Abstract
The integral endoplasmic reticulum (ER)-membrane protein VAP-B interacts with various lipid-transfer/binding proteins containing an FFAT motif through its N-terminal MSP domain. A genetic mutation within its MSP domain, P56S, was identified in familial forms of motor neuron diseases. This mutation induces the formation of insoluble VAP-B(P56S) protein aggregates by an unknown mechanism. In this study, we defined the structural requirements for VAP-B oligomerization and demonstrated their contribution for VAP-B(P56S) aggregation and neurotoxicity. We show that the oligomerization of VAP-B is mainly mediated by its coiled-coil domain and that the GXXXG dimerization motif within the transmembrane domain mediates transmembrane domains self-association but is insufficient to drive VAP-B oligomerization. We further show that the oligomerization of the wild-type VAP-B is independent of its MSP domain. However, we found that the P56S mutation induces conformational changes within the MSP domain and facilitates its propensity to aggregate by exposing hydrophobic patches to the solvent. These conformational changes have no direct effect on FFAT binding. Rather, they enhance VAP-B(P56S) oligomerization driven by the combined contributions of the coiled-coil and the transmembrane domains, thereby preventing accessibility to FFAT-binding site, facilitating the production of VAP-B(P56S)-insoluble aggregates and consequently its neurotoxicity. These results shed light on the mechanism by which VAP-B(P56S) aggregates are formed and induce familial motor neuron diseases.
Keywords: Amyotrophic Lateral Sclerosis (Lou Gehrig Disease), Endoplasmic Reticulum, Genetic Diseases, Lipid Transport, Protein Folding, Aggregates, FFAT Motif, Oligomerization, VAP-B
Introduction
Protein aggregation and inclusion body formation are common pathological features of many neurological disorders including Huntington disease, Alzheimer disease, Parkinson disease, and amyotrophic lateral sclerosis (ALS)3 (1, 2).
ALS is a fatal neurodegenerative disorder involving progressive loss of motor neurons in the cerebral cortex, brain stem, and spinal cord (3). Approximately 10% of the cases are familial, of which 20% are caused by dominantly inherited mutations in the Cu/Zn-superoxide dismutase (SOD) gene (4, 5). Recently, mutations in other genes, including the human VAP-B, have been identified in ALS patients (6–8).
The mutation in VAP-B substitutes a highly conserved proline residue at position 56 by a serine (P56S) and is associated with three forms of human motor neuron diseases: a late-onset spinal muscular atrophy, an atypical ALS type 8, and a typical severe ALS with rapid progression (7, 9). Atypical ALS type 8 is an autosomal dominant slowly progressive disorder characterized by fasciculation, cramps, and postural tremor. It is currently unknown how this P56S mutation induces motor neuron degeneration. Nevertheless, recent studies suggest that the VAP-B(P56S) mutant acts in a dominant fashion through dimerization with the wild-type protein and inactivating the heterodimer and that expression of the VAP-B(P56S) mutant induces the formation of insoluble protein aggregates (7, 10–15). Indeed, co-expression studies suggest that wild-type VAP-B is recruited to the P56S aggregates and that this recruitment leads either to loss of its unfolded protein response-associated activity (11) or to its ability to regulate lipid transport and biosynthesis (10).
The wild-type VAP-B is a type II integral ER-membrane protein that shares high sequence identity (63%) and a similar primary organization with VAP-A. The two VAPs consist of a large N-terminal region facing the cytoplasm and a hydrophobic C terminus that functions as a transmembrane domain (TMD) (16). The cytoplasmic region contains a conserved N-terminal MSP (major sperm protein) domain of 125 amino acids (aa), and a coiled-coil domain (CCD) of ∼50 aa. Proline 56 resides within a highly conserved sequence of 16 aa in the MSP domain in a cis-peptide bond conformation; therefore, the serine substitution induces its cis-to-trans isomerization (10).
VAPs have been implicated in regulation of a wide range of cellular processes (17), including neurotransmitter release (18), intracellular membrane trafficking (19–22), microtubules (MTs) organization and stability (16, 23) (2), the unfolded protein response (11, 15, 24), and lipid transport and metabolism (25–28). Increasing lines of evidence suggest that VAPs regulate cellular lipid homeostasis by direct interaction with lipid-transfer, -binding, or -sensing proteins through their FFAT (two phenylalanines in an acidic tract) motif (25–27, 29). This interaction recruits FFAT motif-containing proteins to the ER membranes and is thought to facilitate lipid transport between the ER, where various lipids are synthesized, to other cellular membranes.
We have previously shown using cross-linking experiments and sucrose-density gradient analysis that VAP-B undergoes dimerization and perhaps further oligomerization (2). Yet, the structural requirements for VAP-B oligomerization were analyzed either by in vitro binding studies, or by using truncated mutants of different structural domains. Accordingly, it was proposed that the MSP, CCD, and the TMD contribute to VAP oligomerization in the following order of importance: TMD > MSP > CCD (30). Further studies have demonstrated a weak interaction between a TMD-truncated mutant and a full-length wild-type VAP-B protein, which was enhanced by the P56S mutation (11, 12). However, this TMD-truncated mutant was localized to the cytosol rather than the ER membranes.
In this study, we used spectroscopic measurements and demonstrate that the P56S mutation induces conformational changes in the MSP domain, leading to an exposure of hydrophobic patches to the solvent and to MSP aggregation. Further studies on the wild-type VAP-B and its P56S mutant in intact cells revealed: (i) that the coiled-coil domain is critical for VAP-B oligomerization, (ii) that the GXXXG dimerization motif within the TMD is sufficient for driving oligomerization of the TMDs but not of the full-length protein, (iii) that the P56S mutation enhances VAP-B oligomerization driven by the coiled-coil and transmembrane domains, thereby impairing FFAT binding, and (iv) that the production of insoluble VAP-B(P56S) aggregates correlates with its neurotoxic effects. These results provide molecular insights into the mechanism underlying VAP-B(P56S) oligomerization and its implication in VAP-B(P56S) pathology.
EXPERIMENTAL PROCEDURES
Antibodies and Chemicals
Monoclonal anti-hemagglutinin (HA) and anti-Myc antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Alexa-488 donkey anti-mouse and anti-rabbit immunoglobulin Gs (IgGs) were purchased from Invitrogen (Carlsbad, CA). Cyanine (Cy)3-conjugated goat anti-rabbit and goat anti-mouse IgGs, as well as Cy5-conjugated goat anti-mouse IgG, were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). Monoclonal anti-α-tubulin and 3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) were purchased from Sigma. All other chemicals were from Sigma.
DNA Constructs
Point mutations within the human VAP-B cDNA were introduced by the QuikChange site-directed mutagenesis kit according to the manufacturer's instructions (Stratagene) and confirmed by DNA sequencing. The following VAP-B point mutants were established: P56S, G235I (I), G235/G239I (II), and K87/M89D (DD). The different VAP-B cDNAs, wild-type and mutants, were subcloned into pCMV-neo-Myc, pCAN-Myc, or pCMV-neo-HA mammalian expression vectors.
Truncated VAP-B mutants were produced by subcloning of corresponding PCR products into the above expression vectors or pEYFP-C1 harboring the A206K mutation (monomeric). The following sense and antisense oligonucleotide primers were used to establish the indicated truncated mutants: ΔMSP (aa 126–243): 5′-CGCGGATCCCCAGCAGAGAATGATAAACC-3′ and 5′-CGGGATATCCTACAAGGCAATCTTCCCAAT-3′; TMD (aa 201–243): 5′-GCCGGATCCACAGTGCAGAGCAACAGCCCC-3 and 5′-CGGGATATCCTACAAGGCAATCTTCCCAAT-3′; MSP (aa 1–125): 5′-CTCAGTGCATATGGCGAAGGTGGAGCAG-3′ and 5′-CGGGATATCCTACAATTCAAACACACATCTAAG-3′. The MSP domain was also subcloned into the pET-14b bacterial expression vector (Novagen) to produce a recombinant protein containing an N-terminal His6 tag.
The ΔCC (aa Δ159–196) mutant was constructed using three PCR steps. The first two PCR products correspond to the 5′ and 3′ DNA sequences upstream and downstream of the CCD. Each of these PCR products consists of complementary sequences flanking the boundaries of the CCD. Annealing of these two PCR products and further amplification with primers corresponding to the 5′ and 3′ sequence of the full-length VAP-B results in the production of the ΔCC mutant. The following set of sense and antisense primers have been used: for 5′ amplification: 5′-CCGGGATCCGCGAAGGTGGAGCA-3′; 5′-CTGTCTTCCTCATCCGACTCAGAGACTTAGACAC-3′; for 3′ amplification: 5′-GTGTCTAAGTCTCTGAGTCGGATGAGGAAGACAG-3′ and 5′-CGGGATATCCTACAAGGCAATCTTCCCAAT-3′. For full-length amplification: 5′-CCGGGATCCGCGAAGGTGGAGCA-3′ and 5′-CGGGATATCCTACAAGGCAATCTTCCCAAT-3′. The cDNA of Myc-tagged wild-type VAP-B, VAP-B(P56S), or the VAP-B(P56S)/ΔCC/II mutant was subcloned into pCSC-SP-PW-IRES/GFP lentiviral vector.
Cell Culture Transfection and Infection
HeLa and HEK293 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. The same medium containing 2 mm l-glutamine was used to grow Neuro2A cells. The cells were transfected with DNA constructs using the calcium phosphate method. Lentiviruses were produced in HEK293T packaging cells as follows: subconfluent (80%) HEK293T cells in 10-cm-diameter tissue culture plates were co-transfected with pMDL-g/p-RRE (HIV gag/pol) packaging plasmid, the rev expression plasmid pRSV-REV, and pCMV VSV-G envelope plasmid together with pCSC-SP-PW-IRES/GFP carrying the cDNA of either Myc-tagged wild-type VAP-B, the P56S mutant, or the P56S/ΔCC/II. Sixteen hours later, the medium was changed and viruses were collected 48 h later. Viral supernatants were filtered through 0.45-μm pore size filters and stored at −80 °C. Neuro2A cells were incubated in lentiviral supernatants containing 25% fresh complete medium and 8 μg/ml Polybrene for 24 h. The infection efficiency was assessed by immunofluorescence analysis and was found to be >90%.
Co-immunoprecipitation
The cells were lysed in lysis buffer (50 mm HEPES, pH 7.5, 150 mm NaCl, 10% glycerol, 1.5 mm MgCl2, 1 mm EGTA, 200 μm NaVO3, 20 mm NaF, 0.5% Triton X-100, and protease inhibitors) and centrifuged at 16,000 × g for 15 min at 4 °C. Proteins were immunoprecipitated from the supernatants using the indicated antibodies. The immunoprecipitates were washed three times with lysis buffer and boiled for 5 min in SDS sample buffer. The samples were then separated on SDS-PAGE and analyzed by Western blotting.
Chemical Cross-linking
Transfected HEK293 cells grown in 60-mm tissue culture plates were incubated with 1% formaldehyde (stock solution, 37%) for 10 min at room temperature. The formaldehyde was then quenched by 125 mm glycine for 5 min. The cells were washed three times with phosphate-buffered saline (PBS), lysed in RIPA buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, 50 mm NaF, 0.5 mm NaVO3, and protease inhibitors), and incubated on ice for 30 min and centrifuged as described below. For Fig. 4A, after washing with PBS, the cells were lysed in lysis buffer (50 mm HEPES, pH 7.5, 150 mm NaCl, 10% glycerol, 1.5 mm MgCl2, 1 mm EGTA, 0.5% Triton X-100, 200 μm NaVO3, 20 mm NaF, and protease inhibitors) containing 0.2% SDS, passed through a 21-gauge syringe ten times, boiled for 5 min, and passed again through the syringe as above. Cell debris was removed by centrifugation at 12,000 rpm for 10 min at 4 °C. The samples were then incubated in SDS-sample buffer and boiled either for 1 min or for 30 min to reverse cross-linking. The samples were separated on SDS-PAGE and analyzed by Western blotting.
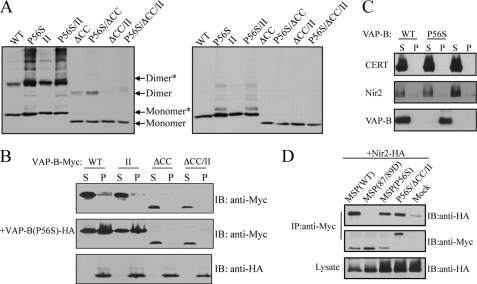
FIGURE 4.
The P56S mutation enhances VAP-B(P56S) dimerization thereby preventing FFAT binding. A, cross-linking of HeLa cells expressing the indicated Myc-tagged VAP-B proteins before (left panel) and after (right panel) cross-linker cleavage. The ability of each mutant to undergo dimerization was assessed by Western blotting using anti-Myc antibody. As shown, the dimerization of the P56S is stronger than the wild-type protein. Densitometric analysis (software: ImageJ) of four different experiments indicated that the ratio between dimer to monomer plus dimer in the WT protein is 47.8 ± 5.7%, whereas in the P56S mutant the ratio is 66.0 ± 4.7%. Likewise, the dimerization of the P56S/ΔCC mutant is stronger than the ΔCC mutant, yet the P56S/ΔCC/II is mainly monomeric. B, recruitment of VAP-B proteins into VAP-B(P56S) aggregates. The indicated Myc-tagged VAP-B proteins were transiently expressed in HeLa cells either alone or together with the HA-tagged VAP-B(P56S) mutant. The distribution of the different VAP-B proteins between the Triton X-100-soluble (S) and insoluble (P) fractions was assessed by Western blotting using the indicated antibodies. C, recruitment of FFAT motif-containing proteins into VAP-B(P56S) aggregates. HA-tagged Nir2 or ceramide transfer protein was transiently expressed in HeLa cells, either alone or together with Myc-tagged wild-type VAP-B or the P56S mutant. Distribution of Nir2 and ceramide transfer protein between the detergent-soluble (S) and insoluble (P) fractions was assessed by Western blotting using anti-HA antibody. D, the P56S mutation does not affect FFAT binding. The indicated Myc-tagged VAP-B mutants were transiently co-expressed with HA-Nir2, and their association was determined by immunoprecipitation with anti-Myc antibody followed by immunoblotting with anti-HA antibody. As shown the MSP domain of the P56S mutant interacts with Nir2 in a similar manner to that of the wild-type domain.
Immunofluorescence
Transfected HeLa cells were grown on coverslips, washed with PBS, and fixed either in methanol for 1 h at −20 °C, or in paraformaldehyde in PBS for 20 min at room temperature, and immunostained as described previously (31). The specimens were analyzed by a confocal laser scanning microscope (Zeiss 510, Carl Zeiss, Jena, Germany) using the 488, 543, 405, and 633 nm excitations for fluorescein, Cy3 epifluorescence, 4,6-diamidino-2-phenylindole, and Cy5, respectively. Images were processed using Adobe Photoshop (Adobe Systems, Mountain View, CA).
Cell Viability and Neurite Outgrowth Quantification
Neuro2A cells were seeded in 6-wells plate at 60% confluency (day 0) and on the following day (day 1) were infected with lentiviruses. 48 h following infection (day 3), the cells were split into 24-well plate for future MTT assays or seeded on polylysine-coated glass coverslips for neurite outgrowth quantification. The growth medium was then replaced by differentiation medium containing Dulbecco's modified Eagle's medium, antibiotics, 2 mm l-glutamine, and 0.2% bovine serum albumin (day 4). At days 5 and 6, 96 and 120 h following infection (24 and 48 h after serum deprivation), cell viability was measured by MTT assay as previously described (32). At day 5, the cells on the glass coverslips were fixed in 4% paraformaldehyde and co-immunostained with polyclonal anti-Myc antibody to detect protein expression and monoclonal anti-α-tubulin antibody to assess full-length neurites. 25 random areas were selected in blind, and z-stack images were acquired using a 63× oil objective. The infected cells were categorized into three different populations; round, cells with short neurites, and differentiated cells possessing long neurites; at least 3-fold longer of the cell body. Average values and standard deviations of two different experiments from different days were calculated.
Biophysical Methods
Fluorescence emission spectra were acquired with a Cary Varian Eclipse instrument, equipped with a Peltier temperature control set for all measurements at 20 °C. Intrinsic aromatic fluorescence emission spectra of proteins (0.05 mg/ml concentration in 50 mm KPi, pH 8, buffer) were acquired between 300 and 400 nm after excitation at 280 nm (excitation slit, 5 nm; emission slit, 10 nm). ANS binding assays were used as an extrinsic fluorescence probe to detect the hydrophobic exposure of the domain (33, 34). A 20-fold molar excess of 1-anilinonaphthalene-8-sulfonic acid (1,8-ANS) was added to the samples (0.05 mg/ml concentration in 50 mm KPi, pH 8, buffer), and ANS fluorescence emission enhancement was evaluated upon excitation at 370 nm, after 60-min incubation in the dark. CD measurements were recorded on a JASCO J-815 spectrometer equipped with a Peltier-thermostatted cell. Far UV-CD spectra of VAP-MSP (0.1 mg/ml protein in 50 mm KPi, pH 8, buffer) were recorded at 20 °C using a 1-mm path length cell (Hellma) and represent an average of ten scans. For CD analysis of thermal transitions, the temperature was raised from 20 to 70 °C at 0.5 °C/min, monitoring the spectroscopic signal at 222 nm (0.4 mg/ml protein in 50 mm KPi, pH 8, buffer). Curve fitting assuming a sigmoid two-state model allowed the determination of the melting temperature (Tm). Dynamic light scattering (DLS) measurements were carried out in a Malvern Zetasizer nano ZS instrument, equipped with a 4-megawatt He-Ne laser (632 nm). Samples were directly filtered through a 0.45-μm filter into a 3-mm light path quartz cuvette (Hellma) prior to analysis. Proteins (0.4 mg/ml in 50 mm KPi, pH 8, buffer) were analyzed at different temperatures, after 5-min incubation at each measuring temperature, between 20 to 70 °C. The operating procedure was set to 17 runs, each being averaged for 17 s. Data were analyzed by using DTS software (Malvern) in respect to the distribution of sizes by volume. Thermal aggregation profiles were determined plotting the intensity of aggregates determined by DLS (>100 nm diameter) as a function of temperature.
RESULTS
The GXXXG Motif in the TMD Is Insufficient to Drive VAP-B Oligomerization
The GXXXG motif, in which two glycine residues are separated by any three amino acids on a helical framework, is recognized as a “dimerization motif,” because it mediates the assembly of two transmembrane helices (35–37). Its presence within the TMD of VAP-B (235GXXXG239) prompted us to investigate its contribution to VAP-B oligomerization using mutagenesis studies and cross-linking experiments in intact cells. Replacing glycine 235 (Gly235) with isoleucine (Ile), either alone (G235I) or in combination with Gly239 (G235I/G239I; hereafter termed II), had no apparent effect on VAP-B homo-oligomerization, as determined by cross-linking experiments (Fig. 1A). These mutations also did not affect oligomerization of the VAP-B K87/M89D (DD) mutant, which fails to interact with the FFAT motif (38). The DD mutant, as well as the DD(I) and DD(II) VAP-B mutants, undergo dimerization similar to the wild-type protein. This implies that dimerization of VAP-B is dependent on intrinsic structural properties of VAP-B proteins, rather than its ability to interact with FFAT motif containing proteins.
FIGURE 1.
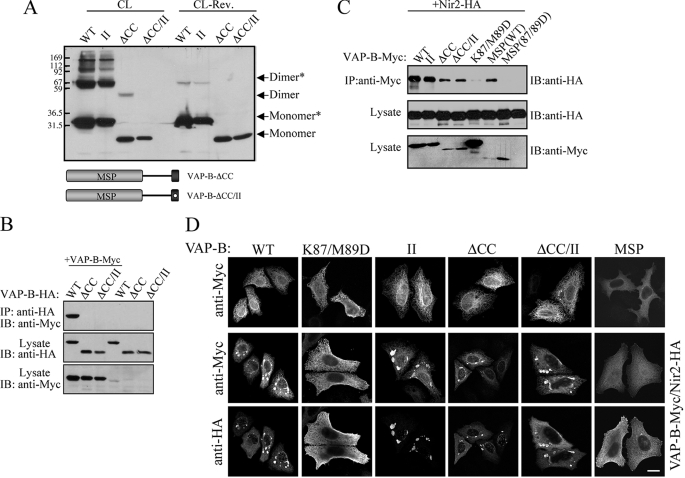
The GXXXG motif mediates self-assembly of the TMD but is insufficient to drive VAP-B oligomerization. A, the oligomerization of Myc-tagged wild-type and VAP-B mutants was assessed following cross-linking (CL) of HEK293 cells expressing the indicated VAP-B proteins, using Western blotting with anti-Myc antibody. Arrows mark the positions of the monomer, dimer, and tetramer. The following VAP-B proteins were examined: wild-type (WT), the G235I (I) mutant, the G235/239I (II) double mutant, the FFAT-binding mutant K87/M89D, the K87D/M89D/G235I; K87/M89D/(I) triple mutant, and the K87D/M89D/G235I/G239I; K87/M89D/(II) mutant. B, the indicated HA-tagged VAP-B proteins were expressed in HEK293 cells either alone or together with the indicated Myc-tagged VAP-B. The interaction between the VAP-B proteins was determined by immunoprecipitation with anti-Myc antibody followed by immunoblotting with anti-HA antibody. C, HEK293 cells expressing the TMD of VAP-B (WT) or the II mutant fused to monomeric YFP (A206K) were cross-linked by formaldehyde, and their oligomerization was assessed by Western blotting using anti-GFP antibody before and after cross-linker cleavage, CL and CL-Rev., respectively. Arrows mark the positions of the monomer and dimer. The weak band that appears above the dimer might represent a complex between YFP-TMD and endogenous VAP-B (or -A). Positions of prestained molecular mass markers, in kDa, are indicated on the left.
We next assessed the ability of the VAP-B(II) mutant to interact with the wild-type VAP-B (hereafter termed WT) or to undergo homo-dimerization by co-immunoprecipitation experiments. As shown in Fig. 1B, the G235/239I (II) mutations had no effect on either VAP-B(WT) or VAP-B(II) binding. These results suggest that the GXXXG motif is either dispensable or insufficient for driving VAP-B oligomerization. To distinguish between these two possibilities, we selectively expressed the TMD of the wild-type and the G235I/G239I (II) mutant fused to monomeric YFP in mammalian cells and assessed their dimerization by cross-linking studies. Only the TMD of VAP-B(WT) was found as a dimer (Fig. 1C), suggesting that the GXXXG motif mediates the assembly of the transmembrane helices in VAP-B and that other structural motifs are involved in oligomerization of the full-length VAP-B protein.
VAP-B Oligomerization Is Independent of the MSP Domain
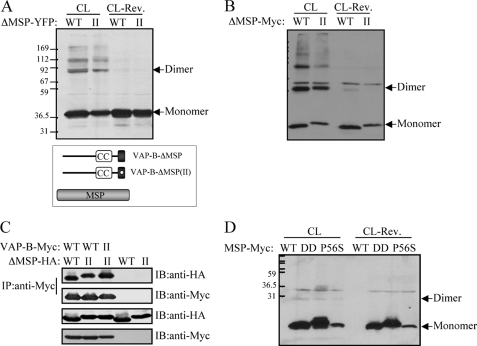
Previous studies using in vitro binding assays have suggested that the MSP domain of VAP-A and/or VAP-B contributes to VAP oligomerization (30). However, the MSP domain of VAP-A has been found as a monomer in solution (38). To examine the contribution of the MSP domain to VAP-B oligomerization, we expressed an MSP-truncated VAP-B mutant (ΔMSP) fused to either monomeric YFP (ΔMSP-YFP, Fig. 2A) or Myc tag (ΔMSP-Myc, Fig. 2B) in mammalian cells, and assessed their dimerization by cross-linking studies. Deletion of the MSP domain had no apparent effect on oligomerization of either the VAP-B(WT) or the VAP-B(II) mutant. Furthermore, co-immunoprecipitation experiments demonstrated that the ΔMSP mutants (VAP-B-ΔMSP and VAP-B-ΔMSP(II)) strongly interacted with either VAP-B(WT) or the VAP-B(II) mutant (Fig. 2C). These results suggest that the MSP has a minor effect on VAP-B oligomerization. This was further supported by cross-linking studies of cells expressing only the MSP domain of either the WT, or the DD or P56S mutants. In all cases, dimerization of the MSP domain was hardly detected (Fig. 2D), suggesting that, under these experimental conditions, the MSP domain of VAP-B does not contribute to VAP-B oligomerization.
FIGURE 2.
The MSP domain does not contribute to VAP-B oligomerization. The indicated VAP-B mutants lacking the MSP domain (ΔMSP) of either the WT or the VAP-B(II) mutant were expressed HEK293 cells. Their self-assembly was assessed following cross-linking by Western blotting using anti-GFP (A) or anti-Myc (B) antibodies. The position of the monomer and the dimer is marked by an arrow. C, the indicated HA-tagged ΔMSP mutants were expressed either alone or together with the indicated Myc-tagged VAP-B proteins, and their interaction was assessed by co-immunoprecipitation studies using anti-Myc antibody for immunoprecipitation and anti-HA antibody for immunoblotting. D, the Myc-tagged MSP domains of the wild-type VAP-B, the K87/M89D (DD) double mutant, or the P56S mutant were expressed in HEK293 cells. Their self-assembly was assessed following cross-linking (CL) by Western blotting using anti-Myc antibody. Cross-linker cleavage: CL-Rev. Note that the expression level of the MSP-P56S mutant is relatively low, implying that it is not as stable as the other mutants. The position of the monomer and the dimer is marked by an arrow. Positions of prestained molecular mass markers, in kDa, are indicated on the left.
The Coiled-coil Domain Has a Major Effect on VAP-B Oligomerization
The α-helical coiled-coil is one of the principal oligomerization motifs in proteins (39). We therefore examined its requirement for VAP-B dimerization using coiled-coil-truncated (ΔCC) mutants of VAP-B(WT) and VAP-B(II) proteins (ΔCC and ΔCC/II, respectively). The mutants were expressed in HEK293 cells, and their oligomerization was assessed following cross-linking. As shown, deletion of the coiled-coil domain from the WT protein gradually attenuated its dimerization, and apparently abolished dimerization of the VAP-B(II) mutant (Fig. 3A). These results suggest that the coiled-coil domain of VAP-B has a major effect on VAP-B oligomerization and together with the GXXXG motif, mediates VAP-B dimerization. These results were further confirmed by co-immunoprecipitation studies: both the ΔCC and the ΔCC/II mutants failed to interact with the VAP-B(WT) protein (Fig. 3B), consistent with their influence on VAP-B homodimerization (Fig. 3A). To ensure that these observations did not hinge upon unusual folding properties of the ΔCC mutants, we examined their ability to interact with the FFAT motif-containing protein Nir2 by co-immunoprecipitation. As shown in Fig. 3C, the ΔCC mutant, as well as the ΔCC/II double mutant, strongly interacted with Nir2. Furthermore, their co-expression with Nir2 in HeLa cells induced the formation of stacked ER-membrane arrays. These ER structures are typically formed by co-expressing VAP proteins with Nir2 (Fig. 3D) (2), or other FFAT motif-containing proteins (40–42).
FIGURE 3.
The coiled-coil domain is critical for VAP-B oligomerization. Deletion of the coiled-coil (ΔCC) domain markedly reduces VAP-B oligomerization and together with the II mutant apparently abolishes VAP-B homo- (A) and hetero-dimerization (B) as determined by cross-linking experiments (A) and coimmunoprecipitation studies (B), respectively. C, deletion of the coiled-coil domain has no effect on Nir2 binding. The indicated Myc-tagged VAP-B proteins were co-expressed with HA-tagged Nir2, and their association was assessed by immunoprecipitation using anti-Myc antibody following by immunoblotting using anti-HA antibody. D, the indicated Myc-tagged VAP-B proteins were expressed in HeLa cells either alone or together with HA-tagged Nir2. The localization of the indicated proteins was determined by indirect immunofluorescence analysis. Representative confocal images demonstrating the formation of stacked ER-membrane arrays upon co-expression with Nir2 are shown in the lower panels. Scale bar, 10 μm.
The P56S Mutation Enhances VAP-B Dimerization but Has No Effect on MSP-FFAT Interaction
We then examined the influence of the coiled-coil domain and the GXXXG motif on the VAP-B(P56S) mutant's oligomerization by cross-linking experiments. As shown in Fig. 4A, the P56S mutant underwent dimerization similar to VAP-B(WT). Densitometric analysis, however, indicated that its dimerization is more efficient than the wild-type protein. Deletion of the coiled-coil domain alone markedly reduced its dimerization, and together with the II mutations, apparently abolished it. Nevertheless, the P56S/ΔCC mutant exhibited stronger dimerization than the ΔCC mutant. Its dimerization was reduced by 22.0 ± 7.6% as compared with the VAP-B(P56S) mutant, whereas the dimerization of the ΔCC mutant was reduced by 58.8 ± 7.2% compared with the wild type, as determined by quantitative densitometric analysis of four different cross-linking experiments. These results suggest that the P56S mutation somehow enhances VAP-B dimerization. It is worth mentioning that the P56S mutation enhanced oligomerization of the full-length protein, but not that of an isolated MSP domain expressed in mammalian cells (Fig. 2D). This suggests that close proximity of adjacent MSP-P56S domains facilitates their dimerization and is consistent with the finding that the P56S/ΔCC/II mutant failed to dimerize. Taken together, these observations suggest that the coiled-coil domain and to a lesser extend the TMD contribute to the oligomerization of both VAP-B(WT) and the P56S mutant. Yet, the MSP domain enhances oligomerization of the P56S mutant.
To further demonstrate the role of these domains for VAP-B(P56S) oligomerization, we examined the ability of VAP-B(P56S) to interact with different VAP-B mutants by assessing their recruitment into insoluble VAP-B(P56S) aggregates. Expression of the P56S mutant in mammalian cells induces the formation of insoluble protein aggregates (supplemental Fig. S1), and its co-expression with VAP-B(WT) resulted in redistribution of the latter from the soluble to insoluble fraction (Fig. 4B). These results suggest that heterodimerization between the WT and the P56S mutant induces the recruitment of VAP-B(WT) protein into P56S-associated aggregates, consistent with previous reports (10–12). Similar results were obtained by co-expressing the VAP-B(P56S) with the VAP-B(II) double mutant, but not with the ΔCC or the ΔCC/II mutants (Fig. 4C). This demonstrates the importance of the coiled-coil domain for VAP-B(P56S) heterodimerization and is consistent with the homodimerization results shown in Figs. 3 and 4A.
We employed the same assay to examine whether FFAT motif-containing proteins are also recruited into the insoluble VAP-B(P56S) aggregates. As shown in Fig. 4C, solubilization of both Nir2 and ceramide transfer protein was unaffected by co-expression with VAP-B(P56S). These results suggested that these FFAT motif-containing proteins are unable to interact with the P56S mutant and are consistent with previous co-immunoprecipitation studies of other FFAT proteins (10). Hence, it could be that the enhanced oligomerization of the MSP domain in the VAP-B(P56S) mutant prevents access to the FFAT-binding site, thereby impairing FFAT binding. Alternatively, the P56S mutation may affect the conformation of the MSP domain and consequently preventing the binding of FFAT proteins. To distinguish between these two possibilities, we examined whether the MSP-P56S domain interacts with Nir2 using co-immunoprecipitation experiments. As shown, the interaction between the MSP-P56S mutant and Nir2 was similar to that of the MSP-WT (Fig. 4D). Furthermore, the P56S/ΔCC/II mutant also interacted with Nir2. These results suggest that the P56S mutation per se does not affect FFAT binding. Rather, the mutation enhances the oligomerization of the MSP domain in the context of the full-length protein, thereby preventing the binding of FFAT proteins.
The P56S Mutation Induces the Exposure of Hydrophobic Patches, Thereby Facilitating MSP Oligomerization
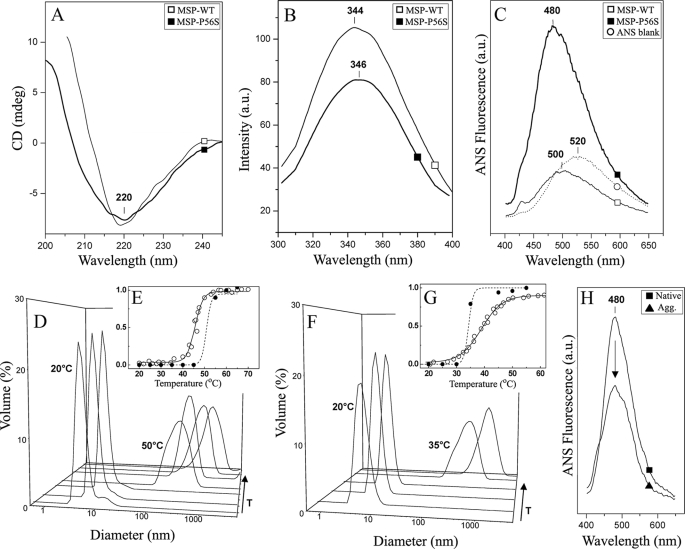
To better understand how the P56S mutation enhances oligomerization of VAP-B(P56S), we characterized the folding properties of recombinant purified MSP domains of the WT and P56S mutant, using different biophysical and spectroscopic methods. Far-UV CD spectroscopy showed spectra typical of a β-sheet-rich protein with a prominent single negative band at 220 nm (43), suggesting that the P56S mutation has no severe impact on the MSP fold (Fig. 5A). This spectroscopic fingerprint agrees with the available structural data for the MSP domain, which comprises seven strands and three α-helices in a immunoglobulin-like β-sandwich topology (38). The fluorescence emission properties of the single Trp106 residue denoted subtle effects of the mutation on tertiary contacts (Fig. 5B). As shown, the emission maxima of the WT and the P56S MSP domains were essentially identical, denoting similar solvent accessibility. However, the increased emission intensity of MSP-P56S suggests a modification of the quenching environment around aromatic residues, compatible with a modified tertiary structure (Fig. 5B). To further evaluate the effect of the P56S mutation on MSP structure, we examined the hydrophobicity of the MSP domain using the fluorophore 1,8-ANS as a reporter for hydrophobic moieties (44, 45). This dye has an emission peak at 520 nm in aqueous solution (Fig. 5C), which may undergo a blue shift down to 480 nm, and a substantial increase in emission, upon interaction with surface hydrophobic patches (44, 45). Incubation of 1,8-ANS with MSP-WT results in a shift of the emission maxima to 500 nm, but no increase in intensity (Fig. 5C). In contrast, a 4-fold increase in the fluorescence intensity and a blue shift to 480 nm were observed upon incubation of 1,8-ANS with MSP-P56S (Fig. 5C). These data indicated that the P56S mutation induces conformational changes within the MSP domain that eventually lead to an exposure of hydrophobic patches accessible to the solvent. Such exposure may affect the hydrophobic interaction network of the domain and consequently its structural packing. Indeed, DLS analysis indicates that the hydrodynamic diameter of MSP-P56S domain (6.2 nm) was 20% higher than that of the MSP-WT (5.1 nm), thereby demonstrating its more expanded conformation. Because the increased exposure of hydrophobic regions could play a role in unspecific interactions, DLS was used to analyze the aggregation propensities of the two MSP variants at different temperatures. As shown in Fig. 5D, the MSP-WT was found as a monomer at a quite wide range of temperatures (20–47 °C). However, aggregation occurred at 49 °C and above, when the protein was already unfolded (Tm(MSP-WT) = 47 °C, Fig. 5E). In contrast, at 35 °C, the average size distribution of MSP-P56S likely represented a population of aggregated proteins (Fig. 5F); under these conditions most of the protein was still folded, as the melting temperature was higher than the aggregation temperature (Tm(MSP-P56S) = 39 °C, Fig. 5G). These results clearly show that the MSP-P56S has an increased propensity to aggregate and that the onset of this process occurs or even starting from native conditions and physiological temperature (Fig. 5G). We then examined if binding of 1,8-ANS would be modified after induction of aggregation events of MSP-P56S at different temperatures. Our results showed that, upon aggregation, the emission was markedly reduced (Fig. 5H). This demonstrated that hydrophobic moieties that were being detected by 1,8-ANS became less accessible to the dye under these conditions, suggesting a possible involvement of these hydrophobic patches in the formation of aggregates. Overall, these results suggest that the P56S mutation induces conformational changes within the MSP domain, resulting in the exposure of hydrophobic surfaces to the solvent. Such hydrophobic patches are likely to facilitate protein-protein interactions in aqueous solution (46, 47), thereby leading to the formation of aggregates under physiological conditions.
FIGURE 5.
Effects of the P56S mutation on the MSP domain. Purified MSP-WT (open square) and MSP-P56S (closed square) were analyzed using different biophysical methodologies: A, far-UV CD (0.1 mg/ml of protein). B, Trp-emission fluorescence (0.05 mg/ml protein). C, 1,8-ANS fluorescence emission (0.05 mg/ml protein). D, DLS distribution of MSP-WT at different temperatures (20, 25, 35, 50, 55, 60, and 65 °C, [protein] = 0.4 mg/ml). E, MSP-WT thermal unfolding curve determined by CD (open circles, Tm = 47 °C) and aggregation profile determined from DLS (closed circles, Tagg > 50 °C). F, DLS distribution of MSP-P56S at different temperatures (25, 25, 30, 35, and 45 °C, [protein] = 0.4 mg/ml). G, MSP-P56S thermal unfolding curve determined by CD (open circles, Tm = 39 °C) and aggregation profile determined from DLS (closed circles, Tagg > 35 °C). H, 1,8-ANS fluorescence emission spectra of native (solid square) and thermally aggregated (solid triangle) MSP-P56S. The spectra were recorded at 20 °C, and the arrow denotes the decrease in the emission of the 1,8-ANS fluorescence observed in the aggregated sample. (0.05 mg/ml of protein). In E and G, the y axis is the fractional change in respect to protein unfolding and aggregation; the solid line is a fit of the unfolding curve to a two-state model, whereas the dashed line on the thermal aggregation profiles is merely represented to guide the eye. See “Experimental Procedures” for detailed conditions.
Oligomerization of VAP-B(P56S) Is Essential for the Production of Insoluble Protein Aggregates
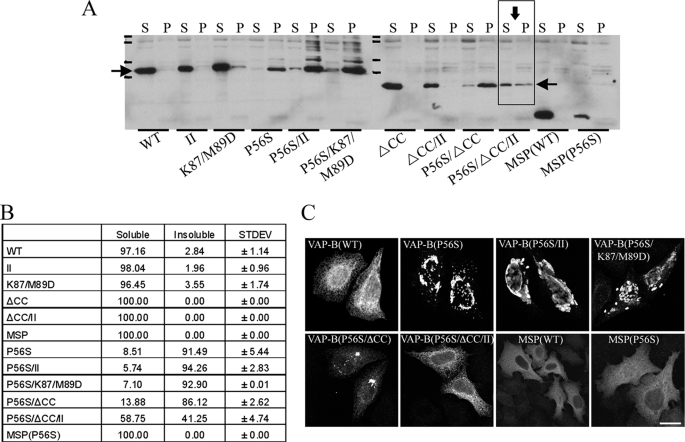
Thus far, we showed that the P56S mutation induces conformational changes of the MSP domain and enhances its oligomerization, and that the TMD and more profoundly the coiled-coil domain are essential for VAP-B(P56S) dimerization. We then examined the contribution of VAP-B(P56S) dimerization to the production of insoluble protein aggregates. For this purpose, we examined the influence of the coiled-coil domain and the GXXXG motif on VAP-B(P56S) solubility using Triton X-100 as the detergent. The indicated VAP-B mutants (Fig. 6A) were transiently expressed in HeLa cells, and their distribution between the Triton X-100-soluble and insoluble fractions was assessed by Western blotting. Solubilization of VAP-B(P56S) was markedly increased by deletion of its coiled-coil domain and concomitant mutation of its GXXXG motif (Fig. 6A). These observations were further confirmed by immunofluorescence analysis. As shown in Fig. 6C, the G235/239I (II) and the K87/M89D (DD) mutations had no detectable effect on the production of VAP-B(P56S)-associated aggregates. Deletion of the coiled-coil domain, however, markedly reduced aggregate size, rendering them barely detectable in cells expressing the VAP-B(P56S)/ΔCC/II mutant. Overall, these results indicated that the oligomerization of VAP-B(P56S) is mediated mainly by the coiled-coil domain and that the GXXXG also contributes to VAP-B(P56S) oligomerization and consequently for the production of insoluble protein aggregates.
FIGURE 6.
Oligomerization of VAP-B enhances the production of VAP-B(P56S)-insoluble aggregates. A, the indicated Myc-tagged VAP-B proteins were expressed in HeLa cells and their distribution between the Triton X-100-soluble (S) and insoluble (P) fractions was assessed by Western blotting using anti-Myc antibody. Densitometric analysis (software: ImageJ) of the Western blot signals was used to estimate the relative distribution of each mutant between the soluble and insoluble fractions (B). C, the P56S/ΔCC/II mutant did not form visible aggregates. The indicated mutants were expressed in HeLa cells, fixed in 4% paraformaldehyde, immunostained with anti-Myc antibody, and analyzed by confocal microscope. Bar, 10 μm.
Insoluble Aggregates of VAP-B(P56S) Protein Induce Neurotoxic Effects
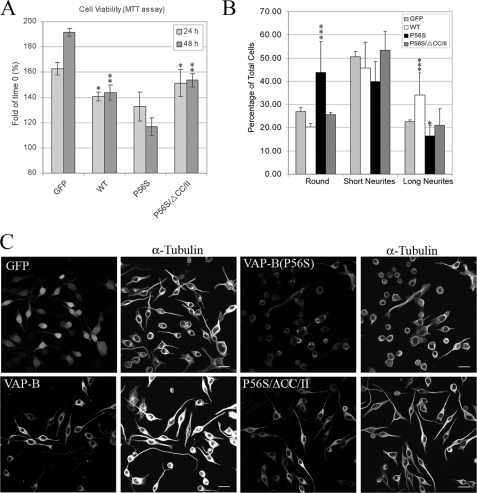
The production of insoluble VAP-B(P56S) protein aggregates was proposed to induce neurotoxicity and eventually neuronal cell death. To demonstrate the correlation between aggregate formation and cell viability, we compared the effect of VAP-B(WT), the P56S mutant, and the P56S/ΔCC/II mutant on Neuro2a (N2a) cell viability using the MTT assay. N2a cells were infected with lentiviruses encoding the indicated VAP-B proteins or with GFP as a control. After 72 h, the cells were incubated with serum-free media to induce neuronal differentiation, and cell viability was assessed 24 or 48 h later. We observed high infection efficiency (∼95%) of the different lentiviruses and comparable protein expression levels (supplemental Fig. S2). Nevertheless, the VAP-B(P56S) mutant reduced N2a cell viability as compared with the WT and the P56S/ΔCC/II mutant (Fig. 7A). Furthermore, the VAP-B(P56S) mutant markedly attenuated neurite outgrowth of N2a cells after serum deprivation (Fig. 7, B and C). Approximately 16% of the cells expressing the P56S mutant underwent differentiation and grew long neurites, exceeding three cell-body diameters, 24 h after serum deprivation. However, most of them (43%) exhibited a rounded morphology. In contrast, only 20 or 25% of the cells expressing the WT or the VAP-B(P56S)/ΔCC/II mutant, respectively, exhibited a rounded morphology. Such inhibition of neurite outgrowth might correspond to axonal degeneration in ALS patients, and has been previously observed for other disease-associated proteins, including SOD1 and Huntingtin mutants (48, 49). Taken together, these results indicate that the P56S mutation markedly affects N2a cell viability and their ability to elongate neurites. Furthermore, these effects on cell viability and neurite outgrowth could barely be detected in P56S/ΔCC/II-expressing cells, demonstrating a direct correlation between VAP-B(P56S) oligomerization, aggregates formation, and its neurotoxic effects.
FIGURE 7.
Disruption of VAP-B(P56S) dimerization attenuates its neurotoxic effects. A, the VAP-B(P56S) mutant affects cell viability. Neuro2a cells were infected with lentivirus encoding the indicated proteins. After 72 h, the cells were incubated in serum-free medium (0.2% bovine serum albumin) to induce neuronal differentiation. MTT assays were performed at time 0, and at 24 and 48 h following serum withdrawal. The relative MTT values obtained at 24 or 48 h compared with time 0 are shown. Data are mean ± S.D. values of triplicates from two independent experiments. Statistical significance was assessed by Student's t test. **, p < 0.0005 and *, p < 0.005, respectively, obtained using Student's t test of comparing P56S mutant versus either the WT protein or the P56S/ΔCC/II mutant for each time point. B, effect of VAP-B(P56S) mutant on neurite outgrowth of Neuro2a cells. Neuro2A cells were infected as above and seeded on polylysine-coated glass coverslips. 72 h after infection, the cells were incubated in serum-free media for 24 h, fixed in 4% paraformaldehyde, and immunostained with anti-Myc antibody (for VAP-B proteins) and with α-tubulin to assess neurite length. The cells were then analyzed by confocal microscopy. Z-stack images of 25 random areas were acquired in blind, and cell morphology was classified into three different categories: round, undifferentiated cells possessing short neurites, and differentiated cells exhibiting long neurites (3-fold of cell body). Approximately 500 cells of each infection were analyzed. The relative distribution of the three phenotypes obtained from two different experiments is shown ± S.D. Representative confocal images of these infections are shown in C. Scale bar, 20 μm. Statistical analysis was performed as above. ***, p values < 0.0005 and *, p < 0.05, respectively, comparing each mutant in each group with GFP.
DISCUSSION
The production of insoluble protein aggregates is a hallmark of many neurodegenerative diseases, such as Parkinson disease, Alzheimer disease, Huntington disease, and ALS. In such diseases, large protein aggregates, known as inclusion bodies, are accumulated within the cells or in extracellular plaques (1). These aggregates frequently contain an abnormal conformation of the disease protein and are often formed through multiple steps of protein conformational changes and accretion (50).
The finding that the VAP-B(P56S) mutant is associated with several familial motor neuron diseases (7, 9, 51), concomitant with its ability to form insoluble protein aggregates, suggests that VAP-B(P56S) pathogenesis shares common features with other aggregation diseases. Nevertheless, the mechanisms by which insoluble VAP-B(P56S) aggregates are formed remain largely unknown.
In this study, we show that oligomerization of VAP-B(P56S) is essential for the production of insoluble protein aggregates and consequently, for VAP-B(P56S) neurotoxicity. By systematic characterization of VAP-B oligomerization in intact cells using cross-linking and co-immunoprecipitation experiments and various VAP-B mutants, we found that the coiled-coil domain is critical for both VAP-B and VAP-B(P56S) oligomerization (Figs. 3A, 3B, and 4A). The TMD also contributes to VAP-B oligomerization through its GXXXG motif (Fig. 1). The MSP domain, on the other hand, had only a minor effect on oligomerization of the wild-type VAP-B (Fig. 2). These results suggest that the TMD does not play a major role in VAP-B oligomerization as was previously proposed (11, 12, 30). Rather, the coiled-coil domain is crucial. The discrepancy between our results and previous reports could be related to the fact that previous studies used truncated TMD mutants, whereas our characterization employed the GXXXG mutant (VAP-B/II), which retains its ER localization. Furthermore, our entire analysis, in contrast to previous studies, avoided using glutathione S-transferase or GFP fusion proteins, which tend to dimerize. We rather used monomeric GFP. Hence, we propose that the different VAP-B domains contribute to VAP-B oligomerization in the following order of importance: CCD > TMD > MSP.
In the case of the VAP-B(P56S) mutant, we clearly observed enhancement of VAP-B(P56S)/ΔCC oligomerization as compared with the VAP-B/ΔCC mutant (Fig. 4A). This suggests that somehow the P56S mutation enhances oligomerization of the full-length protein. Insight into the underlying mechanism of this enhancement was provided by biophysical characterization of an isolated recombinant MSP-P56S domain. This analysis suggested that the P56S mutation induces conformational changes in the domain, leading to the exposure of hydrophobic patches and consequently to MSP aggregation (Fig. 5). However, we could not observe oligomerization of the MSP-P56S domain expressed in mammalian cells (Fig. 2), suggesting that dimerization of this domain will occur only if the two domains are found in close proximity. Such proximity is likely obtained at high protein concentrations and temperatures, and was therefore detected with the recombinant bacterially expressed MSP-P56S domain at temperatures above 35 °C (Fig. 5F).
In the context of the full-length protein expressed in mammalian cells, the proximity between adjacent MSP-P56S domains is accomplished through VAP-B oligomerization driven by the coiled-coil and transmembrane domains. Accordingly, we propose that the coiled-coil and transmembrane domains induce zippering of adjacent MSP-P56S domains possessing exposed hydrophobic patches, thereby sealing the binding of FFAT motif-containing proteins. Consistent with this model, we show that both the MSP-P56S domain and the P56S/ΔCC/II mutant retain their ability to interact with FFAT proteins (Fig. 4D). These results suggest that the P56S mutation by itself has no effect on FFAT binding.
However, the P56S mutation induces conformational changes within the MSP domain and causes alterations in its tertiary structure, a decrease in its overall compactness, and destabilization of the hydrophobic core. The exposure of hydrophobic patches to the solvent appears to enhance the mutated domain's propensity to oligomerize. This oligomerization of an isolated MSP-P56S domain in solution may represent an undirected aggregation driven by hydrophobic interactions. However, when the MSP domain is embedded in the full-length protein and anchored to the ER membrane its ability to undergo self-association driven by hydrophobic interactions is probably more limited and directed by a dimerization-driven mechanism mediated by the coiled-coil and the transmembrane domains. In both circumstances, however, the exposure of hydrophobic patches in MSP-P56S facilitates aberrant oligomerization.
Consistent with our results, changes in surface hydrophobicity have also been observed for SOD1 mutants, and have been proposed to affect SOD1 neurotoxicity by facilitating abnormal filamentous self-association or abnormal interactions with other cellular constituents (52, 53). SOD1 mutants display an abnormal conformation, and here we show, for the first time, that the P56S mutation also causes an abnormal protein conformation, thus sharing a common feature with many conformational neurodegenerative diseases (54, 55).
Although VAP-B(P56S) is not organized into the characteristic fibrillar aggregates known as amyloids (1, 10), its neurotoxicity has been demonstrated in previous studies (10). Here we show that there is a tight correlation between VAP-B(P56S) oligomerization, insoluble aggregate formation, and neurotoxicity (Figs. 4, 6, and 7). The mechanisms by which VAP-B(P56S) mediates its neurotoxic effects are currently unknown. The formation of insoluble aggregates might induce loss-of-function of VAP-B(WT) protein, gain of toxic function, or both. Indeed, previous studies have proposed that the recruitment of WT VAP proteins (A and B) into VAP-B(P56S)-associated aggregates impairs their unfolded protein response-associated activity or affects lipid biosynthesis, thereby inducing neuronal cell death. Here we provide evidence that the P56S mutation induces conformational changes in the MSP domain (Fig. 5), and we therefore propose that gain of toxic functions also plays an important role in VAP-B(P56S) pathology. Gain of toxic functions has also been attributed to the pathology of SOD1 mutants. Thus, multiple mechanisms might contribute to VAP-B(P56S) neurotoxicity involving diverse pathogenic processes, such as impaired lipid transport and metabolism, protein aggregation, mitochondrial dysfunction, and impaired axonal transport. Further studies on VAP-B(P56S) neurotoxicity are expected to provide further mechanistic insights into its pathogenesis.
Supplementary Material
This work was supported by the Minerva foundation with funding from the Federal German Ministry for Education and Research and by Israel Science Foundation Grant 548/08.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- ALS
- amyotrophic lateral sclerosis
- fALS
- familial ALS
- SOD
- Cu/Zn-superoxide dismutase
- TMD
- transmembrane domain
- CCD
- coiled-coil domain
- UPR
- unfolded protein response
- FFAT
- two phenylalanines in an acidic tract
- HA
- hemagglutinin
- Cy
- Cyanine
- MTT
- 3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyl tetrazolium bromide
- PBS
- phosphate-buffered saline
- DLS
- dynamic light scattering
- 1,8-ANS
- 1-anilinonaphthalene-8-sulfonic acid
- N2a
- Neuro2a
- MSP
- major sperm protein
- aa
- amino acid(s)
- CMV
- cytomegalovirus
- GFP
- green fluorescent protein.
REFERENCES
- 1.Ross C. A., Poirier M. A. (2004) Nat. Med. 10, (suppl.) S10–S17 [DOI] [PubMed] [Google Scholar]
- 2.Amarilio R., Ramachandran S., Sabanay H., Lev S. (2005) J. Biol. Chem. 280, 5934–5944 [DOI] [PubMed] [Google Scholar]
- 3.Brown R. H., Jr., Robberecht W. (2001) Semin. Neurol. 21, 131–139 [DOI] [PubMed] [Google Scholar]
- 4.Orrell R. W. (2000) Neuromuscul. Disord. 10, 63–68 [DOI] [PubMed] [Google Scholar]
- 5.Gros-Louis F., Gaspar C., Rouleau G. A. (2006) Biochim. Biophys. Acta 1762, 956–972 [DOI] [PubMed] [Google Scholar]
- 6.Pasinelli P., Brown R. H. (2006) Nat. Rev. Neurosci. 7, 710–723 [DOI] [PubMed] [Google Scholar]
- 7.Nishimura A. L., Mitne-Neto M., Silva H. C., Richieri-Costa A., Middleton S., Cascio D., Kok F., Oliveira J. R., Gillingwater T., Webb J., Skehel P., Zatz M. (2004) Am. J. Hum. Genet. 75, 822–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw P. J. (2005) J. Neurol. Neurosurg. Psychiatry 76, 1046–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishimura A. L., Al-Chalabi A., Zatz M. (2005) Hum. Genet. 118, 499–500 [DOI] [PubMed] [Google Scholar]
- 10.Teuling E., Ahmed S., Haasdijk E., Demmers J., Steinmetz M. O., Akhmanova A., Jaarsma D., Hoogenraad C. C. (2007) J. Neurosci. 27, 9801–9815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanekura K., Nishimoto I., Aiso S., Matsuoka M. (2006) J. Biol. Chem. 281, 30223–30233 [DOI] [PubMed] [Google Scholar]
- 12.Suzuki H., Kanekura K., Levine T. P., Kohno K., Olkkonen V. M., Aiso S., Matsuoka M. (2009) J. Neurochem. 108, 973–985 [DOI] [PubMed] [Google Scholar]
- 13.Tsuda H., Han S. M., Yang Y., Tong C., Lin Y. Q., Mohan K., Haueter C., Zoghbi A., Harati Y., Kwan J., Miller M. A., Bellen H. J. (2008) Cell 133, 963–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ratnaparkhi A., Lawless G. M., Schweizer F. E., Golshani P., Jackson G. R. (2008) PLoS ONE 3, e2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gkogkas C., Middleton S., Kremer A. M., Wardrope C., Hannah M., Gillingwater T. H., Skehel P. (2008) Hum. Mol. Genet. 17, 1517–1526 [DOI] [PubMed] [Google Scholar]
- 16.Skehel P. A., Fabian-Fine R., Kandel E. R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1101–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lev S., Ben Halevy D., Peretti D., Dahan N. (2008) Trends Cell Biol. 18, 282–290 [DOI] [PubMed] [Google Scholar]
- 18.Skehel P. A., Martin K. C., Kandel E. R., Bartsch D. (1995) Science 269, 1580–1583 [DOI] [PubMed] [Google Scholar]
- 19.Soussan L., Burakov D., Daniels M. P., Toister-Achituv M., Porat A., Yarden Y., Elazar Z. (1999) J. Cell Biol. 146, 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weir M. L., Klip A., Trimble W. S. (1998) Biochem. J. 333, 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weir M. L., Xie H., Klip A., Trimble W. S. (2001) Biochem. Biophys. Res. Commun. 286, 616–621 [DOI] [PubMed] [Google Scholar]
- 22.Foster L. J., Weir M. L., Lim D. Y., Liu Z., Trimble W. S., Klip A. (2000) Traffic 1, 512–521 [DOI] [PubMed] [Google Scholar]
- 23.Pennetta G., Hiesinger P. R., Fabian-Fine R., Meinertzhagen I. A., Bellen H. J. (2002) Neuron 35, 291–306 [DOI] [PubMed] [Google Scholar]
- 24.Brickner J. H., Walter P. (2004) PLoS Biol. 2, e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peretti D., Dahan N., Shimoni E., Hirschberg K., Lev S. (2008) Mol. Biol. Cell 19, 3871–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perry R. J., Ridgway N. D. (2006) Mol. Biol. Cell 17, 2604–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawano M., Kumagai K., Nishijima M., Hanada K. (2006) J. Biol. Chem. 281, 30279–30288 [DOI] [PubMed] [Google Scholar]
- 28.Loewen C. J., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., Henry S. A., Levine T. P. (2004) Science 304, 1644–1647 [DOI] [PubMed] [Google Scholar]
- 29.Loewen C. J., Roy A., Levine T. P. (2003) EMBO J. 22, 2025–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimura Y., Hayashi M., Inada H., Tanaka T. (1999) Biochem. Biophys. Res. Commun. 254, 21–26 [DOI] [PubMed] [Google Scholar]
- 31.Litvak V., Dahan N., Ramachandran S., Sabanay H., Lev S. (2005) Nat. Cell Biol. 7, 225–234 [DOI] [PubMed] [Google Scholar]
- 32.Mosmann T. (1983) J. Immunol. Methods 65, 55–63 [DOI] [PubMed] [Google Scholar]
- 33.McClure W. O., Edelman G. M. (1966) Biochemistry 5, 1908–1919 [DOI] [PubMed] [Google Scholar]
- 34.Stryer L. (1965) J. Mol. Biol. 13, 482–495 [DOI] [PubMed] [Google Scholar]
- 35.Russ W. P., Engelman D. M. (2000) J. Mol. Biol. 296, 911–919 [DOI] [PubMed] [Google Scholar]
- 36.Brosig B., Langosch D. (1998) Protein Sci. 7, 1052–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senes A., Engel D. E., DeGrado W. F. (2004) Curr. Opin. Struct. Biol. 14, 465–479 [DOI] [PubMed] [Google Scholar]
- 38.Kaiser S. E., Brickner J. H., Reilein A. R., Fenn T. D., Walter P., Brunger A. T. (2005) Structure 13, 1035–1045 [DOI] [PubMed] [Google Scholar]
- 39.Burkhard P., Stetefeld J., Strelkov S. V. (2001) Trends Cell Biol. 11, 82–88 [DOI] [PubMed] [Google Scholar]
- 40.Lehto M., Hynynen R., Karjalainen K., Kuismanen E., Hyvärinen K., Olkkonen V. M. (2005) Exp. Cell Res. 310, 445–462 [DOI] [PubMed] [Google Scholar]
- 41.Wyles J. P., Ridgway N. D. (2004) Exp. Cell Res. 297, 533–547 [DOI] [PubMed] [Google Scholar]
- 42.Wyles J. P., McMaster C. R., Ridgway N. D. (2002) J. Biol. Chem. 277, 29908–29918 [DOI] [PubMed] [Google Scholar]
- 43.Martin S. R., Bayley P. M. (2002) Methods Mol. Biol. 173, 43–55 [DOI] [PubMed] [Google Scholar]
- 44.Cardamone M., Puri N. K. (1992) Biochem. J. 282, 589–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leal S. S., Gomes C. M. (2007) Proteins 68, 606–616 [DOI] [PubMed] [Google Scholar]
- 46.Kundu B., Guptasarma P. (2002) Biochem. Biophys. Res. Commun. 293, 572–577 [DOI] [PubMed] [Google Scholar]
- 47.Vetri V., Militello V. (2005) Biophys. Chem. 113, 83–91 [DOI] [PubMed] [Google Scholar]
- 48.Takeuchi H., Kobayashi Y., Yoshihara T., Niwa J., Doyu M., Ohtsuka K., Sobue G. (2002) Brain Res. 949, 11–22 [DOI] [PubMed] [Google Scholar]
- 49.Ye C., Zhang Y., Wang W., Wang J., Li H. (2008) Neurosci. Lett. 442, 63–68 [DOI] [PubMed] [Google Scholar]
- 50.Ross C. A., Poirier M. A. (2005) Nat. Rev. Mol. Cell Biol. 6, 891–898 [DOI] [PubMed] [Google Scholar]
- 51.Marques V. D., Barreira A. A., Davis M. B., Abou-Sleiman P. M., Silva W. A., Jr., Zago M. A., Sobreira C., Fazan V., Marques W., Jr. (2006) Muscle Nerve 34, 731–739 [DOI] [PubMed] [Google Scholar]
- 52.Tiwari A., Xu Z., Hayward L. J. (2005) J. Biol. Chem. 280, 29771–29779 [DOI] [PubMed] [Google Scholar]
- 53.Tiwari A., Liba A., Sohn S. H., Seetharaman S. V., Bilsel O., Matthews C. R., Hart P. J., Valentine J. S., Hayward L. J. (2009) J. Biol. Chem. 284, 27746–27758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Pril R., Fischer D. F., van Leeuwen F. W. (2006) Neurobiol. Aging 27, 515–523 [DOI] [PubMed] [Google Scholar]
- 55.Thompson A. J., Barrow C. J. (2002) Curr. Med. Chem. 9, 1751–1762 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.