Abstract
Hypoxia-inducible factor (HIF) has a pivotal role in oxygen homeostasis and cardioprotection mediated by ischemic preconditioning. Its stability is regulated by HIF prolyl 4-hydroxylases (HIF-P4Hs), the inhibition of which is regarded as a promising strategy for treating diseases such as anemia and ischemia. We generated a viable Hif-p4h-2 hypomorph mouse line (Hif-p4h-2gt/gt) that expresses decreased amounts of wild-type Hif-p4h-2 mRNA: 8% in the heart; 15% in the skeletal muscle; 34–47% in the kidney, spleen, lung, and bladder; 60% in the brain; and 85% in the liver. These mice have no polycythemia and show no signs of the dilated cardiomyopathy or hyperactive angiogenesis observed in mice with broad spectrum conditional Hif-p4h-2 inactivation. We focused here on the effects of chronic Hif-p4h-2 deficiency in the heart. Hif-1 and Hif-2 were stabilized, and the mRNA levels of glucose transporter-1, several enzymes of glycolysis, pyruvate dehydrogenase kinase 1, angiopoietin-2, and adrenomedullin were increased in the Hif-p4h-2gt/gt hearts. When isolated Hif-p4h-2gt/gt hearts were subjected to ischemia-reperfusion, the recovery of mechanical function and coronary flow rate was significantly better than in wild type, while cumulative release of lactate dehydrogenase reflecting the infarct size was reduced. The preischemic amount of lactate was increased, and the ischemic versus preischemic [CrP]/[Cr] and [ATP] remained at higher levels in Hif-p4h-2gt/gt hearts, indicating enhanced glycolysis and an improved cellular energy state. Our data suggest that chronic stabilization of Hif-1α and Hif-2α by genetic knockdown of Hif-p4h-2 promotes cardioprotection by induction of many genes involved in glucose metabolism, cardiac function, and blood pressure.
Keywords: Hydroxylase, Hydroxyproline, Hypoxia, Ischemia, Transcription Factors, Hypoxia-inducible Factor, Ischemia-Reperfusion Injury, Prolyl 4-Hydroxylase
Introduction
Hypoxia-inducible transcription factor (HIF),3 which has a pivotal role in the induction of numerous genes involved in the mediation of survival and adaptive responses to hypoxia (1–3), is a heterodimer consisting of an unstable α subunit and a stable β subunit. The stability of the HIF-α subunit isoforms HIF-1α and HIF-2α is regulated by oxygen-dependent prolyl 4-hydroxylation (4–6). HIF-α is synthesized continuously, and its two critical proline residues become hydroxylated under normoxic conditions by a cytoplasmic and nuclear HIF prolyl 4-hydroxylase (HIF-P4H) family (7–9). The 4-hydroxyproline residues thus formed are required for binding of HIF-α to the von Hippel-Lindau E3 ubiquitin ligase complex, resulting in its rapid proteasomal degradation in normoxia (1–3). In hypoxia, this oxygen-dependent hydroxylation is inhibited, and HIF-α escapes degradation, translocates into the nucleus, and forms a functional dimer with HIF-β.
Three HIF-P4H isoenzymes exist in mammals: HIF-P4Hs 1, 2, and 3, also known as prolyl hydroxylase domain-containing proteins 1, 2 and 3; Egl-nine 2, 1, and 3; and HIF prolyl hydroxylases 3, 2, and 1, respectively (7–9). A fourth P4H with an endoplasmic reticulum transmembrane domain is also capable of hydroxylating HIF-α in vitro and in cultured cells, but it remains to be established whether it also participates in the regulation of HIF-α in vivo (10, 11). HIF-P4H-2 is the primary oxygen sensor, because silencing of HIF-P4H-2 alone by RNA interference is sufficient to stabilize HIF-α in cultured cells in normoxia (12, 13). Furthermore, Hif-p4h-2 null mice die during embryonic development because of severe placental and cardiac defects, the latter of which are not due to elevated Hif-α levels, whereas Hif-p4h-1 and Hif-p4h-3 null mice are viable (14). Broad spectrum conditional inactivation of Hif-p4h-2 in mice leads to severe erythrocytosis, hyperactive angiogenesis, and angiectasia (15–17). Heterozygous Hif-p4h-2 deficiency inhibits tumor cell invasion, intravasation, and metastasis in mice by normalizing the endothelial lining and maturation of tumor vessels, resulting in the restoration of tumor oxygenation (18). Hif-p4h-1 and Hif-p4h-3 null mice have no hematopoietic or angiogenic defects, but Hif-p4h-1/Hif-p4h-3 double knock-out leads to moderate erythrocytosis caused by an accumulation of Hif-2α in the liver and activation of the hepatic erythropoietin (Epo) pathway, whereas inactivation of Hif-p4h-2 causes activation of the renal Epo pathway (15, 16). Hif-p4h-1 null mice have lowered oxygen consumption caused by reprogramming of the basal metabolism toward anaerobic energy production, increased hypoxia tolerance, and protection against acute severe ischemia in the skeletal muscle (19). Hif-p4h-3 null mice have abnormal sympathoadrenal development and function, leading to reduced catecholamine secretion and systemic blood pressure (20).
Studies with HIF-P4H-inhibiting small molecule compounds have indicated that pharmacologic HIF activation appears promising as a strategy for treating diseases associated with acute or chronic hypoxia, such as anemia, myocardial infarction, and stroke (1–3, 21). HIF-1α also plays a central role in the tissue protection induced by ischemic preconditioning (IP), i.e. cytoprotective adaptation triggered by brief periods of sublethal ischemia. In the case of the heart, this is evident from data showing that IP-induced acute cardioprotection was lost in heterozygous Hif-1α null mice (22) and in Hif-1α siRNA-treated mice (23) and that acute in vivo stabilization of Hif-1α by administration of the Hif-p4h inhibitor dimethyloxalylglycine or by introduction of Hif-p4h-2 siRNA into the left ventricle of the heart led to cardioprotection (23). Furthermore, intramyocardial injection of Hif-p4h-2 small hairpin RNA led to significant improvement in angiogenesis and contractility following ligation of left anterior descending artery in mice (24). On the other hand, broad spectrum conditional inactivation of Hif-p4h-2 in utero at day 17.5 postfertilization led to congestive heart failure, with enlarged cardiomyocytes and increased contraction band necrosis indicative of early cardiac ischemia (17). It was regarded as possible that these changes might be indirect consequences of the severe polycythemia, volume overload, and hyperviscosity present in these mice, but it was also possible that the cardiomyopathy could have been a direct consequence of chronic, high level Hif activation in the cardiac myocytes (17). When Hif-p4h-2 was inactivated later, at 3 weeks of age, the hearts were only minimally enlarged, retained normal systolic function in vivo, and had only few myocytes showing degenerative changes (25).
We have produced mice in which the Hif-p4h-2 gene is disrupted by a GeneTrap (gt) insertion cassette. Because Hif-p4h-2 knock-out mice die during embryonic development (14), we expected to obtain only Hif-p4h-2+/+ and Hif-p4h-2+/gt mice from the heterozygous matings. Surprisingly, we also obtained Hif-p4h-2gt/gt mice, which appeared to be healthy and fertile and to have a normal lifespan. Analysis of the tissues of these mice showed that varying amounts of wild-type Hif-p4h-2 mRNA were generated from the gene-trapped alleles in different tissues, because of partial skipping of the insertion cassette, the lowest amount of wild-type Hif-p4h-2 mRNA, 8% of that in the wild-type mice, being found in the heart. Because these mice have no polycythemia, they appeared to offer an excellent model for studying the consequences of chronic high level activation of Hif-1α and Hif-2α in the heart.
EXPERIMENTAL PROCEDURES
Generation of Hif-p4h-2gt/gt Mice
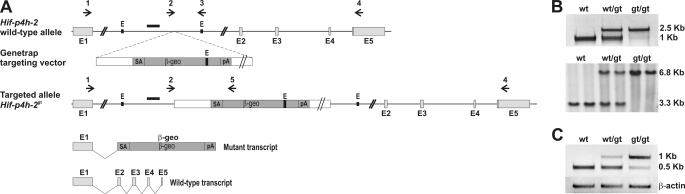
An embryonic stem cell line RRG405 with a GeneTrap targeting vector containing a β-galactosidase reporter in intron 1 of the Hif-p4h-2 gene (Research Consortium BayGenomics) was obtained from the Mutant Mouse Regional Resource Center (University of California, Davis). The cells were expanded and injected into C57BL/6 blastocysts, and the mice were generated. The mice were genotyped by PCR using a primer pair from intron 1 of Hif-p4h-2 (arrows 2 and 3 in Fig. 1A) and a Hif-p4h-2-specific forward primer and a β-galactosidase-specific reverse primer (arrows 2 and 5 in Fig. 1A), producing 1050- and 2500-bp fragments from the wild-type and targeted alleles, respectively. Primer sequences are given in supplemental Table S1. To verify correct targeting, genomic DNA was digested with EcoRI and hybridized with a probe generated by PCR from Hif-p4h-2 intron 1 (black bar in Fig. 1A). All of the animal experiments were performed according to protocols approved by the Provincial State Office of Southern Finland.
FIGURE 1.
Targeted disruption of the mouse Hif-p4h-2 gene. A, an embryonic stem cell line containing a GeneTrap targeting vector in intron 1 of the Hif-p4h-2 gene, disrupting the endogenous transcription and leading to the production of truncated mRNA coding for β-galactosidase, was used to generate a Hif-p4h-2gt/gt mouse line. The arrows represent the primers used in the genotyping by PCR and in RT-PCR. The probe used in Southern blotting is depicted as a black bar. E1–E5, exons 1–5; E, EcoRI; SA, splice acceptor sequence; β-geo, a fusion gene formed from the β-galactosidase and neomycin resistance genes; pA, polyadenylation signal. B, genotyping of the mice by PCR or Southern blotting. A 1050-bp fragment was produced from the wild-type allele with gene-specific primers (arrows 2 and 3 in A), while insertion of the GeneTrap cassette resulted in amplification of a 2500-bp fragment with the Hif-p4h-2 intron 1 forward primer and a β-galactosidase reverse primer (arrows 2 and 5 in A) from ear samples (upper panel). Genomic DNA isolated from embryonic day 12.5 embryos was digested with EcoRI and hybridized with a probe spanning 536 bp of intron 1. It recognized a 3.3-kb fragment in the wild-type allele and a 6.8-kb fragment in the targeted allele (lower panel). C, RT-PCR analysis. Heart RNA samples from wild-type (wt), Hif-p4h-2+/gt (wt/gt), and Hif-p4h-2gt/gt (gt/gt) mice were used as templates. Gene-specific primers from exons 1 and 5 amplifying wild-type cDNA (arrows 1 and 4 in A) produced a 500-bp fragment, whereas a primer set designed to amplify the transcript generated from the targeted allele (arrows 1 and 5 in A) produced a 1000-bp fragment. β-Actin amplification was used as a control for equal template amounts.
RT-PCR, Quantitative Real Time RT-PCR (Q-PCR), and Microarray Analysis
Tissues were quickly dissected immediately after the mice had been sacrificed, snap-frozen in liquid nitrogen, and stored at −70 °C. Total RNA was isolated using TriPure isolation reagent (Roche Applied Science) and further purified with an EZNA total RNA kit (OMEGA Bio-tek), and reverse transcription was performed with an iScript cDNA synthesis kit (Bio-Rad).
Wild-type Hif-p4h-2 mRNA was amplified in RT-PCR using primers from exons 1 and 5 (arrows 1 and 4 in Fig. 1A), and the generation of a transcript containing the GeneTrap cassette was analyzed using primers indicated by arrows 1 and 5 (Fig. 1A). β-Actin was used to verify equal amounts of template in RT-PCR. Q-PCR was performed with either iTaq SYBR Green Supermix with ROX (Bio-Rad) or Brilliant II SYBR Green Q-PCR Master Mix (Stratagene) and the Stratagene MX3005 thermocycler. The sequences for the primers used in RT-PCR and Q-PCR are listed in supplemental Table S1. The sets of primers for Hif-1α, Hif-2α, Epo, vascular endothelial growth factor A (Vegf-a), BCL2/adenovirus E1B interacting protein-3 (Bnip-3), phosphoglycerate kinase-1, angiopoietin-1 and -2 (Ang-1 and -2), Cd73, adenosine receptor A2b, and apelin were according to previously published sequences (14, 23, 26, 27). Quantitect primer assays (Qiagen) were used for lysyl oxidase, myoglobin, glucose transporter-1 and -4 (Glut-1 and -4), plasminogen activator inhibitor-1, DNA damage-inducible transcript 4 (Redd-1), triose phosphate isomerase, enolase-1, aldolase A, pyryvate dehydrogenase kinase-1 and -4 (Pdk-1 and -4), hexokinase-1 and -2, phosphofructokinase-L, lactate dehydrogenase A, peroxisome proliferator activator α (Pparα), and glucokinase-1. The expression data were normalized to β-actin or TATA-binding protein.
Microarray analysis of RNA samples of Hif-p4h-2gt/gt and wild-type hearts on Gene Chip mouse genome 430 2.0 array was carried out according to protocols provided by the chip manufacturer (Affymetrix) in the Gene Analysis Services Core Facility of Biocenter Oulu.
Western Blotting
For Western blotting, snap-frozen tissues were crushed to a powder, lysed in 3 m urea supplemented with 25 mm Tris-HCl, pH 7.5, 75 mm NaCl, and 0.25% Nonidet P-40, and centrifuged. In the case of skeletal muscle, a buffer containing 75 mm Tris-HCl, pH 6.8, 15% SDS, 20% glycerol, 5% mercaptoethanol, and 0.001% bromphenol blue and homogenization with an Ultra-Turrax homogenizer was used to increase solubility. The supernatant protein concentrations were determined by the Bradford method (Bio-Rad protein assay), and the supernatants were resolved by SDS-PAGE and blotted onto Immobilon-P membranes (Millipore). Western blots were blocked with Tris-buffered saline with 5% nonfat dry milk and probed with the following primary antibodies: anti-HIF-1α (NB100-479; Novus Biologicals), anti-HIF-2α (a gift from FibroGen Inc.), anti-HIF prolyl hydroxylase 2 (NB100-2219; Novus Biologicals), anti-HIF prolyl hydroxylase 1 (NB100-310; Novus Biologicals), anti-HIF-β (611078; BD Transduction Laboratories), anti-glucose transporter-1 (NB300-666; Novus Biologicals), anti-pyruvate dehydrogenase kinase-1 (KAP-PK112; Assay Designs), and anti-α-tubulin (B-6199; Sigma-Aldrich). Bound antibodies were detected with horseradish peroxidase-conjugated secondary antibodies (Dako) and ECL detection reagents (Thermo Scientific).
Blood and Serum Analyses
Terminal blood samples from the inferior vena cava of adult mice were drawn into syringes containing EDTA, and complete blood counts were measured with Cell-Dyn Sapphire (Abbott). The samples were allowed to clot overnight at 4 °C followed by centrifugation for 20 min at 1000 × g, and serum Epo and Vegf-a levels were determined with the Quantikine Mouse Epo and Vegf Immunoassay kits (R & D Systems).
Histological Analyses
Dissected hearts were fixed in fresh 4% paraformaldehyde at 4 °C overnight. The specimens were washed with 0.15 m NaCl in 0.02 m phosphate, pH 7.4, and transferred to 70% ethanol, dehydrated, and embedded in paraffin. For the cryosections, paraformaldehyde-fixed specimens were transferred to 15% sucrose at 4 °C for 1 h, followed by overnight incubation in 30% sucrose at 4 °C and embedding in OTC compound (Sakura). 5-μm sections were cut and stained with hematoxylin and eosin for histology. The paraffin and cryosections were stained with rat anti-mouse Pecam-1 (MEC13.3, BD Pharmingen) using the TSA indirect kit (PerkinElmer Life Sciences) and Alexa Fluor 488-conjugated anti-rat antibody (Invitrogen), respectively. The sections were viewed and photographed with either a Leica DM LB2 microscope equipped with a Leica DFC 320 camera or with an Olympus BX51 microscope equipped with an Olympus DP50 camera, and the Pecam-1-stained vessels were quantified from 10 photomicrographs/mouse.
β-Galactosidase activity was detected by X-gal staining. Staining of embryos and adult tissues was performed as described earlier (28) with the following modifications. The embryos or tissue samples were stained overnight at room temperature in 2 mg/ml X-gal, 10 mm K3Fe(CN)6, 10 mm K4Fe(CN)6, 0.01% sodium deoxycholate, 0.02% Nonidet P-40, 5 mm EGTA, 2 mm MgCl2, and 0.1 m potassium phosphate buffer, pH 7.3, and the staining was followed by fixation in 10% formalin. The fixed tissues were processed for paraffin sections, and background was stained with eosin.
Transmission Electron Microscopy
Heart samples were fixed in 1% glutaraldehyde and 4% formaldehyde in 0.1 m phosphate buffer, postfixed in 1% osmiumtetroxide, dehydrated in acetone, and embedded in Epon LX 112 (Ladd Research Industries). Thin sections were cut with a Leica Ultracut UCT ultramicrotome, stained in uranyl acetate and lead citrate, and examined in a Philips CM100 transmission electron microscope. The images were captured using a Morada CCD camera (Olympus Soft Imaging Solutions).
Langendorff Perfusion
The experiments were performed with 2–5-month-old female Hif-p4h-2gt/gt mice and age- and gender-matched controls. The mice were sacrificed by cervical dislocation followed by decapitation. The aorta was immediately cannulated, and perfusion was started in situ with ice-cold Krebs-Henseleit buffer (10 mmol/liter glucose, 118 mmol/liter NaCl, 4.7 mmol/liter KCl, 2.5 mmol/liter CaCl2, 0.5 mmol/liter EDTA, 1.2 mmol/liter MgSO4, 25 mmol/liter NaHCO3, and 1.2 mmol/liter KH2PO4, pH 7.4). The heart was excised and enclosed in a thermostatic chamber, and perfusion was continued with a constant pressure of 100 cm H2O with Krebs-Henseleit buffer maintained at 37 °C and gassed continuously with a mixture of 95% O2 and 5% CO2. Continuous monitoring of left ventricular pressure, measurement of the coronary flow rate, and calculation of the heart rate, left ventricle peak systolic pressure, left ventricle minimum diastolic pressure, and left ventricular developed pressure (LVDP = left ventricle peak systolic pressure − left ventricle minimum diastolic pressure) was performed as described (29). The hearts were subjected to at least 30 min of stabilization perfusion, after which global ischemia was induced by cessation of perfusion for 20 min, followed by reperfusion for 45 min. The venous effluent from the heart was collected in 5-min aliquots, and lactate dehydrogenase washout was measured as described earlier (30).
Metabolite Assays
Hearts from female Hif-p4h-2gt/gt mice and age- and gender-matched controls were subjected to either the stabilization perfusion alone (preischemia) or stabilization perfusion followed by a 20-min global ischemia (end ischemia), after which the hearts were immediately snap-frozen with precooled aluminum clamps, transferred to liquid nitrogen, and stored at −70 °C until further processing. Frozen heart tissue was immersed in 8% perchloric acid in 40% (v/v) ethanol precooled to −20 °C and homogenized immediately with an Ultra-Turrax homogenizer followed by centrifugation for 10 min at 3200 × g. The supernatant was neutralized with 3.75 mol/liter K2CO3 containing 0.5 mol/liter triethanolamine hydrochloride (31). ATP, ADP, CrP, Cr, and lactate in the supernatant were determined by conventional enzymatic methods by measuring the appearance or disappearance of NADH or NADPH with an Aminco DW-2 dual wavelength spectrophotometer (31–34).
Statistical Analyses
The statistical analyses were performed using Student's two-tailed t test. The area under the curve (AUC) during the 45-min post-ischemic period was calculated using the method of summary measures (35). The data are shown as the means ± S.D. or S.E. Values of p < 0.05 were considered statistically significant (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
RESULTS
Hif-p4h-2gt/gt Mice Are Viable and Have No Obvious Phenotypic Abnormalities
We generated mice having their Hif-p4h-2 gene disrupted within intron 1 by a GeneTrap insertion cassette containing a β-galactosidase reporter (Fig. 1A). Genotyping (Fig. 1B) of the pups (n = 470) indicated that 28.9% were Hif-p4h-2+/+, 59.4% Hif-p4h-2+/gt, and 11.7% Hif-p4h-2gt/gt. The Hif-p4h-2gt/gt percentage was lower than expected for normal Mendelian inheritance, but otherwise the Hif-p4h-2gt/gt mice appeared to be healthy and fertile and to have a normal lifespan. Similarly, the Hif-p4h-2gt/gt pup percentage (36%) was reduced in the Hif-p4h-2+/gt and Hif-p4h-2gt/gt intercrosses (n = 50 offspring). The body weights of the Hif-p4h-2gt/gt mice (21.8 ± 1.8 g, n = 14) were ∼88% of those of the wild type (24.8 ± 1.5 g, n = 13) (p < 0.0001). RT-PCR of the heart RNA of Hif-p4h-2gt/gt mice showed that a small amount of wild-type Hif-p4h-2 mRNA was generated from the gene-trapped alleles (Fig. 1C), so that recognition of the splicing acceptor sequence in the 5′ end of the insertion cassette (Fig. 1A) was thus not absolute. A similar phenomenon has been observed also by others using the GeneTrap targeting strategy (36).
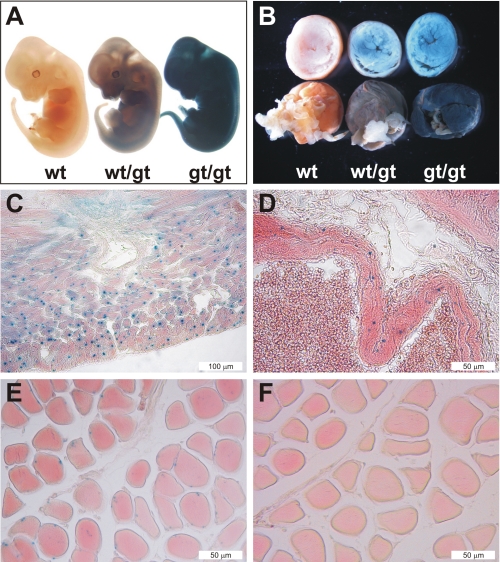
X-gal staining, performed to study the Hif-p4h-2 promoter-driven β-galactosidase expression, was intense and ubiquitous in the Hif-p4h-2gt/gt embryos at embryonic day 12.5 (Fig. 2A). The Hif-p4h-2 gene was also widely expressed in various adult tissues. Intense X-gal staining was seen throughout the whole mount hearts, for example (Fig. 2B). Histological analyses of hearts showed staining in the cardiomyocytes (Fig. 2C), in which both HIF-1α and HIF-2α have been shown to be stabilized under hypoxic conditions in vivo (37). Strong staining was also seen in the smooth muscle cells of the arteries in the lung (Fig. 2D) and in all other tissues studied. Skeletal muscle also showed X-gal staining (Fig. 2E), and no obvious alterations were seen in the morphological structure between the Hif-p4h-2gt/gt and wild-type muscle (Fig. 2, E and F).
FIGURE 2.
Analysis of the expression of Hif-p4h-2 in embryos and adult tissues. Hif-p4h-2 promoter-driven β-galactosidase expression was analyzed by X-gal staining of whole mounts or 5-μm paraffin sections of wild-type (wt), Hif-p4h-2+/gt (wt/gt), and Hif-p4h-2gt/gt (gt/gt) embryos and different tissues from adult mice. Embryonic day 12.5 Hif-p4h-2+/gt and Hif-p4h-2gt/gt embryos (A) and whole mount hearts (B) show ubiquitous X-gal staining with genotype-dependent intensity. C–F, histological analysis demonstrates pronounced X-gal staining in the cardiomyocytes of the Hif-p4h-2gt/gt hearts (C), in the smooth muscle cells of Hif-p4h-2gt/gt pulmonary arterial walls (D), and in myocytes of skeletal muscle (E), whereas no staining was observed in wild-type skeletal muscle (F).
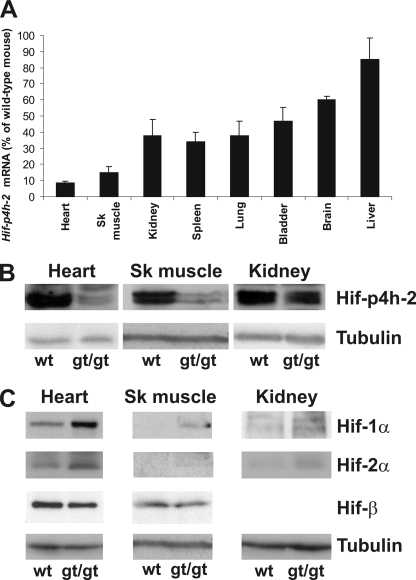
Wild-type Hif-p4h-2 mRNA Is Expressed at Various Levels in Different Hif-p4h-2gt/gt Tissues
The lowest relative amounts of wild-type Hif-p4h-2 mRNA in the various Hif-p4h-2gt/gt tissues versus the Hif-p4h-2+/+ ones, studied by means of Q-PCR (Fig. 3A), were seen in the heart and skeletal muscle, 8 and 15%, respectively, the values being 34–47% in the kidney, spleen, lung, and bladder; 60% in the brain; and 85% in the liver (Fig. 3A). Western blotting showed corresponding decreases in the amounts of Hif-p4h-2 protein in the heart, skeletal muscle, and kidney (Fig. 3B). The reduction in this enzyme level was accompanied by a concordant stabilization of Hif-1α and Hif-2α in these tissues (Fig. 2C).
FIGURE 3.
Analysis of wild-type Hif-p4h-2 mRNA levels and Hif-p4h-2, Hif-1α, and Hif-2α protein levels in selected tissues. A, wild-type Hif-p4h-2 mRNA levels in various Hif-p4h-2gt/gt tissues relative to wild-type tissues were analyzed by Q-PCR with primers amplifying the wild-type cDNA. A significant decrease (p < 0.001, n ≥ 5) in the Hif-p4h-2 mRNA level was seen in all Hif-p4h-2gt/gt tissues studied relative to the wild type, except in the liver. The error bars indicate S.D. B and C, Western blotting of total protein extracts from wild-type (wt) and Hif-p4h-2gt/gt (gt/gt) heart, skeletal (Sk) muscle, and kidney was performed with the antibodies indicated. Tubulin and HIF-1β stainings were used as loading controls.
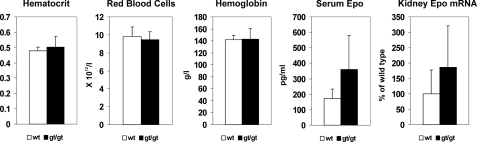
Hif-p4h-2gt/gt Mice Have No Polycythemia
Analysis of blood samples from the Hif-p4h-2gt/gt mice showed no increase in the hematocrit, red blood cell, or hemoglobin values (Fig. 4). The mean serum Epo level was increased ∼2-fold, but this was not statistically significant (p = 0.075, n = 6) (Fig. 4), and there was a trend for an increase in the kidney Epo mRNA level (Fig. 4), but these increases are less than those in mice with conditional broad spectrum Hif-p4h-2 inactivation (16, 17) and were evidently not sufficient to lead to increased erythropoiesis.
FIGURE 4.
Analysis of hematological parameters, serum Epo, and Epo mRNA in the kidney. Hematocrit, the red blood cell count, and hemoglobin level were analyzed in blood samples from four adult Hif-p4h-2gt/gt and age- and gender-matched wild-type mice. Serum Epo levels in six adult Hif-p4h-2gt/gt and age- and gender-matched wild-type mice were analyzed by enzyme-linked immunosorbent assay. The relative amounts of Epo mRNA in kidney samples from eight to ten adult Hif-p4h-2gt/gt and age- and gender-matched wild-type mice were analyzed by Q-PCR. The error bars indicate S.D.
Hearts of Hif-p4h-2gt/gt Mice Show No Signs of Dilated Cardiomyopathy or Increased Angiogenesis
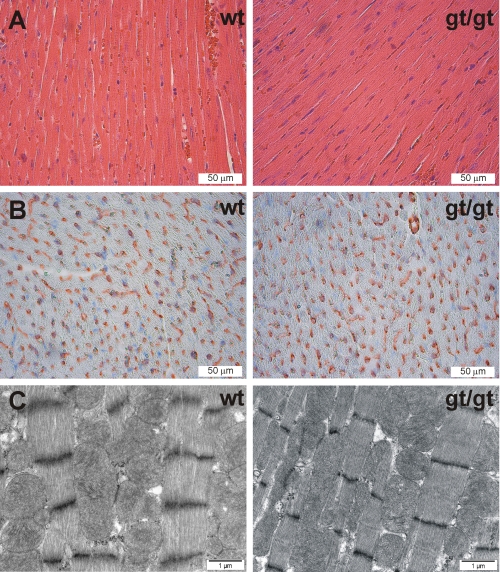
The Hif-p4h-2gt/gt hearts (0.16 ± 0.02 g, n = 11) were similar in weight to those in the wild type (0.18 ± 0.03 g, n = 13). Histological analysis of hematoxylin- and eosin-stained sections of the Hif-p4h-2gt/gt hearts showed no detectable alterations relative to the controls (Fig. 5A). Hearts of the mice with conditional Hif-p4h-2 inactivation in utero at day 17.5 postfertilization have enlarged cardiomyocytes, increased space between the myocardial fibers, and increased contraction band necrosis (17), whereas myocytes with degenerative changes have been found only occasionally when the inactivation occurred at the age of 3 weeks (25). None of these alterations were detected in the Hif-p4h-2gt/gt hearts (Fig. 5A). Likewise, no changes were observed in the number of the blood vessels in anti-Pecam-1-stained sections of the Hif-p4h-2gt/gt hearts (218 ± 26/field, n = 3) relative to the wild type (226 ± 29/field, n = 3) (Fig. 5B), although conditional broad spectrum Hif-p4h-2 inactivation in 6-week-old mice leads to hyperactive angiogenesis in the heart and other organs 6 weeks after the tamoxyfen treatment (15). Statistically significant changes were not found in the serum Vegf-a concentration either (22.0 ± 8.8 pg/ml in wild-type mice, n = 6; and 28.9 ± 16.6 pg/ml in Hif-p4h-2gt/gt mice, n = 6; p = 0.355). Electron microscopy analysis showed no differences in the ultrastructure of the capillaries, myofilaments, or mitochondria of the cardiomyocytes in the Hif-p4h-2gt/gt hearts relative to the wild type (Fig. 5C). Minamishima et al. (25) observed mild mitochondrial swelling in hearts of 10-week-old conditional Hif-p4h-2 null mice 3 weeks after tamoxyfen treatment. The area of mitochondria was also increased in the Hif-p4h-2gt/gt hearts (0.638 ± 0.349 μm2 in wild-type hearts, n = 165; and 0.754 ± 0.418 μm2 in Hif-p4h-2gt/gt hearts, n = 184; p = 0.0056).
FIGURE 5.
Histological analyses of the hearts. A and B, 5-μm paraffin sections of wild-type (wt) and Hif-p4h-2gt/gt (gt/gt) hearts stained with hematoxylin and eosin (A) and Pecam-1 (B). C, transmission electron microscopy of the wild-type (wt) and Hif-p4h-2gt/gt (gt/gt) hearts. The intensity and distance between Z-bands are influenced by the contractile state and sample width and vary within the same samples irrespective of the genotype.
Several HIF Target Genes Are Up-regulated in Hif-p4h-2gt/gt Hearts
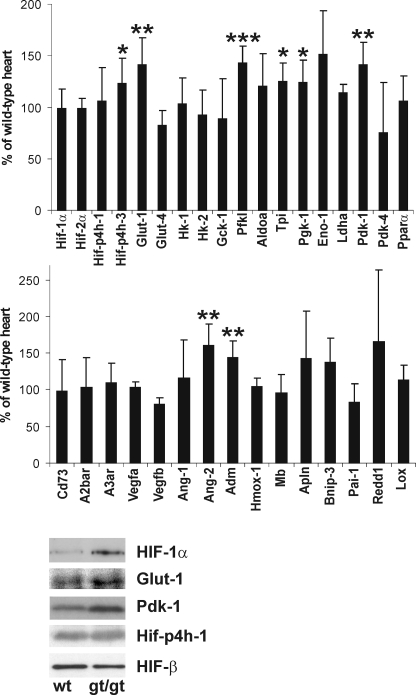
To analyze the consequences of the Hif-p4h-2 deficiency and the resulting HIF-1α and HIF-2α stabilization in the Hif-p4h-2gt/gt hearts, we performed Q-PCR analysis of the mRNA levels of select known HIF targets and some other genes relative to the wild-type hearts. The Hif-1α and Hif-2α mRNA levels were unchanged in Hif-p4h-2gt/gt hearts (Fig. 6), indicating that the increases in the respective protein levels (Fig. 3C) were indeed due to protein stabilization. The level of Hif-p4h-1 mRNA was likewise unchanged, whereas that of Hif-p4h-3 mRNA was increased (Fig. 6), agreeing with data indicating that the latter but not the former is a HIF target (38). Significant increases were found in the mRNA levels of the glucose metabolism genes Glut-1, phosphofructokinase-L, triose phosphate isomerase, and phosphoglycerate kinase-1, and a trend for an increase in the mRNA level of enolase-1 was also found, whereas the mRNA levels of the other glycolysis genes studied were unchanged (Fig. 6). The mRNA level of Pdk-1 was also increased, and significant increases were additionally seen in the mRNA levels of Ang-2 and adrenomedullin. Other genes that showed some trend for increased mRNA expression included those for apelin, which has various beneficial effects on cardiac function and blood pressure, the apoptosis-inducing protein Bnip-3 (p = 0.051), and Redd-1, which reduces protein synthesis by inhibiting the activity of the mammalian target of rapamycin mTOR (2, 27, 39, 40), whereas no changes were found in the levels of several other known HIF targets (Fig. 6). Interestingly, the latter included those for Cd73, the key enzyme for extracellular adenosine generation, and adenosine receptor A2b (Fig. 6), which were markedly increased after the acute stabilization of Hif-1α following left ventricular Hif-p4h-2 siRNA (23), and for Pdk-4 and Pparα, which were increased in the skeletal muscle of the Hif-p4h-1−/− mice (19). The changes in the expression levels of these genes were verified by a microarray analysis of an Affymetrix Gene Chip mouse genome 430 2.0 array with over 39,000 transcripts (data not shown). No further genes with significant changes in expression level were found in the microarray analysis.
FIGURE 6.
Molecular analysis of the preischemic Hif-p4h-2gt/gt hearts. Top and middle panels, Q-PCR analysis of the mRNA levels of Hif-1α, Hif-2α, Hif-p4h-1 and 3, and certain HIF target and other genes in adult preischemic Hif-p4h-2gt/gt hearts relative to age- and gender-matched wild-type hearts (n = 6). Hk-1 and -2, hexokinase-1 and -2; Gck-1, glucokinase-1; Pfkl, phosphofructokinase-L; Aldoa, aldolase A; Tpi, triose phosphate isomerase; Pgk-1, phosphoglycerate kinase-1; Eno-1, enolase-1; Ldha, lactate dehydrogenase A; A2bar, adenosine receptor A2b; A3ar, adenosine receptor A3; Adm, adrenomedullin; Hmox1, heme oxygenase 1; Mb, myoglobin; Apln, apelin; Pai-1, plasminogen activator inhibitor-1; Lox, lysyl oxidase. *, p < 0.05; **, p < 0.01; ***, p < 0.001. The error bars indicate S.D. Bottom panels, Western blotting of total protein extracts from preischemic wild-type (wt) and Hif-p4h-2gt/gt (gt/gt) heart was performed with the antibodies indicated.
To study whether the increased mRNA levels actually led to increased protein levels, we analyzed Glut-1 and Pdk-1 by Western blotting. The levels of both proteins were found to be slightly increased in the Hif-p4h-2gt/gt hearts relative to wild type, whereas no increase was seen in the levels of the Hif-p4h-1 and Hif-β proteins, agreeing with their unchanged mRNA levels (Fig. 6).
Hif-p4h-2gt/gt Mice Show Cardioprotection against Ischemia-Reperfusion Injury
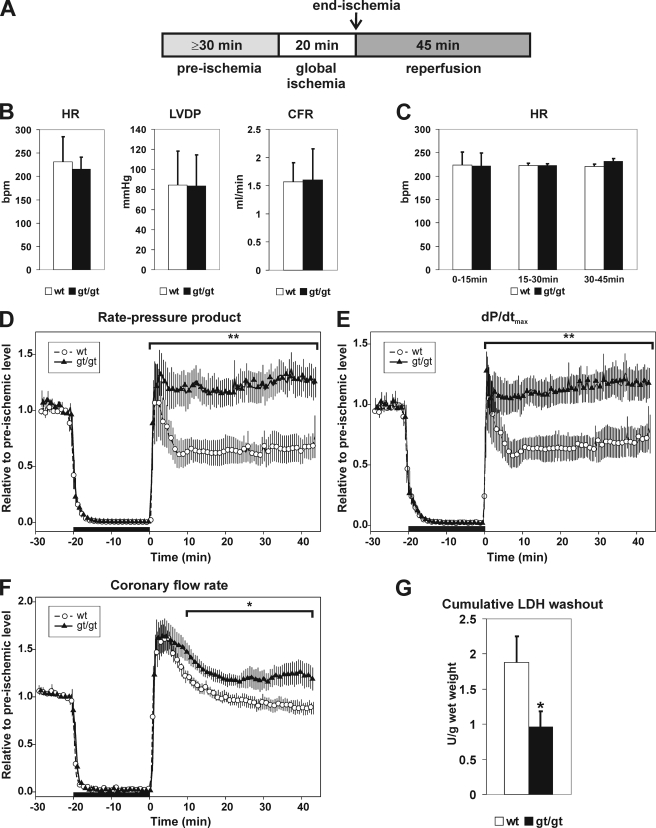
To study whether chronic stabilization of Hif-1α and Hif-2α in the heart provides cardioprotection against ischemia-reperfusion injury, isolated Langendorff-perfused hearts from Hif-p4h-2gt/gt and wild-type mice were subjected to a stabilization perfusion of at least 30 min, followed by global ischemia induced by cessation of perfusion for 20 min and reperfusion for 45 min (Fig. 7A). No differences were observed in the heart rates, LVDP, and coronary flow rates between the wild-type and Hif-p4h-2gt/gt hearts after the stabilization period (Fig. 7B), indicating that no differences exist in their hemodynamic function prior to ischemia.
FIGURE 7.
Analysis of Hif-p4h-2gt/gt hearts in an ex vivo ischemia-reperfusion model. A, hearts from 2–5-month-old female Hif-p4h-2gt/gt (gt/gt) mice and age- and gender-matched controls (wt) were isolated and subjected to Langendorff perfusion as indicated. B, preischemic heart rate (HR), LVDP, and coronary flow rate (CFR) were measured after a minimum 30-min stabilization period. C, post-ischemic heart rates during 0–15, 15–30, and 30–45 min of reperfusion. D–F, global ischemia (black horizontal bar) was induced after the stabilization perfusion (the last 10 min of which is shown) by cessation of perfusion for 20 min. Ischemia was followed by 45 min of reperfusion. The values are the means, and the hatched area depicts ± S.E. Coronary flow rate and left ventricle pressure were continuously recorded, and HR, LVDP, and RPP were calculated. The post-ischemic RPP (D), dP/dtmax (E), and coronary flow rate (F) values were calculated relative to the preischemic mean (mean of RPP, dP/dtmax, and coronary flow rate for the last 3 min of stabilization) in the wild-type (n = 7) and Hif-p4h-2gt/gt (n = 9) hearts. The AUC during the 45-min post-ischemic period was calculated using the method of summary measures. *, p < 0.05; **, p < 0.01. G, lactate dehydrogenase (LDH) levels in the venous effluents collected during the reperfusion were determined, and the cumulative lactate dehydrogenase washout for the first 15 min of reperfusion was calculated (units/g of wet weight) for the wild-type (n = 8) and Hif-p4h-2gt/gt (n = 8) hearts. The error bars indicate S.E.
There was no difference in the heart rates between the Hif-p4h-2gt/gt and wild-type hearts during the reperfusion (Fig. 7C). The rate pressure product (RPP = heart rate × LVDP) and dP/dtmax returned to and even exceeded the preischemic level during reperfusion in the Hif-p4h-2gt/gt hearts, whereas the wild-type hearts reached the preischemic level only momentarily in early reperfusion and stayed at a level of ∼65% of the preischemic one from 5 to 45 min of reperfusion (Fig. 7, D and E). The mechanical recovery was thus significantly improved in the Hif-p4h-2 deficient hearts during the 45-min reperfusion (RPP 29.4 ± 4.9 AUC in wild-type hearts, n = 7; and 55.4 ± 5.8 AUC in Hif-p4h-2gt/gt hearts, n = 9; p = 0.005; and dP/dtmax 30.1 ± 12.8 AUC in wild-type hearts, n = 7; and 56.8 ± 20.0 AUC in Hif-p4h-2gt/gt hearts, n = 9; p = 0.008) Furthermore, the mean end diastolic pressure relative to the preischemic level was slightly lower in the Hif-p4h-2gt/gt hearts than in the wild type throughout the reperfusion, but the difference was not statistically significant. Recovery of the coronary flow rate was significantly improved in the Hif-p4h-2gt/gt hearts (34.4 ± 2.0 AUC in wild-type hearts, n = 7; and 43.6 ± 2.9 AUC in Hif-p4h-2gt/gt hearts, n = 9; p = 0.029) (Fig. 7F). The cumulative release of lactate dehydrogenase during reperfusion was significantly reduced (1.88 ± 0.37 units/g wet weight in wild-type hearts, n = 8; and 0.96 ± 0.23 units/g wet weight in Hif-p4h-2gt/gt hearts, n = 8; p < 0.05) (Fig. 7G). Altogether these data indicate that chronic Hif-p4h-2 deficiency in the heart provides protection against cardiac ischemia-reperfusion injury.
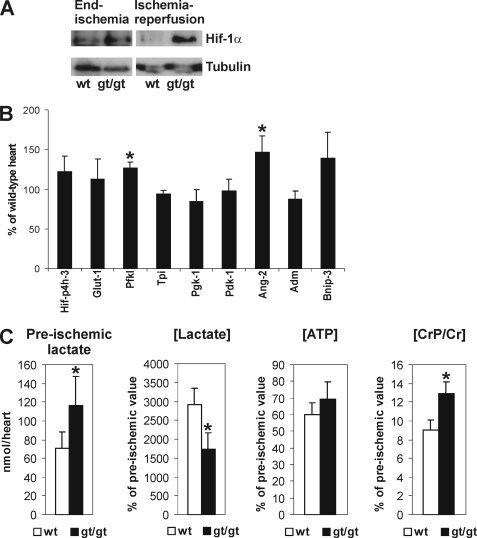
We next analyzed stabilization of HIF-1α at end ischemia and after ischemia-reperfusion (Fig. 8A). HIF-1α was stabilized to a similar extent in both wild-type and Hif-p4h-2gt/gt hearts at the end of the 20-min global ischemia (Fig. 8A). The stabilized HIF-1α was almost completely degraded during the 45-min reperfusion in wild-type hearts, while its amount was unchanged and remained at a high level in Hif-p4h-2gt/gt hearts (Fig. 8A). After ischemia-reperfusion, the mRNA levels of those genes that showed significantly increased expression in the preischemic Hif-p4h-2gt/gt hearts relative to wild type were also increased in wild-type hearts so that differences in the expression levels between wild-type and Hif-p4h-2gt/gt hearts disappeared in all cases except for the phosphofructokinase-L and Ang-2 genes (Fig. 8B). These data clearly suggest that the gene expression changes contributing to the cardioprotection in the Hif-p4h-2gt/gt hearts precede the ischemic insult.
FIGURE 8.
Molecular and metabolic analysis of Hif-p4h-2gt/gt hearts. A, Western blotting of total protein extracts from wild-type (wt) and Hif-p4h-2gt/gt (gt/gt) heart for HIF-1α at end ischemia and after ischemia-reperfusion. Tubulin staining was used as a loading control. B, Q-PCR analysis of the mRNA levels of certain HIF target genes (see Fig. 6) in Hif-p4h-2gt/gt hearts relative to age- and gender-matched wild-type hearts after ischemia-reperfusion (n = 4). The error bars indicate S.D. C, the amount of preischemic lactate (n = 4), and the ratios of the end ischemic versus preischemic [Lactate], [ATP], and [CrP/Cr] (n = 3) in age- and gender-matched Hif-p4h-2gt/gt and wild-type hearts. The error bars indicate S.D.
To study in more detail the cardioprotective mechanism in the Hif-p4h-2gt/gt hearts, we determined the amounts of certain key metabolites in preischemia and at end ischemia. The ATP concentration in the Hif-p4h-2gt/gt hearts at the end of the 20-min global ischemia was 69% of the preischemic value, whereas it was 60% in wild-type hearts (Fig. 8C). No differences in the amount of total ADP were detected in Hif-p4h-2gt/gt and wild-type hearts in preischemia (111.8 ± 24.9 nmol/heart in Hif-p4h-2gt/gt, n = 4; and 109.8 ± 12.6 nmol/heart in wild type, n = 4) or at end ischemia (111.6 ± 13.1 nmol/heart in Hif-p4h-2gt/gt, n = 3; and 105.6 ± 31.4 nmol/heart in wild type, n = 3). The ischemic versus preischemic ratio of CrP/Cr concentration was significantly higher in Hif-p4h-2gt/gt than in wild-type hearts (12.9 ± 1.2% in Hif-p4h-2gt/gt, n = 3; and 9.0 ± 1.1% in wild type, n = 3; p = 0.016) (Fig. 8C). These data indicate that the cellular energy state is retained at a higher level in Hif-p4h-2gt/gt hearts during ischemia. The amount of lactate was significantly increased in the preischemic Hif-p4h-2gt/gt hearts relative to wild type (116.2 ± 31.5 nmol/heart in Hif-p4h-2gt/gt, n = 4; and 71.2 ± 17.4 nmol/heart in wild type, n = 4; p = 0.047) (Fig. 8C), most likely leading to a lower pH in the Hif-p4h-2gt/gt hearts, whereas the lactate levels were equivalent in both genotypes at end ischemia (2.08 ± 0.30 μmol/heart in wild type, n = 3; and 2.03 ± 0.48 μmol/heart in Hif-p4h-2gt/gt, n = 3).
DISCUSSION
We have generated a novel viable Hif-p4h-2 hypomorph mouse line that expresses wild-type Hif-p4h-2 mRNA at varying levels in different organs, with the lowest level, only 8%, being detected in the heart. The data presented here indicate that the Hif-p4h-2 deficiency in the heart leads to chronic stabilization of Hif-1α and Hif-2α with no obvious adverse effects but instead provides cardioprotection against acute ischemia-reperfusion injury observed as improved functional recovery of the heart and reduced infarct size evidenced by decreased lactate dehydrogenase release. Thus, the congestive heart failure found after broad spectrum conditional chronic inactivation of Hif-p4h-2 (17, 25) is likely to be largely an indirect consequence of the severe polycythemia and hyperviscosity observed in these mice. The markedly increased angiogenesis found in the heart and other organs after broad spectrum conditional chronic inactivation of Hif-p4h-2 (15) was not seen in the hearts of our Hif-p4h-2gt/gt mice either. Interestingly, the increased angiogenesis in the conditional Hif-p4h-2 mice was seen even in the brain, where inactivation of the gene occurred only with an efficiency of ∼10%, suggesting that this increased angiogenesis might be mediated by systemic effects such as circulatory angiogenic factors rather than locally produced factors (15). The effectiveness of Hif-p4h-2 inactivation in the heart was 92% in our work, whereas in the work of Takeda et al. (15), it was only 74%, suggesting that even in the heart the increased angiogenesis found in the conditional Hif-p4h-2 mice was not due to locally produced factors. This suggestion is supported by data indicating that there was no up-regulation of the mRNAs for Vegf-a, Vegf-b, and Ang-1 in the heart in either study, whereas the level of serum Vegf-a was markedly increased in the mice studied by Takeda et al. (15) but not in our mice. Furthermore, our mice, unlike those studied by Takeda et al. (15) had significantly increased mRNA levels for Ang-2, which acts as a dose-dependent antagonist for Ang-1 of the receptor tyrosine kinase Tie2 of endothelial cells when Ang-1 is present (41). Mild mitochondrial swelling has been observed in the hearts of conditional Hif-p4h-2 null mice (25). Likewise, an increase in the mitochondrial area (μm2) was seen in the Hif-p4h-2gt/gt hearts, but no signs of ultrastructural damage were detected. An increase in mitochondrial volume is not necessarily a sign of a harmful effect but can be a regulation mechanism because IP has been reported to increase heart mitochondrial volume by ∼25% while providing cardioprotection (42). A similar increase, ∼20%, was seen in the area of Hif-p4h-2gt/gt heart mitochondria relative to wild type.
Our Hif-p4h-2gt/gt mice that had effective preischemic stabilization of both Hif-1α and Hif-2α in the heart showed up-regulation of the mRNAs for Glut-1 and several enzymes of glycolysis, which are all known HIF targets (43), suggesting increased glycolysis. The Hif-p4h-2gt/gt preischemic hearts may additionally have reduced activity of the pyryvate dehydrogenase complex, because the Pdk-1 mRNA level was increased even though the mRNA levels of Pdk-4 and its metabolic regulator Pparα were not increased. This may further enhance glycolysis by preventing pyruvate from entering the citric acid cycle and resulting in reduced oxidative phosphorylation. In the cases of Glut-1 and Pdk-1 we also showed that their increased mRNA levels actually led to increased protein levels. As metabolic level evidence of increased glycolysis, we found a significantly higher amount of lactate in the preischemic Hif-p4h-2gt/gt hearts relative to wild type (Fig. 8C). It has been reported that at equal ATP levels, hearts can withstand ischemia better if the source of ATP production is glycolytic rather than mitochondrial (44). This is likely to be caused by the increased cytosolic [ATP] that keeps the sarcolemmal KATP channel closed and the Na+/K+-ATPase energized and thereby prevents Na+ overload and subsequent Ca2+ overload caused by reversed Na+/Ca2+ exchange (45) and the following mitochondrial damage and decreased functional recovery during reperfusion (44). It has been shown that the skeletal muscle of Hif-p4h-1 null mice is protected against ischemic necrosis (19). Although expression of glycolytic enzymes is not increased in the Hif-p4h-1 null skeletal muscle, the basal metabolism is reprogrammed from oxidative to more anaerobic ATP production by a 1.6–2-fold preischemic up-regulation of Pparα, Pdk-1, and Pdk-4 mRNAs (19).
The mitochondrial permeability transition plays a major role in reperfusion damage, and opening of the mitochondrial permeability transition pore (MPTP) determines the amount of irreversible cardiac damage, which is reflected in infarct size (46). Reduction in adenine nucleotides favors MPTP opening, whereas low pH has been reported to inhibit opening of MPTP (46). The ischemic versus preischemic concentration of ATP remained higher in Hif-p4h-2gt/gt than in wild-type hearts (Fig. 8C), and the ischemic versus preischemic [CrP/Cr], which qualitatively reflects the cellular energy state and the free [ATP]/[ADP], was significantly higher in Hif-p4h-2gt/gt than in wild-type hearts (Fig. 8C). Although the end ischemic lactate levels were equal in the Hif-p4h-2gt/gt and wild-type hearts, probably because lactate accumulation reaches limiting conditions caused by concomitant pH decrease, they are likely to return during reperfusion to the preischemic levels, where the amount of lactate was significantly higher in Hif-p4h-2gt/gt hearts than in wild-type hearts (Fig. 8C). The elevated amount of ATP and the higher amount of lactate, which leads to acidosis and low pH, are thus also likely to contribute to the observed protection against ischemia-reperfusion injury in Hif-p4h-2gt/gt hearts via regulation of MPTP. These together with the above data suggest that the Hif-p4h-2gt/gt hearts are primed for preference toward glycolytic metabolism and thus have an advantage in surviving an ischemic insult. Furthermore, the hemodynamic findings, i.e. significantly increased rate pressure product, LVDP, dP/dtmax, and improved coronary flow rate after ischemia-reperfusion in the Hif-p4h-2gt/gt hearts relative to the wild type (Fig. 7, D–F), all indicate that the recovery of the mechanical performance is better in the Hif-p4h-2 hypomorph hearts than in the wild type, most likely contributing to a better prognosis.
The HIF targets adrenomedullin, a vasodilatory peptide that also increases the tolerance to oxidative stress in cells, and the peptide apelin, with emerging regulatory actions in the heart, protect the myocardium against ischemia-reperfusion injury-induced infarction and apoptosis (27, 40, 47, 48). Because the adrenomedullin mRNA level was significantly increased in the preischemic Hif-p4h-2gt/gt hearts and the apelin mRNA level showed a trend for an increase, it seems likely that increased adrenomedullin and apelin expression may have contributed to the cardioprotection seen here. The Bnip-3 mRNA level also showed a trend for an increase, but the significance of this change is unknown. Although Bnip-3 has been implicated as a pro-apoptotic gene, several reports do not support this assertion and indicate that under hypoxic conditions Bnip-3 plays a role in a survival mechanism (39).
Hif-1α was stabilized to a similar extent in the wild-type and Hif-p4h-2gt/gt hearts at end ischemia, whereas after reperfusion the amounts returned to those seen in preischemic hearts, i.e. Hif-1α was degraded in the wild-type heart but remained stabilized in the Hif-p4h-2gt/gt heart (Fig. 8A). Our results showed that the difference in the expression level of most of the HIF target genes that were up-regulated in the preischemic Hif-p4h-2gt/gt hearts relative to the wild type disappeared after ischemia-reperfusion because of the stabilization of Hif-1α also in the wild-type heart (Fig. 8, A and B). Although Hif-1α was degraded in the wild-type heart during reperfusion, the half-lives of the mRNAs are likely to exceed the 45-min reperfusion time, thus representing the outcome of HIF-1α stabilization during ischemia. Thus, the cardioprotection in the Hif-p4h-2gt/gt hearts must be due to Hif-mediated differences in gene expression already existing before the start of the ischemic insult.
Cardioprotection induced by IP has been shown to be lost in heterozygous Hif-1α null mice with concomitant impairment of reactive oxygen species production, oxidation of the phosphatase, and tensin homologue and phosphorylation of protein kinase B (22). The Hif-1α-dependent acute cardioprotection obtained by IP or by administration of the Hif-p4h inhibitor dimethyloxalylglycine or the introduction of Hif-p4h-2 siRNA into the left ventricle of the heart has been shown to be due to an approximately 10–15-fold induction of cardiac Cd73 and A2br mRNA levels, leading to elevated cardiac adenosine concentrations (23). Our Hif-p4h-2gt/gt mice had no increases in either of these two mRNAs, however, suggesting that the mechanisms involved in chronic cardioprotection are different from that seen in the acute case. Hmox-1 has also been shown to be a cardioprotective Hif-1α-target (49, 50), but the Hif-p4h-2gt/gt mice had no increase in its mRNA level either. An inducible nitric-oxide synthase-dependent pathway has also been suggested to be involved in the attenuation of reperfusion injury after stabilization of Hif-1 in the heart via in vivo Hif-p4h-2 siRNA treatment (51). However, the siRNA used in that study (51) did not target Hif-p4h-2, but instead collagen P4H isoenzyme II that does not act on HIF-1α (5).
Taken together, our data suggest that chronic stabilization of Hif-1α and Hif-2α by genetic knockdown of the Hif-p4h-2 isoenzyme in mouse heart promotes cardioprotection by induction of a number of genes involved in glucose metabolism, cardiac function, and blood pressure. In contrast to the outcome of acute depletion of Hif-p4h-2 where cardioprotection is provided by 10–15-fold induction of cardiac Cd73 and A2br mRNA levels (23), the magnitude of changes in the gene expression levels is relatively low, ∼1.5-fold, in the Hif-p4h-2 hypomorphic hearts, similar to the magnitude of changes seen in Hif-p4h-1 null skeletal muscle (19). It is thus likely that in our model the concerted action of a number of HIF target genes with slightly up-regulated expression is required for the generation of the cardioprotective effect instead of one or a few major highly up-regulated targets. The novel viable Hif-p4h-2 hypomorph mouse line generated here offers a unique model to study the consequences of chronic Hif-p4h-2 deficiency of varying extents in different organs.
The data obtained with HIF-P4H inhibitors in various animal models of anemia and ischemia and in ongoing clinical trials suggest efficacy in a number of therapeutic applications (52). Distinct differences are observed in the inhibitory potency of many small molecule 2-oxoglutarate analogues between the HIF-P4Hs and the other major P4H family, the collagen P4Hs, suggesting that their catalytic sites differ sufficiently to allow the development of specific inhibitors of the two P4H classes (52). Furthermore, differences exist even in the inhibition between the individual HIF-P4H isoenzymes, suggesting that it may be possible to develop specific inhibitors for each HIF-P4H isoenzyme (52).
Supplementary Material
Acknowledgments
We thank T. Aatsinki, R. Juntunen, E. Lehtimäki, R. Polojärvi, and M. Siurua and the Biocenter Oulu Transgenic and EM Core Facilities for excellent technical assistance; Dr. E.-R. Savolainen (Hematology Laboratory, Oulu University Hospital) for the hematological analyses; and Dr. J. Vuoristo (Biocenter Oulu) for the microarray analysis.
This work was supported by Health Science Council Grants 200471 and 202469 (to J. M.) and 120156 (to P. K.) from the Academy of Finland and by funds from the S. Juselius Foundation and FibroGen Inc. (to J. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- HIF
- hypoxia-inducible factor
- P4H
- prolyl 4-hydroxylase
- Epo
- erythropoietin
- IP
- ischemic preconditioning
- gt
- GeneTrap allele
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- RT
- reverse transcription
- Q-PCR
- quantitative real time RT-PCR
- AUC
- area under the curve
- Vegf
- vascular endothelial growth factor
- Bnip-3
- BCL2/adenovirus E1B interacting protein-3
- Ang
- angiopoietin
- Glut
- glucose transporter
- Pdk
- pyryvate dehydrogenase kinase
- Pparα
- peroxisome proliferator activator α
- LVDP
- left ventricular developed pressure
- RPP
- rate pressure product
- MPTP
- mitochondrial permeability transition pore
- siRNA
- small interfering RNA
- CrP
- creatine phosphate
- Cr
- creatine.
REFERENCES
- 1.Kaelin W. G., Jr., Ratcliffe P. J. (2008) Mol. Cell 30, 393–402 [DOI] [PubMed] [Google Scholar]
- 2.Fraisl P., Aragonés J., Carmeliet P. (2009) Nat. Rev. Drug Discov. 8, 139–152 [DOI] [PubMed] [Google Scholar]
- 3.Semenza G. L. (2009) Physiology 24, 97–106 [DOI] [PubMed] [Google Scholar]
- 4.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 5.Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., Kriegsheim Av, Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 6.Yu F., White S. B., Zhao Q., Lee F. S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9630–9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruick R. K., McKnight S. L. (2001) Science 294, 1337–1340 [DOI] [PubMed] [Google Scholar]
- 8.Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P. H., Pugh C. W., Schofield C. J., Ratcliffe P. J. (2001) Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- 9.Ivan M., Haberberger T., Gervasi D. C., Michelson K. S., Günzler V., Kondo K., Yang H., Sorokina I., Conaway R. C., Conaway J. W., Kaelin W. G., Jr. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13459–13464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oehme F., Ellinghaus P., Kolkhof P., Smith T. J., Ramakrishnan S., Hütter J., Schramm M., Flamme I. (2002) Biochem. Biophys. Res. Commun. 296, 343–349 [DOI] [PubMed] [Google Scholar]
- 11.Koivunen P., Tiainen P., Hyvärinen J., Williams K. E., Sormunen R., Klaus S. J., Kivirikko K. I., Myllyharju J. (2007) J. Biol. Chem. 282, 30544–30552 [DOI] [PubMed] [Google Scholar]
- 12.Berra E., Benizri E., Ginouvès A., Volmat V., Roux D., Pouysségur J. (2003) EMBO J. 22, 4082–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Appelhoff R. J., Tian Y. M., Raval R. R., Turley H., Harris A. L., Pugh C. W., Ratcliffe P. J., Gleadle J. M. (2004) J. Biol. Chem. 279, 38458–38465 [DOI] [PubMed] [Google Scholar]
- 14.Takeda K., Ho V. C., Takeda H., Duan L. J., Nagy A., Fong G. H. (2006) Mol. Cell. Biol. 26, 8336–8346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeda K., Cowan A., Fong G. H. (2007) Circulation 116, 774–781 [DOI] [PubMed] [Google Scholar]
- 16.Takeda K., Aguila H. L., Parikh N. S., Li X., Lamothe K., Duan L. J., Takeda H., Lee F. S., Fong G. H. (2008) Blood 111, 3229–3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minamishima Y. A., Moslehi J., Bardeesy N., Cullen D., Bronson R. T., Kaelin W. G., Jr. (2008) Blood 111, 3236–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazzone M., Dettori D., Leite de Oliveira R., Loges S., Schmidt T., Jonckx B., Tian Y. M., Lanahan A. A., Pollard P., Ruiz de Almodovar C., De Smet F., Vinckier S., Aragonés J., Debackere K., Luttun A., Wyns S., Jordan B., Pisacane A., Gallez B., Lampugnani M. G., Dejana E., Simons M., Ratcliffe P., Maxwell P., Carmeliet P. (2009) Cell 136, 839–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aragonés J., Schneider M., Van Geyte K., Fraisl P., Dresselaers T., Mazzone M., Dirkx R., Zacchigna S., Lemieux H., Jeoung N. H., Lambrechts D., Bishop T., Lafuste P., Diez-Juan A., Harten S. K., Van Noten P., De Bock K., Willam C., Tjwa M., Grosfeld A., Navet R., Moons L., Vandendriessche T., Deroose C., Wijeyekoon B., Nuyts J., Jordan B., Silasi-Mansat R., Lupu F., Dewerchin M., Pugh C., Salmon P., Mortelmans L., Gallez B., Gorus F., Buyse J., Sluse F., Harris R. A., Gnaiger E., Hespel P., Van Hecke P., Schuit F., Van Veldhoven P., Ratcliffe P., Baes M., Maxwell P., Carmeliet P. (2008) Nat. Genet. 40, 170–180 [DOI] [PubMed] [Google Scholar]
- 20.Bishop T., Gallagher D., Pascual A., Lygate C. A., de Bono J. P., Nicholls L. G., Ortega-Saenz P., Oster H., Wijeyekoon B., Sutherland A. I., Grosfeld A., Aragones J., Schneider M., van Geyte K., Teixeira D., Diez-Juan A., Lopez-Barneo J., Channon K. M., Maxwell P. H., Pugh C. W., Davies A. M., Carmeliet P., Ratcliffe P. J. (2008) Mol. Cell. Biol. 28, 3386–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myllyharju J. (2008) Ann. Med. 40, 402–417 [DOI] [PubMed] [Google Scholar]
- 22.Cai Z., Zhong H., Bosch-Marce M., Fox-Talbot K., Wang L., Wei C., Trush M. A., Semenza G. L. (2008) Cardiovasc. Res. 77, 463–470 [DOI] [PubMed] [Google Scholar]
- 23.Eckle T., Köhler D., Lehmann R., El Kasmi K., Eltzschig H. K. (2008) Circulation 118, 166–175 [DOI] [PubMed] [Google Scholar]
- 24.Huang M., Chan D. A., Jia F., Xie X., Li Z., Hoyt G., Robbins R. C., Chen X., Giaccia A. J., Wu J. C. (2008) Circulation 118, S226–S233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minamishima Y. A., Moslehi J., Padera R. F., Bronson R. T., Liao R., Kaelin W. G., Jr. (2009) Mol. Cell. Biol. 29, 5729–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rankin E. B., Rha J., Unger T. L., Wu C. H., Shutt H. P., Johnson R. S., Simon M. C., Keith B., Haase V. H. (2008) Oncogene 27, 5354–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ronkainen V. P., Ronkainen J. J., Hänninen S. L., Leskinen H., Ruas J. L., Pereira T., Poellinger L., Vuolteenaho O., Tavi P. (2007) FASEB J. 21, 1821–1830 [DOI] [PubMed] [Google Scholar]
- 28.Wurst W., Gossler A. (2000) in Gene Targeting: A Practical Approach, 2nd Ed. (Joyner A. L. ed) pp. 234–236, IRL Press at Oxford University Press, New York [Google Scholar]
- 29.Karpanen T., Bry M., Ollila H. M., Seppänen-Laakso T., Liimatta E., Leskinen H., Kivelä R., Helkamaa T., Merentie M., Jeltsch M., Paavonen K., Andersson L. C., Mervaala E., Hassinen I. E., Ylä-Herttuala S., Oresic M., Alitalo K. (2008) Circ. Res. 103, 1018–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergmeyer H. U., Bernt E. (1970) in Methoden der Enzymatischen Analyse (Bergmeyer H. U. ed) pp. 533–538, Verlag Chemie, Weinheim [Google Scholar]
- 31.Lamprecht W., Stein P., Heinz F., Weisser H. (1970) in Methoden der Enzymatischen Analyse (Bergmeyer H. U. ed) pp. 1729–1733, Verlag Chemie, Weinheim, Germany [Google Scholar]
- 32.Bernt E., Bergmeyer H. U., Möllering H. (1970) in Methoden der Enzymatischen Analyse (Bergmeyer H. U. ed) pp. 1724–1728, Verlag Chemie, Weinheim, Germany [Google Scholar]
- 33.Lamprecht W., Trautschold I. (1970) in Methoden der Enzymatischen Analyse (Bergmeyer H. U. ed) pp. 2024–2033, Verlag Chemie, Weinheim, Germany [Google Scholar]
- 34.Noll F. (1970) in Methoden der Enzymatischen Analyse (Bergmeyer H. U. ed) pp. 1433–1437, Verlag Chemie, Weinheim, Germany [Google Scholar]
- 35.Matthews J. N., Altman D. G., Campbell M. J., Royston P. (1990) Brit. Med. J. 300, 230–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voss A. K., Thomas T., Gruss P. (1998) Dev. Dyn. 212, 258–266 [DOI] [PubMed] [Google Scholar]
- 37.Jürgensen J. S., Rosenberger C., Wiesener M. S., Warnecke C., Hörstrup J. H., Gräfe M., Philipp S., Griethe W., Maxwell P. H., Frei U., Bachmann S., Willenbrock R., Eckardt K. U. (2004) FASEB J. 18, 1415–1417 [DOI] [PubMed] [Google Scholar]
- 38.Metzen E., Berchner-Pfannschmidt U., Stengel P., Marxsen J. H., Stolze I., Klinger M., Huang W. Q., Wotzlaw C., Hellwig-Bürgel T., Jelkmann W., Acker H., Fandrey J. (2003) J. Cell Sci. 116, 1319–1326 [DOI] [PubMed] [Google Scholar]
- 39.Bellot G., Garcia-Medina R., Gounon P., Chiche J., Roux D., Pouysségur J., Mazure N. M. (2009) Mol. Cell. Biol. 29, 2570–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleinz M. J., Baxter G. F. (2008) Regul. Pept. 146, 271–277 [DOI] [PubMed] [Google Scholar]
- 41.Yuan H. T., Khankin E. V., Karumanchi S. A., Parikh S. M. (2009) Mol. Cell. Biol. 29, 2011–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim K. H., Javadov S. A., Das M., Clarke S. J., Suleiman M. S., Halestrap A. P. (2002) J. Physiol. 545, 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Semenza G. L. (2007) J. Bioenerg. Biomembr. 39, 231–234 [DOI] [PubMed] [Google Scholar]
- 44.Opie L. H., Sack M. N. (2002) J. Mol. Cell. Cardiol. 34, 1077–1089 [DOI] [PubMed] [Google Scholar]
- 45.Schäfer C., Ladilov Y., Inserte J., Schäfer M., Haffner S., Garcia-Dorado D., Piper H. M. (2001) Cardiovasc. Res. 51, 241–250 [DOI] [PubMed] [Google Scholar]
- 46.Halestrap A. P. (2009) J. Bioenerg. Biomembr. 41, 113–121 [DOI] [PubMed] [Google Scholar]
- 47.Burley D. S., Hamid S. A., Baxter G. F. (2007) Heart Fail. Rev. 12, 279–291 [DOI] [PubMed] [Google Scholar]
- 48.Cormier-Regard S., Nguyen S. V., Claycomb W. C. (1998) J. Biol. Chem. 273, 17787–17792 [DOI] [PubMed] [Google Scholar]
- 49.Czibik G., Sagave J., Martinov V., Ishaq B., Sohl M., Sefland I., Carlsen H., Farnebo F., Blomhoff R., Valen G. (2009) Cardiovasc. Res. 82, 107–114 [DOI] [PubMed] [Google Scholar]
- 50.Yoshida T., Maulik N., Ho Y. S., Alam J., Das D. K. (2001) Circulation 103, 1695–1701 [DOI] [PubMed] [Google Scholar]
- 51.Natarajan R., Salloum F. N., Fisher B. J., Kukreja R. C., Fowler A. A., 3rd (2006) Circ. Res. 98, 133–140 [DOI] [PubMed] [Google Scholar]
- 52.Myllyharju J. (2009) Curr. Pharm. Des. 15, 3878–3885 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.