Abstract
The cell catalysts calnexin (CNX) and protein-disulfide isomerase (PDI) cooperate in establishing the disulfide bonding of the HIV envelope (Env) glycoprotein. Following HIV binding to lymphocytes, cell-surface PDI also reduces Env to induce the fusogenic conformation. We sought to define the contact points between Env and these catalysts to illustrate their potential as therapeutic targets. In lysates of Env-expressing cells, 15% of the gp160 precursor, but not gp120, coprecipitated with CNX, whereas only 0.25% of gp160 and gp120 coprecipitated with PDI. Under in vitro conditions, which mimic the Env/PDI interaction during virus/cell contact, PDI readily associated with Env. The domains of Env interacting in cellulo with CNX or in vitro with PDI were then determined using anti-Env antibodies whose binding site was occluded by CNX or PDI. Antibodies against domains V1/V2, C2, and the C terminus of V3 did not bind CNX-associated Env, whereas those against C1, V1/V2, and the CD4-binding domain did not react with PDI-associated Env. In addition, a mixture of the latter antibodies interfered with PDI-mediated Env reduction. Thus, Env interacts with intracellular CNX and extracellular PDI via discrete, largely nonoverlapping, regions. The sites of interaction explain the mode of action of compounds that target these two catalysts and may enable the design of further new competitive agents.
Keywords: Protein/Viral, Protein/Biosynthesis, Protein/Disulfide, Protein/Drug Interactions, Viruses/HIV, HIV Envelope, Calnexin, Protein-disulfide Isomerase
Introduction
As a viral component, the HIV5-envelope glycoprotein (Env) depends on the catalytic machinery of the host cell for its functional expression. Cell catalysts include cellular foldases that increase the rate and yield of folding by catalyzing reactions such as disulfide bond formation and molecular chaperones that bind unfolded proteins to prevent their aggregation (1). Among these catalysts, previous studies have indicated that, as a glycoprotein, the gp160 Env precursor of the mature envelope subunits gp120 and gp41 interacts early in biosynthesis with the transmembrane chaperone calnexin (CNX) and, to a lesser extent, with its lumenal homolog calreticulin (2–4). CNX retains improperly folded glycoproteins in the endoplasmic reticulum via its lectin-binding domain, which recognizes the transient glucosylated tag specific to immature N-glycans. CNX also binds proteins by a glycosylation-independent mechanism (4–8), although the sequence specificity of CNX binding remains unknown. Besides its documented role as a chaperone, evidence shows that glycoprotein interaction with CNX plays a central role in the induction of the correct network of disulfide bonds in conjunction with oxidoreductases, with some glycoproteins requiring only protein-disulfide isomerase (PDI) (9) for correct disulfide bond formation following entry into the CNX cycle (10). In agreement with these observations, Env was also shown to bind PDI (11, 12), and in the context of the redox potential of the endoplasmic reticulum, the interplay between CNX, PDI, and Env results in the establishment of the pattern of disulfide bonds (13).
When mature Env present at the virus surface interacts with the target cell, experimental evidence indicates that its fusogenicity, the capacity to mediate membrane fusion between virus and cell membranes leading to entry of the viral genome into the cell (14), does not depend solely on interaction between viral glycoproteins and cell-surface receptors. We and others have shown that enzymatic activities associated with the lymphoid cell surface play a key role probably by catalyzing the receptor-mediated conformational changes that occur within Env and eventually trigger fusion (15, 16). Among these reactions, cleavage of the Env disulfides by a lymphocyte surface-associated PDI activity, which acts as a reductase in the redox potential conditions of the extracellular environment, is necessary for entry (17–21). However, in contrast to the regions of Env that bind to the viral receptors or to neutralizing antibodies (Abs), the regions of the molecule interacting with such a key protein partner during the fusion reaction have not been investigated.
Using the extensive array of biological reagents available for study of Env functional expression, we have investigated the interaction between the catalysts CNX and PDI and the Env protein substrate. We focused on CNX (Env associates significantly more with CNX than with calreticulin (3)) and PDI because of their emblematic and well documented roles during the early (virus production) and late (virus/cell contact) stages of the acquisition of the Env functional conformation, respectively. In addition, these catalysts are targets for experimental therapeutic strategies directed against HIV based on glycosylation inhibitors in the case of CNX (22–24) or oxidoreductase inhibitors for PDI (16, 25). The prototypic lymphotropic HIV-1Lai Env was chosen as a model ligand for both catalysts because of the availability of many well characterized Abs directed against such T-tropic Envs, enabling us to establish a competitive immunoassay based on their binding to Env associated, or not, with CNX or PDI. Mapping of Abs whose binding site was occluded by either catalyst allowed the identification of their respective sites of interaction with Env.
EXPERIMENTAL PROCEDURES
Reagents
Abs recognize the following EnvHXB2 sequences (sources are detailed in the Acknowledgments): SR2 (mouse) is directed against amino acids (aa) 31–50; 187 2.1 (mouse), aa 101–120; 8/19b (rat), C1+C3; CRA3 (mouse), V2 and C1 area at the base of V2; 11/4C (rat), aa 152–181; SR1 (mouse), aa 160–175; 11/68b (rat), V1/V2+C4; CRA4 (mouse), V2; 213.1 (mouse), aa 252–261; IIIBV3-13 (mouse), aa 309–317; 5F7 (mouse), aa 308–322; 0.5β (mouse), aa 311–324; IIIB-V3-01 (mouse), aa 320–328; IRC 38.1a (rat), aa 429–438/CD4-binding site; IRC 39.13g (rat), CD4-binding site; D7324 (sheep; Aalto, Dublin, Ireland), gp120 C terminus (aa 497–511). The anti-CNX C4731 Ab (rabbit) was from Sigma. The anti-PDI SPA-890 (rabbit) and RL90 (mouse) Abs were from StressGen (Paris, France) and Alexis Biochemicals (Lörrach, Germany), respectively. Peroxidase-coupled anti-species Abs and other immunochemical reagents were from Dakopatts (Glostrup, Denmark). Purified PDI was from Sigma. The thiol reagent 3-(N-maleimidylpropionyl) biocytin (MPB) was obtained from Molecular Probes (Cergy-Pontoise, France) (19). Other reagents were from Sigma. Recombinant soluble CD4 was produced using the baculovirus expression system (26). Soluble gp160LAI was produced in baby hamster kidney (BHK-21) cells using a recombinant vaccinia virus vector encoding a sequence lacking the gp120-gp41 cleavage sites and transmembrane domain (VV TG1163; Transgène SA, Strasbourg, France (27)) and purified using lectin chromatography and high performance liquid chromatography (28).
Coimmunoprecipitation Experiments
Env was expressed in BHK-21 cells using a recombinant vaccinia virus encoding native EnvLai (VV TG9-1; Transgène SA (27)). Two days post-infection (2 plaque-forming units/cell in serum-free medium) and the subsequent culture in Glasgow medium supplemented with 5% fetal calf serum, cells (107) were washed in PBS and lysed on ice (PBS, Igepal 1%, 1 mm phenylmethanesulfonyl fluoride, 10 mm iodoacetamide, 100 mm antitrypsin inhibitor, and 20 μg/ml each leupeptin and aprotinin). Lysates were centrifuged (10,000 × g; 15 min; 4 °C), and the supernatant was further processed.
Coprecipitation of Chaperones Using Anti-Env Abs
The supernatant was incubated with D7324 Ab (30 μg) or irrelevant sheep IgG coupled to Sepharose (3 h; 4 °C). After three washes in lysis buffer, proteins were eluted in loading buffer (2% β-mercaptoethanol, 5% SDS) and analyzed by SDS-PAGE (8%) and Western blotting (WB). The filters were blocked (PBS, 2% casein, 0.5% Tween 20; 30 min) prior to incubation with D7324 (3 μg/ml PBS, 0.5% casein, 0.5% Tween 20; 90 min), C4731 (1:10,000), or RL90 (1:1,000) Abs. Membranes were then probed with peroxidase-coupled anti-species Abs (1:2,500; 60 min) followed by incubation with SuperSignal working solution (Pierce). Chemiluminescence was detected using a Chemigenius apparatus and quantified using GeneTools software (Ozyme, Montigny, F).
Coprecipitation of Cellular Env Using Anti-chaperone Abs
The supernatant was incubated with the anti-CNX C4731 Ab (30 μg; 3 h) or irrelevant rabbit IgG coupled to Sepharose. Alternatively, the supernatant was incubated with the anti-PDI RL90 Ab (1:100) or irrelevant mouse ascites and protein G-Sepharose (25 μl; 3 h). The resulting precipitated samples were then analyzed by SDS-PAGE and WB using D7324, C4731, or RL90 Abs for staining. Membranes were then processed, and chemiluminescence was determined as described.
Analysis of the Interaction Domains of Env with Chaperones
To analyze Env/CNX interaction in cellulo, the panel of anti-Env Abs (see under “Reagents”; 1:3–30 in PBS) was dot-blotted, and nitrocellulose filters were blocked (PBS, 2% casein; 30 min) prior to incubation (90 min) with cell lysates obtained using 0.2% Triton and subsequently diluted in PBS 0.2% casein (2 × 107 BHK-21 cells expressing Env following vaccinia virus TG9-1 infection). After two washes, membranes were incubated with D7324 Ab (3 μg/ml; 90 min) to determine the amount of Env bound to dot-blotted Abs or with C4731 Ab (1:10,000; 90 min) to determine the amount of CNX bound to Env. Irrelevant Abs determined nonspecific binding. After two washes, membranes were probed with peroxidase-coupled anti-species Abs (1:2,500; 60 min) prior to processing and chemiluminescence determination as described above. The interaction of PDI with soluble gp160 was studied in vitro as described in Ref. 29. gp160LAI (5 μg/20 μl of PBS) preincubated with PDI (2 μg in 1 mm GSH) for 45 min was incubated with dot-blotted anti-Env Abs. Env bound to the Abs was quantified using a pool of human HIV+ sera (1:300), and nonspecific binding was assessed using HIV− sera. The amount of PDI associated with Env was determined using SPA-890 (1:1,000), and the nonspecific binding was determined by incubating the filter with gp160 in the absence of PDI.
The effect of glycosaminoglycans on Env interaction with PDI was examined as described previously (29); gp160LAI (1 μg/20 μl of PBS) was incubated with soluble CD4 (500 ng; 45 min; 25 °C), in the presence or absence of heparin (1.5 μg) and heparan sulfate (3 μg) (Sigma). PDI (400 ng in 1 mm GSH) was then added to the samples for 45 min prior to incubation with a pool of human HIV+ sera (1:100 in 300 μl of PBS; 90 min; 4 °C) followed by addition of protein A-agarose (25 μl; 90 min). The use of HIV− sera determined nonspecific binding. The presence of PDI-Env complexes in immunopurified samples was examined by SDS-PAGE and WB using HIV+ sera (1:300) or SPA-890 Ab (1:1,000) for staining. Membranes were then processed, and chemiluminescence was determined as described.
The effect of anti-Env Abs on Env reduction by PDI was examined as follows: gp160Lai was coupled to CNBr-activated Sepharose 4B (GE Healthcare) according to the manufacturer's instructions (3 μg/100 μl suspension). Bound Env (1 μg/200 μl final volume) was incubated with the following Abs: SR1, 11.68, 213.1, 38.1a, and 39.13g (final dilution, 1:4); CRA3, 5F7, and 11/4c (1:10); SR2 and 8/19b (1:30) (these dilutions were similar to those at which mapping was determined). After washing with PBS, the sample was treated using PDI preactivated with GSH under the conditions described above and elsewhere (29). Env sulfhydryls were then labeled using the biotin-associated sulfhydryl reagent MPB prior to washing and incubation with streptavidin-peroxidase, as described previously (19, 29). After four washes in PBS casein (0.25%), Tween (0.05%), color was developed using o-phenylenediamine.
RESULTS
Interaction of Env with CNX and PDI in Cell Lysate
We examined CNX and PDI association with native Env expressed using a recombinant vaccinia vector in mammalian BHK-21 cells. This system is well documented as suitable for the interactions examined here (3, 30) and to produce authentically folded Env antigens capable of fusion between Env-expressing cells and CD4+ lymphocytes (17). We also used this system because the expression level obtained in many other cell lines (e.g. lymphoid cell lines), including following infections with vaccinia virus vectors, was too low to perform the analysis of the binding domains reported below.
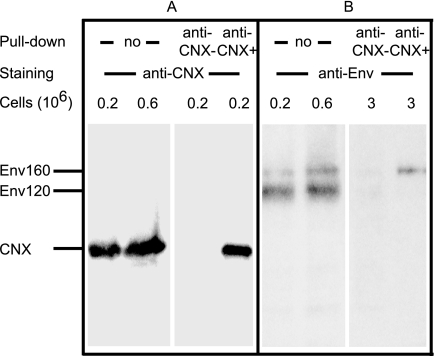
Cell lysates were analyzed in reducing conditions by SDS-PAGE and Western blotting using the anti-gp120 C-terminal Ab D7324 (Fig. 1A). This Ab reacts with Env irrespective of its glycosylation (31) or redox status (19). Both gp120 and gp160 were detected with a gp120:gp160 ratio of ≈5, as determined by reference to a standard curve obtained following SDS-PAGE analysis and WB of increasing amounts of cell lysate (two conditions are shown hereafter) and densitometric analysis of the resulting bands. This observation is in agreement with previous data obtained using this expression system (32). The cell lysate was also incubated with D7324 coupled to Sepharose beads in a “pulldown” assay to recover Env and any associated proteins. After elution from the beads, the sample was analyzed using SDS-PAGE and WB as above (Fig. 1A). Densitometric analysis showed that ≈80% of cell-associated Env was isolated using this procedure, which is consistent with the high efficiency previously noted for large scale Env production (33), and showed that no preferential purification of gp120 or gp160 occurred because a gp120:gp160 ratio ≈5 was maintained. When anti-CNX (Fig. 1B) or anti-PDI (Fig. 1C) Abs were used instead of anti-Env Ab for WB, the presence of both chaperones in the Env eluate pulled down using D7324 was shown. Control procedures using an irrelevant sheep IgG coupled to Sepharose-beads precipitated neither the Env species nor either of the chaperones. Together, these data indicate that both CNX and PDI associate with Env during biosynthesis and that their interaction is sufficiently robust to allow isolation in a pulldown assay. By reference to a standard curve obtained following SDS-PAGE analysis and WB of increasing amounts of cell lysate using anti-chaperone Abs and densitometric analysis, we determined that ≈10 and ≈0.5% of the total pool of CNX and PDI present in the cell lysate were pulled down using anti-Env Abs, respectively (Fig. 1, B and C, and data not shown). We then estimated the total amount of chaperones and Env produced by cells. CNX was quantified following immunoprecipitation (in conditions where about 90% of the cellular species was isolated; see below) and protein quantitation because a commercial source of pure CNX was not available. The levels of PDI and Env were quantified following the establishment of a standard curve using a dot blot with known amounts of pure, commercially sourced, antigen. We found that 106 cells expressed about 1.5 μg of Env (12 pmol), 0.3 μg of CNX (3 pmol), and 0.25 μg of PDI (4 pmol). Taking into account the coprecipitated Env/chaperone ratio above and the yield of precipitation, we estimated the amounts of chaperones interacting with Env and concluded that ≈0.3 pmol of CNX and ≈0.02 pmol of PDI were pulled down when ≈10 pmol of Env was immunoprecipitated. Thus, 1 Env out of 30 and out of 500 was found associated with CNX and PDI, respectively. These estimates relate to the subset of Env involved in a complex with either chaperone at the moment cells were lysed and not the overall rate of interaction with either chaperone during the whole course of Env biosynthesis. Moreover, these values probably underscore the actual Env-chaperone complex formed when lysis occurs as some loosely bound chaperone is undoubtedly lost during cell disruption and the pulldown procedure.
FIGURE 1.
Coprecipitation of CNX and PDI with Env using anti-Env Abs. Lysate of BHK-21 cells (the corresponding cell number is indicated) expressing Env was submitted (no pulldown) to SDS-PAGE analysis and WB using anti-Env (A), -CNX (B), or -PDI (C) Abs. Alternatively, lysate was incubated with irrelevant sheep IgG (anti-Env−) or D7324 Ab (anti-Env+) prior to precipitation, elution using loading buffer, SDS-PAGE, and WB as indicated. A, *, nonspecific species detected by the peroxidase-labeled anti-sheep Ab staining system.
To identify the Env species interacting with chaperones, we incubated cell lysates with anti-chaperone Abs. When anti-CNX Abs were used in the pulldown and immunoblotting assays, we determined by densitometry that ≈90% of the cellular CNX was purified (Fig. 2A). When the Env species coprecipitated with CNX was examined following immunoblotting using D7324, only gp160 was found to associate with CNX (Fig. 2B). Using densitometry and a standard curve, we found that ≈15% of cell-associated gp160 coprecipitated with CNX. This subset of gp160 represented ≈3% of the total immunopurified Env (gp120 and gp160) because the gp120/gp160 ratio was ≈5 (see above).
FIGURE 2.
Coprecipitation of Env with CNX using anti-CNX Ab. Lysate of BHK-21 cells expressing Env was submitted (no pulldown) to SDS-PAGE analysis and WB using anti-CNX (A) or -Env (B) Abs. Alternatively, lysate was incubated with irrelevant rabbit IgG (anti-CNX−) or anti-CNX Ab (anti-CNX+) prior to precipitation, elution using loading buffer, SDS-PAGE, and WB as indicated.
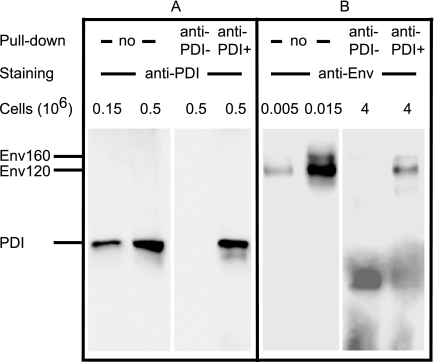
When anti-PDI Abs were used in the pulldown and immunoblotting assays, we observed that ≈90% of PDI was purified (Fig. 3A). Following D7324 immunoblotting, both gp120 and gp160 species were shown to be coprecipitated with PDI (Fig. 3B). Using densitometry and a standard curve, we determined that ≈0.25% of cell-associated Env coprecipitated with PDI.
FIGURE 3.
Coprecipitation of Env with PDI using anti-PDI Ab. Lysate of BHK-21 cells expressing Env was submitted (no pulldown) to SDS-PAGE analysis and WB using anti-PDI (A) or -Env (B) Abs. Alternatively, lysate was incubated with irrelevant mouse IgGs (anti-PDI−) or anti-PDI Ab (anti-PDI+) prior to precipitation, elution using loading buffer and analysis by SDS-PAGE and WB as indicated.
Together with the quantification of each protein precipitated and the yield of precipitation (see above), the results of Figs. 1–3 indicate that ≈0.3 pmol of CNX interacted with ≈0.3 pmol of gp160 and ≈0.02 pmol of PDI interacted with ≈0.02 pmol of Env. Therefore, the average stoichiometry of the Env/chaperone interaction was ≈1:1 in our experimental conditions. In further experiments to examine the strength of the interaction, we observed that the presence of either 0.2% Triton X-100 or 1% Igepal CA-630 during cell lysis and immunoprecipitation led to similar coprecipitation yields but that the presence of 0.1% SDS abolished the observed Env interaction with chaperones (data not shown).
Domains of Env Involved in Contact with CNX
Because the data shown in Figs. 1 and 2 indicate that the recombinant vaccinia virus/BHK-21 expression system enables intracellular Env precursor association with CNX to be monitored, we repeated these conditions to investigate the domains of the glycoprotein involved in contact with each chaperone. We immobilized a panel of anti-Env Abs onto nitrocellulose at a variety of concentrations and then incubated the filter with a lysate of BHK-21 cells expressing Env. Following incubation, the relative capture of Env and CNX by each anti-Env Ab was assessed (Fig. 4).
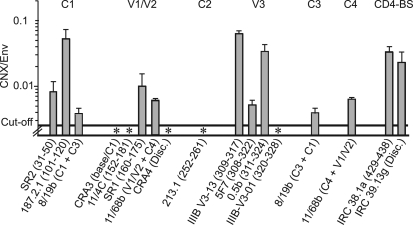
FIGURE 4.
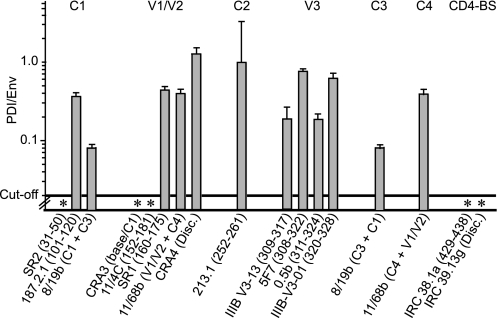
CNX association with Env in cellulo, epitopes involved. Clarified lysate of BHK-21 cells expressing Env was incubated with dilutions of dot-blotted anti-Env Abs. The amount of Env associated with each dot was assessed using D7324 Ab or irrelevant sheep IgG prior to staining and densitometry. The presence of CNX associated with samples bound to the Abs was assessed in parallel using anti-CNX Ab or irrelevant rabbit IgGs. Env and CNX quantification was done using standard curves obtained using known amounts of antigens. For each Ab dilution, the CNX/Env molar ratio was determined (n = 2 experiments). The mean ratio ± S.D. obtained for each Ab is presented. * indicates Abs that did not bind Env complexed with CNX.
Env was quantified using D7324 combined with a standard curve obtained following a dot blot assay using known amounts of Env. The background signal was assessed using an irrelevant sheep IgG. Env reacted with Abs in a dose-dependent manner limited by the level of Ab immobilized on the filter, indicating that Env was in excess in these conditions. The amounts of Ab-bound Env ranged from 0.2 ng (e.g. 187.2.1 Ab at a dilution of 1:30) to 20 ng (e.g. 8/19b; 1:3), and the threshold of Env detection was 0.1 ng (threshold absorbance was defined as two times background absorbance hereafter). Similarly, CNX associated with the Env species immobilized on the filter via anti-Env Abs was quantified as described above using an anti-CNX Ab following determination of the background signal using an irrelevant rabbit IgG. Samples associated with most anti-Env Abs of the panel reacted with the anti-CNX Ab, indicative of an Env/CNX interaction. CNX quantification ranges were from 0.015 ng (e.g. IRC 38.1a; 1:30) to 0.6 ng (IIIB-V3-13; 1:3), and the threshold of detection was 0.003 ng.
Based on the amount of Env and CNX associated with each CNX-positive spot, we calculated the CNX/Env molar ratio and found it to be close to 1:30 for five Abs (187.2.1; IIIB-V3-13; 0.5β; IRC38.1a; IRC39.13g; Fig. 4), in agreement with the ratio of CNX/Env association determined in Fig. 1. The association of CNX to Env immobilized via five Abs (SR2, C1; 8/19b, C1+C3; SR1, V1-V2; 11/68b, V1/V2+C4; 5F7, V3) was weaker than that reported above, whereas the samples associated with five Abs (CRA3, 11/4c, CRA4, 213.1, IIIB-V3-0.1) directed against V1/V2 (34), C2, and the C-terminal flanking region of V3 led to signals below the cutoff of the assay. The threshold of CNX detection and the amount of Env associated with these Abs indicated less than 1 molecule of CNX per 400 Env molecules, suggesting that Env complexed with CNX essentially did not associate with this subset of captured Abs.
Lack of CNX association with Env may result either from an overlap between the Ab epitopes and the CNX interaction domain or from the weak reactivity of these Abs with the subpopulation of Env complexed with CNX due to particular folding. Accordingly, we tested whether these Abs reacted with Env in a conformation-dependent manner. Cells expressing Env were lysed using 0.2% SDS, effectively denaturing the protein (31), and the lysate was diluted 15-fold prior to incubation with the immobilized Ab panel. We observed that SDS-treated Env reacted with Abs in our conditions (data not shown), suggesting that the lack of detection of CNX in the samples immobilized via these Abs was not the result of a particular Env conformation. These data are consistent with the domains of Env involved in CNX interaction being composed, at least in part, of regions of V1/V2, C2, and the C-terminal flanking region of V3.
Domains of Env Involved in Contact with PDI
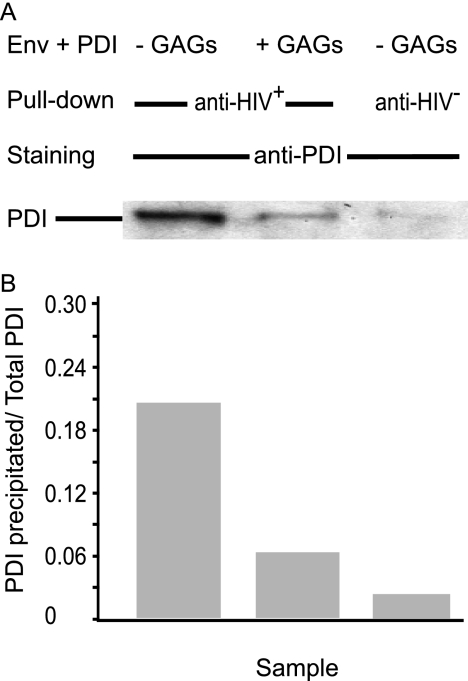
A similar strategy was used to localize the domains of Env interacting with PDI. However, when cellular Env was captured using the panel of Abs described above, we were unable to detect associated PDI (data not shown), consistent with the low levels of association between Env and PDI in cell lysates (cf. Figs. 1 and 3). These observations, along with data from other authors showing a very weak association by pulldown assay between Env and PDI during biosynthesis (12), indicated that the Env/PDI interaction in the biosynthetic pathway was below a level necessary to provide consistent and reproducible measurement using the Ab capture assay described here. As an alternate approach, we used the documented capacity of Env to react with, and to be reduced by, PDI in in vitro (18, 29) conditions that mimic their interaction during HIV/cell contact and that lead to Env reduction (18–20). Env was coincubated with PDI at a molar ratio of 1 (29) and then pulled down with pooled human HIV+ sera; 5 out of 6 pmol of Env present in the assay were purified under these conditions (data not shown and determined as described above), which also coprecipitated 1.2 out of 6 pmol of PDI (Fig. 5). Preincubation of Env with glycosaminoglycans, which inhibit Env reduction by PDI (29), prevented Env/PDI coprecipitation, supporting the specificity of the Env/PDI interaction under these conditions. As a control, we verified that the amount of Env precipitated in the presence or absence of glycosaminoglycans was similar and that the use of HIV− sera did not precipitate detectable amounts of Env (data not shown).
FIGURE 5.
Glycosaminoglycans and PDI association with Env in vitro. Env was incubated with equimolar amounts of PDI in the presence or absence of glycosaminoglycans (GAGs) prior to precipitation using human HIV+ or HIV− sera. The resulting samples were analyzed by SDS-PAGE and WB using anti-PDI Abs (A) prior to densitometric quantification (B) using a standard curve obtained using known amounts of PDI. The ratio “PDI recovered following precipitation/PDI in the assay” is shown.
Env preincubated with PDI as above was then allowed to react with the panel of anti-Env Abs immobilized onto nitrocellulose (Fig. 6). Quantification of the Env bound by these Abs using a dot blot assay with pooled HIV+ sera showed that the antigen bound Abs in a dose-dependent manner ranging from 0.3 ng (187.2.1 Ab; 1:30) to 20 ng (8/19b Ab; 1:3), and the threshold of Env detection was 0.03 ng.
FIGURE 6.
PDI association with Env in vitro, epitopes involved. Env preincubated with PDI was incubated with dilutions of dot-blotted anti-Env Abs. The amount of Env associated with each dot was assessed using HIV+ or HIV− sera prior to staining and densitometry. The presence of PDI associated with samples bound to the Abs was assessed in parallel using anti-PDI Ab, similar experiments using Env but not PDI determining the background signal. Env and PDI quantification was done using standard curves obtained using known amounts of antigens. For each Ab dilution, the PDI/Env molar ratio was determined (n = two experiments). The mean ratio ± S.D. obtained for each Ab is presented. * indicates Abs that did not bind Env complexed with PDI.
Samples associated with most Abs of the panel were also found to react specifically with anti-PDI Ab indicative of an Env/PDI interaction. PDI associated with Env was quantified as described above, and the range was from 0.2 ng of PDI associated with Env bound to 8/19b Ab (1:30) to 4 ng for the Env bound to IIIB-V3-01 Ab (1:3).
Based on the amount of Env and PDI associated with each PDI-positive dot, we calculated the PDI/Env molar ratio for each Ab dilution for each experiment and found it to be close to 1:4 for 10 Abs (187.2.1, 8/19b, SR1, 11/68b, CRA4, 213.1, IIIB-V3-13, 5F7, 0.5β, III-V3-01; Fig. 6), in agreement with the ratio observed when Env was incubated in vitro with PDI and precipitated with HIV+ sera (Fig. 5).
In contrast, we did not detect PDI in samples immobilized via an Ab directed against the C1 domain (SR2), Abs against residues of V2 in the C1 area (CRA3, 11/4C; Ref. 34), and Abs against the CD4-binding site (IRC 38.1a, IRC 39.13g). Taking into account the amount of Env linked to these Abs and the fact that the threshold of PDI detection in our assay was 0.04 ng, we conclude that the corresponding Env population exhibited <1 PDI per ≈50 Envs bound suggesting that Env-PDI complexes cannot be captured effectively via the epitopes concerned. Epitope occlusion could be either because they were located within the binding site for PDI or because they were not present on the PDI-reduced form of Env. To test the latter hypothesis, we studied the binding of this subset of Abs to reduced Env. Env was treated using 1% mercaptoethanol, a condition that fully reduces the antigen (19), prior to iodoacetamide treatment and lyophilization. Treated or mock-treated Env was then diluted and incubated with test Abs. Ab binding was not altered following Env reduction, except for CRA3 and IRC 39.13g. The results of Fig. 6 therefore suggest that the domains of Env interacting with PDI involve C1, the N-terminal flanking region of V1/V2, and the CD4-binding site.
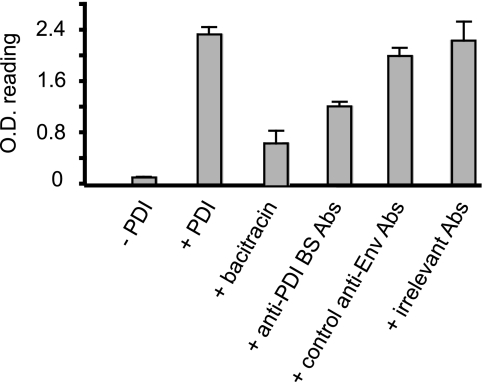
We then hypothesized that Abs interacting with the PDI-binding site could interfere with Env reduction by the reductase. To test this, we designed and validated a new assay where Env immobilized via CNBr-Sepharose beads is reduced by PDI prior to labeling of the resulting Env SH groups by the biotin-coupled thiol reagent MPB. Env-associated MPB was detected using streptavidin-peroxidase. In this assay, Env labeling by MPB was weak in the absence of PDI, consistent with the low level of free thiols associated with mature Env (Fig. 7) (19, 29). Addition of PDI to the Env sample significantly increased MPB labeling, indicative of the generation of free sulfhydryls on Env as a result of disulfide reduction by the catalyst. Bacitracin, a specific and potent PDI inhibitor, strongly inhibited the PDI-induced MPB labeling of Env. These results are in agreement with previous observations that both membrane and soluble PDI similarly reduce Env in vitro and in vivo (18–20, 29). Using this format, we then investigated PDI-induced MPB labeling of Env in the presence of those Abs found to interact with the PDI-binding site. Env labeling was reduced by 50% when the antigen was preincubated with this Ab mixture (namely SR2, CRA3, 11/4c, IRC 38.1a, and IRC 39.13g). Further analysis revealed the following: (a) a mixture composed of 8/19b, SR1, 11.68, 213.1, and 5F7 inhibited Env reduction by about 15%, possibly by limiting Env surface accessibility to PDI (Fig. 7); (b) irrelevant mouse Abs had no effect (Fig. 7); and (c) SR2, CRA3, 11/4c, IRC 38.1a, and IRC 39.13g did not alter reduction when tested separately (data not shown). Together, these data indicate that Abs raised against parts of C1, the N-terminal flank of V1/V2, and the CD4-binding site interfere with Env reduction by PDI by blocking the site of PDI binding to Env.
FIGURE 7.
Effect of anti-Env Abs on reduction by PDI. Env coupled to Sepharose beads was incubated in the presence or absence of PDI. The sulfhydryl groups were then labeled using MPB, a thiol-reactive reagent that was detected using streptavidin-peroxidase and o-phenylenediamine. Bacitracin was used to inhibit Env reduction by PDI, as indicated. Alternatively, Env was incubated with anti-PDI-binding site Abs (a mixture of SR2, CRA3, 11/4c, IRC 38.1a, and IRC 39.13g), other anti-Env Abs (a mixture of 8/19b, SR1, 11.68, 213.1, and 5F7), or irrelevant mouse Abs, as described under “Experimental Procedures,” prior to reduction by PDI, MPB labeling, and processing (n = 3; the mean ratio ± S.D. is presented).
DISCUSSION
Because viruses encode only a few proteins and the range of biochemical reactions they can perform is limited, the completion of the infectious cycle requires the co-opting of host enzymes. This is illustrated by the HIV replication cycle that requires the involvement of several cellular catalysts, from the early steps of the intracellular biosynthetic pathway through particle assembly and release to the ultimate fusion of the virus and cellular membranes (2, 3, 11, 12, 15, 17, 30, 35, 36). We investigated here the relationship of Env with two major cellular partners involved in Env folding and hence in establishing its disulfide network, namely CNX, which functions early during biosynthesis (2, 3), and PDI, which functions both during biosynthesis (12) and following virus binding at the cell surface to produce the fusogenic conformation of the viral envelope (18–20).
Using a vaccinia virus expression system, we show that 15% of gp160 molecules are associated with CNX. In contrast, the gp120 subunit did not interact with CNX. These results, which agree with previous works using recombinant systems or HIV to express Env (2, 3), showed that, as a glycoprotein, the folding of the gp160 precursor is assisted by CNX during the early steps of its biosynthesis in the endoplasmic reticulum (2, 3) where gp120 is essentially absent because Env processing in the Golgi has yet to occur (36).
Subsequently, we undertook a detailed analysis of the Env domains involved in CNX interaction. A competitive binding assay based on the use of a panel of anti-Env Abs immobilized on nitrocellulose was developed to examine the interacting domains. Most Abs were directed against C1-4-, V1-3-, and the CD4-binding site, and they typically bound to mature forms of Env, consistent with the accessibility of their corresponding epitopes on the folded antigen (37). Other Abs tested, mainly directed against V4 and C5, reacted poorly with mature Env in the cell lysate or as a soluble antigen probably because they recognized regions that are hidden on the folded molecule (37). We observed that epitopes within the V1/V2 and C2 domains as well as the C-terminal flanking region of V3 were not available on Env species interacting with CNX. These findings are consistent with an earlier study based on coprecipitation of truncated Env with anti-calreticulin Abs, which indicated that calreticulin binding occurred in the N-terminal half of the molecule (3).
We did not further discriminate between epitope occlusion following lectin-type binding of CNX to Env and occlusion resulting from CNX/protein interaction. Although the use of glycosylation inhibitors may allow such a distinction, the consequence of abnormal glycosylation on Env conformation would more likely interfere with the results obtained. Indeed, we have shown previously that the regions of Env that we found here interacting with CNX also have altered immunoreactivity when the viral protein is expressed in the presence of deoxynojirimycin (38), a glycosidase inhibitor with potent anti-HIV activity (22–24) that arrests Env glycosylation at the stage of glycan processing where CNX is thought to act. Together, these data support a mode of action of deoxynojirimycin in which the drug-altered glycosylation modifies the conformation of the Env domains recognized by CNX, interferes with CNX interaction, and leads to aberrant folding and the subsequent inhibition of fusion at the coreceptor binding step (38).
In the case of PDI, the catalyst associated with both gp120 and gp160 but only 1 molecule in 500 bound the oxidoreductase intracellularly in our experimental conditions. Such weak interaction was unlikely to be caused by low level expression of PDI as its level in lysates was found here to be similar to that of CNX, i.e. about 0.15% of the total cell proteins and in the range reported previously (39). The very weak coprecipitation of intracellular Env with PDI more likely resulted from only transient interaction during biosynthesis, in agreement with the poor association of Env and PDI reported previously (12). It is noteworthy that when mercaptoethanol was omitted prior to SDS-PAGE analysis, we did not detect high molecular weight species reacting with both anti-Env and anti-PDI Abs in cell lysates, suggesting that Env-PDI complexes cross-linked by disulfides are either not produced or are very transient species in our cell system (data not shown).
In addition to PDI interaction with Env during the biosynthetic pathway, PDI is found within the CD4-CXCR4-Env complexes that appear following HIV interaction with the surface of the target lymphoid cell (18–20). Env is reduced within these cell surface complexes by a catalyst that is most probably PDI. Indeed, various compounds considered to be specific PDI inhibitors such as Bacitracin, aT3, and anti-PDI Abs have been found to potently interfere with HIV/cell fusion or Env reduction (17–20, 29). In contrast, Ou and Silver (21), using small interfering RNA to inhibit PDI expression, also observed entry inhibition but to a lesser extent, challenging PDI intervention. The PDI family of proteins continues to grow with roles in oxidative folding, protein degradation, trafficking, calcium homeostasis, and antigen presentation in addition to virus entry (16, 40). Many members of the family contain common features, including antibody epitopes, and are thus poorly distinguishable and generally considered as a single class. As much of the cellular chaperone machinery includes extensive redundancy, a sequence-specific siRNA would be unable to potently block generic PDI activity involved in HIV entry and thus would have only marginal effect. Env reduction can be mimicked using soluble components in vitro (18, 29). We took advantage of these observations and preincubated equimolar mixture of Env and PDI to identify the PDI binding regions on Env. First, we showed that in these in vitro conditions Env was associated with PDI, and this association was inhibited by incubation with glycosaminoglycans that prevent virus entry (41) and interfere with Env reduction by PDI (29), further supporting the relevance of the complex formed in our in vitro assay (29). Second, when the Env/PDI mixture was used with the immobilized panel of Abs, PDI was found to bind Env via regions including the N-terminal flanks of the C1 and V1/V2 domains and the CD4-binding site. This conclusion is supported by the finding that addition of the mixture of the Abs against these sequences inhibited Env reduction by PDI. The extent of inhibition achieved using this set of Abs was similar to that obtained using a mixture of glycosaminoglycans that bind Env and inhibit its reduction (see above and Ref. 29), although below the inhibition of reduction obtained using compounds that specifically target PDI such as Bacitracin (19, 29). Inhibition of Env reduction by the Abs directed against the PDI-binding site is also consistent with their capacity to neutralize HIV (42, 43), reduction of Env by PDI being a prerequisite to the Env-mediated membrane fusion process (18–20). In contrast, we did not find binding of PDI to the V3 loop despite reports indicating that its basic nature may favor interaction with the acidic C terminus of PDI (9, 16, 25) and observations showing the susceptibility of the disulfide at the base of V3 to PDI (44). It is therefore possible that although PDI binds Env at the domains we have identified, final catalysis is distal to them, in agreement with the separate domains of PDI involved in substrate binding and catalysis (9). Recently, four glycosaminoglycan-binding domains have been mapped on gp120, in the V2 and V3 loops, C terminus (residues 420–435), and within the CD4-induced bridging sheet (45). Our mapping of the PDI-binding sites to similar sequences offers a plausible mode of action for the observed glycosaminoglycan inhibition of PDI-mediated Env reduction (29) and the subsequent blocking of HIV cell fusion (41): interference with PDI binding to its contact sites on Env. This explanation is supported by the observation that glycosaminoglycans prevented Env/PDI coprecipitation (Fig. 5).
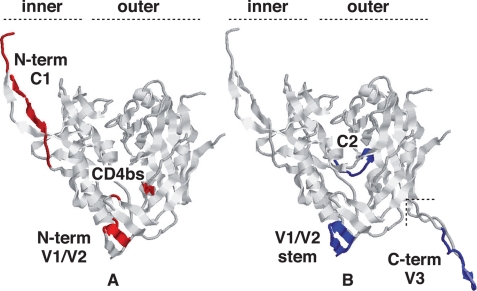
In addition to providing explanations of the mode of action of compounds with anti-HIV activity, deoxynojirimycin, which targets CNX, and glycosaminoglycans, which target PDI, our mapping of the regions interacting with intracellular CNX and extracellular PDI is also consistent with current Env structural data. The structure of the liganded and unliganded forms of the gp120 subunit, largely deleted of the variable regions to enable crystallization, reveals significant changes in the inner domain and bridging sheet in response to CD4 binding (46, 47). Cryoelectron tomography studies have recently confirmed these conformational changes following CD4 association by reporting the three-dimensional structure of trimeric and HIV-associated native Env pre- and post-CD4 binding (48). The interaction of viral Env to cell surface PDI is a post-CD4 binding event (16–20), and the PDI-binding sites identified here on Env are among the regions that become accessible following HIV attachment to the target cell, namely the C1, the N-terminal region of V1/V2, and the CD4-binding site sequences (Fig. 8A) (49). Of note, our mapping is also consistent with the reports showing that heparin mediates inhibition of HIV entry at a post-CD4 binding step by occupying the same sites (45) and that it interferes with both Env reduction by PDI (29) and coprecipitation of Env and PDI (Fig. 5). By contrast, the sites identified as binding CNX are largely buried in mature gp120 (Fig. 8B) (46–48). Our data are therefore consistent with published reports showing CNX acting in the endoplasmic reticulum on the partially unfolded gp160 prior to oligomerization and with PDI acting at the virus surface on the folded, assembled, and CD4-interacting molecule (2, 3, 16).
FIGURE 8.
Location of the interaction sites for PDI and CNX in the mature gp120 structure. The gp120 structure shown was rendered from the recently released structure (Protein Data Bank code 3JWO (50)) that includes an extended N terminus when compared with earlier structures. The V3 loop in B was rendered from the single structure reporting it (Protein Data Bank code 2B4C) and then grafted where shown. The rendering is in the canonical view of gp120 (45) with the inner and outer domains of the molecule indicated. The definitive epitopes for the Abs that blocked chaperone binding are indicated, where available. PDI-reactive sites are largely accessible on the mature molecule (A), and those reacting with CNX are buried or face inward (B), consistent with access only prior to folding. The sites are mutually exclusive except for the V1/V2 region where precise residues cannot be indicated as they are not present in any solved structure. The diagram is illustrative, and the actual structure of the molecule during interaction with either chaperone remaining unknown.
We have described the first mapping of the binding sites for CNX and PDI on HIV Env. The data suggest that each chaperone binds a distinct footprint on the molecule. As a result of the key role of PDI during virus entry, deciphering the Env/PDI interaction surfaces may have therapeutic impact because it designates additional targets for the development of competing agents with antiviral potential.
Acknowledgments
We are indebted to Dr. M. P. Kieny and Transgene S.A. for the gift of recombinant vaccinia virus vectors and to the scientists who contributed to this work via the National Institute for Biological Standards and Controls (United Kingdom) by giving the following antibodies (M. Page (CRA3 and CRA4); C. Thiriard and C. Bruck (187.2.1 and 213.1); J. Cordell and C. Dean (ICR38.1a and ICR39.13g); A. von Brunn (5F7); K. Takatsuki (0.5β); C. Shotton and C. Dean (11/4C, 8/19b, and 11/68b); J. Laman (IIIB-V3-01 and IIIB-V3-13); and S. Ranjbar (SR1 and SR2)).
This work was supported by Agence Nationale de Recherche sur le Sida (to E. F., R. B., and I. M. J.) and the United Kingdom Medical Research Council (to I. M. J.).
- HIV
- human immunodeficiency virus
- Ab
- antibody
- BHK
- baby hamster kidney
- CNX
- calnexin
- Env
- envelope glycoprotein
- PBS
- phosphate-buffered saline
- PDI
- protein-disulfide isomerase
- V
- variable domain
- WB
- Western blot
- aa
- amino acid
- MPB
- 3-(N-maleimidylpropionyl) biocytin.
REFERENCES
- 1.Nagradova N. (2007) Curr. Protein Pept. Sci. 8, 273–282 [DOI] [PubMed] [Google Scholar]
- 2.Dettenhofer M., Yu X. F. (2001) J. Biol. Chem. 276, 5985–5991 [DOI] [PubMed] [Google Scholar]
- 3.Otteken A., Moss B. (1996) J. Biol. Chem. 271, 97–103 [DOI] [PubMed] [Google Scholar]
- 4.Caramelo J. J., Parodi A. J. (2008) J. Biol. Chem. 283, 10221–10225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ihara Y., Cohen-Doyle M. F., Saito Y., Williams D. B. (1999) Mol. Cell 4, 331–341 [DOI] [PubMed] [Google Scholar]
- 6.Wanamaker C. P., Green W. N. (2005) J. Biol. Chem. 280, 33800–33810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Free R. B., Hazelwood L. A., Cabrera D. M., Spalding H. N., Namkung Y., Rankin M. L., Sibley D. R. (2007) J. Biol. Chem. 282, 21285–21300 [DOI] [PubMed] [Google Scholar]
- 8.Brockmeier A., Brockmeier U., Williams D. B. (2009) J. Biol. Chem. 284, 3433–3444 [DOI] [PubMed] [Google Scholar]
- 9.Ferrari D. M., Söling H. D. (1999) Biochem. J. 339, 1–10 [PMC free article] [PubMed] [Google Scholar]
- 10.Jessop C. E., Tavender T. J., Watkins R. H., Chambers J. E., Bulleid N. J. (2009) J. Biol. Chem. 284, 2194–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Land A., Braakman I. (2001) Biochimie 83, 783–790 [DOI] [PubMed] [Google Scholar]
- 12.Land A., Zonneveld D., Braakman I. (2003) FASEB J. 17, 1058–1067 [DOI] [PubMed] [Google Scholar]
- 13.Leonard C. K., Spellman M. W., Riddle L., Harris R. J., Thomas J. N., Gregory T. J. (1990) J. Biol. Chem. 265, 10373–10382 [PubMed] [Google Scholar]
- 14.Pierson T. C., Doms R. W. (2003) Curr. Top. Microbiol. Immunol. 281, 1–27 [DOI] [PubMed] [Google Scholar]
- 15.Avril L. E., Di Martino-Ferrer M., Barin F., Gauthier F. (1993) FEBS Lett. 317, 167–172 [DOI] [PubMed] [Google Scholar]
- 16.Fenouillet E., Barbouche R., Jones I. M. (2007) Antioxid. Redox. Signal. 9, 1009–1034 [DOI] [PubMed] [Google Scholar]
- 17.Fenouillet E., Barbouche R., Courageot J., Miquelis R. (2001) J. Infect. Dis. 183, 744–752 [DOI] [PubMed] [Google Scholar]
- 18.Gallina A., Hanley T. M., Mandel R., Trahey M., Broder C. C., Viglianti G. A., Ryser H. J. (2002) J. Biol. Chem. 277, 50579–50588 [DOI] [PubMed] [Google Scholar]
- 19.Barbouche R., Miquelis R., Jones I. M., Fenouillet E. (2003) J. Biol. Chem. 278, 3131–3136 [DOI] [PubMed] [Google Scholar]
- 20.Markovic I., Stantchev T. S., Fields K. H., Tiffany L. J., Tomiç M., Weiss C. D., Broder C. C., Strebel K., Clouse K. A. (2004) Blood 103, 1586–1594 [DOI] [PubMed] [Google Scholar]
- 21.Ou W., Silver J. (2006) Virology 350, 406–417 [DOI] [PubMed] [Google Scholar]
- 22.Jones I. M., Jacob G. S. (1991) Nature 352, 198. [DOI] [PubMed] [Google Scholar]
- 23.Fenouillet E., Gluckman J. C. (1992) Virology 187, 825–828 [DOI] [PubMed] [Google Scholar]
- 24.Fischer P. B., Collin M., Karlsson G. B., James W., Butters T. D., Davis S. J., Gordon S., Dwek R. A., Platt F. M. (1995) J. Virol. 69, 5791–5797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryser H. J., Flückiger R. (2005) Drug Discov. Today 10, 1085–1094 [DOI] [PubMed] [Google Scholar]
- 26.Morikawa Y., Overton H. A., Moore J. P., Wilkinson A. J., Brady R. L., Lewis S. J., Jones I. M. (1990) AIDS Res. Hum. Retroviruses 6, 765–773 [DOI] [PubMed] [Google Scholar]
- 27.Kieny M. P., Lathe R., Rivière Y., Dott K., Schmitt D., Girard M., Montagnier L., Lecocq J. P. (1988) Protein Eng. 2, 219–225 [DOI] [PubMed] [Google Scholar]
- 28.Jones D. H., McBride B. W., Roff M. A., Farrar G. H. (1995) Vaccine 13, 991–999 [DOI] [PubMed] [Google Scholar]
- 29.Barbouche R., Lortat-Jacob H., Jones I. M., Fenouillet E. (2005) Mol. Pharmacol. 67, 1111–1118 [DOI] [PubMed] [Google Scholar]
- 30.Otteken A., Earl P. L., Moss B. (1996) J. Virol. 70, 3407–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papandreou M. J., Idziorek T., Miquelis R., Fenouillet E. (1996) FEBS Lett. 379, 171–176 [DOI] [PubMed] [Google Scholar]
- 32.Fenouillet E., Jones I., Powell B., Schmitt D., Kieny M. P., Gluckman J. C. (1993) J. Virol. 67, 150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore J. P., McKeating J. A., Jones I. M., Stephens P. E., Clements G., Thomson S., Weiss R. A. (1990) AIDS 4, 307–315 [DOI] [PubMed] [Google Scholar]
- 34.Moore J. P., Sattentau Q. J., Yoshiyama H., Thali M., Charles M., Sullivan N., Poon S. W., Fung M. S., Traincard F., Pinkus M., Drobey G., Robinson J., Ho D., Sodrosky J. (1993) J. Virol. 67, 6136–6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryser H. J., Levy E. M., Mandel R., DiSciullo G. J. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 4559–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfeiffer T., Zentgraf H., Freyaldenhoven B., Bosch V. (1997) J. Gen. Virol. 78, 1745–1753 [DOI] [PubMed] [Google Scholar]
- 37.Wyatt R., Kwong P. D., Desjardins E., Sweet R. W., Robinson J., Hendrickson W. A., Sodroski J. G. (1998) Nature 393, 705–711 [DOI] [PubMed] [Google Scholar]
- 38.Papandréou M. J., Barbouche R., Guieu R., Kieny M. P., Fenouillet E. (2002) Mol. Pharmacol. 61, 186–193 [DOI] [PubMed] [Google Scholar]
- 39.Roth R. A., Koshland M. E. (1981) Biochemistry 20, 6594–6599 [DOI] [PubMed] [Google Scholar]
- 40.Appenzeller-Herzog C., Ellgaard L. (2008) Biochim. Biophys. Acta 1783, 535–548 [DOI] [PubMed] [Google Scholar]
- 41.Rider C. C. (1997) Glycoconj. J. 14, 639–642 [DOI] [PubMed] [Google Scholar]
- 42.McKeating J. A., Thali M., Furman C., Karwowska S., Gorny M. K., Cordell J., Zolla-Pazner S., Sodroski J., Weiss R. A. (1992) Virology 190, 134–142 [DOI] [PubMed] [Google Scholar]
- 43.McKeating J. A., Shotton C., Cordell J., Graham S., Balfe P., Sullivan N., Charles M., Page M., Bolmstedt A., Olofsson S., Kayman S. C., Wu Z., Pinter A., Dean C., Sodrosky J., Weiss R. A. (1993) J. Virol. 67, 4932–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Billington J., Hickling T. P., Munro G. H., Halai C., Chung R., Dodson G. G., Daniels R. S. (2007) J. Virol. 81, 4604–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crublet E., Andrieu J. P., Vivès R. R., Lortat-Jacob H. (2008) J. Biol. Chem. 283, 15193–15200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwong P. D., Wyatt R., Robinson J., Sweet R. W., Sodroski J., Hendrickson W. A. (1998) Nature 393, 648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen B., Vogan E. M., Gong H., Skehel J. J., Wiley D. C., Harrison S. C. (2005) Nature 433, 834–841 [DOI] [PubMed] [Google Scholar]
- 48.Liu J., Bartesaghi A., Borgnia M. J., Sapiro G., Subramaniam S. (2008) Nature 455, 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J., Sen J., Rong L., Caffrey M. (2008) J. Biol. Chem. 283, 32644–32649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pancera M., Majeed S., Ban Y. E., Chen L., Huang C. C., Kong L., Kwon Y. D., Stuckey J., Zhou T., Robinson J. E., Schief W. R., Sodroski J., Wyatt R., Kwong P. D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1166–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]