Abstract
The effects of UV irradiation on herpes simplex virus type 1 (HSV-1) gene expression and DNA replication were examined in cell lines containing mutations inactivating the XPA gene product required for nucleotide-excision repair, the DNA polymerase η responsible for translesion synthesis, or the Cockayne syndrome A and B (CSA and CSB) gene products required for transcription-coupled nucleotide excision repair. In the absence of XPA and CSA and CSB gene products, virus replication was reduced 106-, 400-, and 100-fold, respectively. In DNA polymerase η mutant cells HSV-1 plaque efficiency was reduced 104-fold. Furthermore, DNA polymerase η was strictly required for virus replication at low multiplicities of infection but dispensable at high multiplicities of infection. Knock down of Rad 51, Rad 52, and Rad 54 levels by RNA interference reduced replication of UV-irradiated HSV-1 150-, 100-, and 50-fold, respectively. We find that transcription-coupled repair efficiently supports expression of immediate early and early genes from UV-irradiated HSV-1 DNA. In contrast, the progression of the replication fork appears to be impaired, causing a severe reduction of late gene expression. Since the HSV-1 replisome does not make use of proliferating cell nuclear antigen, we attribute the replication defect to an inability to perform proliferating cell nuclear antigen-dependent translesion synthesis by polymerase switching at the fork. Instead, DNA polymerase η may act during postreplication gap filling. Homologous recombination, finally, might restore the physical and genetic integrity of the virus chromosome.
Keywords: DNA Recombination, DNA Repair, DNA Replication, DNA Nucleotide-excision Repair, Herpesvirus, Viral Replication
Introduction
Herpes simplex virus has a linear double-stranded genome, which, upon entry into the eukaryotic nucleus, circularizes and becomes replicated by a replisome consisting of six virus-encoded proteins (1–4). The herpes simplex virus replisome is composed of a DNA polymerase made up from the UL30 and UL42 gene products, a helicase-primase complex encoded by the UL5, UL8, and UL52 genes and a single-stranded DNA-binding protein ICP8, which is a product of the UL29 gene (4, 5). The replisome is loaded on the origins of replication oriS and oriL by a sequence-specific superfamily II DNA helicase termed OBP or UL9 protein (6–9), and it is capable of processive leading strand DNA synthesis coupled to discontinuous synthesis of lagging strand intermediates (5). Processive polymerization is dependent on the UL42 protein, which is a monomer folded as the proliferating cell nuclear antigen (PCNA)2 protomer but with no sequence similarity to the cellular processivity factor (10). The UL30 subunit is a proofreading family B DNA polymerase (11). It appears that the overall replication fidelity is high, and existing genetic variability may be created by recombination between a limited number of strains rather than random replication errors (12–14). The contribution of the DNA polymerase to replication fidelity has been thoroughly examined, but cellular mechanisms contributing to the genetic stability of herpesviruses have, with few exceptions, not been extensively looked at (15–18). It has been noted that several cellular repair proteins co-localize with viral replication proteins in a limited number of replication foci (19–21). In cells, genomic stability is maintained by an intricate interplay among the replication machinery, repair proteins, and factors controlling the cell cycle (22, 23). Viruses are known to interact and interfere with these mechanisms to promote their own replication (24). Such interactions may depend on specific molecular interactions with the viral replication machinery, and the outcome may thus vary considerably. Lytic replication of herpes simplex virus DNA has several distinguishing features. First, the viral replisome is structurally different from its cellular counterpart. Second, herpesvirus replication is independent of cell cycle. Third, the herpes simplex virus chromosome has no regular chromatin structure during replication (25). Together, these considerations might indicate that not all repair pathways are readily available to the virus and, conversely, that not all repair proteins are required for repair and recombination to be carried out. Furthermore, because the virus genome is being continuously rereplicated, there will be little time for repair between successive rounds of replication. It is, therefore, possible that the virus may be more vulnerable to certain types of DNA damage and that rapid successive rounds of replication may increase the probability for replication forks to encounter unrepaired lesions and generate an error catastrophe. To counteract such consequences, the virus must adapt to existing cellular mechanisms for repair and recombination to ensure survival.
As a first step toward clarifying the role of cellular repair systems during herpesvirus replication, we have looked at repair of UV-induced lesions in herpes simplex virus type 1 (HSV-1) DNA and how they affect the progression of the viral replication cycle. The herpes simplex virus genome does not encode enzymes suitable for handling UV-damaged DNA. In the eukaryotic cell, however, several mechanisms contribute to repair of UV-damaged DNA (26). Nucleotide excision repair in both the global genome and the transcription-coupled repair modes plays the predominant role as witnessed by the diseases xeroderma pigmentosum and Cockayne syndrome. In addition, translesion synthesis by DNA polymerase η has been shown to be significant (27–29). In the latter instance, DNA polymerase η may either act at the replication fork as part of a tool belt provided by the PCNA trimer, or it may fulfill its role during gap-filling postreplicative repair (30, 31). Finally, unrepaired single-stranded breaks, occurring as a consequence of disrupted replication or incomplete repair, may be converted to double-stranded breaks that will require homologous recombination for repair (32).
Here, we have analyzed the contribution of these repair pathways to replication of UV-damaged HSV-1 DNA. We find that both global genome and transcription-coupled nucleotide excision repair has the essential role. Interestingly, DNA polymerase η is strictly required for replication of UV-damaged HSV-1 at low multiplicities of infection (m.o.i.). On the other hand, at high m.o.i., homologous recombination involving Rad51, Rad52, and Rad54 proteins plays a significant role. The latter observation suggests that viable genomes can be reconstructed from defective genomes. These studies provide the first functional insights into cellular mechanisms contributing to genomic stability of herpesviruses and demonstrate that HSV-1 may serve as a readily accessible model system for mechanistic studies of DNA repair and recombination.
EXPERIMENTAL PROCEDURES
Cells, Viruses, and Plaque Assays
MRC5, XP12 (XPA), XP30 (XPV), and XP30eGFPη, which is stably transfected with a plasmid encoding the wild type DNA polymerase η, were kindly provided by Alan R. Lehmann (University of Sussex, UK) and propagated in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) (33). 600 μg/ml G418 was added to XP30eGFPη growth medium for selective pressure. Cockayne syndrome A and B mutant cell lines, GM16094 and GM16095, were from Coriell Cell Repositories and propagated as above. BHK-21 cells were cultured in Glasgow minimum essential medium (Invitrogen) supplemented with 10% fetal bovine serum. HSV-1 (Glasgow strain 17 syn+), UV-irradiated HSV-1 (UV/HSV-1), and calicheamicin-treated HSV-1 (calicheamicin/HSV-1) titers were determined by plaque assay on the indicated cell line, as previously described (2).
Viral DNA Damage
100 μl of HSV-1 virus on a 35-mm dish was mock-irradiated or irradiated (UV/HSV-1) with 1600 J/m2 UV for 6 s at 254 nm using an UV Cross-linker CL-1000 (UVP, Upland, CA). For production of calicheamicin/HSV-1, HSV-1 viruses were mock-treated or treated with 0.1 and 1 μm calicheamicin (kindly provided by Ola Hammarsten, University of Gothenburg, Sweden) for 1 h on ice, followed by overnight incubation with 1 mm dithiothreitol on ice.
Growth Curve Analysis
MRC5 and XP30 monolayers were infected with HSV-1 and UV/HSV-1 at a m.o.i. of 5 or 0.1 plaque-forming units (pfu)/cell at 37 °C. After 1 h, the inocula were removed, and the cells were washed extensively with Dulbecco's modified Eagle's medium, followed by incubation with Dulbecco's modified Eagle's medium containing 2% fetal bovine serum. High and low m.o.i. supernatants were harvested at the indicated time points, clarified by centrifugation, and stored at −80 °C. Titers were determined by plaque assay on BHK-21 cells.
RNA Interference Assay
Validated siRNAs Hs_RAD51_7 and Hs_RAD54_7, and FlexiTube siRNAs Hs_RAD52_5, Hs_RAD52_6, Hs_RAD52_7, and Hs_RAD52_8 were purchased from Qiagen. MRC5 monolayers at 30% confluence on 24-well plates were transfected with 150 pmol of Rad51, Rad52 (a pool containing 37.5 pmol of each flexitube siRNA), or Rad54 siRNAs by using Oligofectamine as described before (2). At 72 h after transfection, the cells were infected with HSV-1 or UV/HSV-1 at an m.o.i. of 5 pfu/cell for 24 h. Titers were determined by plaque assay on BHK-21 cells.
Protein Analysis
At 72 h after transfection, MRC5 monolayers were treated with SDS lysis buffer (0.5% SDS, 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EDTA, and Complete protease inhibitors mix (Roche Applied Science)) and immunoprecipitated, as described previously (2), using mouse monoclonal antibodies to human Rad51 (ab213) or Rad54 (ab11055) or rabbit polyclonal antibodies to human Rad52 (ab18264). The immunoprecipitates as well as β-actin were visualized by Western blotting using the antibodies above and an antibody against a synthetically produced β-cytoplasmic actin N-terminal peptide (ab6276). All antibodies were purchased from Abcam. For detection of ICP8 and glycoprotein C, the indicated cell monolayers were infected with HSV-1 or UV/HSV-1 at an m.o.i. of 5 pfu/cell for 12 h followed by Laemmli lysis buffer treatment and Western blot analysis, using rabbit polyclonal antibodies to HSV-1 ICP8 and mouse monoclonal antibodies to HSV-1 glycoprotein C (kindly provided by Tomas Bergström, University of Gothenburg, Sweden). All protein bands were detected with the Super Signal West Pico Chemiluminescence kit (Pierce).
Immuno-Dot Blot Analysis
MRC5, XP12, and XP30 cells were infected with HSV-1 or UV/HSV-1 at an m.o.i. of 5 pfu/cell for 1 h, 3 h, or 6 h, and, at each time point, total DNA was isolated by using the QIAamp DNA blood mini kit (Qiagen). All samples were denatured by boiling for 10 min, followed by incubation with an equal volume of 1 m NaOH at room temperature for 20 min. The DNA samples were next dot-blotted onto a Hybond-N+ membrane (GE Healthcare) by using the Bio-Dot apparatus (Bio-Rad) and according to the manufacturer's instructions. The membrane was then blocked in phosphate-buffered saline and 0.1% Tween 20 (PBST) containing 5% milk at room temperature for 1 h, followed by incubation with a primary anti-thymine dimer monoclonal antibody (1:5000 in PBST; MC-062, Kamiya Biomedical Company), at room temperature for 2 h. After washing, the membrane was incubated with a secondary goat anti-mouse IgG conjugated to horseradish peroxidase (1:10,000 in PBST) at room temperature for 1 h. Signals were detected as above.
RESULTS
Nucleotide Excision Repair, Translesion Synthesis, and Homologous Recombination Contribute to Repair of UV-damaged HSV-1 DNA
To monitor repair of UV-damaged HSV-1, we used unirradiated cell lines mutated in essential repair proteins. In those instances where suitable mutant cell lines were not available, we have treated cells with siRNA to knock down the levels of Rad51, Rad52, and Rad54 proteins. Our standardized conditions for UV treatment of virus caused an ∼10-fold reduction in plaque number on wild type MRC5 cells.
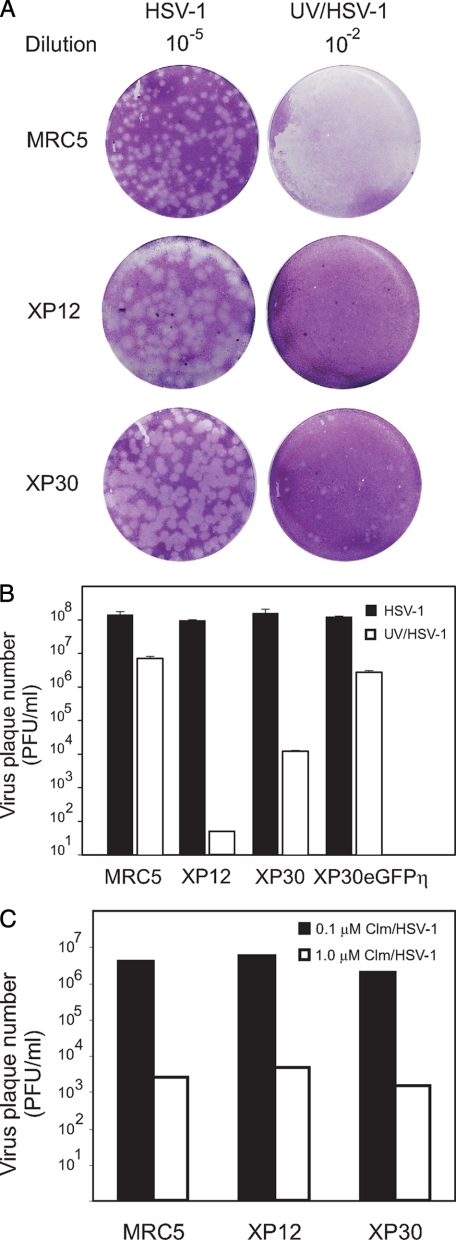
To study effects of UV irradiation on virus replication in mutant cells, we first serially diluted and plated UV-irradiated HSV-1 on normal and mutant human cell lines (Fig. 1). Nonirradiated virus replicated equally well on wild type MRC5 cells, XP12, mutated in the XPA protein, an essential component of nucleotide excision repair, and XP30 cells, mutated in DNA polymerase η (Fig. 1A). The variation in plaque size for unirradiated virus, seen in Fig. 1A, reflects differences in cell morphology rather than kinetics of virus replication. In stark contrast, we found that the plaque efficiency of UV-irradiated virus was reduced 104-fold on XP30 cells and at least 106-fold on XP12 cells (Fig. 1B). UV irradiation also caused reduced plaque size on XP30 cells. The replication-defect seen in XP30 cells could be restored by stable expression of eGFP-DNA polymerase η in XP30 cells (Fig. 1B) (33). We also examined virus treated with calicheamicin, a drug capable of inducing a high proportion of double-stranded breaks in DNA (34) (Fig. 1C). MRC5, XP12, and XP30 cells all supported replication to the same extent, thus confirming that the replication defects in the mutant cell lines were specifically due to the exposure of virus to UV irradiation.
FIGURE 1.
Replication of UV-irradiated HSV-1 is impaired in cells deficient in nucleotide excision repair and DNA polymerase η. A, nonirradiated HSV-1 and UV-irradiated HSV-1 (UV/HSV-1) diluted 105- and 102-fold, respectively, and plated on wild type MRC5, XP12, deficient in the nucleotide excision repair XPA gene product, and XP30 cells, lacking DNA polymerase η. B, HSV-1 and UV/HSV-1 plaque efficiency on MRC5, XP12, XP30, and XP30 cells stably expressing DNA polymerase η as a GFP fusion protein, XP30eGFPη. C, plaque efficiency of HSV-1 and HSV-1 treated with calicheamicin (Clm/HSV-1) on MRC5, XP12, and XP30 cells. The numbers are the averages derived from three independent experiments.
To study the replication defects further, we next examined the production of infectious virus particles at low and high m.o.i. on XP30 cells. This analysis was not performed on XP12 cells because UV-irradiated virus did not grow on this cell line. We found that the yield of virus on XP30 cells after low m.o.i. infection was reduced 105-fold by UV irradiation. Possibly, lesions persisting in single-stranded regions of DNA remain unrepaired and prevent further synthesis of viable progeny. It is also likely that, in the absence of DNA polymerase η, the surviving viruses become heavily mutagenized, resulting in reduced replication efficiency. This indicates that DNA polymerase η is essential for replication of UV-damaged HSV-1 at low m.o.i. (Fig. 2A). Remarkably, the replication defect was almost abolished at high m.o.i., suggesting that a mechanism requiring multiple independent viral genomes was able to rescue viable progeny (Fig. 2B).
FIGURE 2.
Growth curve analyses of HSV-1 and UV-irradiated HSV-1 on MRC5 and XP30 cells. A, cells were infected at a m.o.i. of 0.1 pfu/cell. B, cells were infected at a m.o.i. of 5 pfu/cell. Titers were determined by plaque assay on BHK-21 cells as described under “Experimental Procedures.” The numbers are the averages derived from three independent experiments.
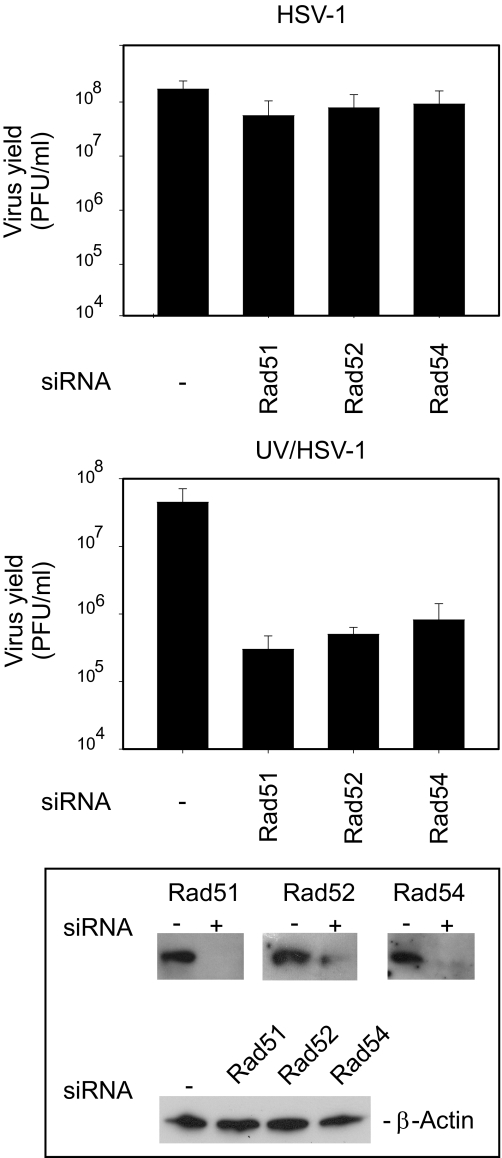
We, therefore, surmised that the most plausible mechanism capable of restoring viable genomes at high m.o.i. would be homologous recombination. Because the enzymes required in this instance are likely to be of cellular origin, we made use of siRNA against mRNA encoding Rad51, Rad52, and Rad54 proteins. We found that siRNA had little if any effect on replication of nonirradiated HSV-1 in MRC5 cells (Fig. 3, top panel). However, when UV-irradiated virus was used, we observed 150-, 100-, and 50-fold reductions on virus yields from cells treated with siRNA against Rad51, Rad52, and Rad 54, respectively (Fig. 3, middle panel). We also verified an efficient knockdown of Rad51, Rad52, and Rad54 protein levels in siRNA-treated cells prior to virus infection (Fig. 3, bottom panel). The exact mechanism by which these proteins contribute to replication of UV-irradiated DNA is unknown, but it is plausible that viable virus genomes may be reconstructed from fragments produced during repeated rounds of DNA replication.
FIGURE 3.
Effects of RNA interference against Rad51, Rad52 and Rad54 on HSV-1 and UV/HSV-1 replication in MRC5 cells. Cells were treated with siRNA for 72 h followed by infection with HSV-1 (top panel) and UV/HSV-1 (middle panel), for 24 h. Titers were determined by plaque assay on BHK-21 cells as described under “Experimental Procedures.” The numbers are the averages derived from three independent experiments. The bottom panel represents a Western blot analysis of Rad51, Rad52, and Rad54 following siRNA treatment.
Kinetics of the Removal of Thymine Dimers from UV-irradiated HSV-1 DNA during the Viral Replication Cycle
To evaluate the physiological effects of UV-induced lesion in DNA on the viral life cycle, we have first monitored the kinetics of DNA repair. We made use of antibodies directed toward thymine dimers. The antibodies do not bind nonirradiated DNA immobilized on membranes, but readily detect lesions in UV-irradiated DNA (Fig. 4). DNA from infected cells was isolated and subjected to a dot blot analysis. We found that, in all cells, high levels of UV-damaged DNA was detected already at 1 h after infection, reflecting rapid uptake and transport of viral DNA to the nucleus. We also found that, in MRC5 cells, DNA repair was activated very early, leading to a 60% decrease of thymine dimers at 3 h after infection (Fig. 4). In XP12 cells, devoid of nucleotide excision repair, damaged DNA persisted for at least 6 h (Fig. 4). Our results suggest that nucleotide excision repair has almost immediate access to viral DNA once it has been delivered into the nucleus. Surprisingly, lesions remained to a large extent unrepaired also in XP30 cells (Fig. 4). This observation suggests that DNA polymerase η acts very early, already at 3 h after infection, during the replication cycle. It also indicates that replication of damaged DNA, in the absence of DNA polymerase η, generates intermediates that are inaccessible for nucleotide excision repair.
FIGURE 4.
Repair of thymine dimers in UV-irradiated HSV-1 DNA. MRC5, XP30, and XP12 cells were infected with HSV-1 and UV/HSV-1 at a m.o.i. of 5 pfu/cell for the indicated times. Upper panel, total DNA was isolated and analyzed by immuno-dot blot using an anti-thymine dimer antibody (see “Experimental Procedures”). Lower panel, relative amounts of thymine dimers determined after quantification of the chemiluminescent signals on exposed films using a Fuji FLA-7000 bioimaging analyzer. Time p.i., time after infection.
Transcription-coupled Nucleotide Excision Repair Is Required for Expression of Early Genes, and Translesion Synthesis by DNA Polymerase η Is Needed for Replication-dependent Expression of Late Genes
We next wanted to investigate whether or not the reduction in virus replication could be explained by inhibition of gene expression or DNA replication. The infectious cycle of HSV-1 is characterized by tight coordination of controlled gene expression and DNA replication (1, 35). The powerful transcription activator VP16 activates a limited number of immediate early α genes. ICP4, one of the proteins encoded by these genes, is strictly required for transcription of the early β genes which, in turn, encode the enzymes required for DNA synthesis. Finally, true late γ genes are only transcribed once DNA replication has been initiated and allowed to proceed unperturbed. Thus, these properties of the HSV-1 replication cycle allow us to distinguish effects of UV irradiation on gene expression from effects on DNA replication by looking at expression, for example, of the early gene encoding ICP8 and the late gene encoding glycoprotein C.
We first found that at high m.o.i., in wild type MRC5 cells, early gene expression was equally efficient at 12 h after infection with either irradiated or nonirradiated virus (Fig. 5A). In contrast, replication-dependent expression of glycoprotein C was severely impaired in cells infected with irradiated virus, which correlates well with the low production of virus particles observed at this time point (Fig. 2B). In XP12 cells, lacking nucleotide excision repair, neither early nor late gene expression occurred at 12 h after infection with irradiated virus (Fig. 5A).
FIGURE 5.
Effects of UV irradiation on HSV-1 gene expression. Cells were infected with HSV-1 and UV/HSV-1 at a m.o.i. of 5 pfu/cell for 12 h followed by Western blot analysis of the early gene product ICP8 and the late gene product glycoprotein C, in the precursor pgC and the mature gC forms (38). A, MRC5, XP12, and XP30 cells were used to examine effects of nucleotide excision repair and translesion synthesis by DNA polymerase η on gene expression. B, CSA and CSB cells were used to analyze effects of transcription-coupled nucleotide excision repair on gene expression.
The role of transcription-coupled nucleotide excision repair was then examined by looking at expression of ICP8 in cells containing mutations responsible for Cockayne syndromes A and B (36). The CSA protein is associated with a cullin4a containing E3 ubiquitin ligase, and the CSB protein is a member of the SWI2/SNF2 family of ATP-dependent chromatin-remodeling factors (37). We first looked at the plaque efficiency of irradiated and nonirradiated HSV-1 on these cell lines. The results showed that UV-irradiated virus produced about 400- and 100-fold fewer plaques on CSA and CSB cells, respectively, compared with nonirradiated HSV-1 (results not shown). Analysis of ICP8 expression revealed that although it was unaffected by UV irradiation in wild type MRC5 cells, it was significantly reduced in CSA and CSB cells (Fig. 5B). This suggests that transcription-coupled nucleotide excision repair promotes expression of immediate early and early genes very efficiently during replication of UV-irradiated HSV-1. The results also indirectly infer that the major reason for reduced production of virus particles is reduced DNA replication in wild type cells.
To examine the significance of DNA polymerase η on replication of UV-irradiated HSV-1 at high m.o.i., we looked at replication-dependent expression of the late gene encoding glycoprotein C in XP30 cells lacking functional DNA polymerase η. We found that, in contrast to robust expression of the early gene product ICP8, the expression of both the precursor protein, pgC, and the mature glycosylated processed form of glycoprotein C, gC, was abolished (38) (Fig. 5A). Because glycoprotein C expression can be detected in wild type MRC5 cells infected with UV-irradiated virus, our results suggest that DNA polymerase η contributes to DNA replication and subsequent late gene expression. It is worth noting that, although translesion synthesis by DNA polymerase η is essential for replication of UV-irradiated HSV-1 DNA at low m.o.i., it does not seem to be a very efficient process, as reflected by the low levels of glycoprotein C expression in wild type cells at high m.o.i.. As further discussed below, replication of UV-irradiated virus DNA may generate aberrant replication intermediates that might be difficult to handle.
DISCUSSION
The studies presented in this report demonstrate an intimate functional relationship between cellular proteins involved in repair and recombination of viral DNA exposed to UV-irradiation and the herpes simplex virus replication cycle. In brief, our results suggest that transcription-coupled nucleotide excision repair promotes expression of immediate early and early genes (Fig. 6A). Once early replication proteins are produced, the replication cycle progresses into the DNA synthesis phase. Our studies indicate that DNA polymerase η must be active on replicating virus genomes already at 3 h after infection and that it is essential for copying UV-damaged template DNA. In wild type cells infected with irradiated virus, DNA replication appears inefficient inasmuch as late gene expression is impaired. Probably, the progression of replisome is impaired once it hits a lesion. In eukaryotic cells, translesion synthesis coupled to polymerase switching, and template switching may assist replisome progression (39). Both mechanisms rely on Rad6-Rad18-dependent monoubiquitination of PCNA. However, the herpesvirus replisome is fully functional in the absence of PCNA (5). Instead, the viral encoded UL42 protein, which has a three-dimensional structure resembling a PCNA protomer but no sequence similarity, serves a similar role by increasing the processivity of the HSV-1 DNA polymerase (4, 40). Furthermore, HSV-1 UL42 acts as a monomer as opposed to PCNA, which is a trimer (10). It is thus possible that the proper interactions between DNA polymerase η and the viral replication fork cannot be established. However, DNA polymerase η may still be active in postreplication gap-filling repair (Fig. 6B). These considerations may help to explain why the products of DNA replication made in the complete absence of DNA polymerase η appear to be inaccessible for nucleotide excision repair. In this instance, one might imagine that once a viral replisome hits a lesion it falls apart; DNA polymerase is left at the site of the lesion, and the UL5/8/52 helicase is allowed to proceed, generating extensive stretches of single-stranded DNA. The observations that the UL5/8/52 helicase-primase assisted by the single-stranded DNA-binding protein ICP8 can efficiently bypass a 1-2-intrastrand d(GpG) cross-link but the UL30 DNA polymerase would stall at the same lesion lend credibility to this suggestion (41, 42). In wild type cells, however, postreplication gap filling may provide nucleotide excision repair with the appropriate substrates, allowing the replication cycle to continue (Fig. 6B). With these considerations in mind, we would like to suggest that the herpesvirus replisome and the cellular replication machinery may be affected in different ways by DNA damage.
FIGURE 6.
Model for repair of UV lesions during HSV-1 replication. A, transcription-coupled nucleotide excision repair (TCR) promotes expression of immediate early and early genes prior to DNA replication. Postreplication gap filling by DNA polymerase η is required for copying UV-damaged template DNA before late gene expression can start. Homologous recombination (HR) may act to repair double-stranded breaks resulting from incomplete replication. Global genome repair is likely to be active throughout the entire infectious cycle. B, postreplication gap filling by DNA polymerase η may create substrates for nucleotide excision repair (NER). C, simplified model illustrates how multiple rounds of DNA synthesis, initiated before DNA repair aided by DNA polymerase η has been completed, may result in double-stranded breaks. Because HSV-1 has three origins of DNA replication, multiple fragments may be generated especially at high m.o.i. These fragments may be substrates for homologous recombination acting to restore the physical integrity of the virus chromosome.
Homologous recombination serves to promote virus replication at high m.o.i., conditions under which DNA polymerase η appears to become dispensable. We favor the idea that an increased number of single-stranded gaps may give rise to double-stranded breaks upon reinitiated DNA synthesis (Fig. 6C). Homologous recombination may not only restore the physical integrity of the virus chromosome but also restore the genetic integrity by bringing together a complete set of fully functional genes, a result that would only be possible under conditions in which the cell has been infected with several virus particles. It is worth noting that although homologous recombination in cells is allowed only between sister chromatids, no such limitations appear to exist for virus chromosomes.
It has long been known that HSV-1 DNA, and probably all other productively replicating herpesviruses, participate in homologous recombination stimulated by double-stranded breaks (43). Different mechanisms have been proposed. For example, HSV-1 proteins, such as the single-stranded DNA-binding protein ICP8, may be active in recombination (44). In fact, an in vitro system for recombination-dependent DNA synthesis requiring ICP8, HSV-1 helicase-primase, and HSV-1 DNA polymerase has been described (45). Using a different approach, we have demonstrated that linear plasmids transfected into cultured cells may undergo homologous recombination and subsequently become replicated by the HSV-1 DNA replisome (46). The latter study suggested that homologous recombination was independent of viral gene functions and that it was likely to be carried out by cellular proteins. We also previously noted that expression of an ATPase-defective version of Rad51 acts as a transdominant inhibitor of recombination between HSV-1 tsS and tsK mutant viruses, resulting in reduced yield of virus with a wild type genotype (47). Furthermore, it was recently observed that siRNA-mediated knockdown of Rad51 caused an approximate 5-fold reduction in Epstein-Barr virus lytic replication (48). Here, we find that replication of UV-damaged HSV-1 DNA is reduced 50–150-fold by siRNA-mediated knockdown of Rad54, Rad52, and Rad51 proteins demonstrating a direct role in HSV-1 recombination repair. We conclude that the cellular apparatus for homologous recombination may act efficiently on HSV-1 DNA and promote recombination. However, our results do not exclude a direct role for HSV-1 replication proteins in similar reactions.
It is apparent that the herpes simplex virus replication cycle and the cellular mechanisms for controlling and executing the DNA damage response are coordinated, causing HSV-1 to make use of, and even depend on, certain repair pathways while down-regulating other branches of the DNA damage response. For example, the DNA ligase IV/XRCC4 complex is required for circularization of linear genomes (2). The ATM kinase is also activated during HSV-1 replication (49, 50). This phenomenon is dependent on viral gene expression because UV-inactivated virus and HSV-1 amplicons fail to activate the ATM kinase. Whether or not viral DNA synthesis is required for activation remains an open question because treatment of infected cells with inhibitors of DNA synthesis only has modest effects on activation of ATM. HSV-1 may also down-regulate repair pathways. ICP0-dependent degradation of DNA-PKcs has been observed in some cell lines (51). HSV-1 also seems to disarm the ATR-dependent DNA-damage response and exclude γH2AX and hyperphosphorylated RPA from viral replication compartments. As a consequence, hyperphosphorylated RPA and the ATR partner ATRIP become relocated to intranuclear VICE domains (52). Also, Mre11 is lost during HSV-1 replication (53).
It now seems possible to make use of HSV-1 as a model system to study molecular mechanisms involved in DNA damage sensing and repair independently from cell cycle regulation and chromatin structure. Because an active HSV-1 replication fork can be reconstituted in vitro with purified components (5), the possibility of studying coupling of DNA replication with repair and recombination using purified enzymes is within reach. In addition, interactions between virus replication and cellular repair systems may influence the efficiency of antiviral treatments and also contribute to the emergence of resistance to antiviral compounds.
Acknowledgment
We thank Dr. Alan Lehmann for generously supplying cells and reagents.
This work was supported by grants from the Swedish Cancer Foundation, the Swedish Research Council, and the Sahlgrenska University Hospital Läkarutbildningsavtal.
- PCNA
- proliferating cell nuclear antigen
- m.o.i.
- multiplicity of infection
- HSV-1
- herpes simplex virus type 1
- XP
- xeroderma pigmentosum
- pfu
- plaque-forming unit
- siRNA
- small interfering RNA
- CSA
- CSB, Cockayne syndrome A and B, respectively.
REFERENCES
- 1.Knipe D. M., Howley P. M. (eds) (2007) Fields Virology, 5th Ed, pp. 2501–2601, Lippincot Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2.Muylaert I., Elias P. (2007) J. Biol. Chem. 282, 10865–10872 [DOI] [PubMed] [Google Scholar]
- 3.Strang B. L., Stow N. D. (2005) J. Virol. 79, 587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehman I. R., Boehmer P. E. (1999) J. Biol. Chem. 274, 28059–28062 [DOI] [PubMed] [Google Scholar]
- 5.Falkenberg M., Lehman I. R., Elias P. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3896–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elias P., O'Donnell M. E., Mocarski E. S., Lehman I. R. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 6322–6326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olivo P. D., Nelson N. J., Challberg M. D. (1989) J. Virol. 63, 196–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruckner R. C., Crute J. J., Dodson M. S., Lehman I. R. (1991) J. Biol. Chem. 266, 2669–2674 [PubMed] [Google Scholar]
- 9.Olsson M., Tang K. W., Persson C., Wilhelmsson L. M., Billeter M., Elias P. (2009) J. Biol. Chem. 284, 16246–16255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuccola H. J., Filman D. J., Coen D. M., Hogle J. M. (2000) Mol. Cell 5, 267–278 [DOI] [PubMed] [Google Scholar]
- 11.Liu S., Knafels J. D., Chang J. S., Waszak G. A., Baldwin E. T., Deibel M. R., Jr., Thomsen D. R., Homa F. L., Wells P. A., Tory M. C., Poorman R. A., Gao H., Qui X., Seddon A. P. (2006) J. Biol. Chem. 281, 18193–18200 [DOI] [PubMed] [Google Scholar]
- 12.Cavanaugh N. A., Urban M., Beckman J., Spratt T. E., Kuchta R. D. (2009) Biochemistry 48, 3554–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norberg P., Bergström T., Rekabdar E., Lindh M., Liljeqvist J. Å. (2004) J. Virol. 78, 10755–10764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowden R., Sakaoka H., Donnelly P., Ward R. (2004) Infect. Genet. Evol. 4, 115–123 [DOI] [PubMed] [Google Scholar]
- 15.Song L., Chaudhuri M., Knopf C. W., Parris D. S. (2004) J. Biol. Chem. 279, 18535–18543 [DOI] [PubMed] [Google Scholar]
- 16.Jiang C., Komazin-Meredith G., Tain W., Coen D. M., Hwang C. B. C. (2009) J. Virol. 83, 7573–7580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tain W., Hwang Y. T., Lu Q., Hwang C. B. C. (2009) J. Virol. 83, 7194–7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogani F., Chua C. N., Boehmer P. E. (2009) J. Biol. Chem. 284, 16784–16790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilcock D., Lane D. P. (1991) Nature 349, 429–431 [DOI] [PubMed] [Google Scholar]
- 20.Taylor T. J., Knipe D. M. (2004) J. Virol. 78, 5856–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilkinson D. E., Weller S. K. (2004) J. Virol. 78, 4783–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harper J. W., Elledge S. J. (2007) Mol. Cell 28, 739–745 [DOI] [PubMed] [Google Scholar]
- 23.Jackson S. P., Bartek J. (2009) Nature 461, 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaurushiya M. S., Weitzman M. D. (2009) DNA Repair 8, 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knipe D. M., Cliffe A. (2008) Nat. Rev. Microbiol. 6, 211–221 [DOI] [PubMed] [Google Scholar]
- 26.Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., Ellenberger T. (2006) DNA Repair and Mutagenesis, American Society for Microbiology, Washington, D. C. [Google Scholar]
- 27.Masutani C., Kusumoto R., Yamada A., Yuasa M., Araki M., Nogimori T., Yokoi M., Eki T., Iwai S., Hanaoka F. (2000) Cold Spring Harb. Symp. Quant. Biol. 65, 71–80 [DOI] [PubMed] [Google Scholar]
- 28.Lehmann A. R., Niimi A., Ogi T., Brown S., Sabbioneda S., Wing J. F., Kannouche P. L., Greene C. M. (2007) DNA Repair 6, 891–899 [DOI] [PubMed] [Google Scholar]
- 29.Friedberg E. C., Lehmann A. R., Fuchs R. P. P. (2005) Mol. Cell 18, 499–505 [DOI] [PubMed] [Google Scholar]
- 30.Waters L. S., Minesinger B. K., Wiltrout M. E., D'Souza S., Woodruff R. V., Walker G. C. (2009) Microbiol. Mol. Biol. Rev. 73, 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shachar S., Ziv O., Avkin S., Adar S., Wittschieben J., Reissner T., Chaney S., Friedberg E. C., Wang Z., Carell T., Geacintov N., Livneh Z. (2009) EMBO J. 28, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.San Filippo J., Sung P., Klein H. (2008) Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 33.Kannouche P., Broughton B. C., Volker M., Hanaoka F., Mullenders L. H. F., Lehmann A. R. (2001) Genes Dev. 15, 158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elmroth K., Nygren J., Mårtensson S., Ismail I. H., Hammarsten O. (2003) DNA Repair 2, 363–374 [DOI] [PubMed] [Google Scholar]
- 35.Honess R. W., Roizman B. (1974) J. Virol. 14, 8–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cleaver J. E., Lam E. T., Revet I. (2009) Nat. Rev. Genet. 10, 756–768 [DOI] [PubMed] [Google Scholar]
- 37.Groisman R., Kuraoka I., Chevallier O., Gaye N., Magnaldo T., Tanaka K., Kisselev A. F., Harel-Bellan A., Nakatami Y. (2006) Genes Dev. 20, 1429–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Compton T., Courtney R. J. (1984) J. Virol. 49, 594–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prakash S., Johnson R. E., Prakash L. (2005) Annu. Rev. Biochem. 74, 317–353 [DOI] [PubMed] [Google Scholar]
- 40.Hernandez T. R., Lehman I. R. (1990) J. Biol. Chem. 265, 11227–11232 [PubMed] [Google Scholar]
- 41.Tanguy Le Gac N., Villani G., Boehmer P. E. (1998) J. Biol. Chem. 273, 13801–13807 [DOI] [PubMed] [Google Scholar]
- 42.Arana M. E., Song L., Tanguy Le Gac N., Parris D. S., Villani G., Boehmer P. E. (2004) DNA Repair 3, 659–669 [DOI] [PubMed] [Google Scholar]
- 43.Sarisky R. T., Weber P. C. (1994) J. Virol. 68, 34–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makhov A. M., Sen A., Yu X., Simon M. N., Griffith J. D., Egelman E. H. (2009) J. Mol. Biol. 386, 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nimonkar A. V., Boehmer P. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10201–10206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao X.-D., Elias P. (2001) J. Biol. Chem. 276, 2905–2913 [DOI] [PubMed] [Google Scholar]
- 47.Yao X.-D. (1999) Replication and Recombination in Cells Infected with Herpes Simplex Virus Type 1 Ph.D. thesis, University of Gothenburg [Google Scholar]
- 48.Kudoh A., Iwahori S., Sato Y., Nakayama S., Isomura H., Murata T., Tsurumi T. (2009) J. Virol. 83, 6641–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lilley C. E., Carson C. T., Muotri A. R., Gage F. H., Weitzman M. D. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shirata N., Kudoh A., Daikoku T., Tatsumi Y., Fujita M., Kiyono T., Sugaya Y., Isomura H., Ishizaki K., Tsurumi T. (2005) J. Biol. Chem. 280, 30336–30341 [DOI] [PubMed] [Google Scholar]
- 51.Lees-Miller S. P., Long M. C., Kilvert M. A., Lam V., Rice S. A., Spencer C. A. (1996) J. Virol. 70, 7471–7477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilkinson D. E., Weller S. K. (2006) J. Cell Sci. 119, 2695–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gregory D. A., Bachenheimer S. L. (2008) Virology 373, 124–136 [DOI] [PMC free article] [PubMed] [Google Scholar]