Abstract
Virtually nothing is known about the mechanisms and enzymes responsible for the glycosylation of arabinogalactan proteins (AGPs). The glycosyltransferase 37 family contains plant-specific enzymes, which suggests involvement in plant-specific organs such as the cell wall. Our working hypothesis is that AtFUT4 and AtFUT6 genes encode α(1,2)fucosyltransferases (FUTs) for AGPs. Multiple lines of evidence support this hypothesis. First, overexpression of the two genes in tobacco BY2 cells, known to contain nonfucosylated AGPs, resulted in a staining of transgenic cells with eel lectin, which specifically binds to terminal α-linked fucose. Second, monosaccharide analysis by high pH anion exchange chromatography and electrospray ionization mass spectrometry indicated the presence of fucose in AGPs from transgenic cell lines but not in AGPs from wild type cells. Third, detergent extracts from microsomal membranes prepared from transgenic lines were able to fucosylate, in vitro, purified AGPs from BY2 wild type cells. Susceptibility of [14C]fucosylated AGPs to α(1,2)fucosidase, and not to α(1,3/4)fucosidase, indicated that an α(1,2) linkage is formed. Furthermore, dearabinosylated AGPs were not substrate acceptors for these enzymes, indicating that arabinosyl residues represent the fucosylation sites on these molecules. Testing of several polysaccharides, oligosaccharides, and glycoproteins as potential substrate acceptors in the fucosyl transfer reactions indicated that the two enzymes are specific for AGPs but are not functionally redundant because they differentially fucosylate certain AGPs. AtFUT4 and AtFUT6 are the first enzymes to be characterized for AGP glycosylation and further our understanding of cell wall biosynthesis.
Keywords: Arabidopsis, Cell Wall, Glycoprotein Biosynthesis, Golgi, Mass Spectrometry (MS), Arabinogalactan Proteins, Fucosyltransferases
Introduction
Plants cell wall synthesize many fucose (Fuc)2-containing polymers including xyloglucans (XyGs), rhamnogalacturonan (RG), arabinogalactan proteins (AGPs), and N-glycans (1–3). In most cases, Fuc residues at terminal positions of the polymer are linked via α(1,2) or α(1,3) linkages. Terminal Fuc exists in a methylated form in type A side chains of RG-II; however, in type B side chains, the Fuc is internal and attached to rhamnose via an α(1,4) linkage and bears a glucuronic acid at its C-4 position (2). In animals, Fuc residues are involved in several physiological processes including fertilization, development, host-pathogen interactions, and cancer (4–6). In plants, Fuc is important in regulating cell wall integrity and extensibility in growing tissues. For example, Fuc in XyGs terminates sugar side chains that are attached to a β(1,4)glucan backbone, which in turn binds tightly to the cellulose network of dicot primary cell walls (1). Molecular models predict that such side chains enhance the binding of XyGs to the surface of cellulose microfibrils (7). In addition, XyG subunits containing Fuc possess growth regulating activity (8–10). An Arabidopsis mutant that cannot synthesize fucosylated polymers (mur1) is brittle and shows stunted growth compared with wild type (11). The fact that this mutant recovers sufficiently to complete its life cycle is probably explained in part by the substitution of l-Fuc by l-galactose in XyGs (12) and N-glycans (13). O'Neill et al. (14) reported that the dwarf phenotype of mur1 is due to Fuc reduction that affects the ability of RG-II to form a stable borate-RG-II complex. More recent analysis of the mur1 mutant showed that fucosylated AGPs are required for root cell elongation (15).
Fucosyltransferases (FUTs) catalyze the transfer of Fuc from GDP-Fuc to various substrate acceptors. They are classified into seven glycosyltransferase (GT) families in the CAZy (carbohydrate-active enzymes) database. Three of these GT families (GT11, GT23, and GT74) contain α(1,2/6)FUTs for N-glycans, whereas α(1,3/4)FUTs for N-glycans cluster together in the GT10 family. The GT65 and GT68 families contain enzymes involved in protein O-fucosylation in eukaryotes and prokaryotes, whereas the GT37 family contains exclusively plant enzymes. Thus, members of the GT37 family are most likely involved in fucosylation of plant-specific organelles such as the cell wall. The Arabidopsis and rice genomes contain 10 and 17 members of GT37 family, respectively. Among these Arabidopsis genes, which are designated AtFUT1-AtFUT10, only AtFUT1 (At2g03220) has a defined function as a XyG-specific α(1,2)FUT (16, 17). Our working hypothesis is that some GT37 family members are responsible for adding Fuc to various AGPs. AGPs are found in the cell walls, plasma membranes, and extracellular secretions of plants and are extensively glycosylated members of the hydroxyproline-rich glycoprotein family, which function in various aspects of plant growth and development (reviewed in Ref. 18).
Although progress in elucidating the genes responsible for the biosynthesis of several plant cell wall polysaccharides is being made, virtually nothing is known about the mechanisms and enzymes responsible for the glycosylation of AGPs (19). Although the Golgi complex has been shown to be involved in hydroxyproline-rich glycoprotein glycosylation a while ago (20) and an arabinosyltransferase (AraT) activity for extensin was identified in particulate membranes (21), only recently was an Arabidopsis gene At2g35610 (a member of the GT77 family) for this AraT activity identified (22). Fuc is present in AGPs in several dicot plants such as Arabidopsis (15) and radish (23). Linkage analysis, reactivity with eel lectin, and digestion with α(1,2)fucosidase indicate that these fucosyl residues are terminal and attached, via an α-linkage, to the C-2 position of the adjacent arabinofuranosyl (Araf) residue (23, 24). The only report describing an α(1,2)FUT activity for AGPs came from a work on radish root, in which a Ara-α(1,3)Gal-β(1,6)Gal trisaccharide was used as exogenous substrate acceptor to mimic an arabinogalactan polysaccharide in an enzymatic assay (25). In this report, we describe the functional characterization of two putative AGP-α(1,2)FUTs, encoded by the AtFUT4 and AtFUT6 genes, from the GT37 family. Our results demonstrate that both enzymes can fucosylate AGPs in vivo and in vitro and are not active against other cell wall polymer substrates in vitro. These two enzymes are the first to be characterized for AGP glycosylation and further our understanding of cell wall biosynthesis.
EXPERIMENTAL PROCEDURES
Chemicals and Plant Material
GDP-[14C]Fucose (200 mCi/mmol) was from PerkinElmer Life Sciences. Tobacco Bright Yellow 2 (BY2) wild type cell suspension cultures and β-glucosyl Yariv reagent (2 mg/ml) in 1% (w/v) NaCl were kindly provided by Dr. Marcia Kieliszewski (Department of Chemistry and Biochemistry, Ohio University). All of the detergents, α1-acid glycoprotein, Dowex 1-X80 resins were purchased from Sigma. Tamarind XyG, rhamnogalacturonans (from soybean and potato), and arabinan (from sugar beet) were purchased from Megazyme International (Bray, Ireland). N-Acetyllactosamine was from Fisher. PD-10 desalting columns were from GE Healthcare. Eel lectin (Anguilla anguilla) linked to the fluorescent Texas Red was from EY Laboratories Inc (San Mateo, CA). α(1,2)Fucosidase (GK80170, 400 milliunits/mg) was from Glyko (Bicester, UK), and α(1,3/4)fucosidase (E-F134, 2.3 units/mg) from almond meal was from QA-Bio, LLC (Palm Desert, CA). The Bio-gel P2 was from Bio-Rad.
Cloning and Expression of AtFUT4.1 and AtFUT6 Genes in Tobacco BY2 Cells
Full-length cDNA clones of AtFUT4.1 and AtFUT6 were obtained from INRA-CNRGV and the ABRC DNA Stock Center (Columbus, Ohio), respectively.
For the AtFUT4.1 cDNA, an N terminus His-tagged construct was created with a PCR-based strategy using Taq DNA Polymerase (New England BioLabs), a forward primer containing the His6 tag (5′-caccatgcatcatcatcatcatcacACCATCGTCTTTAGTACCTTAC-3′), and a reverse primer (5′-CTATAACTCATCAAAAAGCTTAAGC-3′). After amplification, the PCR product was cloned into pCR4-TOPO vector via a TOPO cloning reaction (Invitrogen). The final construct was sequenced twice by the Ohio University Genomics Facility for verification. The His-AtFUT4.1 construct was released from pCR4-TOPO:His-FUT4.1 by EcoRI digestion, gel-purified using QIAquick gel extraction kit (Qiagen), and ligated with T4 DNA ligase (New England BioLabs) into the Gateway pENTR2B vector (Invitrogen). A diagnostic digest was used to check for the correct orientation of His-FUT4.1. For expression in plants (tobacco BY2 cells), the His-FUT4.1 construct was transferred from pENTR2B to pMDC32 via an LR Clonase reaction using ApaI digestion. The pMDC32 expression vector uses a double 35S cauliflower mosaic virus promoter to drive expression and encodes kanamycin resistance in bacteria and hygromycin resistance in plants (26).
For the AtFUT6 cDNA, the N terminus His-tagged construct was created with a PCR-based strategy using AccuPrime Pfx Polymerase (Invitrogen), a forward primer containing the His6 tag (5′-caccatgcatcatcatcatcatcacACCATGAAGATTCTGCTAACACTA-3′) and a reverse primer (5′-CATTAACTCATCAAATAGCTT-3′). The resulting PCR product was cloned into the pENTR/d-TOPO Gateway vector (Invitrogen). The final construct was sequenced twice for verification. His-FUT6 was then transferred from pENTR/d-TOPO to pMDC32 by an LR Clonase reaction using MluI digestion. All of the plasmid constructs were transformed into DH5α Escherichia coli cells, and plasmids were purified using a QIAprep spin miniprep kit (Qiagen).
Transformation of BY2 Cells
The pMDC32 vector containing His-AtFUT4.1 or His-AtFUT6 cDNAs was introduced into electrocompetent Agrobacterium strain EHA105 by electroporation (Bio-Rad; voltage, 2.4 kV, 200 Ω, 5-ms pulse). Electroporated cells were mixed with 1 ml of LB and incubated at 28 °C for 4 h with agitation (200rpm). Transformed EHA105 were selected on medium containing kanamycin and rifampicin for 48 h at 28 °C and confirmed by PCR. To transform BY2 cells, 100 μl of 24 h EHA105 culture grown in LB containing 200 μm acetosyringone was co-incubated with 5 ml of 4-day-old tobacco BY2 cells for 48 h at 22 °C in the dark. After washing, BY2 cells were spread over plates of NT-1 medium (4.3 g/liter of Murashige and Skoog salts, 30 g/liter of sucrose, 100 mg/liter of KH2PO4, 50 mg/liter of myo-inositol, 1 mg/liter of thiamine, and 0.22 mg/liter of 2,4-dichlorophenoxyacetic acid, pH 5.6) containing 0.6% Gelrite, hygromycin (20 mg/liter), and cefotaxime (250 mg/liter) for 4 weeks at 22 °C in the dark for microcalli generation.
Transgenic BY2 cell suspension cultures were generated from individual transformed calli and maintained by subculturing every 3 weeks (10 ml of suspension cells into 100 ml of fresh medium supplemented with 50 mg/ml hygromycin). Wild type BY2 cells were cultured in media without antibiotic.
Transient Expression in Nicotiana tabacum Leaves
Cloning of AtFUT6-GFP was done as described above using the following primer sequences: forward, 5′-CACCATGAAGATTCTGCTAACACTA-3′, and reverse, 5′-CATTAACTCATCAAATAGCTT-3′. The pMDC83 expression vector that encompasses the GFP gene was used. The sialyltransferase (ST)-YFP construct was a gift from Dr. Yumiko Sakuragi (Department of Plant Biology and Biotechnology, Copenhagen University, Denmark). The AtFUT6-GFP and ST-YFP genes along with P19 were co-incubated for 1 h at room temperature and co-infiltrated into leaves of N. tabacum. The epidermal cell layers were observed for fluorescence after 2 days in the dark. Fluorescence was observed using a Leica TCS SP2 AOBS confocal laser-scanning microscope. For co-localization of GFP and YFP in epidermal cells, a 488- and 514-nm argon ion laser was used. Fluorescence signals were separated using an acoustico-optical beam splitter; the emission signal was 495–510 nm for GFP and 540–580 nm for YFP. Appropriate controls were performed to exclude the possibility of cross-talk between the two fluorochromes before image acquisition.
Purification and Dearabinosylation of Arabinogalactan Proteins from Tobacco BY2 Cells
AGPs were purified by precipitation with Yariv reagent according to the procedures described in Ref. 27. Dearabinosylation of AGPs was carried out using mild acid conditions as described in Ref. 28. Dearabinosylated AGPs-BY2WT were solubilized in water to a final carbohydrate concentration of 1%(w/v), before use as acceptor (100 μg) in enzyme assays. The removal of Ara residues was confirmed by monosaccharide composition analysis.
Preparation of Golgi-enriched Membranes and Detergent Solubilization
Microsomal membrane preparation and enzyme solubilization were carried out essentially as described in Ref. 29. Detergent extracts were prepared using 0.5%(w/v) digitonin (Sigma), and all of the detergent extracts were adjusted to a final protein concentration of ∼2 mg/ml before use in the enzyme assay.
AGP-Fucosyltransferase Assay
The fucosyltransferase enzyme assay was performed essentially as described earlier (17).
Fucosidase Treatments and Gel Filtration Fractionation
Two α-fucosidases were used: α(1,2)fucosidase II and α(1,3/4)fucosidase from almond meal. Reactions consisted of 10 μg of AGPs (1000–4000 cpm) being incubated in the presence of ∼800 milliunits of fucosidase in the appropriate buffer (recommended by the manufacturers) for 24 h at 37 °C. The reactions were stopped by boiling at 100 °C for 10 min, and insoluble material was removed by centrifugation (10 min, 10,000 rpm). Supernatants were fractionated by gel filtration on a 90 × 1.5-cm Bio-gel P2 column (Bio-Rad) that was eluted by gravity with degassed water containing 0.02% sodium azide. Fractions (2.3 ml) were collected, and the [14C]radiolabel content was monitored for each fraction. Untreated reaction products were used as controls. Fucose, xylo-oligosaccharides (DP2–6), and XyG-oligosaccharides (DP7–9) were used as standards to calibrate the column.
Monosaccharide Analysis by High pH Anion Exchange Chromatography (HPAEC)
Both total acid hydrolysis and fractionation of the monosaccharides on a CarboPac PA20 column (Dionex) were carried out as described earlier in Ref. 29. The standards used are known monosaccharides treated under the same conditions.
Electrospray Ionization Mass Spectrometry (ESI-MS) Analysis
The samples were desalted and analyzed by electrospray on an Esquire 6000 mass spectrometer system (Bruker Daltonics, Fremont, CA). Freeze dried samples containing 0.1–0.5 mg of carbohydrate were dissolved in 0.5 ml of 50% isopropanol in water. The sample flow rate was 3 μl/min. The source voltage was set at 4 kV. The capillary temperature was set at 300 °C. The Cap Exit voltage was set at 350 V, and the dry gas was nitrogen (purity 99.3%). Helium was introduced into the system to an estimated pressure of 4 × 10−6 mbar to improve trap efficiency.
Staining with Eel Lectin Cross-linked to Texas Red
Tobacco BY2 cells were stained with eel lectin according to the manufacturer's recommendations. Briefly, the cells were harvested and washed in phosphate-buffered saline (10 mm phosphate buffer, pH 7.4, containing 137 mm NaCl and 2.7 mm KCl). After incubation (∼1 × 106 BY2 cells in 1 ml of diluted eel lectin) for 5 h at room temperature (24 °C), the cells were washed and observed under a Motic BA400 EPI-Fluorescence Upright Biological Microscope using Texas Red/Cy3.5 filter set. The image was captured by Moticam 2300 software from Motic Instruments Inc. and processed by the Motic Images Plus 2.0 program (New York, NY).
RESULTS
AtFUT4 and AtFUT6 Genes as Putative Fucosyltransferases for AGPs
Our hypothesis is that AGP-FUTs should cluster in the GT37 family along with XyG-FUT (AtFUT1) because fucosyl residues in AGPs and XyGs are α(1,2)-linked and added by an inverting mechanism, consistent with the GT37 family. Additional supporting evidence comes from analysis of Arabidopsis mur1 mutant roots, which show 40% less Fuc in their AGP and 50% less reactivity with eel lectin, which recognizes specifically terminal Fuc residues, compared with wild type (15). Furthermore, expression patterns of Arabidopsis GT37 family members showed that only AtFUT1, AtFUT4, and AtFUT6 genes are highly expressed in roots (30). Thus, in roots, it is likely that AtFUT4 and AtFUT6 genes are responsible for AGP fucosylation, whereas the AtFUT1 gene functions in XyG fucosylation. Walls from leaves of mur1 plants have an even more drastic reduction in Fuc content (less than 2% of the wild type) (31). Because AtFUT4 along with AtFUT1 are the only two GT37 members highly expressed in leaves (30), it is likely that AtFUT4 is responsible for AGPs fucosylation in leaves. Taken together, these results indicate that AtFUT4 and AtFUT6 likely encode AGP-FUTs.
TAIR data bases have two entries for the AtFUT4 gene, labeled AtFUT4.1 (Atg2g15390.2) and AtFUT4.2 (At2g15390.1), and one entry for AtFUT6 (At1g14080). The AtFUT4.1 has structural similarity with AtFUT6 and most other members of the GT37 family. Both AtFUT4.1 and AtFUT4.2 entries are supported by expressed sequence tags in public data bases, and translation of their DNA sequences gave proteins with a slight difference at the N terminus, but all three sequences have the canonic motifs of the GT37 family (17). In this study, we used the AtFUT4.1 sequence.
Tobacco BY2 Cells Expressing AtFUT4.1 and AtFUT6 Genes Are Stained with Eel Lectin
To evaluate the biochemical function of AtFUT4.1 and AtFUT6 in vivo, the AtFUT4.1 and AtFUT6 genes were expressed in tobacco BY2 cells to determine whether these cells would produce AGPs with α-linked terminal l-Fuc residues, which could be detected by eel lectin. BY2 cells were chosen as the expression system for two reasons: (i) tobacco BY2 and Arabidopsis suspension cells produce similar AGPs (32), and (ii) AGPs in both BY2 and Arabidopsis suspension cells lack Fuc (32), making them ideal substrates for AGP-FUTs.
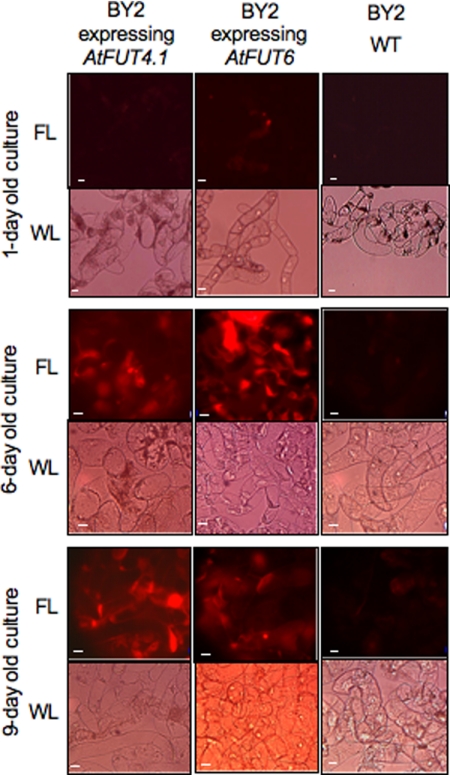
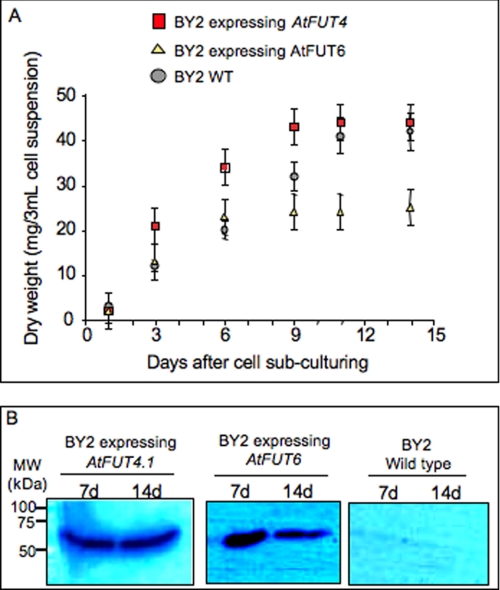
Initially, 1-, 6-, and 9-day-old transgenic and wild type (WT) cells were screened by staining with eel lectin linked to a fluorescent dye (Texas Red) to determine whether they produced fucosylated polymers in their cell walls. Only transgenic cells from 6- and 9-day-old cultures showed strong staining (Fig. 1), indicating that the expression of AtFUT4.1 and AtFUT6 resulted in the incorporation of α-linked terminal Fuc residues onto some cell wall polymers. The 9-day-old cells expressing AtFUT4.1 had stronger red staining compared with younger cultures, whereas cells expressing AtFUT6 showed stronger staining at 6 days (Fig. 1). Both BY2 transgenic cell lines produced a single band with a size of 60 kDa compatible with the predicted sizes of the His-AtFUT4.1 (∼61kDa) and His-AtFUT6 (59kDa) proteins (Fig. 2).
FIGURE 1.
Detection of eel (A. anguilla) lectin epitope in tobacco BY2 cells. Eel lectin (anti-H agglutinin) linked to a fluorescent dye (Texas Red) was used to stain (5 h) tobacco BY2 cells from 1-, 6-, and 9-day-old cultures of WT or transgenic cells expressing the AtFUT4.1 or AtFUT6 genes. The cells were observed under fluorescent light (FL) and white light (WL) as indicated. Scale bars, 20 μm.
FIGURE 2.
Expression of His-tagged versions of AtFUT4.1 and AtFUT6 proteins in tobacco BY2 cells. A, dry weight of the cells was determined after 1, 3, 6, 9, 11, and 14 days of culture to monitor growth stages of the cells. The growth was steady until 9 days of culture and then stabilized, indicating a separation of cell division stage (3–9 days) from cell elongation stage (9–14 days). B, 7- and 14-day-old cultures of BY2 WT cells or transgenic BY2 cells were screened by Western blotting using anti-His6 tag antibodies (Clontech). The cells were harvested and directly ground in liquid nitrogen before extraction with SDS-PAGE loading buffer, and then the proteins were separated on 10% gels. The protein bands detected had an estimated size of ∼60 kDa in agreement with the predicted size of AtFUT4.1 (∼61 kDa) and AtFUT6 (∼59 kDa) proteins. Marker sizes (kDa) are indicated on the left. MW, molecular mass; d, days.
AtFUT4.1 and AtFUT6 Are Fucosyltransferases Specific to AGPs
To confirm that the observed staining with eel lectin was due to the incorporation of α-Fuc residues onto AGPs, purified AGPs were prepared from transgenic and WT cell cultures using Yariv reagent and analyzed for monosaccharide composition. As expected, all of the purified AGPs contained galactose (Gal) and arabinose (Ara) as the two main monosaccharides, representing ∼50 and ∼35 mol %, respectively (Table 1). The most noticeable difference between AGPs from WT (AGPs-BY2WT) and transgenic (AGPs-BY2:F6 and AGPs-BY2:F4) cultures was the presence of Fuc (Table 1). Using HPAEC separation, Fuc was estimated to be ∼1.2 and 0.5 mol % in 7-day-old cells expressing AtFUT6 and AtFUT4.1, respectively. AGPs-BY2:F4 also showed a higher xylose content compared with AGPs-BY2WT or AGPs-BY2:F6.
TABLE 1.
Neutral monosaccharide content of purified AGPs from transgenic tobacco BY2 cells expressing AtFUT4.1 or AtFUT6 and WT cells
Monosaccharide content was determined after fractionation by HPAEC. The values are the averages of at least three experiments from two biological replicates. The standard deviations are indicated.
| BY2 WT | BY2 expressing AtFUT4.1 | BY2 expressing AtFUT6 | |
|---|---|---|---|
| mol % | mol % | mol % | |
| Galactose | 51 ± 2 | 47 ± 2 | 56 ± 2 |
| Arabinose | 36 ± 2 | 37 ± 2 | 32 ± 3 |
| Glucose | 13 ± 1 | 12 ± 2 | 10 ± 2 |
| Xylose | 0.7 ± 0.1 | 4 ± 0.5 | 0.8 ± 0.1 |
| Fucose | 0 | 0.5 ± 0.1 | 1.2 ± 0.2 |
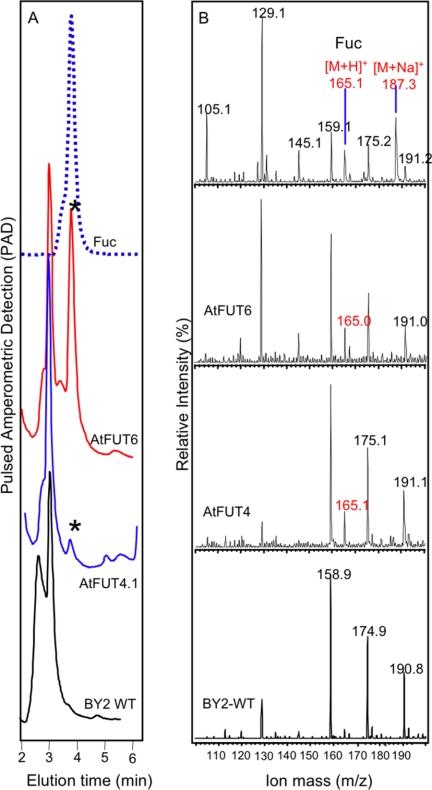
ESI-MS analysis provided further confirmation of fucosylated AGPs in the transgenic cultures. Following acid hydrolysis of transgenic and WT AGPs, the released sugars were fractionated by HPAEC. Fractions eluting between 2 and 6 min, which encompasses the elution time of a Fuc standard, were collected and analyzed by ESI-MS. The ion m/z of 165 ([M+H]+), characteristic of Fuc, was detected in transgenic, but not in WT samples, indicating that both genes encode FUTs that incorporate Fuc into AGPs (Fig. 3).
FIGURE 3.
Determination of the presence of Fuc in purified AGPs by HPAEC and ESI-MS. Purified AGPs from tobacco BY2 cells expressing the AtFUT4.1 or AtFUT6 genes or from WT cells were subjected to acid hydrolysis (2 m trifluoroacetic acid), and the resulting monosaccharides were fractionated by HPAEC on a CarboPac PA20 column (A). Fractions eluting between 2 and 6 min were collected, and the peak corresponding to Fuc is indicated by an asterisk in A. HPAEC fractions were then analyzed by ESI-MS (B). The ion at m/z 165 ([M+H]+) confirmed the presence of Fuc in the purified AGPs from transgenic BY2 cells and not in AGPs from the BY2 WT cells. Purified l-Fuc was used as standard.
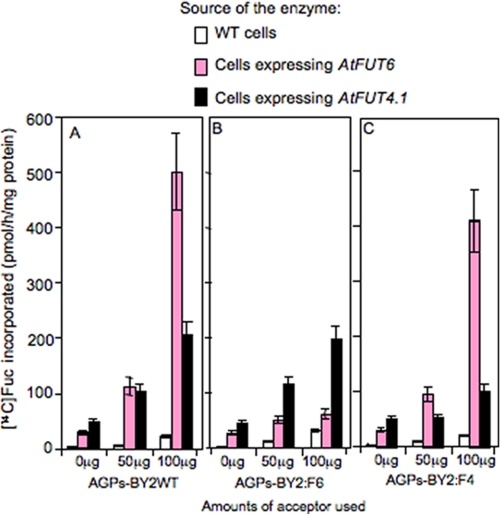
Next, the FUT activity of both enzymes was measured in vitro, using GDP-[14C]Fuc and digitonin (0.5%)-solubilized extracts from microsomal membranes prepared from transgenic and WT cells. Here detergent-soluble extracts from the transgenic cells were able to incorporate substantial amounts of [14C]Fuc onto AGPs-BY2WT (used as acceptors) in a concentration-dependent manner (Fig. 4A). Detergent extracts from WT cells did not catalyze this transfer. Similarly, control reactions (GDP-[14C]Fuc in the presence of AGPs-BY2WT alone or in the presence of protein extracts alone) did not show any [14C]Fuc incorporation. These data indicate that [14C]Fuc incorporation was AGP-dependent and due to the expression of AtFUT4.1 and AtFUT6.
FIGURE 4.
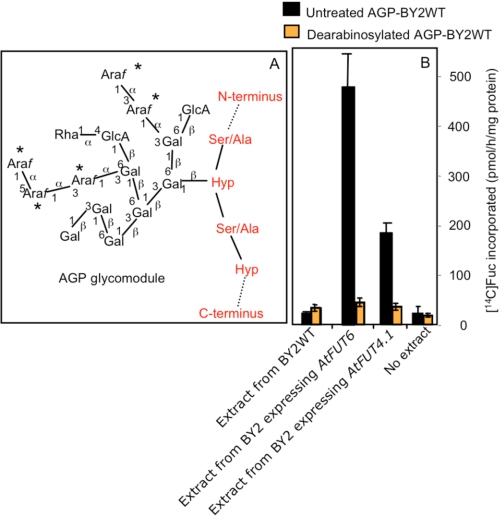
Fucosyltransferase activity in detergent extracts from tobacco BY2 cells expressing the AtFUT4.1 or AtFUT6 gene and from BY2 WT cells. Fucosyltransferase activity was monitored by measuring [14C]Fuc transfer onto AGPs from GDP-[14C]Fuc as described under “Experimental Procedures.” Enzyme sources were prepared from microsomal membranes solubilized with 0.5% digitonin from WT cells and transgenic cells. A–C show the fucosyltransferase activity resulting from using AGP substrate acceptors purified from wild type (AGPs-BY2WT), from cells expressing the AtFUT6 gene (AGPs-BY2:F6), and from cells expressing the AtFUT4.1 gene (AGPs-BY2:F4), respectively. All of the AGPs were used at 0, 50, and 100 μg/reaction. Each value is an average based on at least three experiment sets. The error bars represent the S.E.
To verify that both enzymes were specific to AGPs, several molecules, known to act as substrates for FUTs or contain Fuc, were tested as acceptors: XyG, RG, N-acetyllactosamine, and glycoproteins. Two types of RGs were used; one contained Fuc (RG from soybean), and the other one did not (RG from potato). N-Acetyllactosamine and α1-acid glycoprotein were used as acceptors for α(1,3)/α(1,2)FUT activities for N-glycans, and tamarind XyG was used as an acceptor for XyG-FUT activity. The data presented in Table 2 compared [14C]Fuc incorporated onto these substrates after incubation with detergent extracts from transgenic and WT BY2 cells. As judged by the comparably low [14C]Fuc incorporation obtained for all these wall polymers, none of these polymers (except AGPs-BY2WT) served as effective substrate acceptors for AtFUT4.1 and AtFUT6. These results indicate that both AtFUT4.1 and AtFUT6 are FUTs specific to AGPs.
TABLE 2.
Substrate specificity of AtFUT4.1 and AtFUT6 enzymes
Each acceptor (100 μg) was tested for [14C]Fuc incorporation in the presence of extracts from transgenic cell lines expressing AtFUT4.1 or AtFUT6 genes. Fucosyltransferase assays were conducted as described in Ref. 17. Extracts from BY2 WT cells were used as a control. The values are the averages of at least three experiments from two biological replicates. The values are the amounts of [14C]Fuc incorporated (pmol/h/mg of protein). The standard deviations are indicated.
| Acceptor added (100 μg) | Source of enzyme extract |
||
|---|---|---|---|
| BY2 WT | BY2 expressing AtFUT6 | BY2 expressing AtFUT4.1 | |
| None | 2 ± 1 | 6 ± 1 | 7.5 ± 2 |
| AGPs-BY2WT | 12 ± 3 | 485 ± 10 | 210 ± 5 |
| RGI from potato | 80 ± 9 | 100 ± 5 | 87 ± 6 |
| RG from soybean | 109 ± 5 | 94 ± 8 | 95 ± 2 |
| Arabinan | 67 ± 9 | 65 ± 6 | 66 ± 5 |
| Tamarind XyG | 49 ± 9 | 39 ± 5 | 37 ± 2 |
| N-Acetyllactosamine | 122 ± 10 | 111 ± 9 | 107 ± 5 |
| α1-acid glycoprotein | 4 ± 1 | 3.5 ± 1 | 3 ± 1 |
Interestingly, when AGPs-BY2WT were used in the FUT assay, extracts containing AtFUT4.1 were less than 50% as active compared with extracts containing AtFUT6 (Fig. 4A). This low activity of AtFUT4.1 could be attributed to low expression levels of the enzyme or to a difference in the specificity. To investigate this hypothesis, purified AGPs-BY2:F6 and AGPs-BY2:F4 were used as substrate acceptors in the presence of extracts containing AtFUT4.1 or AtFUT6 proteins. With AGPs-BY2:F6 as substrate, very low [14C]Fuc incorporation was obtained with extracts from cells expressing AtFUT6, but substantial incorporation (comparable with AGPs-BY2WT) was observed with extracts from cells expressing AtFUT4.1 (Fig. 4B). Reciprocally, purified AGPs-BY2:F4 were effective acceptors for extracts containing AtFUT6 protein (comparable with AGPs-BY2WT) but were not very effective substrates for extracts containing AtFUT4.1 protein (Fig. 4C). In addition, the level of AtFUT4.1 expression was comparable with AtFUT6 as indicated by Western blot analysis (Fig. 1). These data indicate that AtFUT4.1 and AtFUT6 differentially fucosylate AGPs, most likely on different Ara residues on AGPs.
AtFUT4.1 and AtFUT6 Transfer the Fucose onto Arabinosyl Residues in an α(1,2)-type Linkage
To examine whether [14C]Fuc is transferred onto Ara branches of AGPs, purified AGPs-BY2WT were treated with mild acid treatment at 100 °C to remove Ara residues (28) and used as acceptors. The efficiency of the acid treatment was assessed by the change in the ratio of Gal to Ara in untreated and dearabinosylated AGPs from 1.7:1 to 30:1, respectively (data not shown). When used as acceptors with extracts containing AFUT4.1 or AtFUT6, [14C]Fuc incorporation was minimal and similar to negative control reactions lacking these FUTs or an extract. In contrast, untreated AGPs-BY2WT (positive controls) were good acceptors (Fig. 5). These results demonstrate that fucosylation sites reside on Ara residues of AGPs.
FIGURE 5.
A simplified representation of an AGP glycomodule (A), and the effect of arabinose (Ara) removal on the ability of these glycomodules to act as acceptors in FUT assay (B). A, the simplified structure of arabinogalactan polymers in BY2 AGPs was adapted from Ref. 32. The Araf residues susceptible to mild acid treatment are indicated by asterisks. B, fucosyltransferase assays were performed using as acceptors mild untreated and acid-treated AGPs-BY2WT (100 μg) in the presence of GDP-[14C]Fuc and detergent extracts from BY2 WT cells or transgenic BY2 cell lines expressing the AtFUT4.1 or AtFUT6 genes. The reactions containing no protein extracts were used as negative controls. Each value is an average based on at least three experimental sets. The error bars represent the S.E.
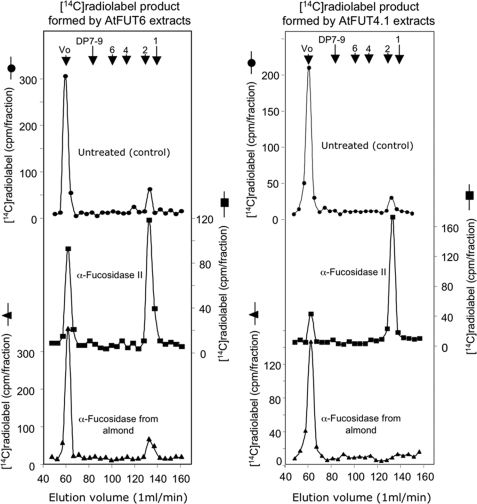
The linkage between Fuc and Ara was further characterized using two α-fucosidases, namely α-fucosidase II that removes terminal Fuc residues specifically α-linked to the C-2 position of the adjacent sugar and an α-fucosidase from almond meal that removes terminal Fuc residues specifically α-linked to the C-3 or C-4 positions of the adjacent sugar. When the [14C]fucosylated AGPs-BY2WT generated by AtFUT6 extracts were treated with α-fucosidase II and fractionated on a Biogel P2 column, more than 60% of the radiolabel was released at the total volume as a DP1 sugar (Fig. 6). The identity of this released sugar was confirmed as [14C]Fuc by HPAEC. However, similar experiments using α-fucosidase from almond meal did not release [14C]Fuc (Fig. 6). Similar results were obtained using [14C]fucosylated AGPs-BY2WT generated by AtFUT4.1 extracts (Fig. 6). Taken together with the dearabinoslylation data, these results indicated that AtFUT4.1 and AtFUT6 incorporate terminal α-l-Fuc residues on the C-2 positions of adjacent Ara residues of AGPs.
FIGURE 6.
Bio-gel P2 fractionation of the [14C]Fuc-labeled AGPs-BY2WT after treatment with α(1,2)fucosidase II or α(1,3/4)fucosidase from almond meal as described under “Experimental Procedures.” The [14C]Fuc-labeled AGPs-BY2WT were formed by detergent extracts from microsomal membranes of BY2 cells expressing AtFUT4.1 or AtFUT6 genes. Bio-gel P2 columns were eluted with degassed water, and the elution volumes of dextran (Vo) (average molecular weight 500,000) and sugars with degree of polymerization (DP) of 1, 2, 4, 6, and 7–9 are indicated with arrows at the top.
AtFUT6 Is Localized to the Golgi Complex
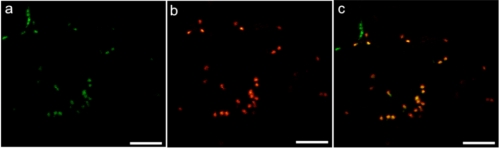
To investigate whether AtFUT6 is localized in the Golgi, AtFUT6-green fluorescent protein (AtFUT6-GFP) fusion protein was constructed and transiently co-expressed with a Golgi marker protein, α(2,6)sialyltransferase (33), fused to yellow fluorescent protein (ST-YFP) in tobacco leaves. AtFUT6-GFP was observed in punctate structures typical of a Golgi-localized staining pattern (Fig. 7a). The same staining pattern was also obtained with ST-YFP (Fig. 7b), and both staining patterns overlapped (Fig. 7c), consistent with the likely Golgi localization of AtFUT6 protein. Because of technical issues, we did not succeed in determining localization of AtFUT4.1.
FIGURE 7.
Subcellular localization of AtFUT6-GFP in N. tabacum leaves. AtFUT6-GFP and ST-YFP (a Golgi marker) fusion proteins were transiently co-expressed in tobacco plant leaves. Fluorescence spots are seen following the expression of AtFUT6-GFP (a) and ST-YFP (b). Fluorescence spots observed for both AtFUT6-GFP and ST-YFP are co-localized as shown in the overlay (c), suggesting a Golgi localization of the AtFUT6 protein. The scale bars are 8 μm.
DISCUSSION
Despite the fact that plants contain substantial amounts of AGPs, there is an embarrassing lack of knowledge on the enzymology of AGP biosynthesis, and none of the biosynthetic enzymes have been cloned. In the present work, we describe the characterization of the biochemical function of two Arabidopsis genes, AtFUT4 and AtFUT6, as α(1,2)FUTs specific for AGPs. These two genes along with eight others are members of the GT37 family. The only GT37 member fully characterized previously is the XyG-specific α(1,2)FUT encoded by the AtFUT1 gene (16, 17). Using a multifaceted strategy, we demonstrated that both AtFUT4.1 and AtFUT6 specifically fucosylate AGPs via an α(1,2) linkage onto Ara residues. In addition, our localization study supported earlier work (20, 21) on the role of the Golgi apparatus in the glycosylation of hydroxyproline-rich glycoproteins.
The in vitro FUT assay also revealed that the two FUT genes are not functionally redundant. Because AtFUT4.1 could fucosylate AGPs-BY2:F6, and AtFUT6 could fucosylate AGPs-BY:F4 and neither of the enzymes could use AGPs from their own cells (Fig. 4), we propose that the two enzymes transfer Fuc residues onto two different Ara residues on AGP molecules (enzyme specificity dictated by the fucosylation sites). Tan et al. (32) have shown that the expression of a synthetic gene encoding an [Ala-Hyp]51 peptide in tobacco BY2 cells produced and secreted a mixture of glycoproteins having Hyp residues substituted with arabinogalactan (AG) polysaccharides (DP13–26) and containing terminal Ara residues α(1,3)- or α(1,5)-linked. A simplified presentation of this AG polysaccharide structure is depicted in Fig. 5A; this figure shows that AGs can contain up to five Ara residues as side chains that could be potential sites for fucosylation. Thus, AtFUT4.1 and AtFUT6 likely specify FUT activities that transfer Fuc onto different Ara residues in these side chains. The difference in the enzyme activity between AtFUT4.1 and AtFUT6 could be explained by the difference in the amounts of AGPs with various Ara side chains.
Because of the difference in eel lectin staining patterns of transgenic cells (Fig. 1), it is tempting to speculate that the substrate acceptors for AtFUT4.1 and AtFUT6 have subtle differences in their AG polymers, as a result of the action of AraTs. For example, the expression of various α(1.3)- or α(1,5)-AraT genes during BY2 cell growth could generate different fucosylation sites (i.e. substrates) for AGP-FUTs. The GT77 family contains at least three putative AraTs (22, 34) and may also contain AraT for AGPs. Misawa et al. (25) have used a trisaccharide with terminal Araf residues α(1,3)-linked to Gal as an exogenous acceptor for radish AGP-FUT. One of the Arabidopsis AGP-FUT proteins described here could have this activity, and the other AtFUT protein might be specific to a terminal Araf α(1,5)-linked to an Araf or to any internal Araf in the side chains. Verification of this hypothesis is needed when purified acceptors are available.
In conclusion, this work demonstrated that the AtFUT4 and AtFUT6 genes encode α(1,2)FUTs specific for AGPs. Furthermore, our data indicate that these two AGP-AtFUTs are not functionally redundant on a biochemical level and thus may have different physiological roles. The present study has increased our understanding of the AGP biosynthesis, specifically with respect to glycosylation. Identification of these genes also opens new avenues to examine other AGP glycosyltransferases that may be in complexes with these FUTs and to probe AGP function. Indeed, analysis of Arabidopsis mutants for these two genes should help to elucidate AGP function and the contribution of specific carbohydrate epitopes to cell wall integrity and plant growth.
This work was supported by National Research Initiative Competitive Grants 2008-35318-04563 and 2008-35318-04572 from the United States Department of Agriculture National Institute of Food and Agriculture.
- Fuc
- fucose
- AGP
- arabinogalactan protein
- GT
- glycosyltransferase
- XyG
- xyloglucan
- RG
- rhamnogalacturonan
- FUT
- fucosyltransferase
- AraT
- arabinosyltransferase
- ESI-MS
- electrospray ionization trap mass spectrometry
- GFP
- green fluorescent protein
- YFP
- yellow fluorescent protein
- ST
- sialyltransferase
- HPAEC
- high pH anion exchange chromatography
- WT
- wild type
- Gal
- galactose
- Ara
- arabinose
- AG
- arabinogalactan.
REFERENCES
- 1.Carpita N. C., Gibeaut D. M. (1993) Plant J. 3, 1–30 [DOI] [PubMed] [Google Scholar]
- 2.O'Neill M., Albersheim P., Darvill A. (1990) in Methods in Plant Biochemistry (Dey P. M. ed) Vol. 2, pp. 415–441, Academic Press, London [Google Scholar]
- 3.Johnson K. D., Chrispeels M. J. (1987) Plant Physiol. 84, 1301–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchet M. A., Odom E. W., Vasta G. R., Amzel L. M. (2002) Nat. Struct. Biol. 9, 628–634 [DOI] [PubMed] [Google Scholar]
- 5.Hakomori S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10231–10233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shida K., Misonou Y., Korekane H., Seki Y., Noura S., Ohue M., Honke K., Miyamoto Y. (2009) Glycobiology 19, 1018–1033 [DOI] [PubMed] [Google Scholar]
- 7.Levy S., York W. S., Stuike-Prill R., Meyer B., Staehelin L. A. (1991) Plant J. 1, 195–215 [PubMed] [Google Scholar]
- 8.Augur C., Yu L., Sakai K., Ogawa T., Sinaÿ P., Darvill A. G., Albersheim P. (1992) Plant Physiol. 99, 180–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDougall G. J., Fry S. C. (1990) Plant Physiol. 93, 1042–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warneck H. M., Fulton D. C., Seitz H. U., Fry S. C. (1997) Planta 204, 78–85 [Google Scholar]
- 11.Reiter W. D., Chapple C. C., Somerville C. R. (1993) Science 261, 1032–1035 [DOI] [PubMed] [Google Scholar]
- 12.Zablackis E., York W. S., Pauly M., Hantus S., Reiter W. D., Chapple C. C., Albersheim P., Darvill A. (1996) Science 272, 1808–1810 [DOI] [PubMed] [Google Scholar]
- 13.Rayon C., Cabanes-Macheteau M., Loutelier-Bourhis C., Salliot-Maire I., Lemoine J., Reiter W. D., Lerouge P., Faye L. (1999) Plant Physiol. 119, 725–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Neill M. A., Eberhard S., Albersheim P., Darvill A. G. (2001) Science 294, 846–849 [DOI] [PubMed] [Google Scholar]
- 15.van Hengel A. J., Roberts K. (2002) Plant J. 32, 105–113 [DOI] [PubMed] [Google Scholar]
- 16.Perrin R. M., DeRocher A. E., Bar-Peled M., Zeng W., Norambuena L., Orellana A., Raikhel N. V., Keegstra K. (1999) Science 284, 1976–1979 [DOI] [PubMed] [Google Scholar]
- 17.Faik A., Bar-Peled M., DeRocher A. E., Zeng W., Perrin R. M., Wilkerson C., Raikhel N. V., Keegstra K. (2000) J. Biol. Chem. 275, 15082–15089 [DOI] [PubMed] [Google Scholar]
- 18.Showalter A. M. (2001) Cell Mol. Life Sci. 58, 1399–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farrokhi N., Burton R. A., Brownfield L., Hrmova M., Wilson S. M., Bacic A., Fincher G. B. (2006) Plant Biotechnol. J. 4, 145–167 [DOI] [PubMed] [Google Scholar]
- 20.Gardiner M., Chrispeels M. J. (1975) Plant Physiol. 55, 536–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owens R. J., Northcote D. H. (1981) Biochem. J. 195, 661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gille S., Hänsel U., Ziemann M., Pauly M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14699–14704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsumuraya Y., Ogura K., Hashimoto Y., Mukoyama H., Yamamoto S. (1988) Plant Physiol. 86, 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsumuraya Y., Hashimoto Y., Yamamoto S., Shibuya N. (1984) Carbohydr. Res. 134, 215–228 [Google Scholar]
- 25.Misawa H., Tsumuraya Y., Kaneko Y., Hashimoto Y. (1996) Plant Physiol. 110, 665–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curtis M. D., Grossniklaus U. (2003) Plant Physiol. 133, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schultz C. J., Johnson K. L., Currie G., Bacic A. (2000) Plant Cell 12, 1751–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kieliszewski M., de Zacks R., Leykam J. F., Lamport D. T. (1992) Plant Physiol. 98, 919–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng W., Chatterjee M., Faik A. (2008) Plant Physiol. 147, 78–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarria R., Wagner T. A., O'Neill M. A., Faik A., Wilkerson C. G., Keegstra K., Raikhel N. V. (2001) Plant Physiol. 127, 1595–1606 [PMC free article] [PubMed] [Google Scholar]
- 31.Reiter W. D., Chapple C., Somerville C. R. (1997) Plant J. 12, 335–345 [DOI] [PubMed] [Google Scholar]
- 32.Tan L., Qiu F., Lamport D. T., Kieliszewski M. J. (2004) J. Biol. Chem. 279, 13156–13165 [DOI] [PubMed] [Google Scholar]
- 33.Saint-Jore C. M., Evins J., Batoko H., Brandizzi F., Moore I., Hawes C. (2002) Plant J. 29, 661–678 [DOI] [PubMed] [Google Scholar]
- 34.Egelund J., Obel N., Ulvskov P., Geshi N., Pauly M., Bacic A., Petersen B. L. (2007) Plant Mol. Biol. 64, 439–451 [DOI] [PubMed] [Google Scholar]