Abstract
Fibroblast growth factor 1 (FGF1) has been suggested to have an important role in cell growth, proliferation, and neurogenesis. Human FGF1 gene 1B promoter (−540 to +31)-driven green fluorescence (F1BGFP) has been shown to monitor endogenous FGF1 expression. F1BGFP could also be used to isolate neural stem/progenitor cells from embryonic, neonatal, and adult mouse brains or to isolate glioblastoma stem cells (GBM-SCs) from human glioblastoma tissues. Here, we present evidence that transcription factor RFX1 could bind the 18-bp cis-elements (−484 to −467) of the F1B promoter, modulate F1BGFP expression and endogenous FGF1 expression, and further regulate the maintenance of GBM-SCs. These observations were substantiated by using yeast one-hybrid assay, electrophoretic mobility shift assay, chromatin immunoprecipitation assay, gain- and loss-of-function assays, and neurosphere assays. Overexpression of RFX1 was shown to down-regulate FGF-1B mRNA expression and neurosphere formation in human glioblastoma cells, whereas RNA interference knockdown of RFX1 demonstrated the opposite effects. Our findings provide insight into FGF1 gene regulation and suggest that the roles of FGF1 and RFX1 in the maintenance of GBM-SCs. RFX1 may negatively regulate the self-renewal of GBM-SCs through modulating FGF-1B and FGF1 expression levels by binding the 18-bp cis-elements of the F1B promoter.
Keywords: Gene/Promoters, Gene/Regulation, DNA-binding Protein, DNA-Protein Interaction, Gene Expression, FGF1, RFX1, Glioblastoma Stem Cells, Neural Stem Cells, Neurosphere
Introduction
The human FGF1 gene is over 120 kb long and contains three protein-coding exons as well as a long 3′-untranslated region. It also contains at least four upstream untranslated exons, designated 1A, 1B, 1C, and 1D, which are alternatively spliced to the first protein-coding exon (1–3). This tissue-specific expression of the four mRNAs generated by alternative splicing relies on the use of different promoters (2, 4). Thus, FGF-1A transcript predominates in the human kidney (2), and FGF-1C and -1D transcripts predominate in vascular smooth muscle cells and fibroblasts (5). FGF-1B is the major transcript within the human brain (2) and retina (6). A previous study also showed that most malignant gliomas express FGF1 utilizing the 1B promoter (3). Moreover, the expression of FGF-1B mRNA is restricted to sensory and motor nuclei in the brain stem, subventricular zone, spinal cord, and other areas that are known to be abundant for neural stem/progenitor cells (NSPCs)2 (7, 8).
NSPCs are defined on the basis of the ability to self-renew and their potential to differentiate into neurons, astroglias, and oligodendrocytes in vitro (9–13). It has been suggested that glioblastoma stem cells (GBM-SCs) share many properties with normal NSPCs (14–17). However, GBM-SCs are highly tumorigenic in mice and display aberrant proliferative capacities (14). NSPCs and GBM-SCs can be isolated using fluorescence-activated cell sorting (FACS) with a specific cell surface marker such as CD133 (18, 19) or GFP expression driven by NSPC-specific promoters, e.g. SOX1 (20, 21), SOX2 (22, 23), NESTIN (24–26), and FGF-1B (27) in the serum-free culture supplemented with FGF2 (10, 12, 28) or FGF1 (29). NSPCs and GBM-SCs are examined to determine whether they could expand to form neurospheres. The capacity to form neurospheres is defined as self-renewal (30–32). Neurosphere assay has been suggested as a standard to evaluate the self-renewal ability of NSPCs and GBM-SCs from different origins, such as human fetal brain (33, 34) or glioblastoma tissues (27), glioblastoma cell lines (35, 36), and developing mouse brains (11, 37).
The 540-bp (−540 to +31) sequence upstream of the 1B transcription start site (F1B) has been demonstrated to drive the expression of luciferase (4), green fluorescence protein GFP reporter genes in cultured cells (27, 29), and the SV40 large T-antigen in transgenic mice (8). We recently demonstrated that the F1BGFP reporter could be used to isolate NSPCs with self-renewal and multipotent capacities from human glioblastoma tissues in developing (E11.5, E14.5, E17.5, and P1) or adult mouse brains (27, 29). Furthermore, we showed that F1BGFP-selected NSPCs from mouse brains were able to repair the damaged sciatic nerve of paraplegic rats (38, 39).
The regulatory factor protein of the X-box (RFX) family is characterized by a highly conserved DNA-binding domain (DBD) and consists of seven members in mammals (RFX1–7) (40). The RFX family is conserved throughout the evolution in eukaryotic species and contains one member each from yeast (41), Caenorhabditis elegans (daf-19) (42, 43), two members from Drosophila (dRfx and dRfx2) (44, 45), and seven members each from mouse and human. These RFX proteins feature a characteristic 76-amino acid DBD with a wing-helix structure (46). The function of RFX1, the prototype of the RFX family, is not yet clear. RFX1 is expressed in various tissues, with especially high amounts in mammalian brain (40). RFX1 is expressed in the neurons of rat brain and contributes to the regulation of the expression of the neuron specifically expressed glutamate transporter type 3 (47). These results suggest a role of RFX1 in the nervous system. Knock-out of Rfx homologue in C. elegans leads to severe sensory defects (42). In addition, the Drosophila RFX homologue is necessary for ciliated sensory neuron differentiation (45).
In this study, we present evidence that transcription factor RFX1 could bind the 18-bp cis-elements (−484 to −467) of the F1B promoter, modulate F1BGFP and FGF1 expression levels, and further regulate the self-renewal ability of GBM-SCs. Overexpression of RFX1 could down-regulate FGF-1B mRNA expression and neurosphere formation, whereas RNAi knockdown of RFX1 demonstrated the opposite effects. Our findings provide insights into FGF1 gene regulation and suggest the role of RFX1 in the maintenance of GBM-SCs. RFX1 may negatively regulate the self-renewal of GBM-SCs through modulating FGF-1B and FGF1 expression levels by binding the 18-bp cis-elements of the FGF-1B promoter.
EXPERIMENTAL PROCEDURES
Biological Data Base
FGF-1A promoter (−826 to +77), FGF-1B promoter (−540 to +31), FGF-1C promoter (−1601 to +88), and FGF-1D promoter (−985 to +40) sequences (3) were analyzed using the MatInspector program (48) with matrix library 6.3 in the matrix group of vertebrates.
Yeast One-hybrid Assay
To test the DNA-binding ability of the DNA-binding domain of RFX1 (amino acids 441–512), we generated a target reporter construct for library screening (49). Four tandem repeats of the 18-bp (−484 to −467) sequences were ligated and subcloned into the upstream region of the minimal promoter of either pHISi-1 or placZi reporter plasmids. The resultant plasmids were then transformed and integrated into the yeast genome of YM4271 to generate a dual reporter yeast strain designated YM4271/p1B18H1/p1B18Z. The cDNA of RFX1 DBD, which is highly homologous to other RFX proteins, was inserted in sense or antisense orientation behind the activation domain of pGAD10, with these designated pGAD10-RFX1-DBD(+) and pGAD10-RFX1-DBD(−), respectively. Subsequently, these plasmids were transformed into YM4271/p1B18H1/p1B18Z, and the transformants were selected in SD/−Leu/−His medium with 45 mm 3-aminotriazole. In this experiment, pGAD10 and pGAD10-RFX1-DBD(−) were used as negative controls. The process of yeast one-hybrid was according to the manufacturer's protocol (Clontech).
F1BGFP Reporter
Nucleotides −540 to +31 of the 1B promoter of the human FGF1 gene (4) and nucleotides 5171–2533 of the SV40 immediate early gene were cloned into the SmaI-BamHI sites of pGL2-Basic (Promega) and designated pF1BTag (8). We also cloned the nucleotides −540 to +31 of the human FGF-1B promoter into the pEGFP1 (Clontech) vectors to construct the pF1BGFP reporter (27, 29). All constructs were verified by DNA sequencing. pF1BGFP was prepared using the EndoFree plasmid maxi kit (Qiagen Inc., Chatsworth, CA). For generation of U-1240 MG/F1BGFP(+) cells (27), human glioblastoma U-1240 MG cells were plated in 60-mm tissue culture dishes (BD Labware) to achieve 60–80% confluence by day 2. On day 2, cells were transfected with 10 μg of pF1BGFP using GeneJuice transfection reagent (Merck). Percentage of F1BGFP-positive cells was analyzed by using flow cytometry according to the procedures described elsewhere (27, 29, 50). A total of 1 × 104 cells were gated on a dot plot forward side scatter on the x axis and side scatter on the y axis. The gated cells were evaluated on a histogram displaying FL1 (GFP) on the x axis and side scatter on the y axis.
Cell Culture and Transfection
Human glioblastoma cells, U-1240 MG, were cultured in minimal essential media (Invitrogen) supplemented with 10% calf serum, 100 units/ml penicillin, and 500 μg/ml streptomycin (Invitrogen) at 37 °C as described previously (4). U-1240 MG/F1BGFP cells were further cultured in culture medium containing 100 μg/ml G418. KT98 cells were derived from brain tumors of transgenic mice that expressed the large T-antigen driven by the F1B promoter (8, 27). KT98 cells were cultured in Dulbecco's modified Eagle's medium/F-12 nutrient mixture (1:1) supplemented with 10% fetal bovine serum (Invitrogen), 100 units/ml penicillin, and 500 μg/ml G418 at 37 °C. The pHA-RFX1 construct was provided by Dr. Shaul (51, 52). Cells were transfected with pHA-RFX1-wt using the GeneJuice transfection reagents according to the manufacturer's instructions.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was performed using the EZ ChIP kit (Upstate Biotechnology, Inc., Lake Placid, NY) according to the manufacturer's description. Briefly, U-1240 MG cells were cross-linked with 1% formaldehyde in the medium for 5 min at room temperature, and this reaction was stopped by adding glycine to a final concentration of 125 mm. Subsequently, cells were rinsed twice with phosphate-buffered saline, scraped in phosphate-buffered saline, pelleted at 700 × g at 4 °C for 5 min, and lysed in SDS/lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris, pH 8.1). DNA was fragmented into around 200-bp pieces using the sonicator (Sonicor, Deer Park, NY). Sheared chromatin was diluted 10 times and precleared with protein G-agarose at 4 °C for 1 h with rotation. After pelleting the protein G-agarose at 4000 × g for 1 min, 10 μl of the supernatant was removed as 1% input group and saved until the reverse cross-linking step. Each reaction mixture was reacted with 5 μg of polyclonal anti-RFX1 (I-19X) and anti-RFX1 (D-19X) (Santa Cruz Biotechnology, Santa Cruz, CA) antibodies, an anti-acetyl-histone H3 (Upstate, 06-599B) antibody for positive control, and nonspecific rabbit IgG (PP64B) (Upstate) as a negative control. The immunoprecipitated products were washed sequentially with low salt immune complex wash buffer, high salt immune complex wash buffer, LiCl immune complex wash buffer, and twice with TE buffer. The chromatin was eluted from the agarose by incubating with elution buffer (1% SDS, 100 mm NaHCO3), and the DNA-protein complexes were reversely cross-linked by high salt solution containing 200 mm NaCl at 65 °C for at least 5 h. To eliminate contaminations of proteins and RNAs, the mixture was treated with 10 μg of RNase A at 37 °C for 30 min and then treated with protease K for 2 h at 45 °C. Finally, the precipitated DNA was recovered using the spin column provided in the ChIP kit and eluted with 50 μl of elution buffer. PCR was conducted using TaqDNA polymerase (Roche Applied Science). 2 μl of the precipitated DNA was used as template. The sequences of the primers used in the ChIP assay were as follows: amplicon A, 5′-ACAGGGTTTCACAACTGGACATAA-3′ and 5′-CCAGATTCCCCCCCTCCTA-3′ with the amplicon size of 186 bp; amplicon B, 5′-GCAGGGATGCCAGATGACA-3′ and (5′-TGTGTGAGCCGAATGGACTTC-3′ with the amplicon size of 166 bp; amplicon C, 5′-TCAGGGTTTTGGTAGGGTGGTA-3′ and (5′-GATGTGGGTGTGGATAGTGTATGTG-3′ with the amplicon size of 177 bp; GAPDH, 5′-TACTAGCGGTTTTACGGGCG-3′ and 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′ with the amplicon size of 166 bp.
Preparation of Nuclear Extracts
Nuclear extracts were prepared using NE-PER nuclear and cytoplasmic extraction reagent (Pierce) according to the manufacturer's instructions. Briefly, 1 × 106 cells were trypsinized, followed by lysing in 100 μl of CERI buffer. After the lysates were vortexed for 15 s and incubated on ice for 10 min, 5.5 μl of CERII was added. The lysates were vortexed for 5 s, incubated on ice for 1 min, and vortexed again for 5 s. The nuclei were pelleted at 16,000 × g at 4 °C for 5 min, and the cytoplasmic extracts were removed. Nuclei were resuspended in 25 μl of nuclear extraction buffer and vortexed for 15 s. The nuclei were extracted on ice and vortexed for 15 s every 10 min, for a total of 40 min. The extracts were centrifuged at 16,000 × g at 4 °C for 5 min, and the supernatants were collected as nuclear extracts. Protein concentration was determined by the Bradford method using bovine serum albumin as a standard (Bio-Rad).
Electrophoretic Mobility Shift Assay (EMSA)
Binding reaction containing 20 μl of binding buffer (10 mm Tris, 50 mm KCl, 1 mm dithiothreitol, and 5 mm MgCl2, 1 μg of poly(dI·dC), 10 μg of nuclear extracts, 200-fold excess of cold competitors, and 20 fmol of 5′-biotin-labeled oligonucleotide probes) was added sequentially and incubated at room temperature for 20 min. The reaction mixture was separated on 4% native polyacrylamide gel at 50 V for 8–10 h. The resolved probes were transferred onto nylon membranes (Hybond-N+ nylon transfer membrane, RPN303B, Amersham Biosciences) at 600 mA for 2.5 h. Biotin-labeled probe on the membranes was detected using the streptavidin-horseradish peroxidase conjugate and the chemiluminescent substrate provided in the chemiluminescent nucleic acid detection module (Pierce) according to the manufacturer's instructions; subsequently, the membranes were exposed to x-ray film. For EMSA supershift assay, 2 μg of anti-RFX1 (I-19X, Santa Cruz Biotechnology) polyclonal antibodies were added after adding the nuclear extracts and incubated for 15 min. Finally, the probe was added and incubated further for 15 min. The sequences of probes and cold competitors used in the EMSA experiment were as follows: 26 bp, 5′-ACGACCTGCTGTTTCCCTGGCAACTC-3′) AP-1, 5′-CGCTTGATGAGTCAGCCGGAA-3′; 18 bp, 5′-CTGTTTCCCTGGCAACTC-3′; 18-bp mut, 5′-CTTTTTCCCTTTCAACTC-3′; MAP1A, 5′-CGGCGTTGCCATGGAGACAACTCCG-3′; PyEP, 5′-GGCCAGTTGCCTAGCAACTAATAC-3′; m26 bp, 5′-ACAACCAGTTGTTTCCCTGGTGACAG-3′; and m18 bp, 5′-TTGTTTCCCTGGTGACAG-3′ (Protech Technology, Taipei, Taiwan). The above oligonucleotides were incubated with respective complementary oligonucleotides in 10 mm Tris, 1 mm EDTA, 50 mm NaCl, pH 8.0, reaction buffer at 95 °C for 5 min, and then the temperature was decreased by 1 °C/s to 4 °C to anneal the complementary oligonucleotides.
Reverse Transcription-PCR
For expression analysis of human and mouse FGF-1B transcripts, RNA extracted from the U-1240 MG and KT98 cells was primed with oligo(dT) and reverse-transcribed using SuperScriptII reverse transcription (Invitrogen). Each cDNA transcribed from 500 ng of RNA was amplified using specific primer pairs with TaqDNA polymerase (Roche Applied Science) under the conditions of initial denaturing at 95 °C for 10 min, followed by 30 cycles denaturing at 95 °C for 15 s and extension at 60 °C for 30 s, and finally extension at 60 °C for 1 min for completing the polymerization. The primer used in the PCR were as follows: human FGF-1B, 5′-TGAGCGAGTGTGGAGAGAGGTA-3′ and 5′-GCTGTGAAGGTGGTGATTTCC-3′ with amplicon size of 114 bp; and mouse Fgf-1B, 5′-CCGTCTTGTGATAAAGTGGAGTGA-3′ and 5′-CAGCAAGCAGCGGTGGTA-3′ with amplicon size of 81 bp. Quantitative PCR analysis was performed using an ABI prism 7500 HT sequence detection system (Applied Biosystems). We used the SYBR Green method to analyze the expression levels of RFX1, FGF-1B, and FGF1. The RFX1 primers are 5′-AGACCGGCGTTCCTACTCA-3′ and 5′-GGGGCACTTGGATGTTGGT-3′)with amplicon size of 129 bp. The FGF1 primers are 5′-ACAAGGGACAGGAGCGAC-3′ and 5′-TCCAGCCTTTCCAGGAACA-3′ with amplicon size of 63 bp.
Western Blot Analyses
Ten micrograms of nuclear protein fraction was separated on an 8% SDS-PAGE and transferred onto Immobilon-polyvinylidene difluoride (Millipore, Bedford, MA) in a transfer buffer (6.2 mm boric acid, pH 8.0). Blots were incubated initially with blocking buffer (5% bovine serum albumin) for 1 h at room temperature and then with specific primary antibodies against GFP, hemagglutinin, and β-actin (Santa Cruz Biotechnology). Primary antibodies had been diluted (1:200) with Tris-buffered saline/Tween 20 (TBS-T) containing 5% bovine serum albumin and 0.01% sodium azide. After antibody incubation, the blots were washed with TBS-T for 1 h and incubated with anti-goat IgG conjugated with horseradish peroxidase (Santa Cruz Biotechnology) for 1 h at room temperature. After washing the secondary antibodies (1:2000) with TBS-T, immunodetection was performed, using an enhanced chemiluminescence kit for Western blot detection (Amersham Biosciences).
RNAi Experiments
Small interfering RNA knockdown experiments were performed with stealth RNAi (Invitrogen). Stealth RNAi for each RFX is as follows: RFX1-RNAi-I (HSS109204); RFX1-RNAi-II (HSS109205); RFX1-RNAi-III (HSS109206); FGF1-RNAi (HSS142002); nonspecific RNAi (Stealth RNAi negative control duplex, medium GC duplex); and stealth RNAi GFP reporter control (GFP-RNAi). U-1240 MG or U-1240 MG/F1BGFP(+) cells were used in RNAi knockdown experiments. Cells were transfected with small interfering RNA against RFX1 using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer's instructions. Three different RNAi (I, II, and III) against RFX1 were tested, and representative results for RNAi knockdown using RFX1-RNAi-III (HSS109206) were shown in the experiments of U-1240 MG/F1BGFP(+) cells.
Neurosphere Assay
Neurosphere formation has been considered as an indicator for the self-renewal capacity of NSPCs and GBM-SCs. In brief, U-1240 MG/F1BGFP(+) cells were washed with basal medium and seeded at a maximal density of 1 × 104 cells in 60-mm Petri dishes (Falcon Industries, Oxnard, CA) with 5 ml of neurosphere medium (NS medium) as follows: Dulbecco's modified Eagle's medium HG/F-12 supplemented with B27 (Invitrogen), 50 ng/ml epidermal growth factor, 20 ng/ml FGF2, 10 ng/ml leukemia inhibitory factor, and 5 μg/ml heparin (19, 68). Subsequently, cells were cultured in 5% CO2 in a 37 °C incubator. The spheres (diameter larger than 50 μm) were counted directly under microscope after 7 days in vitro.
Statistical Analyses
Data are expressed as means ± S.E. One-way analysis of variance was used for comparison of multiple groups. The data were considered statistically significant at p < 0.05.
RESULTS
Identification of RFX1 as a Binding Candidate of the 18-bp Sequence of FGF-1B Promoter
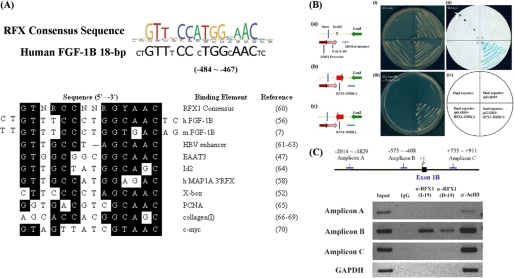
To find the candidate transcription factors that bind to the 18-bp sequence, we analyzed the human FGF-1B promoter region from −540 to +31 using MatInspector (Genomatix) (48). Two putative RFX1-binding sites, namely X-box, located within the 18-bp sequence (−484 to −467) were identified and are similar to the imperfect palindromic RFX1 consensus sequence, which contains a 6-bp half-site. Each sequence in the two complementary strands is separated by a spacer region from 0 to 3 bp, as reported previously (46). To investigate the regulation of the FGF-1B promoter, the homologous sequence information of the 18 bp in mouse was extracted from the University of California, Santa Cruz, genome browser using the track of vertebrate multiple alignment. An alignment of the RFX consensus sequence (5′-GTNRC(C/N)N0–3RGYAAC-3′, where N is any nucleotide, R is a purine, and Y is a pyrimidine), together with the putative RFX1-binding sequence within the 18 bp of the FGF-1B promoter in the human and mouse homologous site, and other published RFX-binding sequences, is shown in Fig. 1A. This result suggests that putative RFX-binding sites in human FGF-1B and mouse Fgf-1B promoters are highly similar to the consensus sequence and other RFX-binding sequences.
FIGURE 1.
A, alignment of the 18-bp sequence (nucleotides −484 to −467) of the human FGF-1B and mouse Fgf-1B promoter with other RFX1-binding sites reveals strong similarity to the RFX consensus binding site. The putative RFX1-binding elements in the 18-bp sequences of the human FGF-1B and mouse Fgf-1B promoter, or other known RFX1-binding sequences, were aligned with the consensus sequence using ClustalW2, a multiple sequence alignment tool. The results are shown above, with the nucleotides consistent with the consensus sequence shown as white letters on a black background. R, Y, and N represent a purine, a pyrimidine, and any nucleotide, respectively (7, 47, 52, 56, 60–70). PCNA, proliferating cell nuclear antigen. B, DNA-binding domain of RFX1 is responsible for the binding to the 18-bp cis-acting elements of the F1B promoter. Panels a–c, constructions of pRFX1-DBD(+) and pRFX1-DBD(−). Panel i, yeast transformants in SD/−Leu medium. Panel ii, yeast transformants in SD/−Leu medium examined by β-galactosidase assay. Panel iii, yeast transformants selected in SD/−Leu-His + 45 mm 3-aminotriazole (3-AT) medium. Panel iv, schematic illustration of each yeast transformant using the dual yeast reporter YM4271/pF1BH1/pF1B18Z in the indicated regions. C, chromatin immunoprecipitation assay confirms that RFX1 could bind the 18-bp sequence in human glioblastoma U-1240 MG cells. Schematic representation of the localization of the primer-amplified region on the FGF1 gene genomic sequences is shown. The exons and introns are shown as black boxes and lines, respectively. The sequences containing the 18-bp sequence are precipitated by the anti-RFX1 antibody (I-19, against the C-terminal region; D-19, against the internal region) and positive control antibody, as well as by the anti-acetyl H3 (α-AcH3) antibody, but not by the negative control antibody, IgG. In addition, the sequences containing the GAPDH promoter region, amplicon A, and amplicon C were not precipitated by the anti-RFX1 antibody.
RFX1 DNA-binding Domain Binds the 18-bp Sequence in Yeast One-hybrid Assay
To functionally verify the computational prediction, we used yeast one-hybrid assay to evaluate the DNA-binding ability of RFX1 DBD, which is highly homologous to other RFX proteins. We constructed the GAL4 fusion proteins of RFX1-DBD in sense orientation (+) and in antisense orientation (−) for yeast one-hybrid screening (Fig. 1B). The dual reporter yeast strain YM4271/p1B18H1/p1B18Z, as described under “Experimental Procedures,” was used in this assay. From our results, we observed that only pGAD10-RFX1-DBD(+) could activate the 18-bp sequence of the F1B promoter to give rise to viable clones in the histidine-deficient selection medium (Fig. 1B, panel iii). In addition, the viable clones could produce β-galactosidase and turn blue within 1 h in the β-galactosidase assay (Fig. 1B, panel ii). These results suggest that the fusion of RFX1-DBD with the GAL4 activation domain could activate, in the yeast one-hybrid assay, the minimal promoter containing four tandem repeats of the 18-bp sequence (Fig. 1B).
RFX1 Binds the 18-bp Sequence in Human Glioblastoma U-1240 MG Cells
To verify whether RFX1 binds to the 18 bp in cultured cells, we performed the ChIP assay using anti-RFX1-specific antibody to precipitate the chromatin in U-1240 MG cells. Three primer pairs were designed to examine the precipitated DNA; the localization of three amplicons amplified by the primers are shown in Fig. 1C. The sequence containing the 18 bp, located in the amplicon B, was precipitated by two different anti-RFX1 antibodies and the positive control antibody, anti-acetyl-H3, but not the negative control antibody, IgG. In addition, the sequences containing the amplicon A, amplicon C, and GAPDH promoter region were not precipitated by the anti-RFX1 antibodies (Fig. 1C). These results further demonstrated that RFX1 bound the 18 bp of F1B promoter in U-1240 MG cells.
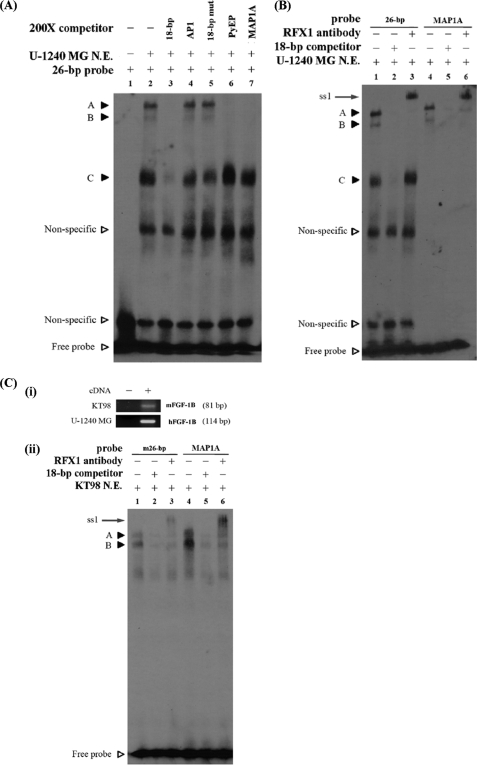
RFX1 Binds the 18-bp Sequence in EMSA Experiments
To examine whether RFX1 could bind to the putative binding site, we performed EMSA experiments with nuclear extracts isolated from human U-1240 MG cells and mouse KT98 cells; both cell lines express the FGF-1B transcript (Fig. 2C, panel i). Several complexes were observed when incubating U-1240 MG nuclear extracts with the 26-bp probe (nucleotides −492 to −467) of the FGF-1B promoter (Fig. 2A, lane 2). The specificity of complex formation was analyzed by the EMSA competition assay; three specific complexes were competed away by the 200-fold excess of the cold 18-bp probe (Fig. 2A, lane 3) but not the nonrelevant cold probe (Fig. 2A, lane 4), AP-1. These three specific complexes were designated as A, B, and C, respectively. The oligonucleotides, MAP1A or PyEP, containing the known RFX1-binding site could diminish the specific complexes A and B (Fig. 2A, lanes 6 and 7). In contrast, the oligonucleotides, 18-bp mut (Fig. 2A, lane 5), with mutated RFX1-binding core sequence could not compete with the wild type 18-bp probe for the binding. These results suggest that the 18-bp sequence contains the RFX1-binding site.
FIGURE 2.
A, EMSA competition assay demonstrates that the 18-bp sequence contains the RFX1-binding site. EMSA experiments using 26 bp (−492 to −467) as a probe and 10 μg of U-1240 MG nuclear extracts revealed several complexes (lane 1). The specificity of complexes formation was confirmed by 18-bp (−484 to −467) competition. Three specific complexes, which were competed by the 18 bp but not the nonrelevant competitor, AP-1, were designated as complexes A, B, and C, respectively (lanes 3 and 4). The oligonucleotides, PyEP or MAP1A, containing the known RFX1-binding site could diminish the specific complexes A and B (lanes 6 and 7) but not the oligonucleotides, 18-bp mut (lane 5), with mutated RFX1-binding core sequence. B, EMSA supershift assay reveals the in vitro interaction between RFX1 and the 18 bp in nuclear extracts (N.E.) isolated from human U-1240 MG cells. EMSA experiment was carried out using MAP1A, which contains the RFX1-binding site and human 26-bp probes. Both complexes A and B were formed with the two probes (lanes 1 and 4), diminished by human 18-bp competition (lanes 2 and 5), and supershifted by anti-RFX1 antibody (lanes 3 and 6). The result of EMSA supershift assay directly demonstrated the in vitro interaction between 18 bp and RFX1. The bands supershifted by anti-RFX1 antibody are designated as ss1. C-i, expression analyses of FGF-1B transcript in human U-1240 MG and mouse KT98 cells. Reverse transcription-PCR experiments conducted using primer specific for mouse or human FGF-1B transcripts, respectively, demonstrated the expression of FGF-1B transcripts in U-1240 MG and KT98 cells. C-ii, EMSA supershift assay reveals the in vitro interaction between RFX1 and the m18 bp in nuclear extracts isolated from mouse KT98 cells. EMSA experiment was carried out using MAP1A, which contains the RFX1-binding site, and mouse 26-bp probes. Both complexes A and B were formed with these two probes (lanes 1 and 4), diminished by mouse 18-bp competition (lanes 2 and 5), and supershifted by anti-RFX1 antibody (lanes 3 and 6). The result of EMSA supershift assay directly demonstrated the in vitro interaction between mouse 18 bp and RFX1. The bands supershifted by anti-RFX1 antibody are designated as ss1.
To investigate if RFX1 is present in the complex formation, the EMSA supershift assays were carried out with the anti-RFX1 antibody. The complexes A and B formed on the 26 bp were similar to the MAP1A probe, and the two complexes were also diminished by 18-bp competition and supershifted by the anti-RFX1 antibodies in the experiments conducted using U-1240 MG and KT98 nuclear extracts (Fig. 2, B and C). The results above demonstrated that the interaction between RFX1 and FGF-1B 18-bp cis-elements is conserved in evolution between mouse and human.
Regulatory Effects of RFX1 on FGF-1B mRNA, F1BGFP, Endogenous FGF1 Expression Levels and Neurosphere Formation in U-1240 MG and U-1240 MG/F1BGFP Cells
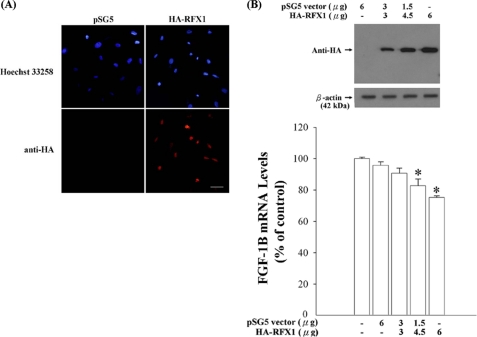
RFX1 is a dual function transcription factor that activates and represses gene expression depending on the promoter contexts. To investigate the regulatory effect of RFX1 on the FGF-1B mRNA expression, we transfected the U-1240 MG cells with pHA-RFX1 plasmid. Representative results of the overexpressed proteins are shown in Fig. 3, A and B. Overexpressed RFX1 was observed in the nucleus of transfected cells (Fig. 3A). The expression level of FGF-1B mRNA was decreased in a dose-dependent manner when RFX1 was overexpressed (Fig. 3B). Knockdown of RFX1 expression was observed using two of three Stealth human RFX1-RNAi sequences (Fig. 4A); and the mRNA expression of FGF-1B was significantly up-regulated when RFX1 was knocked down (Fig. 4B). Of note, knockdown of RFX1 or FGF1 by specific RNAi could have significant effects on endogenous FGF1 gene expression in a time-dependent manner (Fig. 5).
FIGURE 3.
RFX1 regulates FGF-1B mRNA expression in human glioblastoma U-1240 MG cells. A, after pHA-RFX1 transfection for 24 h, the overexpressed RFX1 protein was detected by immunocytochemistry staining and Western blot using anti-hemagglutinin antibody. Representative images are shown. Scale bar, 200 μm. B, overexpression of RFX1 was shown to down-regulate the FGF-1B mRNA expression in a dose-dependent manner. The FGF-1B mRNA expression was analyzed by quantitative PCR and normalized to G3PDH. The result of FGF-1B mRNA expression level is shown as means ± S.E., n = 3; *, p < 0.05.
FIGURE 4.
Knockdown of RFX1 and up-regulation of FGF-1B gene expression by RFX1 Stealth RNAi. A, treatment of human RFX1-RNAi-I and -III for 72 h could efficiently knock down the endogenous RFX1 expression. B, furthermore, FGF-1B mRNA expression level was elevated upon RFX1 knockdown. Control, cells treated with Lipofectamine RNAiMAX. The result of FGF-1B mRNA expression levels was shown as means ± S.E., n = 3; *, p < 0.05; **, p < 0.01 versus control.
FIGURE 5.
Knockdown of RFX1 could up-regulate endogenous FGF1 expression. A, RFX1 mRNA expression level was decreased by RFX1-RNAi-III treatment. B, schematic structure of the human FGF1 gene. Exons are labeled and shown boxed. Locations of PCR primers are indicated by arrows. 1B and F1 indicate the primer pair for FGF-1B and FGF1, respectively. The primer sequences are shown under “Experimental Procedure.” C, FGF1 expression level was significantly elevated by RFX1-RNAi-III treatment in a time-dependent manner. D, FGF1 expression level was significantly decreased by FGF1-RNAi treatment. The results of endogenous RFX1 and FGF1 expression levels normalized with internal control GAPDH expression were shown as means ± S.E.; *, p < 0.05; **, p < 0.01 versus 0 h.
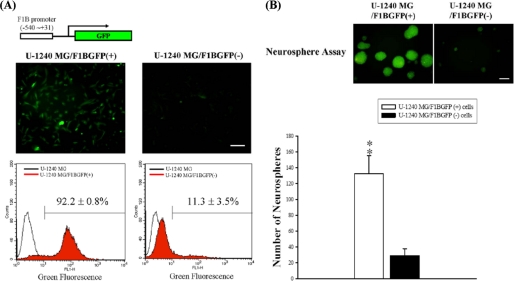
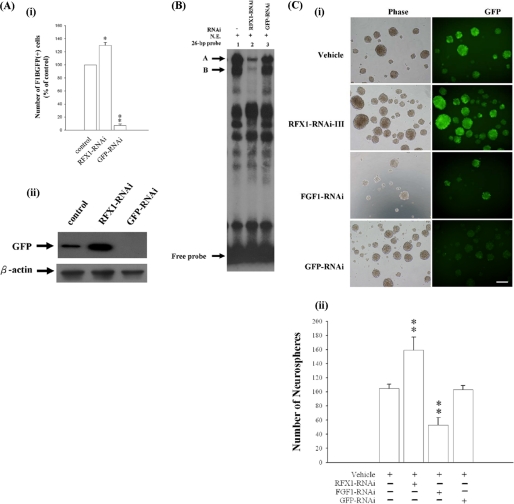
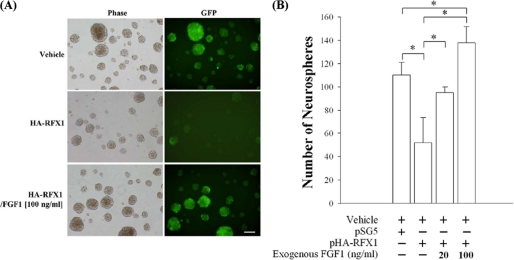
We further transfected U-1240 MG cells with F1BGFP reporter and sorted U-1240 MG/F1BGFP(+) and -(−) cells by FACS (Fig. 6A). We have previously shown that F1BGFP(+) cells from human glioblastoma tissues and mouse brains generate more neurospheres than F1BGFP(−) cells (27, 29). In this study, we showed that U-1240 MG/F1BGFP(+) cells also generated more neurospheres than U-1240 MG/F1BGFP(−) (Fig. 6B), indicating the self-renewal property of F1BGFP-selected GBM-SCs. Using U-1240 MG/F1BGFP(+) cells, we previously demonstrated that mRNA expression of FGF-1B exhibited a positive correlation with the intensity of green fluorescence driven by the F1B promoter (27, 29). Here, we further used U-1240 MG/F1BGFP(+) cells to study the effects of RFX1-RNAi on F1B promoter activation. Knockdown of RFX1 could significantly increase the percentage of GFP(+) cells in U-1240 MG/F1BGFP(+) cells (Fig. 7A, panel i); GFP-RNAi served as a positive control and could significantly decrease the GFP intensity in U-1240 MG/F1BGFP(+) cells (Fig. 7A, panel i). Western blot analysis confirmed the GFP expression (Fig. 7A, panel ii); and EMSA (Fig. 7B) analyses further demonstrated that the RFX1-RNAi treatment decreased most of the RFX1-bound complexes (Fig. 7, A and B). Concomitantly, knockdown of RFX1 increased the number of neurospheres. GFP-RNAi served as a negative control in the neurosphere assay (Fig. 7C). Furthermore, knockdown of endogenous FGF1 significantly decreased the number of neurospheres generated by U-1240 MG/F1BGFP(+) cells (Fig. 7C), suggesting that endogenous FGF1 expression is important for neurosphere formation. We further observed that overexpression of RFX1 could decrease the number of neurospheres (Fig. 8) and that exogenous FGF1 could significantly rescue the inhibitory effect of RFX1 on neurosphere formation.
FIGURE 6.
Comparison of neurosphere formation between U-1240 MG/F1BGFP(+) and U-1240 MG/F1BGFP(−) cells. A, U-1240 MG/F1BGFP(+) and U-1240 MG/F1BGFP(−) cells were sorted by FACS. B, representative images of green fluorescent neurospheres with different treatment are shown. Scale bar, 100 μm. U-1240 MG/F1BGFP(+) generated more neurospheres than U-1240 MG/F1BGFP(−) cells, n = 3; **, p < 0.01.
FIGURE 7.
Knockdown of RFX1 could increase the percentage of F1BGFP(+) cells and neurosphere formation. A-i, U-1240 MG/F1BGFP(+) cells were treated with RFX1-RNAi-III (30 nm) and GFP-RNAi (30 nm) for 72 h, and the percentage of F1BGFP(+) cells was measured by flow cytometry. Data are shown as means ± S.E., n = 3, *, p < 0.05; **, p < 0.01. A-ii, Western blot analysis confirms that F1BGFP expression is up-regulated by RFX1-RNAi and down-regulated by GFP-RNAi. B, EMSA analysis for the effects of RFX1-RNAi-III on complex formation. N.E., nuclear extracts. C-i, representative images of neurospheres with different treatments are shown. Scale bar, 100 μm. C-ii, quantification of neurospheres generated by U-1240 MG/F1BGFP(+) cells treated with FGF1-RNAi, RFX1-RNAi-III, and GFP-RNAi, n = 4; **, p < 0.01.
FIGURE 8.
Exogenous FGF1 treatment could rescue the decrease of neurosphere formation. A, representative images of neurospheres with different treatments are shown. Scale bar, 100 μm. B, neurospheres generated by U-1240 MG/F1BGFP(+) after pSG5 empty vector and pHA-RFX1 transfection. FGF1 (20 ng/ml or 100 ng/ml) were treated in the neurosphere medium. n = 3; *, p < 0.05.
DISCUSSION
FGF1 and FGF2 are the best characterized members of the FGF family; they are important in many biological processes, including cell growth, proliferation, and neurogenesis (53, 54). The transcription factor that regulates FGF2 expression has been cloned and studied (55). In search of the mechanism that regulates FGF1 expression, we observed 18-bp cis-elements in the RR2 region of the FGF-1B promoter (56). Using this sequence, we were able to identify a transcription factor, RFX1, that regulates FGF1 expression in cells. This is the first transcriptional regulator for FGF1 that has been shown to bind the 18-bp cis-element in the FGF-1B promoter. Given the significance of FGF1 in growth control, tumor formation, and neurogenesis, identification of such a transcription factor brings us closer to understanding FGF1-dependent cellular processes.
Human FGF1 gene expression is regulated by the following four tissue-specific promoters: FGF-1A, FGF-1B, FGF-1C, and FGF-1D (3). Among these, FGF-1B is the specific promoter that was utilized in normal brain cells and glioblastoma cells. It is interesting and important to know whether RFX1 also regulates other FGF1 promoters in U-1240 MG cells; therefore, we analyzed all FGF1 promoters using the MatInspector program and found one palindromic RFX1-binding site in the FGF1-B promoter (−483 to −465), but not in any of the other three FGF1 promoters. This observation further supports the view that RFX1 binds specifically to the 18-bp cis-elements in the FGF-1B promoter.
Overexpression of RFX1 could significantly decrease FGF1 expression over 20% in 10% serum-supplemented medium (Fig. 3B); it is interesting to note that the effect of RFX1 overexpression significantly decreased neurosphere formation by 50% in serum-free neurosphere medium and could be rescued by exogenous FGF1 treatment (Fig. 8). The difference of the inhibitory effect of RFX1 overexpression between 10% serum culture conditions and serum-free neurosphere medium may be due to the fact that different culture conditions were applied to GBM-SCs in serum-free neurosphere medium.
In our results, RFX1 had little effect on FGF1 at 24 h and required at least 48 h for significant effects (Fig. 5). One possible explanation is that when RFX1 mRNA was decreased by RNAi at 24 h, the RFX1 proteins may still be present in the cells and require more time for the RFX1 protein levels to be reduced by RNAi. As we have shown, RFX1 mRNA levels were further reduced by 70% at 48 h, and this could significantly induce FGF1 expression.
RFX1 is detected ubiquitously, especially in the highest amount in mammalian brain (57). It has recently been implicated in the regulation of nervous specific gene activation. The two binding sites within exon 1 of the MAP1A gene were identified to be bound by RFX1 and RFX3 as homodimers or heterodimers and are important for effective non-neuronal gene repression (58). The promoter activity of neuronal glutamate transporter EAAT3, which regulates the glutamate neurotransmission, is activated by RFX1; and EEAT3 protein expression is increased in response to RFX1 overexpression (47). Phylogenetic analysis on the DNA-binding domain of RFX family reveals that the subgroup, including RFX1, RFX2, and RFX3 in mammals, is highly similar to the dRFX and daf-19 (59). The interactive roles of RFX1, RFX2, and RFX3 in the regulation of the FGF1 gene promoter are therefore worthy of further investigation. We have an ongoing study to characterize other RFX transcription factors (RFX2 and RFX3) that could also bind the 18-bp cis-elements of the F1B promoter and be detected only in the nucleus of U-1240 MG/F1BGFP(+) cells and not in U-1240 MG/F1BGFP(−) cells. Our preliminary data suggest that RFX2 and RFX3 are the activating factors for FGF1 expression. In this study, we mainly showed the suppression effect of RFX1 on FGF1 gene expression.
In conclusion, we identified RFX1 as a transcription suppressor of the FGF1 gene promoter and provided insights into FGF1 gene regulation. This is the first transcriptional regulator for FGF1 that has been shown to bind the 18-bp cis-element in the FGF-1B promoter. Given the significance of FGF1 in growth control, tumor formation, and neurogenesis, identification of such a transcription factor brings us closer to understanding FGF1-dependent cellular processes.
Acknowledgments
We appreciate the generous gift of pHA-RFX1 constructs from Dr. Yosef Shaul (Department of Molecular Genetics, Weizmann Institute of Science, Israel). We thank Su-Liang Chen, Don-Ching Lee, and Chao-Yang Hsiao for excellent technical assistance and discussions.
This work was supported by the National Science Council, Taiwan, and the National Health Research Institutes, Taiwan.
- NSPC
- neural stem/progenitor cell
- ChIP
- chromatin immunoprecipitation
- DBD
- DNA-binding domain
- EMSA
- electrophoretic mobility shift assay
- RFX
- regulatory factor of X-box
- FACS
- fluorescence-activated cell sorting
- RNAi
- RNA interference
- GFP
- green fluorescence protein
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
REFERENCES
- 1.Payson R. A., Canatan H., Chotani M. A., Wang W. P., Harris S. E., Myers R. L., Chiu I. M. (1993) Nucleic Acids Res. 21, 489–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Myers R. L., Payson R. A., Chotani M. A., Deaven L. L., Chiu I. M. (1993) Oncogene 8, 341–349 [PubMed] [Google Scholar]
- 3.Chiu I. M., Touhalisky K., Baran C. (2001) Prog. Nucleic Acids Res. Mol. Biol. 70, 155–174 [DOI] [PubMed] [Google Scholar]
- 4.Myers R. L., Ray S. K., Eldridge R., Chotani M. A., Chiu I. M. (1995) J. Biol. Chem. 270, 8257–8266 [DOI] [PubMed] [Google Scholar]
- 5.Chotani M. A., Touhalisky K., Chiu I. M. (2000) J. Biol. Chem. 275, 30432–30438 [DOI] [PubMed] [Google Scholar]
- 6.Myers R. L., Chedid M., Tronick S. R., Chiu I. M. (1995) Oncogene 11, 785–789 [PubMed] [Google Scholar]
- 7.Alam K. Y., Frostholm A., Hackshaw K. V., Evans J. E., Rotter A., Chiu I. M. (1996) J. Biol. Chem. 271, 30263–30271 [DOI] [PubMed] [Google Scholar]
- 8.Chiu I. M., Touhalisky K., Liu Y., Yates A., Frostholm A. (2000) Oncogene 19, 6229–6239 [DOI] [PubMed] [Google Scholar]
- 9.Reynolds B. A., Tetzlaff W., Weiss S. (1992) J. Neurosci. 12, 4565–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynolds B. A., Weiss S. (1992) Science 255, 1707–1710 [DOI] [PubMed] [Google Scholar]
- 11.Rietze R. L., Reynolds B. A. (2006) Methods Enzymol. 419, 3–23 [DOI] [PubMed] [Google Scholar]
- 12.Gage F. H., Ray J., Fisher L. J. (1995) Annu. Rev. Neurosci. 18, 159–192 [DOI] [PubMed] [Google Scholar]
- 13.Weiss S., Dunne C., Hewson J., Wohl C., Wheatley M., Peterson A. C., Reynolds B. A. (1996) J. Neurosci. 16, 7599–7609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galli R., Binda E., Orfanelli U., Cipelletti B., Gritti A., De Vitis S., Fiocco R., Foroni C., Dimeco F., Vescovi A. (2004) Cancer Res. 64, 7011–7021 [DOI] [PubMed] [Google Scholar]
- 15.Singh S. K., Clarke I. D., Terasaki M., Bonn V. E., Hawkins C., Squire J., Dirks P. B. (2003) Cancer Res. 63, 5821–5828 [PubMed] [Google Scholar]
- 16.Yuan X., Curtin J., Xiong Y., Liu G., Waschsmann-Hogiu S., Farkas D. L., Black K. L., Yu J. S. (2004) Oncogene 23, 9392–9400 [DOI] [PubMed] [Google Scholar]
- 17.Ignatova T. N., Kukekov V. G., Laywell E. D., Suslov O. N., Vrionis F. D., Steindler D. A. (2002) Glia 39, 193–206 [DOI] [PubMed] [Google Scholar]
- 18.Corti S., Nizzardo M., Nardini M., Donadoni C., Locatelli F., Papadimitriou D., Salani S., Del Bo R., Ghezzi S., Strazzer S., Bresolin N., Comi G. P. (2007) Exp. Neurol. 205, 547–562 [DOI] [PubMed] [Google Scholar]
- 19.Abdouh M., Facchino S., Chatoo W., Balasingam V., Ferreira J., Bernier G. (2009) J. Neurosci. 29, 8884–8896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aubert J., Stavridis M. P., Tweedie S., O'Reilly M., Vierlinger K., Li M., Ghazal P., Pratt T., Mason J. O., Roy D., Smith A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, Suppl. 1, 11836–11841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson C. E., Crawford B. E., Stavridis M., Ten Dam G., Wat A. L., Rushton G., Ward C. M., Wilson V., van Kuppevelt T. H., Esko J. D., Smith A., Gallagher J. T., Merry C. L. (2007) Stem Cells 25, 1913–1923 [DOI] [PubMed] [Google Scholar]
- 22.Suh H., Consiglio A., Ray J., Sawai T., D'Amour K. A., Gage F. H. (2007) Cell Stem Cell. 1, 515–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D'Amour K. A., Gage F. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, Suppl. 1, 11866–11872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma B. F., Liu X. M., Xie X. M., Zhang A. X., Zhang J. Q., Yu W. H., Zhang X. M., Li S. N., Lahn B. T., Xiang A. P. (2006) Neuroreport 17, 377–381 [DOI] [PubMed] [Google Scholar]
- 25.Imayoshi I., Ohtsuka T., Metzger D., Chambon P., Kageyama R. (2006) Genesis 44, 233–238 [DOI] [PubMed] [Google Scholar]
- 26.Park J. H., Ahn J. I., Kim S. Y., Park K. S., Lee Y. D., Yamaguchi M., Chung H. J. (2007) Neurosci. Lett. 421, 185–190 [DOI] [PubMed] [Google Scholar]
- 27.Hsu Y. C., Lee D. C., Chen S. L., Liao W. C., Lin J. W., Chiu W. T., Chiu I. M. (2009) Dev. Dyn. 238, 302–314 [DOI] [PubMed] [Google Scholar]
- 28.Kuhn H. G., Winkler J., Kempermann G., Thal L. J., Gage F. H. (1997) J. Neurosci. 17, 5820–5829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee D. C., Hsu Y. C., Chung Y. F., Hsiao C. Y., Chen S. L., Chen M. S., Lin H. K., Chiu I. M. (2009) Mol. Cell. Neurosci. 41, 348–363 [DOI] [PubMed] [Google Scholar]
- 30.Galli R., Gritti A., Vescovi A. L. (2008) Methods Mol. Biol. 438, 67–84 [DOI] [PubMed] [Google Scholar]
- 31.Cordey M., Limacher M., Kobel S., Taylor V., Lutolf M. P. (2008) Stem Cells 26, 2586–2594 [DOI] [PubMed] [Google Scholar]
- 32.Chojnacki A., Weiss S. (2008) Nat. Protoc. 3, 935–940 [DOI] [PubMed] [Google Scholar]
- 33.Poltavtseva R. A., Revishchin A. V., Aleksandrova M. A., Korochkin L. I., Viktorov I. V., Sukhikh G. T. (2003) Ontogenez. 34, 211–215 [PubMed] [Google Scholar]
- 34.Nakamura Y., Yamamoto M., Oda E., Yamamoto A., Kanemura Y., Hara M., Suzuki A., Yamasaki M., Okano H. (2003) Lab. Invest. 83, 479–489 [DOI] [PubMed] [Google Scholar]
- 35.Sherry M. M., Reeves A., Wu J. K., Cochran B. H. (2009) Stem Cells 27, 2382–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pollard S. M., Yoshikawa K., Clarke I. D., Danovi D., Stricker S., Russell R., Bayani J., Head R., Lee M., Bernstein M., Squire J. A., Smith A., Dirks P. (2009) Cell Stem Cell 4, 568–580 [DOI] [PubMed] [Google Scholar]
- 37.Singec I., Knoth R., Meyer R. P., Maciaczyk J., Volk B., Nikkhah G., Frotscher M., Snyder E. Y. (2006) Nat. Methods 3, 801–806 [DOI] [PubMed] [Google Scholar]
- 38.Lin Y. L., Jen J. C., Hsu S. H., Chiu I. M. (2008) Surg. Neurol. 70, Suppl. 1, 9–18 [DOI] [PubMed] [Google Scholar]
- 39.Hsu S. H., Su C. H., Chiu I. M. (2009) Artif. Organs 33, 26–35 [DOI] [PubMed] [Google Scholar]
- 40.Aftab S., Semenec L., Chu J. S., Chen N. (2008) BMC Evol. Biol. 8, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emery P., Durand B., Mach B., Reith W. (1996) Nucleic Acids Res. 24, 803–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swoboda P., Adler H. T., Thomas J. H. (2000) Mol. Cell 5, 411–421 [DOI] [PubMed] [Google Scholar]
- 43.Senti G., Swoboda P. (2008) Mol. Biol. Cell 19, 5517–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laurençon A., Dubruille R., Efimenko E., Grenier G., Bissett R., Cortier E., Rolland V., Swoboda P., Durand B. (2007) Genome Biol. 8, R195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dubruille R., Laurençon A., Vandaele C., Shishido E., Coulon-Bublex M., Swoboda P., Couble P., Kernan M., Durand B. (2002) Development 129, 5487–5498 [DOI] [PubMed] [Google Scholar]
- 46.Gajiwala K. S., Chen H., Cornille F., Roques B. P., Reith W., Mach B., Burley S. K. (2000) Nature 403, 916–921 [DOI] [PubMed] [Google Scholar]
- 47.Ma K., Zheng S., Zuo Z. (2006) J. Biol. Chem. 281, 21250–21255 [DOI] [PubMed] [Google Scholar]
- 48.Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. (2005) Bioinformatics 21, 2933–2942 [DOI] [PubMed] [Google Scholar]
- 49.Iwama A., Pan J., Zhang P., Reith W., Mach B., Tenen D. G., Sun Z. (1999) Mol. Cell. Biol. 19, 3940–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ducrest A. L., Amacker M., Lingner J., Nabholz M. (2002) Nucleic Acids Res. 30, e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katan-Khaykovich Y., Shaul Y. (2001) Eur. J. Biochem. 268, 3108–3116 [DOI] [PubMed] [Google Scholar]
- 52.Katan-Khaykovich Y., Shaul Y. (1998) J. Biol. Chem. 273, 24504–24512 [DOI] [PubMed] [Google Scholar]
- 53.Eckenstein F. P., Andersson C., Kuzis K., Woodward W. R. (1994) Prog. Brain Res. 103, 55–64 [DOI] [PubMed] [Google Scholar]
- 54.Nurcombe V., Ford M. D., Wildschut J. A., Bartlett P. F. (1993) Science 260, 103–106 [DOI] [PubMed] [Google Scholar]
- 55.Ueba T., Kaspar B., Zhao X., Gage F. H. (1999) J. Biol. Chem. 274, 10382–10387 [DOI] [PubMed] [Google Scholar]
- 56.Ray S. K., Yang X. Q., Chiu I. M. (1997) J. Biol. Chem. 272, 7546–7555 [DOI] [PubMed] [Google Scholar]
- 57.Feng C., Xu W., Zuo Z. (2009) Biochem. Biophys. Res. Commun. 386, 715–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakayama A., Murakami H., Maeyama N., Yamashiro N., Sakakibara A., Mori N., Takahashi M. (2003) J. Biol. Chem. 278, 233–240 [DOI] [PubMed] [Google Scholar]
- 59.Durand B., Vandaele C., Spencer D., Pantalacci S., Couble P. (2000) Gene 246, 285–293 [DOI] [PubMed] [Google Scholar]
- 60.Lubelsky Y., Reuven N., Shaul Y. (2005) Mol. Cell. Biol. 25, 10665–10673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blake M., Niklinski J., Zajac-Kaye M. (1996) J. Virol. 70, 6060–6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reith W., Ucla C., Barras E., Gaud A., Durand B., Herrero-Sanchez C., Kobr M., Mach B. (1994) Mol. Cell. Biol. 14, 1230–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siegrist C. A., Durand B., Emery P., David E., Hearing P., Mach B., Reith W. (1993) Mol. Cell. Biol. 13, 6375–6384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang K. R., Nemoto T., Yokota Y. (2007) J. Biol. Chem. 282, 26167–26177 [DOI] [PubMed] [Google Scholar]
- 65.Liu M., Lee B. H., Mathews M. B. (1999) J. Biol. Chem. 274, 15433–15439 [DOI] [PubMed] [Google Scholar]
- 66.Xu Y., Sengupta P. K., Seto E., Smith B. D. (2006) J. Biol. Chem. 281, 9260–9270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sengupta P., Xu Y., Wang L., Widom R., Smith B. D. (2005) J. Biol. Chem. 280, 21004–21014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sengupta P. K., Fargo J., Smith B. D. (2002) J. Biol. Chem. 277, 24926–24937 [DOI] [PubMed] [Google Scholar]
- 69.Sengupta P. K., Ehrlich M., Smith B. D. (1999) J. Biol. Chem. 274, 36649–36655 [DOI] [PubMed] [Google Scholar]
- 70.Chen L., Smith L., Johnson M. R., Wang K., Diasio R. B., Smith J. B. (2000) J. Biol. Chem. 275, 32227–32233 [DOI] [PubMed] [Google Scholar]