Abstract
In the camphor monooxygenase system from Pseudomonas putida, the [2Fe-2S]-containing putidaredoxin (Pdx) shuttles electrons between the NADH-dependent putidaredoxin reductase (Pdr) and cytochrome P450cam. The mechanism of the Pdr·Pdx redox couple has been investigated by a variety of techniques. One of the exceptions is x-ray crystallography as the native partners associate weakly and resist co-crystallization. Here, we present the 2.6-Å x-ray structure of a catalytically active complex between Pdr and Pdx C73S/C85S chemically cross-linked via the Lys409Pdr-Glu72Pdx pair. The 365 Å2 Pdr-Pdx interface is predominantly hydrophobic with one central Arg310Pdr-Asp38Pdx salt bridge, likely assisting docking and orienting the partners optimally for electron transfer, and a few peripheral hydrogen bonds. A predicted 12-Å-long electron transfer route between FAD and [2Fe-2S] includes flavin flanking Trp330Pdr and the iron ligand Cys39Pdx. The x-ray model agrees well with the experimental and theoretical results and suggests that the linked Pdx must undergo complex movements during turnover to accommodate P450cam, which could limit the Pdx-to-P450cam electron transfer reaction.
Keywords: Electron Transport System, Flavoproteins, Iron-Sulfur Protein, Protein Cross-linking, Protein Structure, Crystal Structure, Putidaredoxin, Putidaredoxin Reductase
Introduction
In Pseudomonas putida, the camphor monooxygenase system uses NADH as a source of electrons and consists of three soluble proteins: FAD-containing putidaredoxin reductase (Pdr,3 45.6 kDa), [2Fe-2S]-containing putidaredoxin (Pdx, 11.4 kDa), and cytochrome P450cam (P450cam, 46.6 kDa) (1). Pdx receives reducing equivalents from Pdr in two one-electron steps and delivers them one at a time to P450cam. Acting as a shuttle, Pdx forms transient electron transfer (ET) complexes with its redox partners during turnover (2–6). The x-ray structures of all components of the camphor monooxygenase have been determined (7–10), but neither Pdr·Pdx nor Pdx·P450cam native complexes have been crystallized thus far.
The mechanism of interaction and interprotein ET in the Pdr·Pdx pair has been the focus of several research groups including ours. Significant progress has been made in the general understanding of how the Pdr·Pdx complex is formed and functions. In particular, both two- and one-electron reduced species of Pdr were identified as catalytically competent redox intermediates (3, 4, 11); ionic and hydrophobic interactions as well as steric complementarity were proven to contribute to molecular recognition between the partners (4, 12, 13) and involve Tyr33, Asp38, Arg66, Glu72, and Cys73 of Pdx (14–16); and, based on the x-ray structures of the flavo- and iron-sulfur proteins (8, 10, 17), a computer model for the Pdr·Pdx ET complex was generated and experimentally tested (16). However, the precise manner of the Pdr-Pdx interaction, the docking sites, and interface residues remained unknown. The most direct way to obtain this information would be determination of the x-ray structure of the Pdr·Pdx complex, but, despite our multiple attempts, the native proteins resisted co-crystallization. This could in part be due to weak association (Kd = 66 μm) (16) and quick inactivation of wild type Pdx in solution (8).
To overcome this problem, we attempted to produce a functionally active Pdr·Pdx conjugate. Having screened various cross-linking agents, reaction conditions, and Pdx mutants, we prepared a catalytically competent complex between wild type Pdr and Pdx C73S/C85S covalently linked by 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide (EDC) via the Lys409Pdr-Glu72Pdx salt bridge (6). The double mutant of Pdx was chosen for a functional analysis because substitution of cysteines 73 and 85 with serine, a highly isosteric analogue, only mildly affects the electron-accepting ability of the iron-sulfur cluster but significantly increases protein stability by preventing the intermolecular disulfide bridge formation and loss of the metal cluster (10). The covalent complex effectively catalyzes reduction of small redox-active molecules, receiving electrons from Pdr, as well as cytochrome c and P450cam, receiving electrons from Pdx. Importantly, the fused partners reduce cytochrome c 150-fold more effectively than the intact Pdr·Pdx pair, whereas ET to P450cam from the linked Pdx is partially impaired due to interference of the attached Pdr. In the presence of a large excess of P450cam (P450cam:Pdx ≥ 30), however, linked Pdr and Pdx transfer electrons to the hemoprotein more efficiently than the freely diffusible pair. Tethered Pdx can also serve as an effector and support the camphor hydroxylation reaction with no uncoupling of ET from hydroxylation and, thus, induces the same type of functionally important structural changes in P450cam as free Pdx. The kinetic behavior of P450cam reduction and camphor hydroxylation by the covalent complex, neither of which is saturated at high P450cam concentrations, suggests that the fused Pdx spends a short time in an orientation favorable for ET to P450cam and that suboptimal orientation of the iron-sulfur protein may limit turnover.
Having produced a highly purified, homogeneous and catalytically active Pdr·Pdx C73S/C85S cross-link, we were able to crystallize it and analyze by x-ray crystallography. The 2.6-Å crystal structure of the complex described here provides a detailed view on the Pdr-Pdx interface and cofactor geometry, suggests a FAD-to-[2Fe-2S] ET pathway, explains previously reported experimental results, and enables structural comparisons with the homologous redox pairs.
MATERIALS AND METHODS
Preparation and Crystallization of the Cross-linked Pdr·Pdx Complex
Protein purification and preparation of the EDC-linked Pdr·Pdx C73S/C85S complex were carried out as reported previously (6, 8, 18). The fusion protein was crystallized at room temperature by a hanging-drop vapor diffusion method. Droplets contained 4 μl of 4 mg/ml protein in 30 mm HEPES, pH 7.4, and 2 μl of a reservoir solution of 1.3 m malonate, pH 6.4. Crystals grew overnight and belonged to the orthorhombic P212121 space group with the unit cell dimensions of 67.6 Å × 103.4 Å × 167.7 Å and two molecules/asymmetric unit. Thirty percent glycerol was used as a cryoprotectant.
X-ray Data Collection
Diffraction data were collected at −170 °C on the Advanced Light Source beamline 5.0.2 at the Lawrence Berkley National Laboratory. Data were indexed, integrated, and scaled with DENZO and SCALEPACK (19) (Table 1). Subsequent data manipulation was performed with the CCP4 software suite (20).
TABLE 1.
Data collection and refinement statistics
| Space group | P212121 |
|---|---|
| Unit cell (Å) | |
| a | 67.6 |
| b | 103.4 |
| c | 167.7 |
| Molecules/unit | 2 |
| Resolution range (Å) | 49.5–2.60 |
| Total observations | 139,651 |
| Unique reflections | 36,524 |
| Completeness (%) | 98.6 (99.9)a |
| I/σ (I) | 25.3 (5.6) |
| Rsym (%) | 5.3 (17.9) |
| R/Rfreeb (%) | 24.4/27.2 |
| No. of residues | 1,038 |
| No. of water molecules | 62 |
| Average B-factor (Å2) | 75.8 |
| Root mean square deviations | |
| Bonds (Å) | 0.015 |
| Angles (°) | 1.4 |
a Numbers in parentheses are for the highest resolution bin.
b Rfree was calculated from a subset of 5% of the data that were excluded during refinement.
Structure Determination and Refinement
The Pdr·Pdx complex structure was solved by molecular replacement with PHASER (20) using Pdr and Pdx C73S/C86S molecules as search models (Protein Data Bank (PDB) codes 1Q1R and 1OQQ, respectively). The final model was refined with O (21) and CNS (22) and consists of residues 2–420Pdr and 1–104Pdx in complex 1, 7–418Pdr and 1–103Pdx in complex 2, and 62 water molecules. Refinement statistics are summarized in Table 1. Coordinates have been deposited into the RCSB Protein Data Bank with the PDB ID code 3LB8. Figures were prepared with PyMOL (32).
RESULTS AND DISCUSSION
Overall Structure of the Pdr·Pdx Complex
Crystals of the Pdr·Pdx complex have a yellow-brown color, indicating the presence of both flavin and iron-sulfur cofactors, whereas electrophoretic analysis of the dissolved crystals confirmed intactness of the covalent link (supplemental Fig. 1SA). Although the crystals contain two Pdr·Pdx complexes in the asymmetric unit, which we will refer to as complexes 1 and 2, during molecular replacement searches only solutions for two Pdr molecules were found but none for Pdx. Electron density maps calculated based on the molecular replacement phases were of sufficient quality to identify two metal cluster positions, stronger for complex 1 and weaker for complex 2 (supplemental Fig. 1S, B–D), which enabled Pdx molecules to be positioned manually. The resulting structure was refined to R and Rfree of 24.4 and 27.2%, respectively (Table 1).
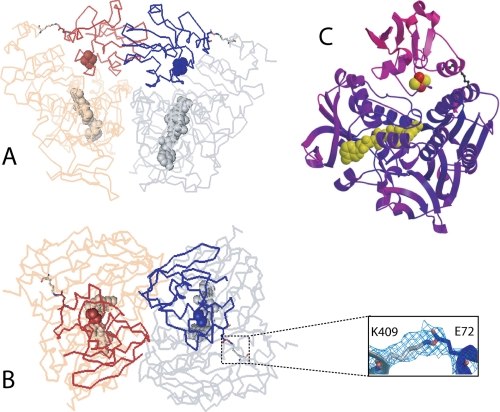
Different views at the crystallographic dimer can be seen in Fig. 1, A and B. In each complex, Pdx is docked at the groove above the si side of the isoalloxazine ring, the active site of Pdr. In agreement with our previous study (6), the electron density map clearly shows that Lys409Pdr and Glu72Pdx are covalently linked by EDC (Fig. 1B). Notably, Pdx in the crystal lattice is fully exposed to the solvent channel and makes no contacts other than with the attached Pdr (supplemental Fig. 2S). This may explain why Pdx molecules have high thermal motion and are poorly defined (Fig. 1C). Also, the lack of spatial constrains indicates that the Pdr·Pdx orientation observed in the crystal is the one trapped by EDC. As better defined overall, complex 1 was used for structural analysis outlined below.
FIGURE 1.
A and B, two views at the crystallographic Pdr·Pdx dimer. The 2 Fo − Fc electron density around the Lys409Pdr-Glu72Pdx covalent bond in complex 1 is contoured at 1.0 σ. Pdr molecules are rendered in gray and beige; Pdx is in dark blue and brown. FAD and the [2Fe-2S] cluster are in cpk representation. C, ribbon diagram of complex 1 colored according to the B-factor. Deep purple and magenta correspond to the B-factor values of 20.0 and 150.0, respectively.
Pdr-Pdx Interface
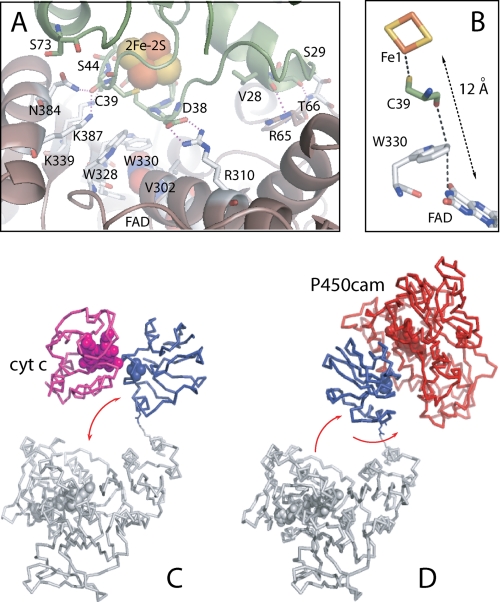
The Pdr-Pdx interface is relatively small and constitutes approximately 365 Å2 of the surface area in each protein. Among 20 interface residues in Pdr, there are 9 hydrophobic, 4 polar, and 5 positively charged. In Pdx, 14 interacting residues include 4 hydrophobic, 4 polar, and 3 charged. Hence, the protein-protein interface provides the basis for both hydrophobic and polar interactions. Besides the cross-linked Lys409Pdr-Glu72Pdx pair, there is only one bidentate Arg310Pdr-Asp38Pdx salt bridge (Fig. 2A). For many redox couples, electrostatic forces play an important role in formation and stabilization of productive ET complexes as they provide long range steering for the initial protein-protein association, optimize the partner orientation for maximal activity, and control solvent access to the interface. Given that substitution of Asp38Pdx with Asn, Ala, or Ile decreases the Pdr-to-Pdx ET by 50–70% and diminishes the binding affinity between the partners (6, 14–16), we conclude that the centrally located Arg310Pdr-Asp38Pdx charge-charge pair is the key element that guides docking and positions Pdr and Pdx optimally for ET.
FIGURE 2.
A, Pdr-Pdx interface. Only selective side chains of Pdr (brown/gray) and Pdx (green) are displayed. Redox centers are in cpk representation. Dotted lines indicate charge-charge and H-bonding interactions. B, FAD-to-[2Fe-2S] ET pathway predicted by HARLEM (24). C and D, hypothetical complexes of the cross-linked Pdr and Pdx with cytochrome c and P450cam, respectively. Pdx movements required for docking with the electron acceptors are shown by arrows.
That electrostatic interactions are important for Pdr-Pdx association follows also from our molecular dynamics simulation of the Pdr·Pdx complex derived from the crystallographic complex 1 (described in detail in the supplemental molecular dynamics simulation), showing that the Arg310Pdr-Asp38Pdx and Lys409Pdr-Glu72Pdx ion pair interactions remain stable throughout the 15-ns molecular dynamics trajectory even if the intermolecular cross-link is not treated as a covalent bond. In accord with the mutagenesis data (6, 15, 16, 31), this suggests that the salt bridge-forming residues may be crucial for establishing and stabilization of the Pdr·Pdx ET complex.
Other polar interactions include five peripheral hydrogen bonds between the main/side chain atoms of Val28, Ser29, and Ser44 of Pdx and Arg65, Thr66, Lys339, Asn384, and Lys387 of Pdr (Fig. 2A), whereas hydrophobic Val302, Pro303, Leu306, Trp328, Trp330, Pro380, and Phe383 of Pdr and Val28, Tyr33, Leu36, and Met70 of Pdx, surrounding the redox centers, contribute to nonpolar contacts. The very limited number of specific charge-charge and H-bonding interactions suggests that the Pdr-Pdx association is assisted by hydrophobic forces and steric complementarity, as proposed previously (4, 12).
One of the mutated residues, Ser73Pdx, is located at the edge of the protein-protein interface, only 3.0 Å away from the Asn384Pdr side chain (Fig. 2A). Thus, the residue at position 73 not only modulates the Pdx redox potential (the C73S mutation decreases E1/2 by 28 mV) (16) but also defines how close Pdx and Pdr can approach each other. Because substitution of Cys73 with either smaller Gly and Ser or bulky Arg affects Pdr-Pdx association and interprotein ET (8, 15), we conclude that the surface cysteine is one of the residues that finely tunes interactions at the interface. Another substituted residue, Ser85Pdx, is solvent-inaccessible and cannot directly influence Pdr-Pdx association but may affect the Pdr-to-Pdx ET rate (10) by altering the environment of the metal cluster as it neighbors the iron-ligating Cys86. It is of interest to mention also that the C-terminal Trp106Pdx, which has no significant affect on the Pdr·Pdx complex formation and interprotein ET (6, 16), is not seen in the structure.
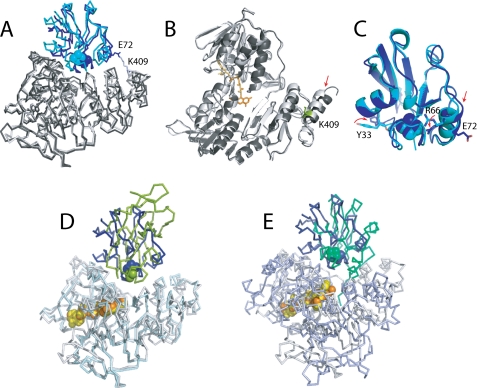
Judging from the “out” conformation of the Cys45-Ala46 peptide bond in Pdx and the overall conformation of the upper [2Fe-2S]-binding loop, resembling that observed in the sodium dithionite-reduced Pdx (17), we conclude that the metal cluster was reduced by synchrotron radiation (23). It is possible that the [2Fe-2S] reduction triggered movements of the Tyr33 and Arg66 side chains (Fig. 3C). As part of the 32Ile-Asp38 peptide that precedes the metal-binding loop and participates in the redox reorganization, Tyr33 undergoes a large positional shift upon Pdx oxidoreduction (up to 2 Å), occupying multiple conformations and forming a hydrogen bond between the tyrosine hydroxyl and the Asp9 carboxyl in the reduced iron-sulfur protein (17). On the other hand, the positively charged side chain of Arg66, mainly nonbonded in oxidized Pdx, becomes H-bonded with the main or side chains atoms of Ser42, Ala43, or Ser44, partially compensating for the negative charge and stabilizing the reduced form of Pdx (17). The conformational changes in Tyr33 and Arg66 may be important not only for regulation of the redox properties of Pdx but also for Pdr-Pdx interaction because, as our mutagenesis study revealed, the Tyr33 and Arg66 substitutions have the largest effect on the kinetics of Pdr-to-Pdx ET (16). To clarify whether or not conformational perturbations in Tyr33 and Arg66 affect orientation of Pdx in the Pdr·Pdx complex, determination of the crystal structure with the fully oxidized Pdx would be necessary.
FIGURE 3.
A, structural comparison of the crystallographic (gray/blue) and computer-generated (black/cyan; model 2 (16) Pdr·Pdx complexes. B and C, ribbon diagrams of backbone overlays of intact and cross-linked Pdr and Pdx, respectively. Displacement of α-helices in Pdr and Pdx caused by the Lys409Pdr-Glu72Pdx covalent link and positional changes in redox-sensitive Tyr33Pdx and Arg66Pdx is indicated by arrows. D and E, structural overlay between the Pdr·Pdx complex (gray/blue/orange) and x-ray models of native BphA4-BphA3 (light cyan/green/yellow, PDB code 2YVJ) and EDC-cross-linked Adr·Adx (light blue/sage/yellow, PDB code 1E6E), respectively. Only the flavoproteins were used for the least-squares superposition.
Redox Cofactor Geometry and Electron Transfer Path
The edge-to-edge distance and the interplanar angle for the FAD and [2Fe-2S] cofactors observed in the crystal structure are 12.0 Å and 75°, respectively. The best electron transfer pathway from the flavin N3 atom to Fe1 of the metal cluster predicted by HARLEM (24) proceeds through the FAD flanking Trp330Pdr and the iron ligand Cys39Pdx (Fig. 2B and supplemental Fig. 3S). This is the most direct route with an average packing density of 0.61 and a decay component of 1.63 that could provide a maximum ET rate of 2.7 × 105 s−1 (supplemental Table 1S). The theoretical Pdr-to-Pdx ET rate is 3 orders of magnitude higher that the experimental value of 235 s−1 (16). One possible reason for this difference could be a large reorganizational energy for the reductive step in Pdx, 2.3 eV (25), due to a major nuclei, dipole, and charge rearrangement taking place during the metal cluster reduction (17).
Movements Required for the Linked Pdx during Turnover
As reported earlier (6), the Pdr·Pdx covalent complex retains substantial electron transferring activities. This means that the ET site of Pdx, which we assume is the closest approach of the iron-sulfur cluster to the molecular surface, must be able to access both P450cam and cytochrome c even while Pdx is covalently tethered to Pdr. Consistent with these observations, the covalent link is at the edge of the complex near the surface so it is sterically quite feasible for Pdx to undergo significant positional changes to allow the electron-accepting partners to access the ET site. To deliver electrons to cytochrome c, for instance, 20–30-Å movements of Pdx along the vertical plane would be sufficient (Fig. 2C). The high turnover number for the cytochrome c reduction reaction catalyzed by the fused Pdr·Pdx couple (5,700 min−−1) (6) suggests that these simple movements do not limit the Pdx-to-cytochrome c ET. In contrast, to dock productively with the considerably larger P450cam, Pdx would have to turn additionally by 130–180° to avoid steric clashing (Fig. 2D). The necessity of such complex movements may be the reason for an impaired exchange of reducing equivalents between the linked Pdx and P450cam (6).
Comparison of the Crystallographic and Computer-generated Pdr·Pdx Models
Structural comparison reveals striking similarity between the crystallographic and computer-generated Pdr·Pdx complexes (Fig. 3A) (16). The detected conformational differences are likely caused by the covalent bonding that pulled the Lys409-containg C-terminal α-helix, normally flexible in Pdr (10), and the Glu72-containing α-helix in Pdx toward each other (indicated by arrows in Fig. 3, B and C). As a consequence, the iron-sulfur protein in the covalent complex is rotated by a few degrees and shifted by approximately 5.0 Å toward the Pdr C terminus. Other than that, there are only slight changes in the Pdr structure caused by the Pdx attachment: the root mean square deviation between the 404 Cα positions of the superimposed cross-linked and intact Pdr is only 0.72 Å. The higher root mean square deviation for the 103 Cα atoms of the respective Pdx molecules (0.86 Å) may be the result of both the covalent linkage and photoreduction of the metal cluster. Markedly, despite positional and conformational differences, electronic coupling parameters, pathways, and ET rates predicted for the computer-generated and x-ray models are nearly identical (supplemental Table 1S; see also Table 2 in Ref. 16).
Comparison of the Pdr·Pdx and Homologous Redox Pairs
To date, structural information on two redox pairs closely related to Pdr and Pdx is available. The first couple is an oxygenase-coupled ferredoxin reductase (BphA4) and a Rieske-type ferredoxin (BphA3) from Pseudomonas sp. (26). BphA4 has the same fold as Pdr and displays higher affinity for BphA3 when the flavin is reduced, because of which it was possible to co-crystallize the native proteins under reduced conditions (27). Unlike Pdx, BphA3 undergoes minor redox-linked structural changes that include the peptide bond flip in one of the [2Fe-2S]-binding residues (27). The superimposed Pdr·Pdx and BphA3-BphA4 complexes are shown in Fig. 3D. Although Pdx and BphA3 belong to two different types of ferredoxins and are structurally distinct, the manner of their interaction with the flavoprotein partners is nearly the same. Moreover, the interface between the Pseudomonas sp. proteins also consists of a small hydrophobic patch surrounded by polar interactions, with the shortest FAD-[2Fe-2S] distance of 9.3 Å and Trp320BphA4 and His66BphA3, corresponding to Trp330Pdr and Cys39Pdx, respectively, comprising the ET path (27).
Cytochrome P450-coupled adrenodoxin reductase (Adr) and adrenodoxin (Adx) are mammalian homologues of Pdr and Pdx. Unlike Pdr, Adr is a membrane-bound protein with a differently folded C-terminal domain and a highly positively charged active site (10, 28), whose interaction with acidic Adx is electrostatically guided (29). In the x-ray structure of the EDC-cross-linked Adr·Adx complex, the flavoprotein undergoes slight domain reorientation to accommodate Adx, and the resulting 580-Å2 interface is composed of predominantly polar and charged residues, with a large number of interprotein hydrogen bonds and salt bridges and the edge-to-edge FAD-[2Fe-2S] distance of 9.7 Å (30). Despite the functional and structural dissimilarities, binding topology and redox center geometry in the Pdr·Pdx and Adr·Adx complexes are remarkably similar (Fig. 3E).
The common manner of the BphA4-BphA3, Adr-Adx, and Pdr-Pdx interaction and good correlation between the x-ray data and experimental and theoretical results suggest that the Pdr·Pdx structure is biologically relevant. Although there might be some differences between the reported structure and a solution complex formed between the native partners due to mutational, cross-linking, and photoreduction effects, the general mode of the Pdr-Pdx interaction is likely to be preserved in the crystallographic complex, and hence, it can provide the structural framework for mechanistic studies on this and related redox couples.
Supplementary Material
Acknowledgment
We thank Dr. H. Li for assistance with synchrotron data collection.
This work was supported, in whole or in part, by National Institutes of Health Grants GM67637 (to I. F. S.) and GM33688 (to T. L. P.). This work was based on research conducted at the Advanced Light Source, which is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Dept. of Energy under Contract DE-AC02-05CH11231.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1S–3S, Table 1S, a molecular dynamics simulation, and additional references.
The atomic coordinates and structure factors (code 3LB8) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Pdr
- putidaredoxin reductase
- Pdx
- putidaredoxin
- P450cam
- camphor-hydroxylating cytochrome P450 from P. putida
- ET
- electron transfer
- EDC
- 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide
- PDB
- Protein Data Bank
- BphA4 and BphA3
- ferredoxin reductase and Rieske-type ferredoxin from Pseudomonas sp., respectively
- Adr
- adrenodoxin reductase
- Adx
- adrenodoxin.
REFERENCES
- 1.Katagiri M., Ganguli B. N., Gunsalus I. C. (1968) J. Biol. Chem. 243, 3543–3546 [PubMed] [Google Scholar]
- 2.Hintz M. J., Mock D. M., Peterson L. L., Tuttle K., Peterson J. A. (1982) J. Biol. Chem. 257, 14324–14332 [PubMed] [Google Scholar]
- 3.Roome P. W., Peterson J. A. (1988) Arch. Biochem. Biophys. 266, 41–50 [DOI] [PubMed] [Google Scholar]
- 4.Sevrioukova I. F., Hazzard J. T., Tollin G., Poulos T. L. (2001) Biochemistry 40, 10592–10600 [DOI] [PubMed] [Google Scholar]
- 5.Purdy M. M., Koo L. S., Ortiz de Montellano P. R., Klinman J. P. (2004) Biochemistry 43, 271–281 [DOI] [PubMed] [Google Scholar]
- 6.Churbanova I. Y., Poulos T. L., Sevrioukova I. F. (2010) Biochemistry 49, 58–67 [DOI] [PubMed] [Google Scholar]
- 7.Poulos T. L., Finzel B. C., Howard A. J. (1987) J. Mol. Biol. 195, 687–700 [DOI] [PubMed] [Google Scholar]
- 8.Sevrioukova I. F., Garcia C., Li H., Bhaskar B., Poulos T. L. (2003) J. Mol. Biol. 333, 377–392 [DOI] [PubMed] [Google Scholar]
- 9.Smith N., Mayhew M., Holden M. J., Kelly H., Robinson H., Heroux A., Vilker V. L., Gallagher D. T. (2004) Acta Crystallogr. Sect. D 60, 816–822 [DOI] [PubMed] [Google Scholar]
- 10.Sevrioukova I. F., Li H., Poulos T. L. (2004) J. Mol. Biol. 336, 889–902 [DOI] [PubMed] [Google Scholar]
- 11.Roome P. W., Peterson J. A. (1988) Arch. Biochem. Biophys. 266, 32–40 [DOI] [PubMed] [Google Scholar]
- 12.Aoki M., Ishimori K., Fukada H., Takahashi K., Morishima I. (1998) Biochim. Biophys. Acta 1384, 180–188 [DOI] [PubMed] [Google Scholar]
- 13.Aoki M., Ishimori K., Morishima I. (1998) Biochim. Biophys. Acta 1386, 168–178 [DOI] [PubMed] [Google Scholar]
- 14.Aoki M., Ishimori K., Morishima I. (1998) Biochim. Biophys. Acta 1386, 157–167 [DOI] [PubMed] [Google Scholar]
- 15.Holden M., Mayhew M., Bunk D., Roitberg A., Vilker V. (1997) J. Biol. Chem. 272, 21720–21725 [DOI] [PubMed] [Google Scholar]
- 16.Kuznetsov V. Y., Blair E., Farmer P. J., Poulos T. L., Pifferitti A., Sevrioukova I. F. (2005) J. Biol. Chem. 280, 16135–16142 [DOI] [PubMed] [Google Scholar]
- 17.Sevrioukova I. F. (2005) J. Mol. Biol. 347, 607–621 [DOI] [PubMed] [Google Scholar]
- 18.Sevrioukova I. F., Poulos T. L. (2002) J. Biol. Chem. 277, 25831–25839 [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 20.CCP4 (1994) Acta Crystallogr. Sect. D 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 21.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. Sect. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 22.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. Sect. D 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 23.Corbett M. C., Latimer M. J., Poulos T. L., Sevrioukova I. F., Hodgson K. O., Hedman B. (2007) Acta Crystallogr. Sect. D 63, 951–960 [DOI] [PubMed] [Google Scholar]
- 24.Kurnikov I. V. (2000) HARLEM, Version 1.0, Department of Chemistry, University of Pittsburgh, Pittsburgh, PA [Google Scholar]
- 25.Kuznetsov V. Y., Poulos T. L., Sevrioukova I. F. (2006) Biochemistry 45, 11934–11944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimbara K., Hashimoto T., Fukuda M., Koana T., Takagi M., Oishi M., Yano K. (1989) J. Bacteriol. 171, 2740–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Senda M., Kishigami S., Kimura S., Fukuda M., Ishida T., Senda T. (2007) J. Mol. Biol. 373, 382–400 [DOI] [PubMed] [Google Scholar]
- 28.Ziegler G. A., Vonrhein C., Hanukoglu I., Schulz G. E. (1999) J. Mol. Biol. 289, 981–990 [DOI] [PubMed] [Google Scholar]
- 29.Grinberg A. V., Hannemann F., Schiffler B., Müller J., Heinemann U., Bernhardt R. (2000) Proteins 40, 590–612 [DOI] [PubMed] [Google Scholar]
- 30.Müller J. J., Lapko A., Bourenkov G., Ruckpaul K., Heinemann U. (2001) J. Biol. Chem. 276, 2786–2789 [DOI] [PubMed] [Google Scholar]
- 31.Aoki M., Ishimori K., Morishima I., Wada Y. (1998) Inorg. Chim. Acta 272, 80–88 [Google Scholar]
- 32.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.