, Germany); anti-human phospho-ERK1/2, anti-phospho-p38, anti-human ERK1/2, and anti-human p38 antibodies were from Cell Signaling (Danvers, MA); and anti-HA was from Roche (Basel, Switzerland).
Cell Lines, Culture, and Transfection
The MEC1 cell line was purchased in DSMZ (ACC 497). This cell line was originally obtained from the peripheral blood of a patient with chronic lymphocytic leukemia (CLL). After culturing for more than 4 years, this cell line was shown to be a good model for investigating the biology of CLL (20). These cells were cultured in Iscove's Modified Dulbecco's medium (IMDM, Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mm glutamine, 50 μg/ml penicillin/streptomycin (Invitrogen), and 100 μg/ml normocin (Amaxa, Khöl, Germany) at 37 °C in a humidified atmosphere containing 5% CO2. The HeLa cervical cancer cell line was cultured in DMEM containing a mixture of antibiotics (100 units of penicillin, 0.1 mg/ml streptomycin, and 0.25 μg/ml fungizone) and supplemented with 10% heat-inactivated FBS. Cells were maintained as monolayer cultures and subcultured every 3–4 days.
HeLa cells were transfected with the hCNT3-cHApcDNA3.1 or hCNT3-pEGFP-C1 plasmids, developed in our laboratory (21), using calcium phosphate precipitation. For ATRA and TGF-β1 experiments, HeLa cells were assayed 48 h after transfection, and MEC1 cells 24 h after seeding.
Nucleoside Transport Assay
Nucleoside transport was measured in HeLa cells as previously described (22). The substrate of interest ([3H]uridine, [3H]guanosine, or [3H]cytidine) was added at a final concentration of 1 μm (specific activity, 1 μCi/nmol), except for kinetic analyses, in which the substrate concentration was varied from 0.5 to 500 μm, in the presence of either 137 mm NaCl or 137 mm choline chloride. The uptake medium also contained 5.4 mm KCl, 1.8 mm CaCl2, 1.2 mm MgSO4, and 10 mm HEPES (pH 7.4). Incubation was stopped after 1 min incubation (linear initial velocity conditions) by washing the monolayers three times in 2 ml of a cold buffer composed of 137 mm NaCl and 10 mm Tris (hydroxymethyl) aminomethane-HEPES (pH 7.4). Cells were then dissolved in 1 ml of Triton X-100, and radioactivity measurements were determined.
Nucleoside uptake was measured in MEC1 cells using a rapid filtration method adapted from a technique previously described by our laboratory (23). Cells were washed and resuspended in either a NaCl or a choline chloride buffer, as previously reported. Uptake assays were started by mixing cell suspensions with an equal volume of buffer, supplemented with a radionucleoside at a specific activity of 4000 dpm/pmol. Nucleosides were routinely used at a concentration of 1 μm. When the selected incubation times had elapsed, aliquots were taken and added to ice-cold 0.4-ml needle Eppendorf tubes, containing an upper buffer phase, an intermediate oil layer (dibutylphthalate/bis-3,5-trimethylhexyl) phthalate [3:2, v/v]), and a 10% HClO4/25% glycerol solution at the bottom. The tube was immediately centrifuged (15,000 × g for 60 s), thus enabling cells to be separated from the incubation medium and pelleted into the HClO4 layer. d-[14C]Mannitol (at a specific activity of 4000 dpm/pmol) was included in the incubation medium to assess the amount of extracellular medium trapped in the bottom acid layer. To ensure that pelleted cells were fully recovered for radioactivity counting, tubes were blade cut at the oil layer level, releasing the bottom part into scintillation counting vials. Double counting allowed discrimination between the transported substrate (a tritiated nucleoside) and the extracellular marker (d-[1-14C]mannitol). Aliquots were sampled for protein determination, according to Bradford (Bio-Rad) and for radioactivity measurements. Uridine uptake increased linearly during the first 3 min, thus a 2-min time point was used for all subsequent transport measurements in MEC1 cells.
In the presence of Na+ ENTs and CNTs are functional, although only CNTs require this ion for substrate translocation. Thus, sodium-dependent nucleoside (CNT-related) activity was determined by subtracting uptake rates measured in the choline chloride medium (almost exclusively related to ENT1 and ENT2 activities) from measurements obtained in the sodium chloride medium.
RNA Isolation and Quantitative RT-PCR
Total RNA extraction, RNA retrotranscription, and real-time RT-PCR were performed as previously described (8). Quantitative real-time PCR amplification of ULBP3, TGF-β1, and RhoA were performed with primers and probes from Applied Biosystems (Foster City, CA). Oligonucleotides used for hCNT3 amplification were designed and were as follows: 5′-GCACACTCAAACTGCTCCACC-3′ and 5′-GGGCTCTGTGAAAGTTCAGC-3′. The relative mRNA level of each target transporter gene, normalized to that of β-glucuronidase (GUS) expression level, was expressed in arbitrary PCR units.
Biotinylation of Plasma Membrane Proteins
hCNT3-cHApcDNA3.1-transfected HeLa (HA-hCNT3 HeLa) cells were washed in PBS containing MgCl2 and CaCl2 (1 mm each) and incubated with 500 μm Sulfo-NHS-Biotin (Pierce) in the same buffer for 30 min at 4 °C with gentle shaking. After biotinylation, cells were washed once in PBS and twice with 100 mm glycine to quench the reaction. After an additional wash with PBS supplemented with 2% (v/v) 2-mercaptoethanol, cells were lysed with a buffer containing 50 mm NaPO4, 1% (v/v) Triton X-100, 300 mm NaCl, and protease inhibitors, and centrifuged at 16,000 × g for 30 min at 4 °C. The supernatants, corresponding to total protein extracts (H), were collected and quantified. Equal amounts of protein from each sample (500 μg) were incubated with 30 μl of streptavidin-agarose beads (Sigma-Aldrich) for 90 min with constant rotation at 4 °C. Complexes were centrifuged at 25,000 × g for 5 min at 4 °C, and supernatants (corresponding to intracellular proteins) and pellets (plasma membrane proteins complexed with streptavidin beads) were obtained. Beads were washed twice in PBS/500 mm NaCl and once in PBS alone. The precipitated complexes were resuspended in sample buffer supplemented with 2-mercaptoethanol and heated to 37 °C for 30 min. After centrifugation at 14,000 × g for 2 min, the supernatants were collected for Western blot analysis.
Western Blot Analysis
Mec1 and HeLa cells (1 × 106) were cultured at 37 °C in medium containing either ATRA or TGF-β1. At specific time points, cells were collected and pelleted. Pellets were washed with cold saline and immediately resuspended in loading buffer (50 mm Tris-HCl pH 7.5, 150 mm NaCl, 1% Nonidet P40, 5 mm sodium pyrophosphate, 50 mm NaF, 1 mm sodium orthovanadate, protease inhibitor mixture (Roche)). Extracts were then centrifuged for 5 min at 13,200 × g and samples of the supernatants containing 20 μg of protein were either boiled for 5 min (ERK and p38) or heated at 37 °C for 30 min (hCNT3). Samples were then separated by SDS-PAGE on standard 12% gels and transferred to Immobilon-P membranes (Millipore). Membranes were then incubated with specific primary antibodies for 1 h, washed with PBS-Tween, and then incubated with horseradish peroxidase-conjugated secondary antibodies. Immunoreactive bands were detected by enhanced chemiluminescence (ECL, Amersham Biosciences).
RhoA Suppression by RNA Interference
Two short interfering RNAs (siRNAs) directed against human RhoA (S758 and S759 from Applied Biosystems-Ambion) were tested at different concentrations by RT-PCR to determine their efficiency in suppressing RhoA expression. HeLa cells grown in 60-mm Petri dishes. Cells were transfected with hCNT3-cHApcDNA3.1 plasmid using calcium phosphate precipitation, and 14 h later the same cells were transfected with siRNAs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). As a control, hCNT3-cHApcDNA3.1- HeLa cells were transfected in parallel with a silencer negative control (AM4613 from Applied Biosystems-Ambion). RNA was extracted and transport assays were performed 24 h later.
Visualization of GFP-hCNT3 Protein
HeLa cells were transfected as explained previously and treated either with ATRA or TGF-β1 for the indicated times. Cells were washed in PBS, Ca2+-Mg2+ and fixed with 3% paraformaldehyde (PFA), 0.06 m sucrose. Images were obtained using a Leica TCS SL laser scanning confocal spectral microscope (Leica Microsystems) with argon and HeNe lasers attached to Leica DMIRE2 inverted microscope lasers. Images were captured by excitation at 488 nm and emission at 508 nm.
Statistical Analysis
Data were expressed as means ± S.E. The significance of differences between two groups was measured using Student's t test. A p value < 0.05 was considered statistically significant. Statistical significance (***, p < 0.001; **, p < 0.01; *, p < 0.05) denotes significant difference between treatments.
RESULTS
ATRA Increases Sodium-dependent Nucleoside Transport Activity in MEC1 Cells
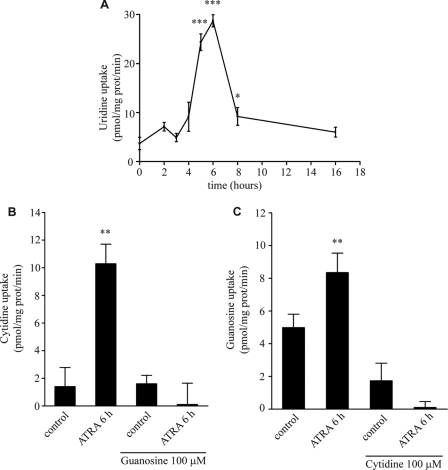
The effect of ATRA on nucleoside transport activity was evaluated in the CLL lymphocyte-like MEC1 cell line. We monitored the time course of sodium-dependent nucleoside activity in MEC1 cells after incubation with a non-toxic ATRA concentration (10 μm) (Fig. 1A). As reported for primary CLL cells, unstimulated MEC1 cells exhibited predominantly an equilibrative (ENT-related) nucleoside transport activity, and poor sodium-dependent transport activity (8). However, ATRA increased this sodium-dependent nucleoside transport activity without affecting equilibrative transport. Peaks in sodium transport activity were recorded 5–6 h after treatment with ATRA.
FIGURE 1.
Effect of ATRA on sodium-related nucleoside transport activity. A, time course of sodium-dependent [3H]uridine uptake was monitored in MEC1 cells cultured with ATRA (10 μm) for the indicated times. Sodium-dependent cross-inhibition of [3H]cytidine and [3H]guanosine transport was monitored in MEC1 cells cultured with ATRA (10 μm) for 6 h. B, inhibition of [3H]cytidine transport with guanosine (100 μm). C, inhibition of [3H]guanosine transport with cytidine (100 μm). Data are expressed as means ± S.E. of quadruplicate measurements from three independent cultures. Statistical significance (*, p < 0.05; **, p < 0.01; ***, p < 0.001) denotes significant difference relative to ATRA-untreated cells.
We next characterized the sodium-dependent nucleoside transport activity induced by ATRA using either [3H]guanosine (transported by hCNT2 and hCNT3) or [3H]cytidine (transported by hCNT1 and hCNT3) as the substrate. The sodium-coupled uptake of both substrates was increased after ATRA treatment, indicating hCNT3 involvement (Fig. 1, B and C). The occurrence of an hCNT3-type transport activity was further confirmed by cis-inhibition studies in which [3H]cytidine and [3H]guanosine uptake were cross-inhibited by each other's substrate. These data are also consistent with hCNT3 being the only sodium-coupled transporter involved in the ATRA-induced activity.
Characterization of ATRA Effects on hCNT3 Transport Activity
We next determined whether the increase in hCNT3-related activity induced by ATRA treatment was caused by transcriptional induction of the SLC28A3 gene. To this end, we evaluated the ability of ATRA to modulate hCNT3 mRNA expression, using quantitative RT-PCR. No significant differences were found between untreated and ATRA-treated MEC1 cells during the time period in which hCNT3 activity was up-regulated (supplemental Fig. S1).
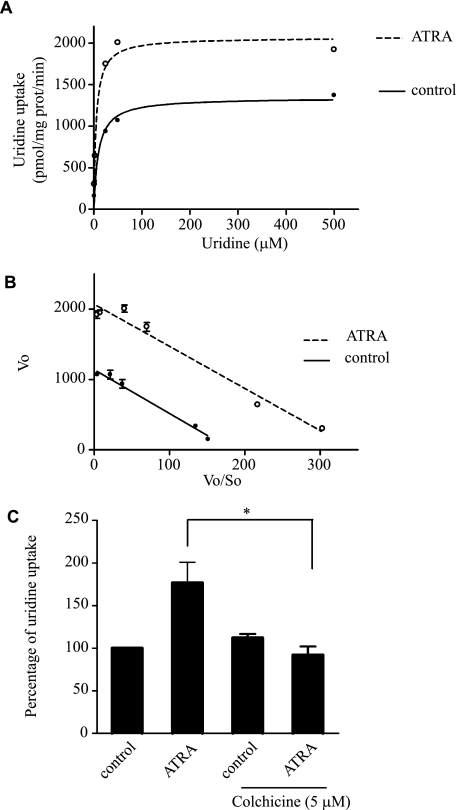
Given that the increase in hCNT3 activity was not associated with induction of hCNT3 expression, we sought to determine if this effect was due to an increase in hCNT3 protein at the plasma membrane, which would result in a Vmax effect, or to an increase in the affinity of hCNT3 for its substrates, which would be consistent with a Km effect. To accurately measure the effect of ATRA on hCNT3-associated kinetic parameters, we used HeLa cells transfected with an hCNT3-HA-encoding cDNA (HA-hCNT3-HeLa) in which ATRA induction of hCNT3-related activity was highly reproducible. As shown in Fig. 2, A and B, ATRA induced nearly a 2-fold increase in the Vmax (Vmax ATRA = 2069 ± 80 pmol/mg prot/min versus Vmax control = 1139 ± 31 pmol/mg prot/min; n = 3, p < 0.05), without significantly affecting the apparent Km values for uridine (6.0 ± 0.5 μm and 6.2 ± 0.36 μm for control and ATRA-treated cells, respectively; n = 3). These results are consistent with an increase in the amount of hCNT3 protein at the plasma membrane. To verify this, we measured hCNT3-related activity in MEC1 cells in the presence of colchicine, which inhibits microtubule polymerization by binding to tubulin. This agent significantly attenuated ATRA-induced hCNT3 activation (Fig. 2C). Furthermore, to discard any possible effect of colchicine on ATRA sensing, we evaluated the expression of ULBP3 (known to be up-regulated in CLL after ATRA treatment) either in the presence or in the absence of colchicine. We found that ATRA significantly increased ULBP3 mRNA after 90 min exposure up to 3.5-fold (supplemental Fig. S2A), and this up-regulation was not affected by the presence of colchicine (supplemental Fig. S2B). Based upon this observation we think that colchicine is not disrupting ATRA sensing. Taken together, these data indicate that the increase in hCNT3 activity observed with ATRA treatment is most consistent with an increase in hCNT3 protein at the plasma membrane.
FIGURE 2.
Characterization of ATRA effects on hCNT3 transport activity. Kinetics were characterized by evaluating the sodium-dependent [3H]uridine uptake over a range of substrate concentrations (1–500 μm). A, hCNT3-mediated uridine uptake conformed to Michaelis-Menten kinetics for HeLa with or without ATRA treatment (10 μm, 6 h). B, Eadie-Hofstee regressions were used to estimate the kinetic parameters, Km and Vmax, for transport. C, sodium-dependent [3H]uridine uptake was monitored in MEC1 cells incubated with colchicine in the presence or absence of ATRA. Results are based on quadruplicate measurements from three independent experiments. Data (means ± S.E.) are shown as the percentage increase in uridine uptake over control (non-treated cells) values (*, p < 0.05).
Protein Synthesis Is Required for hCNT3 Induction by ATRA
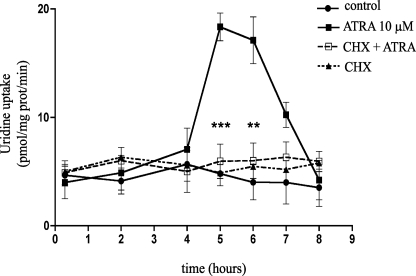
Changes in the amount of protein at the plasma membrane might reflect a relatively rapid trafficking phenomenon. However, as shown above, the increase in hCNT3 transport activity induced by ATRA was delayed (i.e. peaked after 5–6 h), but apparently did not involve a change in the hCNT3-encoding gene transcription rate. We thus considered the possibility that ATRA-induced up-regulation of hCNT3 requires the synthesis of a mediator molecule. To evaluate this possibility, we analyzed the effects of cycloheximide, an inhibitor of protein biosynthesis, on the ATRA-induced up-regulation of hCNT3 activity. Incubation with cycloheximide abrogated the increase in ATRA-induced hCNT3 activity (Fig. 3); cycloheximide alone had no effect on basal uptake. Thus, protein synthesis is required for the induction of hCNT3 activity by ATRA.
FIGURE 3.
Effect of protein synthesis inhibition on ATRA-induced hCNT3 transport activity. The time course of sodium-dependent [3H]uridine transport was measured in cycloheximide (10 μg/ml)-treated MEC1 cells in the presence or absence of ATRA (10 μm, 6 h). Results are expressed as the mean ± S.E. of quadruplicate estimations from three independent determinations. Statistical significance (***, p < 0.001; **, p < 0.01) denotes significant difference relative to ATRA-treated cells.
TGF-β1 Is Involved in hCNT3 Induction by ATRA
Many effects of retinoids have been linked to the production of TGF-β1 and it is known that CLL cells produce TGF-β1 (24, 25). Accordingly, we investigated TGF-β1 involvement by evaluating the effects of modulating TGF-β1 levels on the ATRA-induced increase in hCNT3 activity and changes in TGF-β1 expression with ATRA treatment.
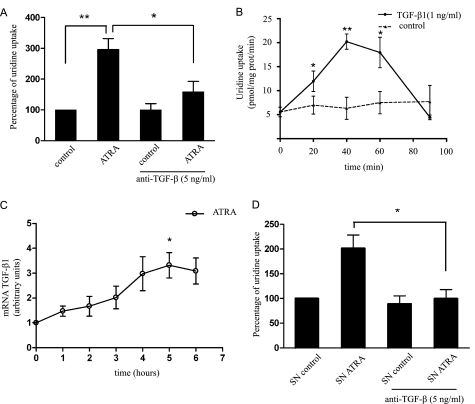
Sodium-dependent [3H]uridine transport measurements performed in the presence of a neutralizing antibody against TGF-β1 showed that inhibition of TGF-β1 significantly decreased ATRA-induced hCNT3 activity (Fig. 4A) without affecting hENT-related transport. We then analyzed the effects of exogenous TGF-β1 addition on hCNT3 activity in a time course assay. TGF-β1 increased hCNT3 activity in MEC1 cells (Fig. 4B); peaks in hCNT3 activity were recorded 40–60 min after TGF-β1 addition, with maximum activation occurring after 40 min.
FIGURE 4.
Role of TGF-β1 in ATRA-induced hCNT3 activity. A, sodium-dependent [3H]uridine transport was measured in MEC1 cells incubated with or without ATRA (10 μm) for 6 h in the presence (or absence) of a neutralizing anti- TGF-β1 antibody. Values are expressed as the percentage increase in uridine uptake over control (non-treated cells) values. B, sodium-dependent [3H]uridine transport was measured in MEC1 cells incubated with (or without) exogenous TGF-β1 (1 ng/ml) for the indicated times. Results are expressed as the mean ± S.E. of quadruplicate estimations from five independent determinations. C, TGF-β1 mRNA levels in MEC1 cells were determined by quantitative RT-PCR and expressed relative to an endogenous reference gene (β-glucuronidase) as described under “Experimental Procedures.” Relative CT values, in arbitrary units, are shown. D, sodium-dependent [3H]uridine transport was evaluated in MEC1 cells cultured for 40 min in the presence of supernatant from untreated MEC1 (SN control) cells or cells treated with ATRA (10 μm) for 5.5 h (SN ATRA). In the indicated cases, a neutralizing antibody against TGF-β1 was added. Results are expressed as the mean ± S.E. of quadruplicate estimations from three independent determinations. Data are shown as the percentage increase in uridine uptake over control (non-treated cells) values. Statistical significance (*, p < 0.05 and **, p < 0.01) denotes significant difference relative to control cells.
Next we analyzed the effects of ATRA on TGF-β1 expression and found that ATRA did increase TGF-β1 mRNA levels (Fig. 4C). We also measured TGF-β1 release by collecting supernatants from cells incubated with ATRA for 5.5 h and then culturing naive cells with these supernatants for 40 min in the presence or absence of TGF-β1 neutralizing antibodies. As shown in Fig. 4D, the supernatants from ATRA-treated cells induced an increase in sodium-dependent activity in a manner similar to that of TGF-β1; co-administration of the neutralizing antibody abrogated this increase in hCNT3 activity. Collectively, these data show that ATRA induces the expression and secretion of TGF-β1, and further demonstrate that this cytokine is responsible, at least in part, for the increase in hCNT3-related activity induced by ATRA.
ATRA and TGF-β1 Up-regulate hCNT3-related Transport Activity by Modulating the Subcellular Localization of hCNT3 Protein
Sodium-dependent [3H]uridine transport activity in MEC1 cells was measured after treatment with TGF-β1 as evaluated for ATRA, in the presence of colchicine or cytochalasin B, which shortens actin filaments by blocking monomer addition at the fast-growing end of polymers. Both agents significantly inhibited TGF-β1-induced transport activity (supplemental Fig. S3, A and B for colchicine and cytochalasin B, respectively), indicating that hCNT3 up-regulation involves changes in cytoskeleton dynamics that could lead to an increase in hCNT3 protein levels at the plasma membrane.
To confirm our hypothesis that ATRA and TGF-β1 alter the localization of hCNT3, we used HA-hCNT3-HeLa cells as a model system. This approach not only yields reproducible results, as noted above but it also allows us to accurately estimate exogenously expressed hCNT3 at the plasma membrane, something that was not possible for endogenous hCNT3 using available anti-hCNT3 antibodies. These experiments were run in parallel with nucleoside transport activity experiments. As expected, as HeLa do not express hCNT3, these cells did not show endogenous hCNT3-related transport activity. Transiently transfected cells expressing HA-hCNT3 showed a basal transport activity of 564 ± 95 pmol/mg prot/min. After either ATRA (10 μm, 6 h) or TGF-β1 (1 ng/ml, 40 min) treatment, HA-hCNT3 activity increased up to 998 ± 10 5 and 1218.2 ± 158 pmol/mg prot/min, respectively.
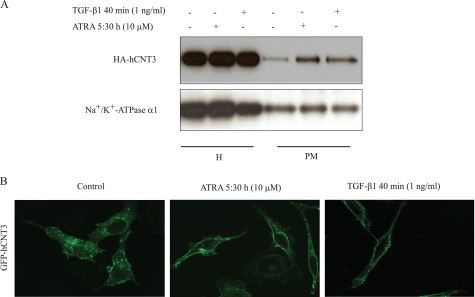
To address the study of subcellular localization of hCNT3, HA-hCNT3-HeLa cells were biotinylated and precipitated with streptavidin; hCNT3 was then detected in streptavidin precipitates by Western blotting using an anti-HA antibody. As shown in Fig. 5A, treatment with ATRA or TGF-β1 significantly enriched HA-hCNT3 in the plasma membrane fraction, without affecting the amount of protein in total cell extracts (H).
FIGURE 5.
Effect of ATRA and TGF-β1 on hCNT3 subcellular localization. A, HeLa cells were transiently transfected with HA-hCNT3 and after 48 h cell extracts (H) and plasma membrane fractions (PM) from HeLa cells incubated in the presence or absence of ATRA (10 μm, 5:30 h) or TGF-β1 (1 ng/ml, 40 min) were prepared and analyzed by Western blotting using an anti-HA-hCNT3 antibody, as described under “Experimental Procedures.” A representative Western blot from three independent experiments is shown. The α1 subunit of Na+/K+-ATPase, a specific plasma membrane protein, was also evaluated as a control of plasma membrane protein. B, HeLa cells were grown on coverslips and transfected with GFP-hCNT3. After 48 h, cells were treated either with ATRA for 5:30 h or with TGF-β1 for 40 min. At that time cells were fixed and analyzed by confocal microscopy. A representative group of cells from three independent experiments is shown.
Densitometric analysis of the HA-hCNT3 protein amounts (relative to the α1 Na+/K+-ATPase subunit) showed an increase in plasma membrane abundance of 2.53 ± 0.31- and 3.4 ± 0.48-fold after ATRA and TGF-β1 treatment, respectively, which correlates to some extent with the above reported changes in hCNT3 biological activity. Evaluation of the α1 subunit of Na+/K+-ATPase, a known plasma membrane protein confirmed that the abundance of this pump subunit remained unaltered after treatments.
The observation that ATRA did not increase hCNT3 mRNA or hCNT3 protein levels, but did increase plasma membrane hCNT3 localization and related activity is consistent with the hypothesis that this retinoid increases hCNT3 activity through a post-translational mechanism that involves trafficking to the plasma membrane. These results indicate that treatment with ATRA or TGF-β1 promotes the trafficking of cytoplasmic vesicles containing pre-existing hCNT3 to the plasma membrane. Additional evidence of hCNT3 trafficking comes from confocal microscopy images (Fig. 5B), showing re-localization of GFP-hCNT3 from intracellular compartments to the plasma membrane either after ATRA or TGF-β1 treatment.
Signaling Pathways Involved in the Induction of hCNT3 Activity by ATRA and TGF-β1
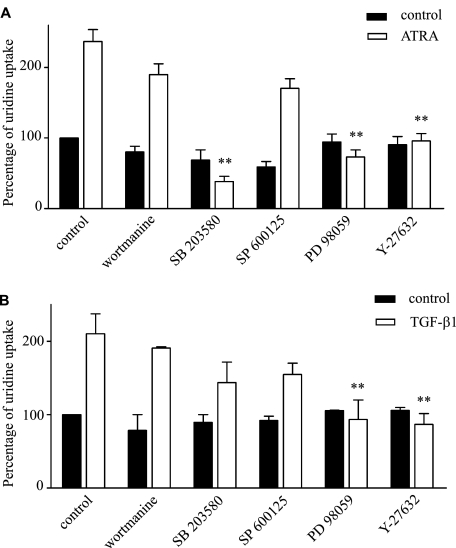
The rapid effects of TGF-β1 on hCNT3-related activity indicate that new gene expression is not required. Given that mitogen-activated protein kinases (MAPKs) are known to be involved in both ATRA and TGF-β1responses (26–29), we next sought to determine whether p38, ERK1/2, and/or JNK have a role in ATRA- and/or TGF-β1-induced hCNT3 activation. Using SB203580, SP600125, and PD98059 to inhibit p38, JNK and ERK1/2, respectively, we found that ATRA-induced hCNT3-related activity was abolished by both SB203580 and PD98059 (Fig. 6A), whereas TGF-β1-dependent hCNT3 induction was inhibited only by PD98059 (Fig. 6B). In each case, hENT-related transport activity was not affected. This indicates that ATRA induces p38 (and ERK1/2) activation, which acts upstream of TGF-β1, and that the effect of TGF-β1 depends only on the activation of ERK1/2.
FIGURE 6.
Signaling pathways involved in the induction of hCNT3 activity by ATRA and TGF-β1. Sodium-dependent [3H]uridine uptake was assayed in MEC1 cells pre-incubated for 15 min with the inhibitors, wortmannin (50 nm), SB203580 (5 μm), SP600125 (10 μm), PD98059 (5 μm), or Y-27632 (5 μm), and then incubated with (A) ATRA (10 μm, 6 h) or (B) TGF-β1 (1 ng/ml, 40 min). Results are based on quadruplicate measurements from three independent experiments. Data (means ± S.E.) are shown as the percentage increase in uridine uptake over control (non-treated cells) values. Statistical significance (**, p < 0.01) denotes significant difference relative to (A) ATRA or (B) TGF-β1-treated cells.
Previous reports have shown that ATRA and TGF-β1 can also activate phosphatidylinositol-3-kinase (PI3K) (28, 30, 31), and depending on the cell line, TGF-β1 can also rapidly activate Rho-like GTPases (28, 32). To determine whether the increased hCNT3 activity induced by ATRA and TGF-β1 was dependent on PI3K activation, we preincubated cells with wortmannin, a PI3K inhibitor. Y-27632, an inhibitor of the RhoA effector kinase, p160ROCK, was used to evaluate the involvement of Rho GTPases. We found that both ATRA and TGF-β1 depended on p160ROCK for the induction of hCNT3 activity, but neither required PI3K activity (Fig. 6, A and B).
Involvement of p38 and ERK1/2 in ATRA- and TGF-β1-induced hCNT3 Activity in MEC1 Cells
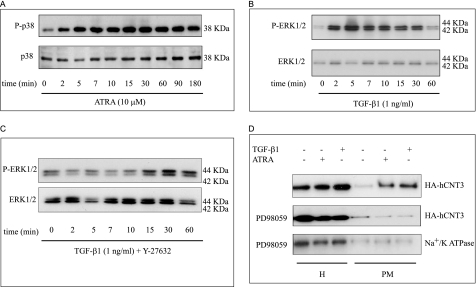
To further investigate the involvement of MAPKs in the action of ATRA and TGF-β1, we evaluated p38 and ERK1/2 activation in MEC1 cells stimulated with ATRA or TGF-β1 by assaying protein extracts for phosphorylated p38 and ERK1/2 by Western blotting. Fig. 7A shows that ATRA rapidly stimulated p38 phosphorylation; by contrast, ATRA did not induce ERK1/2 phosphorylation (not shown). TGF-β1 induced ERK1 and ERK2 phosphorylation within 2–30 min without affecting the total amount of unphosphorylated ERK1 or ERK2 (Fig. 7B). Moreover, the p160ROCK inhibitor Y-27632 abolished TGF-β1-induced ERK1/2 phosphorylation (Fig. 7C). For both, ERK1/2 and p38, quantification after normalizing versus total protein amounts revealed a peak of phosphorylation at 5 min after treatments of about 8–10 fold above non-treated cell values.
FIGURE 7.
Involvement of p38 and ERK1/2 pathways in ATRA- and TGF-β1-induced hCNT3 activity. MEC1 cells were cultured with or without TGF-β1 (1 ng/ml) or ATRA (10 μm) and analyzed by Western blotting for (A) p38 phosphorylation induced by ATRA and (B, C) ERK-phosphorylation induced by TGF-β1, in the absence or presence of Y-27632, respectively. Reactions were stopped at the indicated times by adding cold PBS. Western blotting was performed as described under “Experimental Procedures” using antibodies against phospho-p38 (P-p38) or phospho-ERK (P-ERK). The same membrane was probed with anti-p38 (p38) and anti-ERK (ERK) antibodies to compare the total amount of protein in each sample. D, HeLa cells were transiently transfected with HA-hCNT3. Forty-eight hours later, cells were preincubated with the ERK1/2 inhibitor, PD989059 (5 μm) before adding TGF-β1 (1 ng/ml, 40 min) or ATRA (10 μm, 5:30 h). HA-hCNT3 protein expression total cell extracts (H) and in biotinylated plasma membrane (PM) fractions was analyzed by Western blotting. The α1 subunit of Na+/K+-ATPase was also evaluated as a control. A representative Western blot from three independent experiments is shown.
To further analyze the role of p38 activation in the action of ATRA, we evaluated the levels of TGF-β1 mRNA by real-time RT-PCR in cells preincubated with PD98059, SB203580, or Y-27632 prior to the addition of ATRA. As we have already demonstrated, ATRA increased the levels of TGF-β1 mRNA (∼2.5-fold at 4 h), and this effect was abrogated only by inhibition of p38 activation (supplemental Fig. S4). Taken together, these data indicate that p38 is involved in mediating the induction of TGF-β1 mRNA transcription by ATRA and implicate RhoA and ERK1/2 in the response to TGF-β1.
We next verified the involvement of ERK1/2 in ATRA- and TGF-β1-induced increases in hCNT3 activity by incubating HA-hCNT3-HeLa cells in the absence or presence of PD98059, and evaluating the subcellular localization of hCNT3. hCNT3 was detected by Western blotting in streptavidin precipitates, as described above, using an antibody against HA. As shown in Fig. 7D, inhibition of ERK1/2 abrogated the plasma membrane localization of hCNT3 induced by ATRA or TGF-β1 without altering the amount of the Na+/K+-ATPase α1 subunit.
Involvement of RhoA in the Induction of hCNT3 Activity by TGF-β1
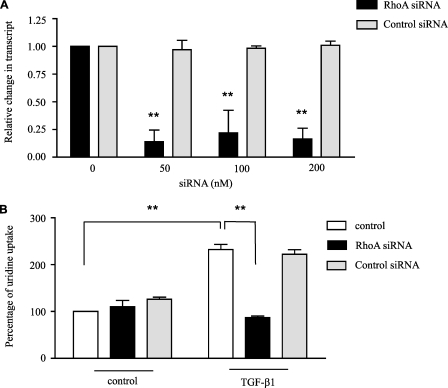
Because inhibiting p160ROCK blocked the induction of hCNT3 activity by TGF-β1, we next studied the role of RhoA on the TGF-β1 effect using a siRNA approach. These experiments were performed in HeLa cells because the ATRA and TGF-β1 effects can be reproduced in these cells which are easier to transfect than the lymphoid cell line, thus making them more suitable for gene silencing. We first validated the conditions for silencing the RhoA gene by transfecting HA-hCNT3-HeLa cells with RhoA-siRNA and evaluating changes in the RhoA transcript level. As a negative control, we used a nonspecific siRNA molecule. As shown in Fig. 8A, transient transfection with the specific RhoA-siRNA optimally suppressed RhoA expression at a concentration of 50 nm; however, the control siRNA had no effect on RhoA expression. We then measured hCNT3-dependent uridine uptake under RhoA-knockdown conditions. As shown in Fig. 8B, hCNT3-related uptake was decreased after siRNA-mediated repression of RhoA in cells exposed to TGF-β1 for 40 min, whereas the control siRNA had no significant effects in the presence or absence of TGF-β1. Taken together, these results indicate that RhoA is required for the observed increase in hCNT3-mediated uridine transport following TGF-β1 treatment.
FIGURE 8.
Involvement of RhoA in the induction of hCNT3 activity by TGF-β1. HeLa cells were transiently transfected with HA-hCNT3 and 14 h later with RhoA siRNA (S758) or control siRNA (AM4613) as described under “Experimental Procedures.” After additional 24 h, mRNA was extracted, quantitative RT-PCR was performed (A) and sodium-dependent [3H]uridine uptake was monitored after culturing cells in the presence or absence of TGF-β1 (1 ng/ml) for 40 min (B). Results are based on quadruplicate measurements from three independent experiments. Data (means ± S.E.) are shown as the percentage increase in uridine uptake over control (non-treated cells) values. Statistical significance denotes significant difference relative to control cells (**, p < 0.01).
DISCUSSION
In this study, we demonstrate that ATRA induces hCNT3 trafficking to the plasma membrane, causing an increase in hCNT3 transporter activity in the CLL lymphocyte-like cell line MEC1. This effect is relatively rapid (i.e. within 5–6 h of treatment) and is not associated with changes in hCNT3-encoding gene transcription. Importantly, the effects of ATRA on hCNT3 are dependent on protein synthesis, and mediated, at least in part, by TGF-β1, which is transcriptionally induced by ATRA treatment.
Reports of post-translational regulation of NT proteins are relatively uncommon. The equilibrative transporter protein hENT1 is rapidly modulated by a PKC-dependent mechanism that likely involves activation of transporters already present at the plasma membrane (9, 33). Regarding CNTs, we have previously shown that CNT2 is under purinergic control in liver cells (22), and its insertion into the plasma membrane is also modulated by bile acids (34). Although previous evidence suggests that ATRA can modulate nucleoside transport activity in the NB4 promyelocytic cell line (16), the mechanism underlying this effect has not been investigated. Thus, to the best of our knowledge, the current study is the first report of post-translational regulation of hCNT3 activity that involves changes in hCNT3 subcellular localization.
Paradoxically, high levels of hCNT3 mRNA and cytosolic hCNT3 protein are associated with clinical resistance to Flu in CLL cells, apparently caused by the absence of hCNT3 at the plasma membrane (11). Similarly, a recent study showed a strong relationship between immunohistochemistry staining of hCNT3 protein and clinical resistance to Flu therapy in CLL patients (10). hCNT3 is located in intracellular structures (10, 11) and according to our observations (not shown) present in secretory vesicles. Our results demonstrate that ATRA can stimulate hCNT3 translocation to the plasma membrane.
Our findings also showed that the induction of hCNT3-related activity in MEC1 cells by ATRA requires activation of p38, which also seems to be involved in the transcriptional induction of TGF-β1. Activation of the p38 pathway by ATRA is consistent with other reports in different cell types (27, 29, 35, 36). It has also been reported that the effects of ATRA in different models might be mediated by increased TGF-β1 levels. For instance, in normal breast cells, ATRA (1 μm) increased TGF-β activity 2- and 5-fold after 3 and 5 days of treatment, respectively (25), and in HL-60 cells, the induction of TGF-β has been shown to be necessary for retinoid-induced growth inhibition (37). Consistent with these reports, our results showed that ATRA-induced hCNT3-related activity was mediated by up-regulation of TGF-β1. In fact, hCNT3 induction was inhibited by cycloheximide and by co-culture with a TGF-neutralizing antibody, both in MEC1 cells exposed to ATRA or to supernatants from ATRA-treated cells.
In agreement with a transcription-independent TGF-β1-mediated effect, our data indicate that TGF-β1 action on hCNT3 is dependent on the rapid activation of RhoA and ERK1/2, consistent with Smad-independent activation of a type II or type I receptor kinase. Previous in vitro studies have shown that TGF-β1 rapidly activates the ERK, JNK, and/or p38 MAPK kinase pathways (38, 39), as well as PI3K. In addition, TGF-β1 can induce activation of the Rho GTPases Rac1, Cdc42, and RhoA (31, 32). RhoA, in particular, is rapidly activated after TGF-β1 stimulation in the human breast adenocarcinoma cell lines MCF-7 and MDA-MB-468 (40) and in normal murine mammary gland (NMuMG) cells (41). Small GTPases also mediate TGF-β1-induced changes in cytoskeletal organization. For example, activation of RhoA and its effector kinase p160ROCK is required for rapid membrane ruffling and lamellipodia formation in response to TGF-β (42).
In our model, Y-27632 abrogated the phosphorylation of ERK1/2, indicating that RhoA activation occurs upstream of ERK1/2 and that it is involved in activating ERK1/2. We also verified that ERK1/2 phosphorylation was a key factor in both ATRA- and TGF-β1-induced trafficking of hCNT3 to the plasma membrane.
Several Rho family members are localized to vesicular compartments, and increasing evidence suggests that they have important roles in the trafficking of vesicles in both the endocytic and exocytic pathways. In particular, RhoA and related small GTPases have been shown to affect various membrane trafficking steps (43, 44). Rho proteins may also affect membrane trafficking by altering the phosphatidylinositide composition of membrane compartments, or through interactions with microtubules and microfilaments. Several reports have described a role for RhoA in the process of plasma membrane docking and fusion of vesicles, showing that RhoA is involved in exocytosis of secretory granules in mast cells (45, 46) and translocation of Na+/K+-ATPase in renal epithelial cells (47). Furthermore, RhoA promotes the trafficking of the epithelial Na+ channel (ENaC) to the plasma membrane, likely through effects on the cytoskeleton (48). Similarly, agonist activation of adrenergic receptors increases the abundance of Na+/K+-ATPase at the plasma membrane in a RhoA-dependent manner (49). Our finding that RhoA is involved in ATRA- and TGF-β1-induced trafficking of hCNT3 to the plasma membrane is consistent with these reports.
RhoA-mediated recruitment of hCNT3 at plasma membrane and subsequent manifestation of hCNT3 activity might also be linked to an increase in actin polymerization, as demonstrated for utrophin (50). Our finding that treatment with cytochalasin B blocked the induction of hCNT3-related activity indicates an involvement of F-actin cytoskeletal assembly, and would thus be consistent with such a mechanism.
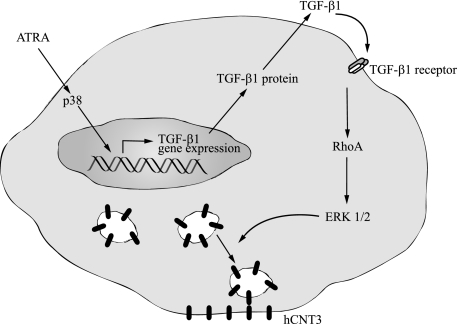
In conclusion, based on the above results, we propose the model summarized in Fig. 9 in which ATRA induces TGF-β1 transcription and secretion through a mechanism that is dependent on p38 activation. TGF-β1 signals through RhoA to activate ERK1/2, resulting in hCNT3 trafficking to the plasma membrane and an increase in hCNT3-related nucleoside transporter activity. Importantly, our results point to a possible role for ATRA in the treatment of CLL, highlighting a potential therapeutic use of retinoic acid in increasing the plasma membrane localization of the hCNT3 protein, which might in turn lead to an increase in fludarabine entrance to cells and enhanced cytotoxicity. In fact preliminary data show that ATRA and the hCNT3 substrate fludarabine, promote a synergistic effect on the programmed cell death of primary CLL cells.4 Moreover, the finding that this effect can be reproduced in non-lymphoid cells transiently expressing hCNT3 supports the view that this mechanism may be more ubiquitous than expected, suggesting that ATRA might be similarly effective in increasing nucleoside-based chemotherapy in solid tumors. Further analysis of the therapeutic potential of combination therapies involving ATRA and nucleoside-derived drugs are warranted.
FIGURE 9.
Proposed mechanism by which ATRA increases hCNT3-related activity. A model for the post-translational activation of hCNT3 transporter function induced by ATRA is shown. ATRA treatment increases the phosphorylation of p38. p38 activation leads to increased TGF-β1 mRNA expression and protein secretion, which induces the activation of the small GTPase, RhoA. RhoA, in turn, activates its effector kinase, p160ROCK, which is required for the activation of the MAPK, ERK1/2. Activation of ERK1/2 is involved in the rapid increase in the abundance of hCNT3 at the plasma membrane, which is ultimately responsible for the observed increase in hCNT3 concentrative transport activity.
Supplementary Material
Acknowledgments
We thank Neus Abella and Maria Calvo from the Confocal Microscopy Facility of Serveis Cientificotècnics (Universitat de Barcelona-IDIBAPS) for support and advice with confocal techniques. This laboratory belongs to the Biomedical Research Institute Network-Liver and Gastrointestinal Diseases (CIBER EHD). CIBER is an initiative of the Instituto de Salud Carlos III (Ministerio de Ciencia e Innovación). Our group is a member of the Network for Cooperative Research on Membrane Transport Proteins (REIT), co-funded by the Ministerio de Educacion y Ciencia, Spain, and the European Regional Development Fund (ERDF) (Grant BFU2007-30688-E/BFI).
This work was supported by Grants SAF2008-00577 (Ministerio de Ciencia e Innovación) and 2005SGR00315 (Generalitat de Catalunya) (to M. P.-A.). This work was also supported by the Acción Transversal en Cáncer (CIBER-Instituto de Salud Carlos III) and by Juan de la Cierva (to P. F.-C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
P. Fernández-Calotti and M. Pastor-Anglada, unpublished observations.
- NT
- nucleoside-specific membrane transporter
- hCNT
- human concentrative nucleoside transporter
- ATRA
- all-trans-retinoic acid
- TGF
- transforming growth factor
- CLL
- chronic lymphocytic leukemia
- PBS
- phosphate-buffered saline
- PI3K
- phosphatidylinositol-3-kinase
- HA
- hemagglutinin
- Flu
- fludarabine
- APL
- acute promyelocytic leukemia.
REFERENCES
- 1.Pastor-Anglada M., Casado F. J., Valdés R., Mata J., García-Manteiga J., Molina M. (2001) Mol. Membr. Biol. 18, 81–85 [DOI] [PubMed] [Google Scholar]
- 2.Baldwin S. A., Beal P. R., Yao S. Y., King A. E., Cass C. E., Young J. D. (2004) Pflugers Arch. 447, 735–743 [DOI] [PubMed] [Google Scholar]
- 3.Chiorazzi N., Rai K. R., Ferrarini M. (2005) N. Engl. J. Med. 352, 804–815 [DOI] [PubMed] [Google Scholar]
- 4.Gamberale R., Geffner J., Arrosagaray G., Scolnik M., Salamone G., Trevani A., Vermeulen M., Giordano M. (2001) Leukemia 15, 1860–1867 [DOI] [PubMed] [Google Scholar]
- 5.Munk Pedersen I., Reed J. (2004) Leuk. Lymphoma 45, 2365–2372 [DOI] [PubMed] [Google Scholar]
- 6.Elter T., Hallek M., Engert A. (2006) Expert Opin. Pharmacother. 7, 1641–1651 [DOI] [PubMed] [Google Scholar]
- 7.Galmarini C. M., Mackey J. R., Dumontet C. (2001) Leukemia 15, 875–890 [DOI] [PubMed] [Google Scholar]
- 8.Molina-Arcas M., Bellosillo B., Casado F. J., Montserrat E., Gil J., Colomer D., Pastor-Anglada M. (2003) Blood 101, 2328–2334 [DOI] [PubMed] [Google Scholar]
- 9.Fernández Calotti P., Galmarini C. M., Cañones C., Gamberale R., Saénz D., Avalos J. S., Chianelli M., Rosenstein R., Giordano M. (2008) Biochem. Pharmacol. 75, 857–865 [DOI] [PubMed] [Google Scholar]
- 10.Tsang R. Y., Santos C., Ghosh S., Dabbagh L., King K., Young J., Cass C. E., Mackey J. R., Lai R. (2008) Modern Pathology 21, 1387–1393 [DOI] [PubMed] [Google Scholar]
- 11.Mackey J. R., Galmarini C. M., Graham K. A., Joy A. A., Delmer A., Dabbagh L., Glubrecht D., Jewell L. D., Lai R., Lang T., Hanson J., Young J. D., Merle-Béral H., Binet J. L., Cass C. E., Dumontet C. (2005) Blood 105, 767–774 [DOI] [PubMed] [Google Scholar]
- 12.Bennett J. M., Catovsky D., Daniel M. T., Flandrin G., Galton D. A., Gralnick H. R., Sultan C. (1976) Br. J. Haematol. 33, 451–458 [DOI] [PubMed] [Google Scholar]
- 13.De Thé H., Lavau C., Marchio A., Chomienne C., Degos L., Dejean A. (1991) Cell 66, 675–684 [DOI] [PubMed] [Google Scholar]
- 14.Chomienne C., Ballerini P., Balitrand N., Daniel M. T., Fenaux P., Castaigne S., Degos L. (1990) Blood 76, 1710–1717 [PubMed] [Google Scholar]
- 15.Warrell R. P., Jr., Frankel S. R., Miller W. H., Jr., Scheinberg D. A., Itri L. M., Hittelman W. N., Vyas R., Andreeff M., Tafuri A., Jabukowski A. (1991) N. Engl. J. Med. 324, 1385–1393 [DOI] [PubMed] [Google Scholar]
- 16.Flanagan S. A., Meckling K. A. (2007) Leuk. Res. 31, 955–968 [DOI] [PubMed] [Google Scholar]
- 17.Flanagan S. A., Meckling-Gill K. A. (1997) J. Biol. Chem. 272, 18026–18032 [DOI] [PubMed] [Google Scholar]
- 18.Flanagan S. A., Meckling K. A. (2003) Cancer Chemother. Pharmacol. 51, 363–375 [DOI] [PubMed] [Google Scholar]
- 19.Freund A., Rössig C., Lanvers C., Gescher A., Hohenlöchter B., Jürgens H., Boos J. (1999) Annals Oncol. 10, 335–338 [DOI] [PubMed] [Google Scholar]
- 20.Stacchini A., Aragno M., Vallario A., Alfarano A., Circosta P., Gottardi D., Faldella A., Rege-Cambrin G., Thunberg U., Nilsson K., Caligaris-Cappio F. (1999) Leuk. Res. 23, 127–136 [DOI] [PubMed] [Google Scholar]
- 21.Errasti-Murugarren E., Molina-Arcas M., Casado F. J., Pastor-Anglada M. (2009) FASEB J. 23, 172–182 [DOI] [PubMed] [Google Scholar]
- 22.Duflot S., Riera B., Fernández-Veledo S., Casadó V., Norman R. I., Casado F. J., Lluís C., Franco R., Pastor-Anglada M. (2004) Mol. Cell. Biol. 24, 2710–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercader J., Gomez-Angelats M., del Santo B., Casado F. J., Felipe A., Pastor-Anglada M. (1996) Biochem. J. 317, 835–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kremer J. P., Reisbach G., Nerl C., Dormer P. (1992) Br. J. Heamatol. 80, 480–484 [DOI] [PubMed] [Google Scholar]
- 25.Yang L., Ostrowski J., Reczek P., Brown P. (2001) Oncogene 20, 8025–8035 [DOI] [PubMed] [Google Scholar]
- 26.Bost F., Caron L., Marchetti I., Dani C., Le Marchand-Brustel Y., Binétruy B. (2002) Biochem. J. 361, 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren X., Li Y., Ma M., Zheng Y., Xu Y., Wang J. (2007) Life Sciences 96, 47–56 [Google Scholar]
- 28.Moustakas A., Heldin C. H. (2005) J. Cell Science 118, 3573–3584 [DOI] [PubMed] [Google Scholar]
- 29.Alsayed Y., Uddin S., Mahmud N. (2001) J. Biol. Chem. 272, 4012–4019 [DOI] [PubMed] [Google Scholar]
- 30.Lewandowski D., Linassier C., Iochmann S., Degenne M., Domenech J., Colombat P., Binet C., Hérault O. (2002) Br. J. Haematol. 118, 535–544 [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y. E. (2009) Cell Res. 19, 128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kardassis D., Murphy C., Fotsis T., Moustakas A., Stournaras C. (2009) FEBS J. 276, 2947–2965 [DOI] [PubMed] [Google Scholar]
- 33.Coe I., Zhang Y., McKenzie T., Naydenova Z. (2002) FEBS Lett. 517, 201–205 [DOI] [PubMed] [Google Scholar]
- 34.Fernández-Veledo S., Huber-Ruano I., Aymerich I., Duflot S., Casado F. J., Pastor-Anglada M. (2006) Biochem. J. 395, 337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai X., Yamasaki K., Shirakata Y., Sayama J., Hashimoto K. (2004) J. Invest. Dermatol. 123, 1078–1085 [DOI] [PubMed] [Google Scholar]
- 36.Glasow A., Prodromou N., Xu K., von Lindern M., Zelent A. (2005) Blood 105, 341–349 [DOI] [PubMed] [Google Scholar]
- 37.Nunes I., Kojima S., Rifkin D. B. (1996) Cancer Res. 56, 495–499 [PubMed] [Google Scholar]
- 38.Yue J., Mulder K. M. (2000) J. Biol. Chem. 275, 30765–30773 [DOI] [PubMed] [Google Scholar]
- 39.Yue J., Mulder K. M. (2000) Methods Mol. Biol. 142, 125–131 [DOI] [PubMed] [Google Scholar]
- 40.Imamichi Y., Waidmann O., Hein R., Eleftheriou P., Giehl K., Menke A. (2005) Biol. Chem. 386, 225–236 [DOI] [PubMed] [Google Scholar]
- 41.Bhowmick N. A., Ghiassi M., Bakin A., Aakre M., Lundquist C. A., Engel M. E., Arteaga C. L., Moses H. L. (2001) Mol. Biol. Cell 12, 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edlund S., Landström M., Heldin C. H., Aspenström P. (2002) Mol. Biol. Cell 13, 902–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ridley A. J. (2001) Traffic 2, 303–310 [DOI] [PubMed] [Google Scholar]
- 44.Symons M., Rusk N. (2003) Curr. Biol. 13, 409–418 [DOI] [PubMed] [Google Scholar]
- 45.Norman J. C., Price L. S., Ridley A. J., Koffer A. (1996) Mol. Biol. Cell 7, 1429–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sullivan R., Price L. S., Koffer A. (1999) J. Biol. Chem. 274, 38140–38146 [DOI] [PubMed] [Google Scholar]
- 47.Maeda A., Amano M., Fukata Y., Kaibuchi K. (2002) Biochem. Biophys. Res. Commun. 297, 1231–1237 [DOI] [PubMed] [Google Scholar]
- 48.Pochynyuk O., Staruschenko A., Bugaj V., Lagrange L., Stockand J. D. (2007) J. Biol. Chem. 282, 14576–14585 [DOI] [PubMed] [Google Scholar]
- 49.Lecuona E., Ridge K., Pesce L., Batlle D., Sznajder J. I. (2003) Mol. Biol. Cell 14, 3888–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonet-Kerrache A., Fortier M., Comunale F., Gauthier-Rouvière C. (2005) Biochem. J. 15, 261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.