Abstract
Heat shock factor-1 (HSF1) is the central regulator of heat-induced transcriptional responses leading to rapid expression of molecular chaperones that protect mammalian cells against proteotoxic stress. The main targets for HSF1 are specific promoter elements (HSE) located upstream of heat shock genes encoding a variety of heat shock proteins, including HSP70, HSP90, HSP27, and other proteins of the network. Herein we report that the zinc finger AN1-type domain-2a gene, also known as AIRAP, behaves as a canonical heat shock gene, whose expression is temperature-dependent and strictly controlled by HSF1. Transcription is triggered at temperatures above 40 °C in different types of human cancer and primary cells, including peripheral blood monocytes. As shown by ChIP analysis, HSF1 is recruited to the AIRAP promoter rapidly after heat treatment, with a kinetics that parallels HSP70 promoter HSF1-recruitment. In transfection experiments HSF1-silencing abolished heat-induced AIRAP promoter-driven transcription, which could be rescued by exogenous Flag-HSF1 expression. The HSF1 binding HSE sequence in the AIRAP promoter critical for heat-induced transcription was identified. Because its expression is induced at febrile temperatures in human cells, AIRAP may represent a new potential component of the protective response during fever in humans.
Keywords: Gene/Promoters, Gene/Regulation, Protein/Heat Shock, Transcription/Regulation, Ubiquitination, AIRAP, HSE, HSF1, HSP70
Introduction
The heat shock response (HSR)2 is a fundamental defense mechanism, which protects living cells against proteotoxic stress, initiating a regulatory cascade for recovery and adaptation (1). This occurs by a nearly instantaneous induction of a set of genes, known as heat shock (HS) genes, leading to expression of cytoprotective heat shock proteins (HSP), which is proportional to the intensity and duration of stress (2, 3).
The HSR is regulated by a family of heat shock transcription factors (HSFs) that are expressed and maintained in an inactive state under non-stress conditions. Among three functionally different HSFs in humans, HSF type 1 (HSF1) mediates signaling of stress-induced stimuli, such as elevated temperatures (4). HSF1 is generally found in the cytoplasm as an inert monomer lacking transcriptional activity; both DNA binding and transcriptional transactivation domains are repressed through intramolecular interactions and constitutive serine phosphorylation (5). Upon exposure to heat shock and other types of stresses, which cause protein damage, HSF1 is derepressed in a stepwise process that involves oligomerization of HSF1 monomers to a trimeric state, localization to the nucleus, inducible phosphorylation and sumoylation, binding of nuclear-localized trimers to DNA, and transcription of HS genes (reviewed in Ref. 4). The main targets for HSF1 are specific promoter elements composed by repeats of the pentameric sequence nGAAn (heat shock elements, HSE) located upstream of HS genes. High rates of transcription are maintained only when HSF1 trimers remain bound to the HSE; when either the stress signal is removed or damaged proteins are no longer generated, the HSR attenuates rapidly, with subsequent conversion of HSF1 back to the monomeric state (6). Inducible acetylation has also been recently shown to negatively regulate DNA binding activity (7).
Heat shock gene promoters that contain HSE elements include molecular chaperones of the HSP70 and HSP90 families, HSP27 and other proteins of the network. In addition to the well known heat shock proteins, HSF binding sites have been also described in genes encoding proteins with non-chaperone function (8).
Herein we report that the zinc finger AN1-type domain 2a gene, also known as AIRAP (arsenite-inducible RNA-associated protein), is a novel human HSF1 target gene. AIRAP has been previously described as a highly conserved gene whose expression can be activated by arsenite and electrophiles but not by other conditions, including heat shock, that perturb protein folding in Caenorhabditis elegans or in mouse cells (9). We now demonstrate that the human AIRAP gene behaves as a canonical heat shock gene, whose expression is temperature-dependent and is strictly controlled by HSF1 through its binding to a specific HSE sequence located in the promoter at position −211 to −202 from the transcription start site.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatments
The production of HeLa cells stably transfected with pSUPER/pcDNA3 (HeLa-pS) and pSUPER-HSF1i/pcDNA3 (HeLa-HSF1i) plasmids was previously described (10). HeLa-pS and HeLa-HSF1i cells were grown at 37 °C in a 5% CO2 atmosphere in Dulbecco's minimum essential medium supplemented with 10% fetal calf serum, 2 mm glutamine, and antibiotics, in the presence of G-418 (400 μg/ml). HaCaT, HCT116, and Jurkat cells (American Type Culture Collection) were maintained in Dulbecco's modified Eagle's medium (HaCaT), McCoy's 5A (HCT116) or RPMI 1640 (Jurkat) medium supplemented with 10% fetal calf serum, 2 mm glutamine, and antibiotics. Human peripheral blood monocytes, isolated and purified from buffy coat of healthy blood donors (kindly provided by Prof. Adorno, Hematology Division, University of Rome Tor Vergata) as described elsewhere (11), were grown for 24 h in RPMI 1640 medium, supplemented with 10% fetal calf serum and antibiotics as indicated above. Murine embryonic fibroblasts were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mm glutamine, and antibiotics. For heating procedures, cells were subjected to heat shock at the indicated temperatures in a precision water bath-W14 (Grant Instruments, Cambridge, UK).
Promoter Cloning, Vector Construction, and Mutagenesis
To generate the AIRAP-PGL3 vector, a pair of gene-specific primers (sense: 5′-CGGGGTACCAGGGTTTCGTCATGTTCACC-3′; antisense: 5′-GCTAGCTAGCCTCAGGTGTCCTCCTCCGTA-3′) was designed to amplify the AIRAP gene promoter region (spanning from −1316 upstream of the gene transcription start site to +160) from human genomic DNA (Promega) by using Pfu Taq polymerase (Promega). The reaction product was analyzed by agarose gel electrophoresis, digested with KpnI and NheI, and inserted upstream of the luciferase gene of the pGL3Basic vector (Promega). The Δ-HSE-AIRAP-PGL3 vector containing the AIRAP promoter region (spanning from −1316 to −232) was generated as described above, by using the following primers: sense: 5′-CGGGGTACCAGGGTTTCGTCATGTTCACC-3′, antisense: 5′-GCTAGCTAGCCGCTTGCTGGGTCACAGT-3′. The AIRAP-PGL3-M26-HSE1 (M26) and AIRAP-PGL3-M210-HSE2 (M210) mutant constructs were generated by using the QuikChange Site-directed Mutagenesis kit following the manufacturer's instructions (Stratagene). Oligos used were: for M26 construct, sense: 5′-CCCCAAGGCCCATACGGCTCTCAGATCCG-3′, antisense: 5′-CGGATCTGAGAGCCGTATGGGCCTTGGGG-3′; for M210 construct, sense: 5′-CGAGAAGCGACCGTACACTTCGAGAAGGG-3′, antisense: 5′-CCCTTCTCGAAGTGTACGGTCGCTTCTCG-3′. To generate the cFlag-tagged AIRAP-pcDNA3 vector, the human AIRAP gene was amplified from the ZFAND2A cDNA (human) clone ID 5263788 (Open Biosystems) by using the following primers: 5′-GGCGGATCCACCATGGAGTTTCCTGATTTGGGGAAG-3′ and 5′-TGCGGTCGACCTACTTATCGTCGTCATCCTTGTAATCCCCAGCTTTGATGGTGGGGCG-3′. The PCR product was digested with BamH1 and Sal1, and inserted into a BamH1/Sal1-cut pcDNA3 vector. The nucleotide sequence of each construct was verified by DNA sequencing. Flag-HSF1-pcDNA3 expression vector was a kind gift from Dr. S. Calderwood, Harvard Medical School.
Cell Transfection and Reporter Assays
All transfections were performed using Lipofectamine Plus reagent (Invitrogen), according to the manufacturer's protocols. For reporter gene experiments, the different AIRAP constructs were co-transfected with a control plasmid (pRL-TK encoding Renilla luciferase, Promega) to normalize transfection efficiency. Transfected cells were grown for 16 h before heat treatment and luciferase activity of quadruplicate samples was measured in a Microplate Luminometer (Wallac-Perkin Elmer) using Dual-Luciferase kit (Promega). AIRAP promoter firefly luciferase activity was normalized to Renilla luciferase activity in the same sample.
Electrophoretic Mobility Shift Assay (EMSA)
Whole cell extracts (15 μg of protein) prepared after lysis in high-salt extraction buffer (12) were incubated with a 32P-labeled HSE DNA probe followed by analysis of DNA binding activity by EMSA. Binding reactions were performed as described (13). Complexes were analyzed by nondenaturing 4% polyacrylamide gel electrophoresis. Quantitative evaluation of HSF-HSE complex formation was determined by Typhoon 8600 imager (Molecular Dynamics, Amersham Biosciences) with the use of ImageQuant (Amersham Biosciences).
Western Blot Analysis
Equal amounts of protein (35 μg/sample) from whole cell extracts were separated by SDS/PAGE, and blotted to nitrocellulose. After blocking with 5% skim milk solution, membranes were incubated with rabbit polyclonal anti-HSF1, anti-ubiquitin (Santa Cruz Biotechnology), and anti-AIRAP (kindly provided by Dr. A. Stanhill, Technion-Israel Institute of Technology) antibodies, or monoclonal anti-HSP70 (Stressgene), anti-Flag M2, and anti-β-actin (Sigma) antibodies followed by decoration with peroxidase-labeled anti-rabbit or anti-mouse IgG, respectively (Super Signal detection kit, Pierce). Quantitative evaluation of proteins was determined by Versadoc 1000 (Bio-Rad) analysis.
RNA Extraction, RT-PCR, and Real-time RT-PCR
Total RNA was prepared using Trizol (Invitrogen) as described in the manufacturer's protocol. For RT-PCR analysis, extracted RNA (1 μg) was digested with 2 units of DNase I (Invitrogen) for 30 min at 37 °C. Samples were reverse transcribed to cDNA with 200 units of Moloney murine leukemia virus reverse transcriptase (RT) (Invitrogen), using 5 μg of random primers (Invitrogen) for 1 h at 45 °C in a total volume of 20 μl. RT was inactivated at 95 °C for 5 min. For each sample, an aliquot of DNase I-digested RNA, without RT, was used as a negative control for PCR amplification. The sequences of AIRAP, AIRAPL, HSP70, β-actin, and GAPDH primers were as follows: AIRAP, sense: 5′-TCATTTTCCATACGCTGCAC-3′, antisense: 5′-CTGTGGTCCAAAGGGTGTCT-3′; AIRAPL, sense: 5′-GTAATGTGCCTGTGCCTGTG-3′, antisense: 5′-CAGCTTGTGCTCTGGAGATG-3′; HSP701A, sense: 5′-CTACAAGGGGGAGACCAAGG-3′, antisense: 5′-TTCACCAGCCTGTTGTCAAA-3′; β-actin, sense: 5′-GCGCTCAGGAGGAGCAAT-3′, antisense: 5′-GCACTCTTCCAGCCTTCC-3′; GAPDH, sense: 5′-GTCATCAATGGAAATCCC-3′, antisense: 5′-GGTGGTGCAGGAGG-3′. Amplification was performed using the following parameters: 94 °C for 45 s, 60 °C for 30 s, and 72 °C for 45 s for 28 cycles for AIRAP, AIRAPL and HSP701A, and 24 cycles for β-actin and GAPDH. PCR products were electrophoresed alongside DNA Molecular Weight Marker IX (Roche Applied Science) in 2% agarose gels and then stained with ethidium bromide. PCR amplification was performed in a thermal cycler GeneAmp 2400 (Applied Biosystems), using Hot-Start Taq polymerase (Qiagen).
Real-time RT-PCR analysis was performed with ABI PRISM 7000 (Applied Biosystem), using RealMasterMix ROX (Eppendorf) to prepare the reaction mixes. Primers used for real-time PCR of human genes were identical to the primers described above for semiquantitative RT-PCR. Primers used for real-time PCR of murine genes were the following: AIRAP, sense: 5′- CTGACTTGGGGAAGCACTGT-3′, antisense: 5′-GCAATTGTGGTCCAGAGGAT-3′; β-actin, sense: 5′-ACTGGGACGACATGGAGAAG-3′, antisense: 5′-TTTGATGTCACGCACGATTT-3′. Relative quantities of selected mRNAs were normalized to β-actin or GAPDH in the same samples.
Chromatin Immunoprecipitation (ChIP) Assay
Cells were fixed by adding formaldehyde (Sigma) to the medium to a final concentration of 1%. After 15 min, cells were washed with ice-cold phosphate-buffered saline (PBS) containing 1 mm phenylmethylsulfonyl fluoride and scraped. After centrifugation, cells were lysed in L1 buffer (50 mm Tris, pH 8.0, 2 mm EDTA, 0.1% Nonidet P-40, 10% glycerol, and proteases inhibitors) and centrifuged for 5′ at 3,000 rpm at 4 °C. After removal of supernatants, nuclei were resuspended in L2 buffer (50 mm Tris, pH 8.0, 1% SDS, 5 mm EDTA), and chromatin was sheared by sonication. After removal of nuclear debris by centrifugation at 13,000 rpm for 5 min at 8 °C, lysates were diluted 10-fold with DB buffer (50 mm Tris pH 8.0, 5 mm EDTA, 200 mm NaCl, 0.5% Nonidet P-40) and then precleared for 3 h using 80 μl of 50% salmon sperm-DNA saturated protein A (ss-proteinA)-agarose beads. Immunoprecipitation was carried out at 4 °C overnight, and immune complexes were collected with ss-protein A-agarose beads. Antibodies utilized included anti-HSF1 (Santa Cruz Biotechnologies) or preimmune rabbit serum as control for nonspecific interaction. After washing three times with high salt WB buffer (20 mm Tris, pH 8.0, 0.1% SDS, 1% Nonidet P-40, 2 mm EDTA, 0.5 m NaCl) and twice with low salt TE buffer (10 mm Tris, pH 8.0, 1 mm EDTA), immunocomplexes were eluted with TE containing 1% SDS. Protein-DNA cross-links were reverted by incubating at 65 °C overnight. After proteinase K digestion, DNA was extracted with phenol-chloroform and precipitated with ethanol using 15 μg of tRNA as carrier. PCR was performed (30 cycles, denaturing at 94 °C for 45 s, annealing at 60 °C for 30 s and extension at 72 °C for 45 s, using the following primers: AIRAP, sense: 5′-CACTTCGAGAAGGGAAGTGG-3′, antisense: 5′-ACCGGAACGCAACATCTC-3′; HSP701A, sense: 5′-CACTCCCCCTTCCTCTCAG-3′, antisense: 5′-TTCCCTTCTGAGCCAATCAC-3′. For quantification of ChIP assays, samples from at least three independent experiments were analyzed by real time PCR. Data were normalized to the input DNA and DNA from untreated cells.
Statistical Analysis
Statistical analysis was performed using the Student's t test for unpaired data. Data are expressed as the mean ± S.D. of duplicate samples. p values of < 0,05 were considered significant.
RESULTS
AIRAP Gene Expression Is Induced by Heat Stress and Is Dependent on the Presence of a Functional HSF1
We have previously generated a stable human cervical carcinoma cell line (HeLa-HSF1i) with HSF1 loss-of-function by using stable RNA-mediated interference (10). By differential microarray analysis of gene expression profiles of HeLa-HSF1i versus HeLa control (HeLa-pS) cells under stress conditions, in addition to the canonical heat shock genes, we identified AIRAP as a new gene whose expression was increased after heat shock selectively in HeLa-pS cells.
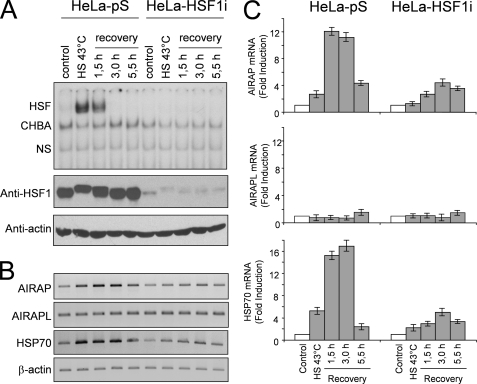
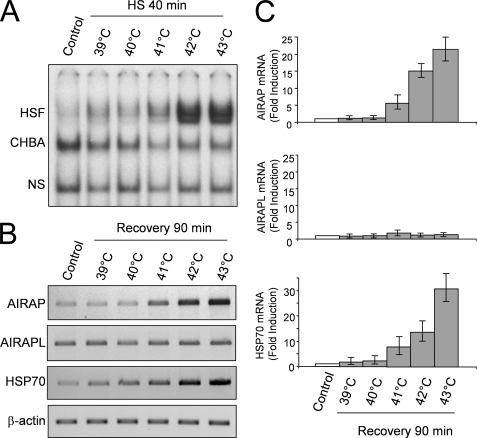
To confirm the results obtained in the microarray analysis, HeLa-pS and HeLa-HSF1i cells were either kept at 37 °C or were subjected to heat shock at 43 °C for 40 min and then allowed to recover at 37 °C. At different times during the recovery period, whole cell extracts were analyzed for HSF1 levels by Western blot and HSF1 DNA binding activity by EMSA; in parallel samples, total RNA was analyzed for AIRAP, the constitutively expressed AIRAP homologue (AIRAPL), HSP70, and β-actin expression by RT-PCR and real-time PCR. As expected, in HeLa-pS cells HSF1 activity was detected at the end of heat shock and continued during the recovery period for 1.5 h, after which time it declined rapidly (Fig. 1A, upper panel). HSF1 activation was followed by sustained AIRAP mRNA expression with kinetics similar to the HSP70 heat shock gene (Fig. 1, B and C). Differently from HeLa-pS cells, neither HSF1 activation nor induction of AIRAP and HSP70 expression, as determined by RT-PCR, were detected at any time in HeLa-HSF1i cells (Fig. 1, A and B). A modest increase in both HSP70 and AIRAP expression in heat-shocked HeLa-HSF1i cells was detected by real-time PCR (Fig. 1C), probably as a consequence of residual HSF1 activity; however, it cannot be excluded that HSF2, that is functional in HeLa-HSF1i cells (10), or other transcription factors could contribute to this activity. No change in the mRNA levels of the constitutively expressed AIRAPL was detected at any time in both HeLa-pS and HeLa-HSF1i cells.
FIGURE 1.
HSF1-mediated induction of AIRAP gene expression by heat shock. A, kinetics of AIRAP gene induction after heat shock. HeLa-pS and HeLa-HSF1i cells were subjected to heat shock (HS) at 43 °C for 40 min. At the end of heat treatment, or at 1.5, 3, and 5.5 h during the recovery period at 37 °C, whole cell extracts were analyzed for HSF1 DNA binding activity by EMSA (top panel). Positions of the HSF DNA binding complex (HSF), constitutive HSE binding activity (CHBA), and nonspecific protein-DNA interaction (NS) are shown. Levels of HSF1 and β-actin were determined in the same samples by Western blot (bottom panels). In parallel samples, total RNA was analyzed for AIRAP, the constitutively expressed AIRAP homologue (AIRAPL), HSP70, and β-actin expression by RT-PCR (B) and real-time PCR (C). For real-time PCR, relative quantities of AIRAP, AIRAPL, and HSP70 RNAs were normalized to β-actin. All reactions were made in duplicates using samples derived from at least three biological repeats. Error bars indicate ± S.D. Fold induction was calculated by comparing the induction of the indicated genes in the treated samples to the relative control which was arbitrarily set to 1.
HSF1 Binds Directly to the AIRAP Promoter in Vivo
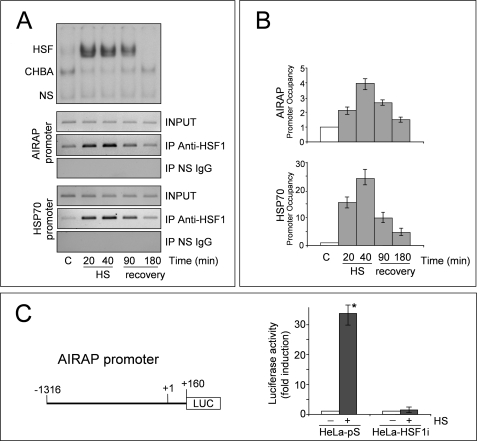
The fact that AIRAP was induced only in HeLa-pS cells suggested that HSF1 might play a direct role in the induction of AIRAP expression. To investigate whether HSF1 is recruited to the AIRAP gene promoter. HeLa-pS cells were subjected to a 40 min heat shock at 43 °C and then allowed to recover at 37 °C. At different times during heat treatment and the recovery period, parallel samples were analyzed for HSF1 DNA binding activity by EMSA (Fig. 2A, upper panel) or for recruitment to the AIRAP and HSP70 promoters by ChIP analysis (Fig. 2A, lower panels). HSF1-coprecipitating DNA was analyzed by PCR (Fig. 2A) and real-time PCR (Fig. 2B) with promoter-specific primers amplifying the AIRAP and HSP70 promoter and the rate of amplification was verified using cross-linked, not immunoprecipitated chromatin. The specificity of chromatin immunoprecipitation was determined by using a control unrelated antibody. Consistently with the DNA binding activity revealed by EMSA (Fig. 2A, upper panel), HSF1 was recruited to the AIRAP promoter during heat treatment and for at least 90 min of the recovery period at 37 °C (Fig. 2A, middle panel). A similar kinetics of HSF1 recruitment was observed also for the HSP70 promoter (Fig. 2A, lower panel).
FIGURE 2.
HSF1 binds directly to the AIRAP promoter in vivo. A, HeLa-pS cells were subjected to heat shock at 43 °C. After 20 and 40 min at 43 °C (HS) or at 1.5 and 3 h during recovery at 37 °C (recovery), whole cell extracts were analyzed for HSF DNA binding activity by EMSA (top panel) and recruitment to the AIRAP and HSP70 promoters by ChIP assay (lower panels). ChIP-enriched DNAs using preimmune serum (IP NS IgG) or anti-HSF1 serum (IP anti-HSF1), as well as input DNAs (INPUT) were prepared, and DNA fragments of the AIRAP gene (−208 to +45) and HSP70 gene (−262 to −70) were amplified by PCR. B, quantification of ChIP assay shown in A. Samples from at least three independent experiments were analyzed by real time PCR. Relative promoter occupancy is expressed as fold induction of control arbitrarily set to a value of 1. Error bars indicate ± S.D. C, reporter analysis of the AIRAP promoter. HeLa-pS and HeLa-HSF1i cells transiently transfected with the AIRAP-PGL3 construct (right panel) were subjected to heat shock (43 °C for 40 min). After 6 h of recovery at 37 °C luciferase activity was determined (left panel). The data, expressed as fold induction of untreated control, represent the mean of quadruplicate samples from two independent experiments. Error bars indicate ± S.D. *, p < 0.01.
Genomic Cloning and Reporter Analysis of the AIRAP Promoter
To verify whether HSF1 binding to the AIRAP promoter effectively and selectively activates AIRAP gene transcription, the upstream region of the AIRAP gene (−1316 to +160) was cloned into the PGL3 reporter vector (AIRAP-PGL3, Fig. 2C) for luciferase reporter activity analysis. HeLa-pS and HeLa-HSF1i cells were transiently transfected with the AIRAP-PGL3 construct; after 24 h, transfected cells were subjected to heat stress at 43 °C for 40 min. Luciferase activity was determined after a 6 h recovery period at 37 °C. As shown in Fig. 2C, heat shock strongly induced reporter activity in HeLa-pS cells, whereas no significant transcriptional activity was detected in HeLa-HSF1i cells, suggesting that HSF1 plays an important role in the control of AIRAP gene expression.
Overexpression of HSF1 Rescues Heat-induced AIRAP Gene Transcription in HeLa HSF1i Cells
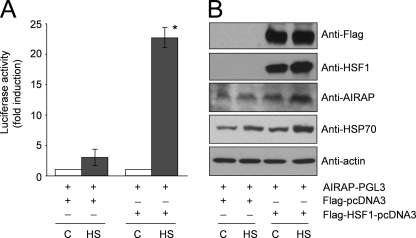
To confirm the role of HSF1 in the transcriptional control of the AIRAP promoter, HeLa-HSF1i cells were co-transfected with AIRAP-PGL3 and Flag-HSF1-pcDNA3 expression vectors, or with AIRAP-PGL3 and the Flag-pcDNA3 empty vector, and after 24 h were incubated at 43 °C for 40 min. After 6 h at 37 °C, whole cell extracts were analyzed for luciferase activity (Fig. 3A), and HSF1, AIRAP, HSP70, and β-actin levels were determined in the same samples by Western blot (Fig. 3B). As shown in Fig. 3B, HeLa-HSF1i cells transfected with the Flag-HSF1 construct express high levels of Flag-HSF1 protein. This result implies that the HSF1-specific shRNAi stably expressed in HeLa-HSF1i cells is able to suppress the basal level of endogenous HSF1, but is unable to prevent exogenous Flag-tagged-HSF1 overexpression. The exogenous Flag-HSF1 protein was found to be functional, being able to restore a canonical heat shock response in HeLa-HSF1i cells, as indicated by reactivation of endogenous HSP70 gene expression after heat shock (Fig. 3B). Concomitant with the ability to induce HSP70 protein expression, the exogenous Flag-HSF1 was able to restore heat shock-driven AIRAP promoter activity in transfected cells (Fig. 3A), as well as to induce endogenous AIRAP protein expression (Fig. 3B). Altogether these results demonstrate that HSF1 is indispensable for heat-induced AIRAP gene expression.
FIGURE 3.
HSF1 overexpression rescues HS-induced AIRAP transcription in HeLa-HSF1i cells. HeLa-HSF1i cells were transiently co-transfected with AIRAP-PGL3 and Flag-HSF1-pcDNA3 expression vectors, or with AIRAP-PGL3 and Flag-pcDNA3 vectors, and after 24 h were subjected to heat shock (43 °C for 40 min) (HS) or left untreated (C). After 6 h of recovery at 37 °C, whole cell extracts were analyzed for luciferase activity (A). The data, expressed as fold induction of untreated control, represent the mean of duplicate samples from a representative experiment of two with similar results. Error bars indicate ± S.D. *, p < 0.05. B, levels of HSF1, Flag-HSF1, AIRAP, HSP70, and β-actin were determined in the same samples by Western blot.
AIRAP Promoter Characterization
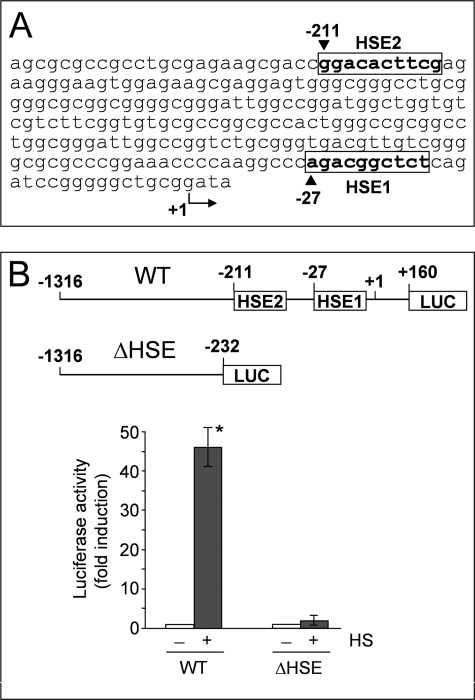
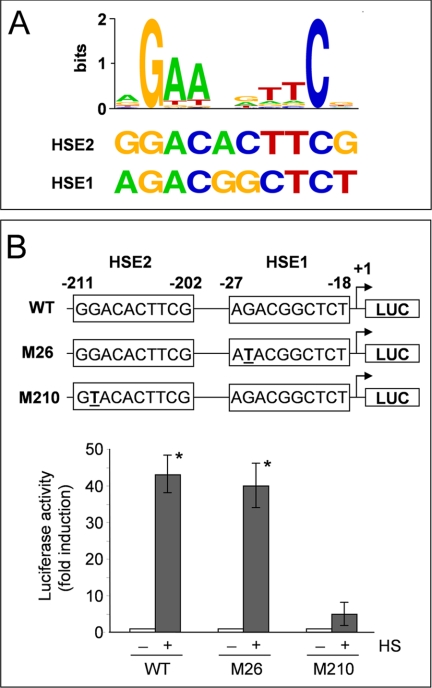
To identify the enhancer elements responsible for HSF1-mediated AIRAP gene expression, the cloned 1316-bp region of the AIRAP promoter was analyzed by TFSEARCH version 1.3 (Yutaka Akiyama, TFSEARCH: Searching Transcription Factor Binding Sites) (14). Two putative HSE located at −27 (HSE1) and −211 (HSE2) from the transcription start site were detected (Fig. 4A). HSE1 presents a minimal dimeric binding site (see Fig. 4) with a score value computed by TFSEARCH similar to that calculated for HSE2 (72.5 and 73.2, respectively). In contrast to HSE1, the minimal dimeric binding site of HSE2 is part of a single contiguous array of seven units of the 5′-nGAAn-3′ consensus motif (Fig. 4, supplemental Fig. S1C, and Ref. 8).
FIGURE 4.
Identification of HSE elements in the AIRAP promoter. A, nucleotide sequence of the 5′-flanking region of the AIRAP gene. Putative HSF1 binding sites (HSE1 and HSE2) localized within this region, identified by TFSEARCH program (ver 1.3), are indicated within boxes. B, AIRAP-PGL3 construct (WT) and AIRAP-PGL3-ΔHSE (ΔHSE) deletion construct (schematically shown in the top of the figure) were transfected in HeLa-pS cells. After 24 h, cells were subjected to heat shock (43 °C for 40 min) (+ HS) or left untreated (− HS). Whole cell extracts were analyzed for luciferase activity after 6 h recovery at 37 °C. The data, expressed as fold induction of untreated control, represent the mean of quadruplicate samples from two independent experiments. Error bars indicate ± S.D. *, p < 0.05.
A construct deleted in the region comprising the two HSE elements (Δ-HSE-AIRAP-PGL3) was then prepared and utilized for reporter analysis. HeLa-pS cells were transfected with the wild-type AIRAP-PGL3 or the Δ-HSE-AIRAP-PGL3 constructs and after 24 h were heat stressed (43 °C for 40 min). Luciferase activity was determined after a 6 h recovery period at 37 °C. As shown in Fig. 4B, deletion of the −232 to +160 region, containing both HSE elements and neighboring sequences, resulted in abrogation of heat-induced reporter activity.
It has been demonstrated that a mutation in the second nucleotide present in the HSE central nGAAn-bp unit (Fig. 5A) causes a strong reduction in HSF1 DNA binding activity in vitro (15). To further define the role of the identified HSEs in heat shock-driven AIRAP induction, a G to T base mutation was inserted in both the HSE1 and the HSE2 elements, and the constructs AIRAP-PGL3-M26-HSE1 (M26) and AIRAP-PGL3-M210-HSE2 (M210), mutated in the HSE1 and the HSE2 elements respectively, were obtained (Fig. 5B, top). The M26, M210 or the wild-type (WT) constructs were transiently transfected in HeLa-pS cells. At 24 h after transfection, cells were subjected to heat shock, and the promoter activity was determined as described above. As shown in Fig. 5B, a G to T base mutation in the HSE1 core sequence did not alter the ability of the AIRAP promoter to respond to heat shock, whereas a similar mutation in the HSE2 element almost completely blocked heat-induced transcription. These data identify the HSE2 sequence present in the AIRAP promoter as a critical element for heat-induced AIRAP transcription.
FIGURE 5.
Characterization of a functional HSE in the AIRAP promoter. A, sequence logo of the consensus motif for HSF1 generated by the program WebLogo (31) using previously known HSF1 binding sites as found in TRANSFAC (14) (top). The sequences of putative HSF binding sites (HSE1 and HSE2) in the AIRAP promoter identified by TFSEARCH are shown (bottom). B, HeLa-pS cells were transfected with the AIRAP-PGL3 construct (WT) and the AIRAP-PGL3-M26HSE (M26), AIRAP-PGL3-M210HSE (M210) point-mutated (G to T) constructs. After 24 h, cells were subjected to heat shock (43 °C for 40 min) (+ HS) or left untreated (− HS). Whole cell extracts were analyzed for luciferase activity after 6 h recovery at 37 °C. The data, expressed as fold induction of untreated control, represent the mean of quadruplicate samples from two independent experiments. Error bars indicate ± S.D. *, p < 0.05. Sequences of WT and point-mutated (M26-HSE1, M210-HSE2) HSEs present in the AIRAP promoter are shown in the top of the figure.
Temperature-dependent AIRAP Gene Expression in Primary and Cancer Human Cells
HSF1 activation by heat is strictly dependent on both the increase in temperature above physiological conditions and the duration of exposure. To investigate the effect of temperature on HSF1 DNA binding activity and AIRAP expression, HeLa-pS cells were either kept at 37 °C or subjected to heat shock at 39, 40, 41, 42, or 43 °C for 40 min. At the end of heat treatment, and after a 90-min recovery period at 37 °C, whole cell extracts were analyzed for HSF1 DNA binding activity by EMSA. In parallel samples levels of AIRAP, AIRAPL, HSP70, and β-actin mRNA were analyzed by RT-PCR and real-time PCR after the 90 min recovery period at 37 °C. As expected the intensity of HSF1 activation was temperature-dependent, and HSF1 DNA binding activity was detected at the end of the heat shock period starting at 41 °C (Fig. 6A). Following exposure to 41 and 42 °C, HSF1 DNA binding activity returned to control levels after a 90 min recovery at 37 °C, whereas after treatment at 43 °C HSF activation was still detected at this time (data not shown). Consistently with HSF1 activation, a temperature-dependent increase in AIRAP mRNA levels was detected starting at 41 °C, reaching maximal levels at 43 °C (Fig. 6, B and C). HSP70 mRNA levels were increased in a temperature-dependent manner, similarly to AIRAP mRNA, whereas no change in the constitutively expressed AIRAP homologue AIRAPL and β-actin levels was detected up to 43 °C.
FIGURE 6.
Temperature-dependent induction of AIRAP expression in HeLa cells. HeLa-Ps cells were subjected to heat stress at 39, 40, 41, 42, and 43 °C for 40 min and then allowed to recover at 37 °C. At the end of heat shock whole cell extracts were analyzed for HSF DNA binding activity by EMSA (A). Positions of the HSF DNA binding complex (HSF), constitutive HSE binding activity (CHBA), and nonspecific protein-DNA interaction (NS) are shown. B, in parallel samples, total RNA extracted after 90 min recovery at 37 °C was analyzed for AIRAP, AIRAPL, HSP70, and β-actin expression by RT-PCR (B) and real-time PCR (C). For real-time PCR, relative quantities of AIRAP, AIRAPL, and HSP70 RNAs were normalized to β-actin. All reactions were made in duplicates using samples derived from at least three biological repeats. Error bars indicate ± S.D. Fold induction was calculated by comparing the induction of the indicated genes in the treated samples to the relative control, which was arbitrarily set to 1.
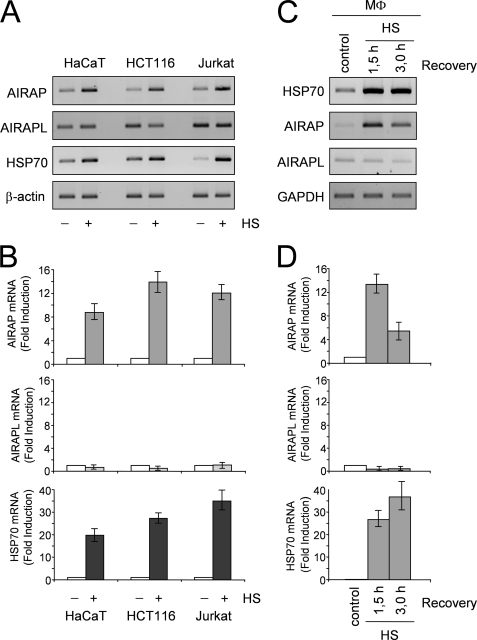
To determine whether AIRAP gene induction by heat represents a general response of human cells to temperature increase, human keratinocytes (HaCaT cells) and two human cancer cell lines of different origin (HCT116 colon carcinoma and Jurkat T-cell lymphoma) were incubated at 43 °C for 40 min and allowed to recover at 37 °C for 90 min. At this time, total RNA was extracted and levels of AIRAP, AIRAPL, HSP70, and β-actin mRNA were analyzed by RT-PCR and real-time PCR. As shown in Fig. 7, A and B, heat treatment resulted in an increase in AIRAP and HSP70 mRNA levels in all cell lines, while no changes were found in AIRAPL levels. Similar results were obtained in human primary blood monocytes subjected to heat stress (43 °C, 40 min) and analyzed at 1.5 and 3 h after recovery at 37 °C (Fig. 7, C and D), indicating that AIRAP expression represents a general response of human cells to temperature increase.
FIGURE 7.
Heat stress induces AIRAP expression in different types of human cells. A, human keratinocytes (HaCaT), colon carcinoma cells (HCT116), and T lymphoblastoid cells (Jurkat) were either kept at 37 °C (− HS) or incubated at 43 °C for 40 min (+ HS) and allowed to recover at 37 °C for 90 min. At this time, total RNA was extracted and levels of AIRAP, AIRAPL, HSP70, and β-actin mRNA were analyzed by RT-PCR (A) and real-time PCR (B). C and D, human peripheral blood monocytes (MΦ) were kept at 37 °C (Control) or subjected to heat stress (43 °C for 40 min) (HS) and allowed to recover at 37 °C. At 1.5 or 3 h recovery at 37 °C, total RNA was extracted and levels of HSP70, AIRAP, AIRAPL, and GAPDH mRNA were analyzed by RT-PCR (C) and real-time PCR (D). For real-time PCR, relative quantities of AIRAP, AIRAPL, and HSP70 RNAs were normalized to β-actin. All reactions were made in duplicates using samples derived from at least three biological repeats. Error bars indicate ± S.D. Fold induction was calculated by comparing the induction of the indicated genes in the treated samples to the relative control, which was arbitrarily set to 1.
DISCUSSION
After its initial discovery in Drosophila as a new puffing pattern of polytene chromosomes induced by heat (16), it became rapidly evident that the HSR is a highly conserved cellular response leading to the rapid expression of cytoprotective heat shock proteins. In mammalian organisms during stress conditions different HSP act in concert constituting the “protein repair machinery” preventing protein denaturation and aggregation that are detrimental to cells, and promoting degradation of irreversibly denaturated cytotoxic proteins (3, 17–18). Under normal conditions, HSP also function as chaperones that assist in protein folding (19). Mammalian genomes encode three homologues of HSF (HSF1, HSF2, and HSF4) regulating HSP expression. Among these HSF1 is considered to be the paralog responsible for regulating the heat-induced transcriptional response (4). HSF2 has also been reported to contribute to inducible expression of heat shock genes through interplay with HSF1 (20).
Whereas it is well established that HSF1 regulates inducible HSP expression, recently it has become evident that the regulation of the mammalian HSR is a more complex phenomenon than previously thought. Microarray and chromatin immunoprecipitation analysis, while confirming that many known heat-inducible genes have HSF1 binding sites in their promoters, have defined another class of genes that recruit HSF1 to their HSE promoter elements but that nevertheless are not induced by heat shock (8). On the other hand, these studies have provided evidence that, in some cases, HSF1 may also control the expression of genes with non-chaperone function (8), such as superoxide dismutase (21), the multiple drug resistance genes (22), lactate dehydrogenase (23), and the T-cell death-associated gene 51 (24).
We have now identified the AIRAP gene as a new HSF1 target gene in human cells. We have shown that the human AIRAP gene behaves as a canonical heat shock gene, whose expression is strictly controlled by HSF1 in a temperature-dependent fashion. Transcription is triggered at temperatures above 40 °C in different types of human cells, including cancer cells of different origin (HeLa cervical carcinoma, HCT116 colon carcinoma, and Jurkat T cell lymphoma), as well as human keratinocytes and primary blood monocytes. Similar results were also obtained in heat stressed human umbilical vein endothelial cells (HUVEC).3 These results indicate that AIRAP induction represents a general response of human cells to heat.
Two putative HSE located at −27 (HSE1) and −211 (HSE2) from the transcription start site were identified in the AIRAP promoter. Deletion in the region comprising the two HSE elements resulted in abrogation of heat-induced AIRAP transcription. Because it has been shown that a mutation in the second nucleotide present in the HSE central nGAAn-bp unit causes a strong reduction in HSF1 DNA binding activity in vitro (15), we have inserted a G to T base mutation in both the HSE1 and the HSE2 elements identified in the AIRAP promoter, and have shown that, whereas a point mutation in the HSE1 element did not alter the ability of the AIRAP promoter to respond to heat shock, a similar mutation in the HSE2 element almost completely blocked heat-induced transcription. These results identify the −211 HSE2 sequence present in the AIRAP promoter as a critical element for heat-induced AIRAP transcription.
ChIP analysis of HSF1 recruitment to the AIRAP promoter after heat stress has shown that HSF1 is recruited to the AIRAP promoter rapidly after heat shock treatment, with a kinetics that parallels the recruitment of the factor to the promoter of canonical HS genes, such as HSP70. Recruitment was followed by AIRAP transcription in HeLa cells, but not in HeLa-HSF1i cells with HSF1 loss-of-function. On the other hand, expression of exogenous Flag-HSF1 was able to restore heat shock-driven AIRAP promoter activity, concomitant with the ability to re-activate endogenous AIRAP and HSP70 expression, in HeLa-HSF1i cells. These data demonstrate that HSF1 plays a fundamental role in the control of heat-induced human AIRAP gene expression.
AIRAP was originally identified as an arsenite-inducible, cysteine- and histidine-rich RNA-associated protein, which is conserved among mammals, Drosophila and C. elegans. In murine cells AIRAP was reported to be selectively induced by arsenite and electrophiles, but not by other conditions that perturb protein folding (9). In fact, oxidative stress induced by H2O2 treatment and tunicamycin-induced stress of the endoplasmic reticulum did not trigger AIRAP expression in NIH3T3 mouse cells or the AIRAP homologue aip-1 in C. elegans (9). In particular, heat shock at 44 °C for 1 h failed to induce AIRAP in NIH3T3 mouse cells (9). We have also found that heat shock at 43 °C for 40 min does not significantly induce AIRAP expression in murine fibroblasts, as compared with human cells (supplemental Fig. S1, A and B). In silico analysis has revealed substantial differences in the murine versus human AIRAP promoter structure, which may account for the differential responses observed. In particular, even though, as revealed by TFSEARCH analysis, the mouse AIRAP promoter contains a putative HSE for HSF1 (mHSE) located at −276 from the transcription start site, differently from the human HSE2 (which presents a single contiguous array of seven 5′-nGAAn-3′ units), mHSE presents only three contiguous units (supplemental Fig. S1C). Considering the strong cooperative nature of HSFs-HSE binding (25), this difference in the number of adjacent 5′-nGAAn-3′ units in the promoter sequences could be crucial in determining the different ability of HSF1 to induce AIRAP in human and murine cells.
In relation to its biological function, AIRAP was identified as an arsenite-inducible component of the 26 S proteasome (26). AIRAP was found to associate tightly with the 19 S cap of the proteasome, altering its biochemical properties to facilitate substrate transit through the particle. In this regard, the AIRAP-containing particles resemble doubly capped hybrid proteasomes with a 19 S on one end and an 11S/REG/PA28 particle on the other (27). Because of these features, that confer to AIRAP-containing proteasomes an enhanced cleavage of peptide substrates, it has been suggested that AIRAP may adapt the cell core protein degradation machinery to counteract proteotoxicity induced by arsenite or other environmental toxins (26). Recently it was discovered that mammals have a second, constitutively expressed, AIRAP-like gene (AIRAPL) that also encodes a proteasome-interacting protein, which shares with AIRAP the property of enhancing peptide accessibility to the proteasome active site (28). The fact that in worms a single gene, aip-1, incorporates features of both the arsenic-inducible AIRAP and the constitutively expressed AIRAPL has suggested that AIRAP and the constitutively expressed AIRAPL represent a relatively late feature of evolution apparently restricted to mammals (28). We have now shown that AIRAP, but not AIRAPL, behaves as a canonical heat shock gene in human cells. Because of the well-known proteotoxic effects of heat, a similar function as a cytoprotective gene, assisting proteasome-mediated misfolded protein degradation during heat stress, could be hypothesized for human AIRAP. This hypothesis is supported by the finding that whereas wild-type HeLa cells exposed to heat shock at 43 °C accumulate polyubiquitinated proteins transiently (up to 90 min and returning to control level by 3 h), in HeLa-HSF1i cells a similar heat treatment leads to a time-dependent increase in polyubiquitinated protein accumulation that persists up to 8 h after heat shock (supplemental Fig. S2A). On the other hand, expression of exogenous AIRAP had only a modest effect on the recovery of heat-induced polyubiquitinated protein accumulation in HSF1-silenced cells (supplemental Fig. S2B), suggesting that if AIRAP is necessary for an optimal response of the cell protein degradation machinery to counteract proteotoxicity, it must function in tight concert with other heat-induced chaperones and non-chaperones to facilitate the clearance of misfolded proteins during the recovery process after heat shock. This is in line with the concept that, whereas chaperone induction constitutes one strand of adaptation to the threat of proteotoxicity, to be effective it has to be complemented by activation of the cell protein degradation machinery (29–30). However, other yet undiscovered functions of AIRAP may intervene during heat stress.
Whereas the function of this novel HSF1 target gene remains to be defined, the fact that AIRAP expression is induced at febrile temperatures in human cells identifies this protein as a new potential component of the protective response during fever in humans.
Supplementary Material
Acknowledgments
We thank Dr. Stuart Calderwood (Harvard University Medical School, Boston, MA) for the Flag-HSF1-pcDNA3 expression vector, and Dr. Ariel Stanhill (Technion-Israel Institute of Technology, Haifa, Israel) for AIRAP antibodies.
This work was supported in part by the Italian Ministry of University and Scientific Research, the Italian Ministry of Public Health (ISS project), and the EU EICOSANOX Project.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
A. Rossi and M. G. Santoro, unpublished results.
- HSR
- heat shock response
- AIRAP
- arsenite-inducible RNA-associated protein
- ChIP
- chromatin immunoprecipitation
- EMSA
- electrophoretic mobility shift assay
- HS
- heat shock
- HSE
- heat shock element
- HSF1
- heat shock factor type 1
- HSP
- heat shock protein
- RT
- reverse transcriptase
- WT
- wild type.
REFERENCES
- 1.Morimoto R. I., Santoro M. G. (1998) Nat. Biotechnol. 16, 833–838 [DOI] [PubMed] [Google Scholar]
- 2.Morimoto R. I. (1998) Genes Dev. 12, 3788–3796 [DOI] [PubMed] [Google Scholar]
- 3.Morimoto R. I. (2008) Genes Dev. 22, 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anckar J., Sistonen L. (2007) Adv. Exp. Med. Biol. 594, 78–88 [DOI] [PubMed] [Google Scholar]
- 5.Holmberg C. I., Tran S. E., Eriksson J. E., Sistonen L. (2002) Trends Biochem. Sci. 27, 619–627 [DOI] [PubMed] [Google Scholar]
- 6.Abravaya K., Phillips B., Morimoto R. I. (1991) Genes Dev. 5, 2117–2127 [DOI] [PubMed] [Google Scholar]
- 7.Westerheide S. D., Anckar J., Stevens S. M., Jr., Sistonen L., Morimoto R.I. (2009) Science 323, 1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trinklein N. D., Murray J. I., Hartman S. J., Botstein D., Myers R. M. (2004) Mol. Biol. Cell 15, 1254–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sok J., Calfon M., Lu J., Lichtlen P., Clark S. G., Ron D. (2001) Cell Stress Chaperones 6, 6–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossi A., Ciafrè S., Balsamo M., Pierimarchi P., Santoro M. G. (2006) Cancer Res. 66, 7678–7685 [DOI] [PubMed] [Google Scholar]
- 11.Elia G., Polla B., Rossi A., Santoro M. G. (1999) Eur. J. Biochem. 264, 736–745 [DOI] [PubMed] [Google Scholar]
- 12.Rossi A., Elia G., Santoro M. G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 746–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossi A., Kapahi P., Natoli G., Takahashi T., Chen Y., Karin M., Santoro M. G. (2000) Nature 403, 103–108 [DOI] [PubMed] [Google Scholar]
- 14.Heinemeyer T., Wingender E., Reuter I., Hermjakob H., Kel A. E., Kel O. V., Ignatieva E. V., Ananko E. A., Podkolodnaya O. A., Kolpakov F. A., Podkolodny N. L., Kolchanov N. A. (1998) Nucleic Acids Res. 26, 362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes M., Xiao H., Lis J. T. (1994) Nucleic Acids Res. 22, 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritossa F. (1962) Experientia 18, 571–573 [Google Scholar]
- 17.Jäättelä M. (1999) Ann. Med. 31, 261–271 [DOI] [PubMed] [Google Scholar]
- 18.Parsell D. A., Lindquist S. (1993) Annu. Rev. Genet 27, 437–496 [DOI] [PubMed] [Google Scholar]
- 19.Dobson C. M. (2003) Nature 426, 884–890 [DOI] [PubMed] [Google Scholar]
- 20.Sandqvist A., Björk J. K., Akerfelt M., Chitikova Z., Grichine A., Vourc'h C., Jolly C., Salminen T. A., Nymalm Y., Sistonen L. (2009) Mol. Biol. Cell 20, 1340–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoo H. Y., Chang M. S., Rho H. M. (1999) J. Biol. Chem. 274, 23887–23892 [DOI] [PubMed] [Google Scholar]
- 22.Vilaboa N. E., Galán A., Troyano A., de Blas E., Aller P. (2000) J. Biol. Chem. 275, 24970–24976 [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y. H., Zhou M., Liu H., Ding Y., Khong H. T., Yu D., Fodstad O., Tan M. (2009) Oncogene 28, 3689–3701 [DOI] [PubMed] [Google Scholar]
- 24.Hayashida N., Inouye S., Fujimoto M., Tanaka Y., Izu H., Takaki E., Ichikawa H., Rho J., Nakai A. (2006) EMBO J. 25, 4773–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao H., Perisic O., Lis J. T. (1991) Cell 64, 585–593 [DOI] [PubMed] [Google Scholar]
- 26.Stanhill A., Haynes C. M., Zhang Y., Min G., Steele M. C., Kalinina J., Martinez E., Pickart C. M., Kong X. P., Ron D. (2006) Mol. Cell 23, 875–885 [DOI] [PubMed] [Google Scholar]
- 27.Cascio P., Call M., Petre B. M., Walz T., Goldberg A. L. (2002) EMBO J. 21, 2636–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yun C., Stanhill A., Yang Y., Zhang Y., Haynes C. M., Xu C. F., Neubert T. A., Mor A., Philips M. R., Ron D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7094–7099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldberg A. L. (2003) Nature 426, 895–899 [DOI] [PubMed] [Google Scholar]
- 30.Höhfeld J., Cyr D. M., Patterson C. (2001) EMBO Rep. 2, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.