Abstract
Rapid advances in genotyping and sequencing technology have dramatically accelerated the discovery of genes underlying human disease. Elucidating the function of such genes and understanding their role in pathogenesis, however, remain challenging. Here, we introduce a genomic strategy to characterize such genes functionally, and we apply it to LRPPRC, a poorly studied gene that is mutated in Leigh syndrome, French-Canadian type (LSFC). We utilize RNA interference to engineer an allelic series of cellular models in which LRPPRC has been stably silenced to different levels of knockdown efficiency. We then combine genome-wide expression profiling with gene set enrichment analysis to identify cellular responses that correlate with the loss of LRPPRC. Using this strategy, we discovered a specific role for LRPPRC in the expression of all mitochondrial DNA-encoded mRNAs, but not the rRNAs, providing mechanistic insights into the enzymatic defects observed in the disease. Our analysis shows that nuclear genes encoding mitochondrial proteins are not collectively affected by the loss of LRPPRC. We do observe altered expression of genes related to hexose metabolism, prostaglandin synthesis, and glycosphingolipid biology that may either play an adaptive role in cell survival or contribute to pathogenesis. The combination of genetic perturbation, genomic profiling, and pathway analysis represents a generic strategy for understanding disease pathogenesis.
Keywords: Metabolic Diseases; Microarray; Mitochondrial DNA; Mitochondrial Metabolism; RNA Interference (RNAi); LRPPRC; Leigh Syndrome, French-Canadian type
Introduction
In recent years, the discovery of genes underlying human disease has progressed rapidly due to the availability of the human genome sequence as well as methods for rapidly genotyping and sequencing. Genome-wide association studies have yielded a plethora of genes associated with common diseases such as cancer and diabetes (1). Integrative genomic approaches made possible by the availability of large scale biological data sets have hastened the discovery of causative genes for rare Mendelian disorders (2). More recent advances in sequencing technologies further promise rapid identification of a multitude of mutant alleles responsible for Mendelian disorders as exemplified by the discovery of DHODH mutations in Miller syndrome (3). Still, pinpointing the exact function of newly discovered genes and the global implications of mutations on cellular function remains extremely challenging. The identification of causative alleles is an important first step in understanding the molecular basis of a disorder; however, determining gene function and characterizing cellular responses to disease-causing mutations are necessary to understand pathogenesis. Although the discovery of disease genes is becoming simpler, the latter step remains a major bottleneck.
One of the first successful applications of genome-wide association studies was Leigh syndrome, French-Canadian type (LSFC)2 (4). LSFC is a rare, monogenic, Mendelian mitochondrial disease that presents as a cytochrome c oxidase (COX) deficiency, common in the Saguenay-Lac-Saint-Jean region of Quebec. Lee et al. (4) mapped the disease to a 2-cm region on chromosome 2 by performing a genome-wide screen for linkage disequilibrium. A follow-up integrative genomic analysis spotlighted LRPPRC, leucine-rich pentatricopeptide repeat motif-containing protein, a high scoring candidate gene whose product is co-expressed with known mitochondrial genes (5). Subsequent sequencing of LRPPRC identified two different causal mutations (5). LRPPRC encodes a 130-kDa RNA-binding protein (6) that localizes primarily to the mitochondria (5, 6). It belongs to a family of pentatricopeptide repeat proteins common to mitochondria and chloroplast that are particularly abundant in plants (7). As a class, pentatricopeptide repeat proteins tend to be sequence-specific RNA-binding proteins with direct roles in RNA editing, processing, splicing, and translation (8).
The precise molecular function of LRPPRC has remained controversial. Consistent with the predicted function of a pentatricopeptide repeat domain-containing protein, Xu et al. (9) have shown that LRPPRC is required for the expression of mitochondrial DNA (mtDNA)-encoded COX subunits CO1 and CO3. However, Cooper et al. (10, 11) proposed a role for LRPPRC in transcriptional activation of nuclear genes via its interaction with PGC-1α. The global effect of loss of LRPPRC on cellular metabolic pathways has not been determined, although previous reports have implicated LRPPRC in hepatic glucose homeostasis and brown fat differentiation (10, 11). Like many rare diseases, unraveling the LSFC pathogenesis and determining the function of LRPPRC are hindered by the limited availability of patient samples. Because LSFC is characterized by a loss of function of LRPPRC, gene silencing by RNAi offers a tractable approach to create cellular models of LSFC.
Here, we use short hairpin RNA (shRNA)-mediated graded knockdown of LRPPRC to construct cellular models of LSFC that recapitulate all of the reported disease phenotypes. To identify mitochondrial and nuclear responses to loss of LRPPRC systematically, we carry out genome-wide expression profiling of these cellular models, followed by gene set enrichment analysis (GSEA) (12). We demonstrate that all mtDNA-encoded mRNA transcripts decrease in proportion to the loss of LRPPRC. In contrast, mtDNA-encoded rRNAs are unaffected, suggesting a specific role for LRPPRC in mt-mRNA homeostasis. In addition, we find up-regulation of key enzymes for fructose and mannose metabolism and of genes for prostaglandin (PG) biosynthesis, as well as down-regulation of genes for glycosphingolipid biosynthesis. These pathways could play a compensatory role in the face of mitochondrial dysfunction, or alternatively may contribute to pathogenesis.
EXPERIMENTAL PROCEDURES
Cell Culture
MCH58 immortalized human skin fibroblasts were cultured in high glucose Dulbecco's modified Eagle's medium (Invitrogen catalog no. 11995) supplemented with 10% fetal bovine serum (Sigma catalog no. F6178) and 1 × penicillin, streptomycin, and glutamine (Invitrogen catalog no. 10378-016). The cell culture medium was further supplemented with 2 μg/ml puromycin and 50 μg/ml uridine to culture cells after lentiviral infection.
Materials
Quantitative real-time PCR (qRT-PCR) assays for mitochondrial transcripts were developed in collaboration with Applied Biosystems (Foster City, CA). Antibodies including CO2, NDUFB8, SDHB, UQCRC2, ATP5A, and VDAC1 were purchased from Mitosciences (Eugene, OR). β-Actin was obtained from Sigma, and LRPPRC antibody was a generous gift from Serafin Piñol-Roma. The cDNA synthesis SuperScript III kit was from Invitrogen (catalog no. 11752). Human U133 plus 2.0 oligonucleotide arrays were purchased from Affymetrix (Santa Clara, CA).
shRNA Lentivirus Production and Infection
Lentiviral vectors for expressing shRNA (pLKO.1) were constructed at the Broad Institute RNAi platform (13). The shRNA sequences are shown in supplemental Table 1. For infection, 200,000 cells were seeded in a 6-well dish. 30 μl of viral supernatant was added to cells for a final volume of 2 ml of medium containing 8 μg/ml Polybrene. The plates were spun at a relative centrifugal force of 805 × g for 30 min at 32 °C, returned to 37 °C, and 24 h after infection were selected for infection using 2 μg/ml puromycin.
qRT-PCR
RNA was isolated from cells using the RNeasy kit (Qiagen). 500 ng of RNA was used as starting material to synthesize first-strand cDNA. 1 ng of genomic DNA, isolated using the DNeasy kit (Qiagen), was used as starting material for the multiplex qRT-PCR assay to quantify mitochondrial DNA as described previously (14). qRT-PCR was performed on cDNA and genomic DNA samples using a 96-well ABI7500 RT-PCR system in 20-μl reactions prepared with 2 × master mix, 20× ABI Taqman assay (supplemental Table 2), and diluted cDNA or genomic DNA samples.
Western Blotting
15–25 μg of protein sample was loaded per lane on a 4–12% BisTris gel followed by electrophoresis at constant voltage (200 V) for 50 min. The separated proteins were blotted onto a polyvinylidene difluoride membrane and blocked for 1 h at room temperature in Tris-buffered saline with 0.1% Tween 20 and 5% BSA (TBST-BSA). Membranes were incubated with primary antibody in TBST-BSA overnight at 4 °C. Primary antibodies were used at the following dilutions: LRPPRC, 1:500; β-actin, 1:10,000; CO2, 1:2,000; NDUFB8, 1:500; SDHB, 1:500; UQCRC2, 1:1,000; ATP5A, 1:10,000; and VDAC1, 1:1,000. Secondary antibody was used at 1:5,000 for 1 h at room temperature. Membranes were developed using WesternLightning Plus-ECL.
Measurement of Cellular Oxygen Consumption and Extracellular Acidification
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) measurements were carried out as described previously (15) with minor modifications. LRPPRC knockdown cells were seeded in XF 24-well cell culture microplates (Seahorse Bioscience) at 30,000 cells/well and incubated at 37 °C and 5% CO2 for 24 h. Prior to measurement, the growth medium was replaced with 925 μl of assay medium (Seahorse Bioscience). The cells were incubated at 37 °C for 60 min in the assay medium prior to measurements. The OCR and ECAR measurements were taken simultaneously every 7 min after a 2-min mix and a 2-min wait period.
Microarray Analysis
Total RNA was extracted from ∼2 million cells with a RNeasy mini kit (Qiagen). 10 μg of RNA was used to synthesize cDNA with a T7-(dT)24 primer and the SuperScript one-cycle cDNA synthesis kit (Affymetrix). cRNA labeling, hybridization to human U133 plus 2.0 oligonucleotide array, washing, and staining were performed as recommended by Affymetrix. Two biological replicates were used for each LRPPRC knockdown cell line, for a total of 14 arrays. The expression data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO series accession number GSE20847. Microarray data were analyzed using GSEA to identify pathways whose expression was altered in concert with the expression of LRPPRC. We used metabolic pathways defined by Edinburgh Human Metabolic Network (16) as well as a set of U133 plus 2.0 probes targeting mtRNA transcripts, nuclear-encoded oxidative phosphorylation (OXPHOS) genes as described previously (14) and a gene set comprising all of the nuclear-encoded mitochondrial proteins as defined in MitoCarta (17).
RESULTS
Engineering and Validating Cellular Models of LSFC
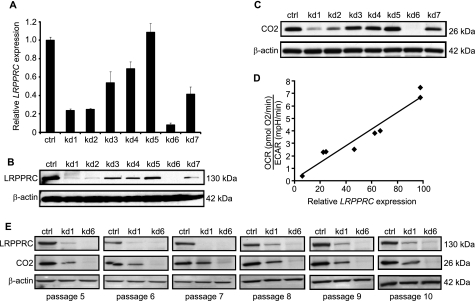
We designed seven shRNAs targeting the LRPPRC cDNA sequence to silence its expression in MCH58 immortalized human skin fibroblasts. We chose diploid human fibroblasts to construct stable cellular models because a number of mitochondrial diseases, including LSFC, express biochemical phenotypes in fibroblasts (supplemental Table 3). We quantified the LRPPRC transcript level by qRT-PCR measurement on RNA extracted from shLRPPRC-infected cells. As shown in Fig. 1A, the extent of knockdown was hairpin-specific, and the LRPPRC transcript level ranged from 8% to 100% in knockdown cell lines. Moreover, depletion of LRPPRC protein was correlated strongly with the pattern of progressive mRNA knockdown (Fig. 1B). A hallmark biochemical feature of LSFC is a reduced level of COX subunits (18). To determine whether our knockdown cell lines faithfully model LSFC, we carried out Western blot analysis for CO2, an mtDNA-encoded subunit of COX, and confirmed that depletion of LRPPRC results in a corresponding and proportional reduction of CO2 (Fig. 1C). To demonstrate further that our knockdown cells recapitulate the physiological defects associated with LSFC, we interrogated oxidative and glycolytic metabolism by assaying for OCR and ECAR, respectively. As expected, decreasing LRPPRC mRNA levels produced a quantitatively correlated decrease in the ratio of OCR to ECAR, indicating a coordinated decrease in respiration and an increase in glycolytic output (Fig. 1D). The knockdown of LRPPRC and the resulting depletion of CO2 protein in lentiviral-shRNA-infected cell lines persisted over the course of 10 passages, approximately 1 month after infection, demonstrating that our cell lines are indeed stable (Fig. 1E).
FIGURE 1.
Using RNAi to engineer cellular models of LSFC. A, LRPPRC mRNA quantified by qRT-PCR on RNA extracted from MCH58 human fibroblasts infected with an empty vector, pLKO.1 (ctrl), or one of seven independent shRNAs targeting LRPPRC. Values are reported as fold change in expression over control (ctrl). Three biological replicates were used per hairpin (error bars represent S.D.). β-Actin expression was used as an endogenous control. B, Western blot detection of LRPPRC protein abundance in LRPPRC knockdown cell lines. C, Western blot detection of CO2 protein abundance in LRPPRC knockdown cell lines. D, correlation between the ratio of OCR to ECAR and LRPPRC expression as measured in A (n ≥ 4). E, Western blots of LRPPRC and CO2 protein abundance over the course of five passages spanning ∼15–30 days of culture after infection with pLKO.1 or one of the two most potent hairpins.
Microarray Profiling of LSFC Cellular Models
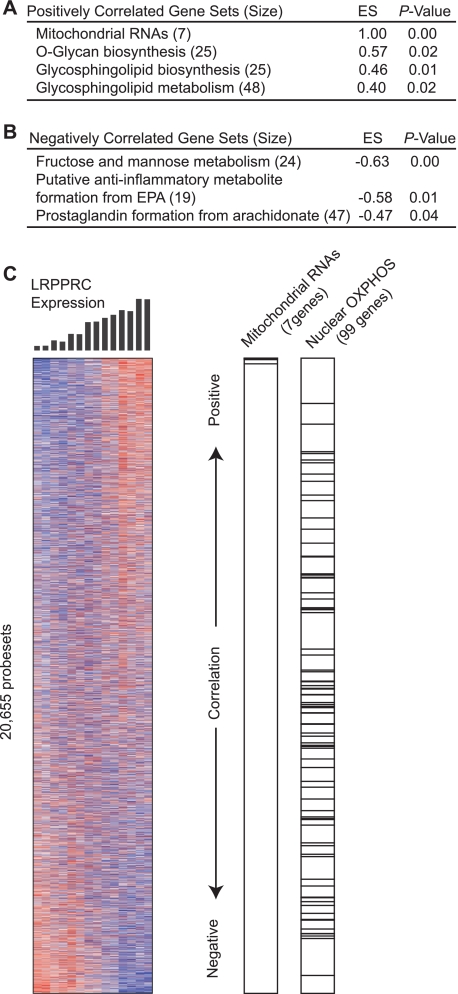
We performed genome-wide expression profiling of the engineered LSFC cell lines to identify systematically biochemical pathways that are altered in response to the depletion of LRPPRC and the consequent loss of mitochondrial respiratory function. The LRPPRC transcript levels determined by microarray analysis were strongly correlated with those obtained by qRT-PCR (Pearson's correlation coefficient = 0.99) (supplemental Fig. 1), confirming the high quality of the microarray data. We performed GSEA on the expression data from the seven engineered LSFC cell lines, each with progressively decreased LRPPRC expression. GSEA identified seven gene sets that are significantly correlated with LRPPRC expression at a nominal p value <0.05 and a false discovery rate <25% (Fig. 2). The gene set corresponding to mtRNAs was correlated strongly and positively with LRPPRC expression and exhibited a nearly perfect enrichment score (ES = 1.00) (Fig. 2, A and C). Among the nuclear-encoded gene sets, three were positively correlated with LRPPRC expression: “O-glycan biosynthesis,” “glycosphingolipid biosynthesis,” and “glycosphingolipid metabolism” (Fig. 2A). The negatively correlated gene sets were “fructose and mannose metabolism,” “putative anti-inflammatory metabolite formation from eicosapentanoic acid,” and “prostaglandin formation from arachidonate” (Fig. 2B). Notably, the nuclear-encoded OXPHOS gene set (ES = −0.25; p = 0.79) (Fig. 2C) and the gene set comprising all nuclear-encoded mitochondrial proteins (ES = 0.16; p = 0.90) did not show significant correlation.
FIGURE 2.
Gene set enrichment analysis of genomic profiles following LRPPRC knockdown. A, list of gene sets that are positively correlated with LRPPRC expression. B, list of gene sets that are negatively correlated with LRPPRC expression. Only gene sets that show p < 0.050 and false discovery rate <0.250 are listed. C, left, heat map showing expression of all 20,655 genes. Genes were ordered by their correlation to the LRPPRC expression profile (shown in black bars), with genes showing the strongest correlation at the top and the genes showing the strongest anticorrelation at the bottom. Red color indicates the highest expression, and blue color indicates the lowest. Right, bar plot indicating genes belonging to mitochondrial RNAs and nuclear OXPHOS genes.
The highest scoring negatively correlated gene set, annotated as fructose and mannose metabolism, consists of genes involved in the synthesis of mannose 6-phosphate, GDP-mannose, and GDP-fucose that are all intermediates in the glycoprotein biosynthesis. The same gene set also includes numerous genes encoding glycolytic enzymes (supplemental Fig. 2A). The increased expression of glycolytic genes may in part represent a biochemical adaptation to compensate for the loss of mitochondrial ATP production, as evidenced by the fact that LRPPRC knockdown cells were more sensitive to the glycolytic inhibitor 2-deoxyglucose (supplemental Fig. 2B). The second- and third-ranking negatively correlated gene sets shared several genes encoding enzymes involved in PG biosynthesis, including the key enzyme cyclooxygenase 2 (supplemental Fig. 3).
Role of LRPPRC in the Regulation of mtDNA-encoded mRNAs
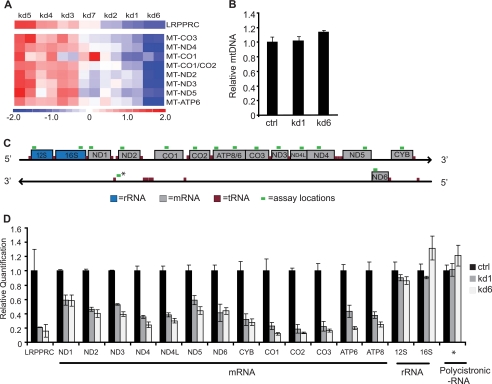
Although it has been shown previously that LRPPRC is necessary for the maintenance of steady-state levels of the mtDNA-encoded subunits of COX (9), we observed reduced expression of almost all of the mtDNA-encoded transcripts, including subunits of respiratory complex I (ND2–ND5), IV (CO1–CO3) and V (ATP6) (Fig. 3A). The reduced expression of these genes was not caused by mtDNA depletion because the two most potent knockdowns, kd1 and kd6, had the same mtDNA content as control cells (Fig. 3B). To determine the precise role of LRPPRC in mitochondrial gene expression, we designed multiple qRT-PCR Taqman assays that specifically probe each of the mtDNA-encoded mRNAs and rRNAs, as well as the polycistronic RNA transcript (Fig. 3C). The qRT-PCR measurement for mRNAs from the two most potent knockdown cell lines showed a significant depletion of each of the 13 mRNAs encoded by the mtDNA (Fig. 3D). However, the expression of the two mtDNA-encoded rRNAs was unaffected by the loss of LRPPRC (Fig. 3D). The mitochondrial mRNAs are generated simultaneously as a polycistronic transcript from the heavy strand (HS) and light strand (LS) promoters (19). However, the mitochondrial rRNAs can be transcribed either as a part of this polycistronic transcript or independently from the HSP1 promoter (19). The differential regulation of mRNAs and rRNAs in LSFC cell lines could either be due to promoter-specific transcriptional activation by LRPPRC, or LRPPRC may act post-trancriptionally on mRNAs. To differentiate between these scenarios, we designed a qRT-PCR assay that specifically probes the polycistronic transcripts (Fig. 3C). We observed no reduction in polycistronic transcript level in the LRPPRC knockdown cells (Fig. 3D), suggesting that LRPPRC acts downstream of the polycistronic transcript to maintain steady-state mRNA levels. Finally, we verified that the reduction in mitochondrial mRNA results in impaired respiratory complex assembly, using a Western blot assay for a labile subunit of each respiratory complex (14). We observed depletion of complex I, III, and IV subunits in the two cell lines with most potent LRPPRC knockdown (supplemental Fig. 4).
FIGURE 3.
LRPPRC is required for the maintenance of mitochondrial mRNAs. A, heat map showing abundance of mitochondrial transcripts in LSFC cell lines as identified by microarray. Two replicates of each knockdown are shown adjacent to each other. B, mtDNA copy number as quantified by qRT-PCR for two most potent knockdowns and the pLKO.1-transduced control (ctrl) cell line. Values are reported as fold change over cells infected with pLKO.1 (n = 3; error bars represent S.D. from mean). C, schematic representation of qRT-PCR Taqman assay locations for mitochondrial transcripts. rRNA are indicated by blue, mRNA by gray, tRNA by red, and assay location by green. * denotes location of assay for polycistronic transcript. D, mitochondrial mRNA, rRNA, and polycistronic RNA transcript were quantified for each of the two most potent knockdowns by qRT-PCR. Values are reported as fold change over control cells; β-actin was used as endogenous control (n = 3; error bars represent S.D.).
DISCUSSION
It has been more than 6 years since LRPPRC was identified as a causative gene for LSFC, yet its precise function has remained unclear. Based on the presence of a pentatricopeptide repeat motif, sequence similarity to the yeast PET309 protein, and the mitochondrial localization of LRPPRC, Mootha et al. (5) postulated that this protein may play a role in mRNA processing. Indeed, a subsequent study on LSFC patient fibroblasts showed a selective decrease in mtDNA-encoded CO1 and CO3 transcripts (9). This finding is consistent with the COX enzyme deficiency in LSFC patients. However, our study shows that LRPPRC is required for the expression of all of the mtDNA-encoded mRNAs and is not specific to COX subunits. This discordance may be due to differences in LRPPRC protein levels, that is, a more severe depletion in our knockdown cellular models compared with a milder decrease in LSFC patients with a point mutation in LRPPRC. Importantly, we have shown that LRPPRC depletion does not alter transcript levels of rRNA (Fig. 3D), suggesting a specific role for LRPPRC in the processing of mitochondrial mRNAs. At present, the exact molecular function of LRPPRC is not known. It is likely that other proteins may interact with LRPPRC to co-stabilize mRNA. In this regard, it is noteworthy that the loss of SLIRP, a RNA-binding protein, results in a remarkably similar pattern of mitochondrial mRNA depletion (14).
Mitochondrial disorders are frequently characterized by a variety of clinical features that are not readily linked to the biochemical defect within the respiratory chain. Our engineered LSFC cell lines offer a powerful tool to discover cellular responses that may help us understand pathogenesis. This approach is effective because LRPPRC protein appears to be limiting in cells: a decrease in its level strongly correlates with mitochondrial mRNA expression (Fig. 3A) and cellular energetics (Fig. 1D). The allelic series of LRPPRC knockdown cell lines offers multiple advantages: it cancels the off-target effects of individual shRNAs; it provides an isogenic background that eliminates genotype specific effects; stable silencing allows us to understand the long term adaptations of the cell to the loss of gene function; and it recapitulates the phenotypes observed in mitochondrial disease where mitochondrial respiration is defective. The transcriptional responses to a graded loss of LRPPRC and reduced mitochondrial function may be either dose-dependent or exhibit a threshold behavior. By analyzing gene expression data based on linear (Pearson) correlation with the LRPPRC expression level, the current analysis is focused on dose-dependent responses; however, our data could be reanalyzed with different similarity metrics to identify genes exhibiting nonlinear responses as well. Of the pathways showing a dose-dependent response, we were particularly interested in gene sets that were negatively correlated with LRPPRC expression because these may represent activation of an adaptive cellular response to the loss of mitochondrial energy-generating capacity. We found three such anticorrelated, statistically significant gene sets (Fig. 2B), representing hexose sugar metabolism and PG biosynthesis. The hexose sugar metabolism gene set included key glycolytic genes such as HK1, HK2, and HK3. Previous studies have shown increased expression of genes involved in glycolysis, including HK1, HK2, LDHB, and PFK in mtDNA mutant cells (20, 21). The up-regulation of these genes may in part facilitate increased ATP production via glycolysis, a necessary adaptation to the loss of mitochondrial respiratory capacity observed in mitochondrial disorders, including LSFC. Not surprisingly, LSFC cells displayed greater sensitivity to a glycolytic inhibitor (supplemental Fig. 2B). Notably, the gene set comprising hexose sugar metabolism includes genes required for the synthesis of GDP-mannose and GDP-fucose, which provide a carbohydrate backbone for glycoprotein synthesis. The increased expression of genes encoding this pathway may reflect alterations in glycoprotein biosynthesis secondary to mitochondrial dysfunction.
The concerted up-regulation of genes involved in the PG biosynthetic pathway was surprising and has not been reported previously in mitochondrial respiratory chain disease. However, recent reports link PG biosynthesis to mitochondrial dysfunction. First, Cillero-Pastor et al. (22) showed that treatment of primary human chondrocytes with the respiratory inhibitors oligomycin or antimycin stimulated PTGS2 expression and PGE2 production. Second, Tomura et al. (23) have shown that acidification of culture media results in an increase in PTGS2 expression and a corresponding increase in PGE2 synthesis. The up-regulation of the PG pathway is also consistent with the retrograde signaling hypothesis, whereby mitochondrial dysfunction results in increased cytosolic Ca2+, which stimulates calcineurin and activates NF-κB (24), a key transcriptional activator of PG biosynthetic genes. The up-regulation of PG biosynthetic genes could be adaptive because PGE2 inhibits apoptosis, up-regulates a number of prosurvival pathways, including phosphatidylinositol-3-OH-kinase/protein kinase B and extracellular signal-regulated kinase, and stimulates VEGF expression through activation of hypoxia-inducible factor 1α (25). Thus, PGE2 biosynthesis could play a cytoprotective role in respiratory compromised cells.
In previous studies, Cooper et al. (10, 11) have shown that LRPPRC interacts with PGC-1α to stimulate the expression of a subset of nuclear-encoded mitochondrial genes in mouse primary hepatocytes and brown fat cells. All shLRPPRC hairpins used in this study are predicted to silence mitochondria-targeted LRPPRC as well as LRPPRC targeted to the nucleus/cytosol. However, our GSEA analysis of LSFC cell lines did not detect any effect on expression of the nuclear-encoded mitochondrial genes that are targets of PGC-1α, including the OXPHOS genes (Fig. 2C). This discrepancy could be due to differences between cell types. Although our data strongly favor a role for LRPPRC in regulating mitochondrial mRNAs, an additional nucleus-specific function cannot be ruled out.
In summary, we report the construction of stable cellular models of LSFC disease in an isogenic background and the discovery of a specific function of LRPPRC in mt-mRNA homeostasis. In addition, we identify nuclear-encoded pathways that could shed light on mitochondrial disease pathogenesis. Our approach overcomes a number of limitations of previous genomic profiling studies conducted on mitochondrial disease cells, where results were confounded by genetic heterogeneity and tissue diversity of samples (26). Our engineered and validated cellular models of LSFC could serve as a valuable resource for understanding mitochondrial disease pathogenesis as well as for drug screening.
Supplementary Material
Acknowledgments
We thank Eric Shoubridge for providing MCH58 cell lines, Olga Goldberger for technical assistance, Oded Shaham for assistance in designing qRT-PCR assays, Serena Silver and Jennifer Grenier for assistance with RNA interference, and Joshua Baughman and Scott Vafai for valuable discussions and comments on this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01DK081457 (to V. K. M.). This work was also supported by grants from the United Mitochondrial Disease Foundation (to V. M. G.), the Broad Institute Scientific Planning and Allocation of Resources Committee, and the American Diabetes Association.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and additional references and Figs. 1–4.
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE20847).
- LSFC
- Leigh syndrome, French-Canadian type
- COX
- cytochrome c oxidase
- LRPPRC
- leucine-rich pentatricopeptide repeat motif-containing protein
- mtDNA
- mitochondrial DNA
- RNAi
- RNA interference
- shRNA
- short hairpin RNA
- PGC-1α
- peroxisome proliferator-activated receptor γ coactivator 1α
- GSEA
- gene set enrichment analysis
- PG
- prostaglandin
- qRT-PCR
- quantitative real-time PCR
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2- (hydroxymethyl)propane-1,3-diol
- OCR
- oxygen consumption rate
- ECAR
- extracellular acidification rate
- ES
- enrichment score.
REFERENCES
- 1.McCarthy M. I., Abecasis G. R., Cardon L. R., Goldstein D. B., Little J., Ioannidis J. P., Hirschhorn J. N. (2008) Nat. Rev. Genet. 9, 356–369 [DOI] [PubMed] [Google Scholar]
- 2.Giallourakis C., Henson C., Reich M., Xie X., Mootha V. K. (2005) Annu. Rev. Genomics Hum. Genet. 6, 381–406 [DOI] [PubMed] [Google Scholar]
- 3.Ng S. B., Buckingham K. J., Lee C., Bigham A. W., Tabor H. K., Dent K. M., Huff C. D., Shannon P. T., Jabs E. W., Nickerson D. A., Shendure J., Bamshad M. J. (2010) Nat. Genet. 42, 30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee N., Daly M. J., Delmonte T., Lander E. S., Xu F., Hudson T. J., Mitchell G. A., Morin C. C., Robinson B. H., Rioux J. D. (2001) Am. J. Hum. Genet. 68, 397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mootha V. K., Lepage P., Miller K., Bunkenborg J., Reich M., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F., Mitchell G. A., Morin C., Mann M., Hudson T. J., Robinson B., Rioux J. D., Lander E. S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 605–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mili S., Piñol-Roma S. (2003) Mol. Cell. Biol. 23, 4972–4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lurin C., Andrés C., Aubourg S., Bellaoui M., Bitton F., Bruyère C., Caboche M., Debast C., Gualberto J., Hoffmann B., Lecharny A., Le Ret M., Martin-Magniette M. L., Mireau H., Peeters N., Renou J. P., Szurek B., Taconnat L., Small I. (2004) Plant Cell 16, 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitz-Linneweber C., Small I. (2008) Trends Plant Sci. 13, 663–670 [DOI] [PubMed] [Google Scholar]
- 9.Xu F., Morin C., Mitchell G., Ackerley C., Robinson B. H. (2004) Biochem. J. 382, 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper M. P., Qu L., Rohas L. M., Lin J., Yang W., Erdjument-Bromage H., Tempst P., Spiegelman B. M. (2006) Genes Dev. 20, 2996–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper M. P., Uldry M., Kajimura S., Arany Z., Spiegelman B. M. (2008) J. Biol. Chem. 283, 31960–31967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C. (2003) Nat. Genet. 34, 267–273 [DOI] [PubMed] [Google Scholar]
- 13.Moffat J., Grueneberg D. A., Yang X., Kim S. Y., Kloepfer A. M., Hinkle G., Piqani B., Eisenhaure T. M., Luo B., Grenier J. K., Carpenter A. E., Foo S. Y., Stewart S. A., Stockwell B. R., Hacohen N., Hahn W. C., Lander E. S., Sabatini D. M., Root D. E. (2006) Cell 124, 1283–1298 [DOI] [PubMed] [Google Scholar]
- 14.Baughman J. M., Nilsson R., Gohil V. M., Arlow D. H., Gauhar Z., Mootha V. K. (2009) PLoS Genet. 5, e1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu M., Neilson A., Swift A. L., Moran R., Tamagnine J., Parslow D., Armistead S., Lemire K., Orrell J., Teich J., Chomicz S., Ferrick D. A. (2007) Am. J. Physiol. Cell Physiol. 292, C125–C136 [DOI] [PubMed] [Google Scholar]
- 16.Ma H., Sorokin A., Mazein A., Selkov A., Selkov E., Demin O., Goryanin I. (2007) Mol. Syst. Biol. 3, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagliarini D. J., Calvo S. E., Chang B., Sheth S. A., Vafai S. B., Ong S. E., Walford G. A., Sugiana C., Boneh A., Chen W. K., Hill D. E., Vidal M., Evans J. G., Thorburn D. R., Carr S. A., Mootha V. K. (2008) Cell 134, 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merante F., Petrova-Benedict R., MacKay N., Mitchell G., Lambert M., Morin C., De Braekeleer M., Laframboise R., Gagné R., Robinson B. H. (1993) Am. J. Hum. Genet. 53, 481–487 [PMC free article] [PubMed] [Google Scholar]
- 19.Bonawitz N. D., Clayton D. A., Shadel G. S. (2006) Mol. Cell 24, 813–825 [DOI] [PubMed] [Google Scholar]
- 20.Heddi A., Stepien G., Benke P. J., Wallace D. C. (1999) J. Biol. Chem. 274, 22968–22976 [DOI] [PubMed] [Google Scholar]
- 21.Behan A., Doyle S., Farrell M. (2005) Mitochondrion 5, 173–193 [DOI] [PubMed] [Google Scholar]
- 22.Cillero-Pastor B., Caramés B., Lires-Deán M., Vaamonde-Garcia C., Blanco F. J., López-Armada M. J. (2008) Arthritis Rheum. 58, 2409–2419 [DOI] [PubMed] [Google Scholar]
- 23.Tomura H., Wang J. Q., Liu J. P., Komachi M., Damirin A., Mogi C., Tobo M., Nochi H., Tamoto K., Im D. S., Sato K., Okajima F. (2008) J. Bone Miner. Res. 23, 1129–1139 [DOI] [PubMed] [Google Scholar]
- 24.Butow R. A., Avadhani N. G. (2004) Mol. Cell 14, 1–15 [DOI] [PubMed] [Google Scholar]
- 25.Greenhough A., Smartt H. J., Moore A. E., Roberts H. R., Williams A. C., Paraskeva C., Kaidi A. (2009) Carcinogenesis 30, 377–386 [DOI] [PubMed] [Google Scholar]
- 26.Reinecke F., Smeitink J. A., van der Westhuizen F. H. (2009) Biochim. Biophys. Acta 1792, 1113–1121 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.