Abstract
The F-box proteins are the substrate recognition subunits of the SCF (Skp1-Cul1-Rbx1-F- box protein) ubiquitin ligase complexes that control the stability of numerous regulators in eukaryotic cells. Here we show that dimerization of the F-box protein Fbx4 is essential for SCFFbx4 (the superscript denotes the F-box protein) ubiquitination activity toward the telomere regulator Pin2 (also known as TRF1). The crystal structure of Fbx4 in complex with an adaptor protein Skp1 reveals an antiparallel dimer configuration in which the linker domain of Fbx4 interacts with the C-terminal substrate-binding domain of the other protomer, whereas the C-terminal domain of the protein adopts a compact α/β fold distinct from those of known F-box proteins. Biochemical studies indicate that both the N-terminal domain and a loop connecting the linker and C-terminal domain of Fbx4 are critical for the dimerization and activation of the protein. Our findings provide a framework for understanding the role of F-box dimerization in the SCF-mediated ubiquitination reaction.
Keywords: Protein Structure, Protein-Protein Interactions, Ubiquitin Ligase, Ubiquitination, X-ray Crystallography
Introduction
Ubiquitin-mediated proteolysis governs the levels of many cellular proteins via a triple-enzyme cascade, involving ubiquitin-activating (E1), -conjugating (E2), and -ligating (E3) activities (1, 2). The E3 ubiquitin ligases recruit both ubiquitin-charged E2 and the targeted substrate so as to confer substrate specificity and mediate ubiquitin transfer from E2 to the substrate (2). RING-based E3s, one of the two major classes of E3s, likely use a zinc-binding RING structural motif to recruit E2 and thus facilitate direct transfer of ubiquitin from E2 onto its substrate (2). The most intensively studied RING E3s are members of the cullin-RING ligase superfamily, which is now recognized as the largest known class of E3 ligases (3–7).
SCF,2 the prototype of the cullin-RING ligases, is built in a modular format that is conserved from yeast to humans (6, 8). In these multisubunit enzymes, an invariable core complex formed by Cul1 (a cullin family member), Rbx1 (a RING domain protein), and Skp1 (an adaptor protein) engages one of a suite of F-box-containing proteins that in turn recruit specific substrates for ubiquitination by an associated E2 enzyme. Structural and mutagenesis studies of SCFs reveal that Cul1 serves as a rigid scaffold on which Rbx1 and Skp1 assemble (9–14). Rbx1 in turn binds an E2 enzyme, whereas Skp1 binds the F-box protein subunit. F-box proteins are the specificity factors in SCF; they interact with Skp1 through the N-terminal ∼40-residue F-box motif to recruit substrates through their variable C-terminal protein-protein interaction domains, including WD40 repeats (Fbxw subfamily), leucine-rich repeats (Fbxl subfamily), and other different or unknown domains (Fbxo subfamily) (15, 16). Recently, a new subfamily within the Fbxo family that recognizes N-linked oligosaccharide, so-called Fbs, has been identified (10, 17). The accumulated evidence suggests that the large number and rich diversity of F-box proteins not only allow the recruitment of numerous, diverse substrates but also position them optimally for the ubiquitination reaction, an important feature of the SCF E3s (13, 14).
The higher order structure of ubiquitin ligases is an important but poorly understood feature of the ubiquitination reaction. For example, a large number of SCF E3s have been shown to dimerize through their F-box subunits, including several Fbxw proteins such as Pop1/Pop2, β-TrCP1/β-TrCP2, Met30, Cdc4, and Fbw7, as well as the Fbxl protein Skp2 (9, 18–24). There is increasing evidence to indicate that F-box protein-mediated dimerization of SCF is important for its ubiquitination activity and may be a key aspect of understanding how ubiquitin transfer occurs (6); however, the underlying mechanism and regulation remain incompletely understood. Notably, all available crystal structures of the F-box-containing proteins contain truncated proteins that are monomeric (6). It has been proposed that the N-terminal region preceding the F-box (named the D domain) is responsible for dimerization of Fbxw proteins (9, 22, 24). Based on the crystal structure of the isolated D domain and small angle x-ray scattering analysis of the dimeric Skp1-Cdc4, a model of the dimeric SCFCdc4 (the superscript denotes the F-box protein) complex has been proposed in which two substrate-binding domains and two E2-binding sites form a coplanar configuration in a suprafacial orientation (22). Hence, atomic structure of a dimeric SCF may provide a framework for understanding how dimerization contributes to the function and regulation of E3s.
The ubiquitin-protein ligase SCFFbx4 was recently identified as the E3 responsible for the ubiquitination and subsequent degradation of the cell cycle regulator cyclin D1 and telomeric DNA-binding protein Pin2 (also known as TRF1) (25, 26). Inactivation of SCFFbx4 appears at least in part to account for overexpression of cyclin D1 in human cancers (26), and mutations in the Fbx4 subunit of SCFFbx4 were found to be associated with human primary esophageal carcinoma (27, 28). On the other hand, overexpression of Fbx4 in human cells leads to degradation of endogenous Pin2 and results in progressive telomere elongation (25). Moreover, several features of the SCFFbx4 ligase make it unique relative to many other SCF ligases. First, Fbx4 belongs to the Fbxo subfamily of F-box proteins that lack recognizable protein-protein interaction motifs in their C termini (16). Second, Fbx4 recognizes its two known substrates, Pin2 and cyclin D1, in two very different manners. Fbx4-mediated ubiquitination of Pin2 is independent of its phosphorylation status (25), whereas that of cyclin D1 requires phosphorylation on its Thr-286 residue, as well as an interaction with αB-crystallin, a small heat-shock protein (26). Fbx4 is the only other known ligase where substrate recognition requires an accessory protein in addition to the F-box component; the other example is the leucine-rich repeat-containing F-box protein Skp2 that utilizes Cks1 for efficient recognition and ubiquitination of p27Kip1 (29, 30). Third, recent studies suggest that Fbx4 forms an active dimeric complex and that the dimerization itself is regulated by phosphorylation of an N-terminal serine (27, 28). The significance of this phosphorylation-dependent dimerization is further highlighted by the discovery of mutations of this serine residue in human cancer (28). As such, Fbx4 dimerization may not only be key to understanding how ubiquitin transfer occurs but may also be a new regulatory mechanism underlying SCFFbx4-mediated ubiquitination.
Here we demonstrate the dimerization-mediated ubiquitination activity of Fbx4 and present the crystal structure of the dimeric Skp1-Fbx4 complex. Our structural, biophysical, and biochemical studies show that: 1) the full-length Fbx4 is monomeric and essentially inactive in ubiquitinating Pin2 in vitro; 2) a loop connecting the linker domain to the C-terminal substrate-binding domain of Fbx4 is crucial for dimerization, and the head-to-tail dimerization configuration of a truncated Fbx4 protein likely plays a role in promoting substrate binding and ubiquitin transfer; 3) the extreme N-terminal region of Fbx4 is important for interaction with Cul1-Rbx1 and may be responsible for regulating Fbx4 dimerization and ubiquitination activity; and 4) Fbx4 possesses a previously uncharacterized type of substrate recognition domain. Collectively, our results delineate a critical role for Fbx4 dimerization in regulating SCFFbx4 ubiquitination activity and support the notion that inactivation of the Fbx4 ligase in human cancer may result from mutations that impair ligase dimerization.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Recombinant human Fbx4 variants and truncated Skp1 (12) were co-expressed as glutathione S-transferase (GST) and His6 fusion proteins, respectively, in Escherichia coli. Co-expression was achieved either by transforming with one plasmid containing a dicistronic message or by transforming with two plasmids containing distinct origins of replication (p15A and pBR322) (31). The Fbx4core construct used in crystallization experiments lacks the N-terminal region preceding the F-box domain (residues 1–54; NTD) and a 21-residue region (residues 150–170; finger) in the linker domain. The Skp1-Fbx4 complexes were purified by glutathione affinity chromatography followed by tobacco etch virus protease cleavage of the GST tag and by anion exchange and gel filtration chromatography. For crystallization, the Skp1-Fbx4core complex was concentrated to 25 mg/ml by ultrafiltration in 25 mm Tris-HCl (pH 8.0), 100 mm NaCl, 5 mm dithiothreitol. Human full-length Pin2 (Pin2FL) and its N-terminal domain (residues 48–268; Pin2NTD) were expressed in E. coli with an N-terminal His6 tag and purified by Ni2+-NTA affinity chromatography, and following cleavage of the tag by tobacco etch virus protease, were purified by anion exchange and gel filtration chromatography. Human E1, UbcH5, and Cul1-Rbx1 were produced as described previously (32).
Crystallization and Structure Determination
The Skp1-Fbx4core complex was crystallized from 1.5–2 m NaCl by the hanging-drop vapor diffusion method at 4 °C. Heavy atom soaks were performed in mother liquor supplemented with 1 mm KAu(CN)2 for 3 h. Crystals were flash-frozen in solutions containing saturated NaCl. Diffraction data were collected at the X3A and X29 beamlines of the National Synchrotron Light Source (NSLS). Data were processed using the HKL2000 suite (33). The crystals contain one Skp1-Fbx4core complex in the asymmetric unit. The structure of the Skp1-Fbx4core complex was determined by the single wavelength anomalous dispersion method using the KAu(CN)2 derivative. Two gold sites were located by SHELXD (34), and phases were calculated with SHARP (35) followed by density modification with SOLOMON (36) and RESOLVE (37). The Skp1 molecule was located in the initial electron density map by molecular replacement using the structure of Skp1 in the Skp1-Skp2 complex (12) as the search model. The entire model was built with the program Coot (38) and improved by several cycles of manual rebuilding and refinement with CNS (39) and REFMAC (36). In the refined model, residues 161–163 of Skp1 and residues 132–135, 242–271, and 384–387 of Fbx4core were not visible in the electron density maps and are likely to be disordered. The final model contains 81.5, 13.9, 4.6, and 0.0% of the amino acids in the most favored, additionally allowed, generously allowed, and disallowed regions of the Ramachandran plot, respectively. All figures depicting structures were prepared using PyMOL (DeLano Scientific LLC).
In Vitro Pin2 Ubiquitination Assay
The ubiquitination assays were performed by incubating E1 (0.5 μm), UbcH5 (2.0 μm), Cul1-Rbx1 (2.0 μm), Pin2FL (17 μm), ubiquitin (205 μm), ATP (10 mm), MgCl2 (20 mm), and the Skp1-Fbx4 wild-type or mutant proteins (2.0 μm) in 50 mm Tris-HCl (pH 8.0), 150 mm NaCl at room temperature for 1 h or as indicated. Reactions were stopped by boiling in SDS-Laemmli buffer and were analyzed by SDS-PAGE. Pin2 and Pin2-ubiquitin-conjugated products were detected by immunoblotting using polyclonal antibodies against a Pin2 N-terminal peptide (Santa Cruz Biotechnology). Pin2FL proteins were phosphorylated by using a 1:40 molar ratio of the GST-tagged Cdk1-cyclin B kinase complex in 50 mm Tris-HCl (pH 8.0), 1 mm ATP, 10 mm MgCl2, 0.2 μCi of [γ-32P]ATP. The phosphorylated Pin2FL was separated from GST-Cdk1-cyclin B by glutathione affinity chromatography followed by G-25 size exclusion chromatography. The ubiquitination reactions were performed as described above except that the Pin2FL used was at 9 μm. The dried gels were exposed to storage phosphor screens, scanned with a Storm 860 molecular imager (GE Healthcare), and quantified using ImageGauge software.
Sedimentation Equilibrium Analysis
Analytical ultracentrifugation measurements were carried out on a Beckman XL-A analytical ultracentrifuge equipped with an An-60 Ti rotor (Beckman Coulter) at 20 °C. Protein samples were dialyzed overnight against 20 mm Tris-HCl (pH 8.0), 200 mm NaCl, 2 mm dithiothreitol and loaded at initial concentrations of 5, 30, and 100 μm for Skp1-Fbx4FL and 30, 100, and 300 μm for Skp1-Fbx4core. Data were acquired at two wavelengths per rotor speed setting and processed simultaneously with a nonlinear least squares fitting routine. Solvent density and protein partial specific volume were calculated according to solvent and protein composition, respectively.
Affinity Pulldown Assays
For reciprocal affinity pulldown assays, the His6-tagged Fbx4 F-box-linker domain (residues 55–149; the wild type and the Val-113 deletion mutant)-Skp1 complex; and the GST-tagged Fbx4 G domain (residues 171–387) were co-expressed in E. coli. Cell lysates were loaded onto either a Ni2+-NTA or a glutathione-Sepharose 4B column at 4 °C. The columns were washed extensively with buffer consisting of 50 mm Tris-HCl (pH 8.0) and 150 mm NaCl. Proteins were eluted with 250 mm imidazole (for Ni2+-NTA) and 15 mm glutathione (for glutathione-Sepharose 4B), respectively. For the wild-type protein, the Ni2+-NTA eluate was then loaded onto a glutathione-Sepharose 4B column and eluted with glutathione. For the GST control, the pulldown assay was performed as described above except that the purified GST and His6-Fbx4 F-box-linker domain-Skp1 complex were used. All proteins were analyzed with SDS-PAGE and Coomassie Blue staining.
For Ni2+-NTA affinity pulldown assays, the indicated proteins (12 μm each) were incubated at room temperature for 10 min in 30 μl of binding buffer consisting of 50 mm Tris-HCl (pH 8.0), 75 mm NaCl, and 10 mm imidazole prior to the addition of 28 μl of Ni2+-NTA resin. After 10 min, the resin was spun down, and the ∼25-μl supernatant, marked as the unbound (U) fraction in Fig. 5B, was removed. The resin was then washed three times with 0.6 ml of binding buffer, and the Ni2+ bound proteins were eluted with 30 μl of buffer consisting of 50 mm Tris-HCl (pH 8.0), 200 mm NaCl, and 600 mm imidazole. One-third of each of the supernatant and eluted fractions (marked as the bound (B) fractions in Fig. 5B) was analyzed by SDS-PAGE and Coomassie Blue staining.
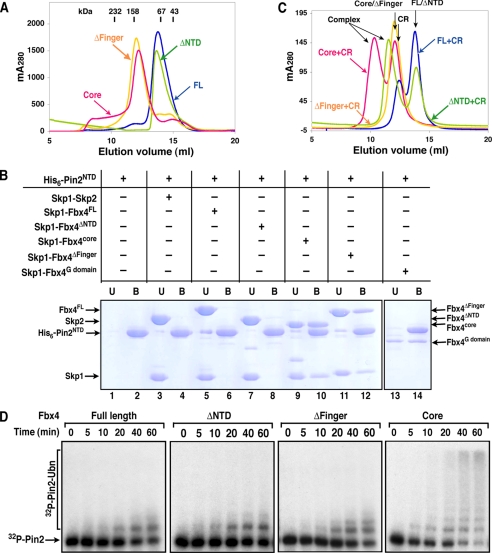
FIGURE 5.
Both the NTD and the finger regulate Fbx4 activity and dimerization. A, overlay of gel filtration chromatography profiles of Skp1-Fbx4FL, Skp1-Fbx4ΔNTD, Skp1-Fbx4ΔFinger, and Skp1-Fbx4core. The retention volumes of proteins of known mass are indicated. mA280, milliunits at A280 nm. B, Ni2+-NTA affinity pulldown assay characterizing the interaction of the His6-tagged Pin2NTD with the Skp1-Fbx4 complexes. Indicated proteins were incubated, and reactions were precipitated with Ni2+-NTA resin. The unbound (U) and eluted bound (B) fractions were analyzed by SDS-PAGE and Coomassie Blue staining. C, superimposed gel filtration profiles of the Skp1-Fbx4-Cul1-Rbx1 (CR) mixtures. The Skp1-Fbx4 complex (Skp1-Fbx4FL, Skp1-Fbx4ΔNTD, Skp1-Fbx4ΔFinger, or Skp1-Fbx4core) was incubated with Cul1-Rbx1 at a 1.5:1 molar ratio before loading onto the gel filtration column. The retention volumes of the four protein complexes and the individual subcomplexes are indicated. D, time course of ubiquitination of the 32P-labeled Pin2FL protein by Fbx4FL, Fbx4ΔNTD, Fbx4ΔFinger, and Fbx4core. 32P-labeled Pin2 and Pin2-ubiquitin conjugates were analyzed by SDS-PAGE followed by autoradiography. Ubn, more than two Ub molecules attached.
RESULTS
Fbx4 Dimerization Is Required for SCFFbx4 Function in Vitro
To characterize the structure-activity relationships of Fbx4, we produced the binary Skp1-Fbx4 complex in E. coli by co-expressing a truncated Skp1 construct (12) with the full-length human Fbx4 (hereafter Fbx4FL). Gel filtration chromatography showed that Skp-Fbx4FL eluted at an apparent molecular mass of ∼75 kDa, comparable with the calculated 61.2 kDa molecular mass for a monomer (Fig. 1A). Sedimentation equilibrium experiments confirmed that Skp-Fbx4FL forms exclusively a monomer (Fig. 1B). To determine the ubiquitination activity of this monomeric Skp1-Fbx4FL complex, we developed an in vitro assay in which purified human E1, UbcH5 E2, Cul1-Rbx1, and Skp1-Fbx4FL were reconstituted to measure the ubiquitination of the purified human Pin2 protein, one of two known substrates of SCFFbx4 (25). Interestingly, the addition of Skp1-Fbx4FL to reaction mixtures only resulted in the formation of low molecular weight Pin2-ubiquitin conjugates (Pin2-Ub1 and Pin2-Ub2) in 60 min, as detected by immunoblotting with a Pin2-specific antibody (Fig. 1C, lane 5). This activity was comparable with those of the control reactions in the absence of F-box protein (lane 3) or in the presence of Skp1-Skp2 (an F-box protein belonging to the Fbxl subfamily) (lane 4). Because αB-crystallin is the accessory protein for the SCFFbx4 ligase to target cyclin D1 (26), we further sought to determine whether αB-crystallin plays a role in Pin2 ubiquitination by SCFFbx4 in our in vitro assay. As shown in Fig. 1C (lane 7), the addition of αB-crystallin did not stimulate the polyubiquitination of Pin2. We conclude that the monomeric full-length Fbx4 protein lacks specific ubiquitination activity toward Pin2 in our in vitro settings. The low molecular weight Pin2-ubiquitin conjugates produced in the Fbx4FL reactions presumably resulted from transient interaction between the ubiquitin-charged E2 and Pin2, which is independent of the activation by SCF (40).
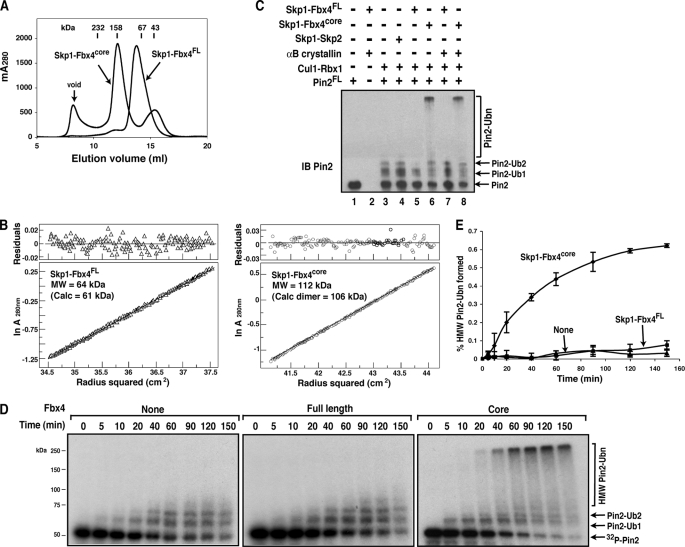
FIGURE 1.
Oligomerization state and ubiquitination activity of the Skp1-Fbx4 complexes. A, overlay of gel filtration chromatography profiles of Skp1-Fbx4FL and Skp1-Fbx4core. The retention volumes of proteins of known mass and the void volume of the Superdex 200 column are indicated. mA280, milliunits at A280 nm. B, representative sedimentation equilibrium data for the Skp1-Fbx4FL (left, 100 μm and 12 krpm) and Skp1-Fbx4core (right, 100 μm and 10 krpm). MW, molecular weight; Calc, calculated; krpm, kilorevolutions per minute. C, in vitro ubiquitination of Pin2FL by Skp1-Fbx4FL and Skp1-Fbx4core in the presence and absence of αB-crystallin. Pin2 and Pin2-ubiquitin conjugates were detected by immunoblotting (IB) with an anti-Pin2 antibody. Ubn, more than two Ub molecules attached. D, time course of ubiquitination of the 32P-labeled Pin2FL protein in the absence and presence of Skp1-Fbx4FL or Skp1-Fbx4core. 32P-labeled Pin2FL and Pin2FL-ubiquitin conjugates were analyzed by SDS-PAGE followed by autoradiography. HMW, high molecular weight. E, quantitative PhosphorImager analysis of the high molecular weight ubiquitinated Pin2FL from D. The ratios of the high molecular weight Pin2/total Pin2 proteins are plotted as a function of reaction time. Data represent the mean ± S.D. for 2–3 independent experiments.
During the course of this study, we designed and produced a series of truncation mutants of Fbx4 by using limited proteolysis and mass spectrometry to identify and eliminate flexible loop regions that might interfere with protein crystallization. We found that the N-terminal region preceding the F-box domain (residues 1–54; NTD) and a 21-residue region (residues 150–170; denoted as finger) of Fbx4 were highly susceptible to limited proteolysis and are presumably unstructured. Unexpectedly, Skp1-Fbx4core (Fbx4 lacking both the NTD and the finger) eluted earlier than Skp1-Fbx4FL on a gel filtration column, with an apparent molecular mass of ∼135 kDa (the calculated molecular mass for a dimer is 106 kDa) (Fig. 1A). Sedimentation equilibrium experiments showed that Skp1-Fbx4core sediments as a discrete dimer in solution (Fig. 1B).
Unlike the monomeric Skp1-Fbx4FL described above, dimeric Skp1-Fbx4core significantly stimulated the formation of high molecular weight Pin2-ubiquitin conjugates (Fig. 1C, lane 6). The addition of αB-crystallin did not result in any stimulatory effect (Fig. 1C, lane 8). To compare the ubiquitination activities of Skp1-Fbx4FL and Skp1-Fbx4core in a more quantitative manner, we used the Pin2 protein phosphorylated by the Cdk2-cyclin B kinase complex so as to label Pin2 with 32P. As shown in Fig. 1, D and E, Skp1-Fbx4core substantially increased both the initial rate of Pin2 ubiquitination and the formation of high molecular weight polyubiquitin conjugates, whereas Skp1-Fbx4FL failed to promote polyubiquitination of Pin2. After 20 min, the Skp1-Fbx4FL reaction yielded ∼23% mono- and diubiquitinated Pin2, a value comparable with that of the reaction without the F-box protein (∼18%). In contrast, the Skp1-Fbx4core reaction took only 5 min to produce similar yields (∼22%). Moreover, the addition of Skp1-Fbx4core greatly increased the rate of the formation of high molecular mass polyubiquitin conjugates (>75 kDa with ≥3 Ub attached). In 150 min, ∼65% of the Pin2 substrate was modified with more than 3 Ub by Skp1-Fbx4core but less than 8% by Skp1-Fbx4FL or ∼5% in the absence of the F-box protein (Fig. 1E). Taken together, our results indicate that monomeric full-length Fbx4 is essentially inactive in ubiquitinating Pin2 in vitro, whereas dimeric Fbx4core efficiently stimulates polyubiquitination of Pin2 in vitro.
Overall Structure of the Dimeric Skp1-Fbx4core Complex
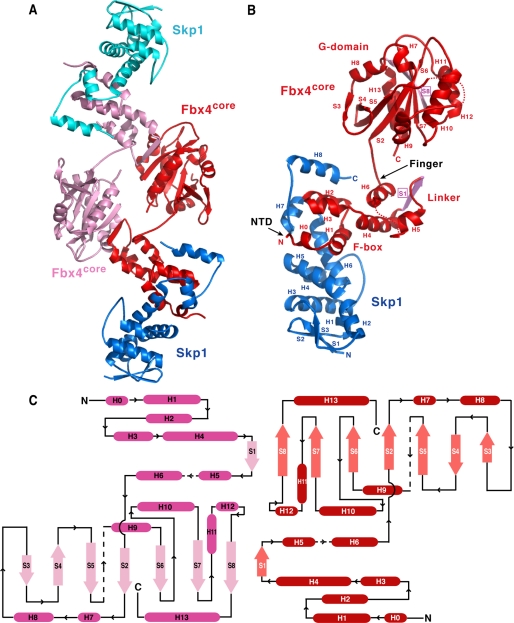
To explore the molecular mechanism of Fbx4 dimerization and activation, we crystallized the binary Skp1-Fbx4core complex and determined its structure at 2.8 Å resolution by the single wavelength anomalous dispersion method (Table 1). The Skp1-Fbx4core complex contains a central Fbx4core homodimer with symmetrically disposed Skp1 molecules on each side, whose overall structure resembles a twisted section sign § (Fig. 2, A and C). The Fbx4 dimer is created by a crystallographic two-fold axis, and two protomers associate in an antiparallel manner such that the N-terminal region of one molecule interacts with the C-terminal region of another. Each Skp1-Fbx4 molecule has an overall elongated structure, with the N-terminal β sheet of Skp1 and the C-terminal substrate-binding domain of Fbx4 located at opposite ends (Fig. 2B). The Fbx4 protomer consists of an N-terminal F-box domain (helices H0, H1–H3); a C-terminal domain (S2–S8 and H7–H13, denoted as the G domain (below)); and a linker domain connecting the N- and C-terminal domains (helices H4–H6, strand S1, as well as H5–H6 and H6–S2 loops) (Fig. 2, B and C). The Fbx4 F-box domain has the same overall fold as in other Fbxl, Fbxw, and Fbs structures (9–13, 41) (supplemental Fig. S1). The binding of the F-box to Skp1 is also very similar to other Skp1-F-box protein complexes (supplemental Fig. S1). Interestingly, the C-terminal domain of Fbx4 adopts a compact α/β fold, which is distinct from the known structures of substrate-binding regions of F-box proteins, reinforcing the notion that Fbx4 represents a new subfamily of substrate recognition proteins in SCF E3s (Fig. 2B).
TABLE 1.
Summary of crystallographic analysis
| Nativea | KAu(CN)2a | |
|---|---|---|
| Data collection | ||
| Space group | P3121 | P3121 |
| Cell dimensions (Å) | ||
| a, b, c (Å) | 92.2, 92.2, 148.1 | 91.9, 91.9, 147.8 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 50.0-2.80 (2.90-2.80) | 50.0-2.95 (3.06-2.95) |
| Rsymb (%) | 8.6 (85.6) | 8.5 (48.9) |
| I/σI | 31.5 (2.2) | 46.4 (4.9) |
| Completeness (%) | 99.8 (100.0) | 99.8 (100.0) |
| Redundancy | 14.3 (14.3) | 11.6 (11.7) |
| Refinement | ||
| Resolution (Å) | 50.0-2.80 | |
| No. of reflections | 18,235 | |
| Rwork/Rfree | 24.9/29.5 | |
| No. of atoms | ||
| Protein | 3,350 | |
| Water | 66 | |
| B-factors | ||
| Protein | 45.62 | |
| Water | 39.98 | |
| r.m.s.c deviations | ||
| Bond lengths (Å) | 0.017 | |
| Bond angles (°) | 1.9 | |
a The diffraction data collected with native and KAu(CN)2-derivatized crystals. Values in parentheses are for the highest-resolution shell.
b Rsym = ΣhΣj|Ih,j − Ih|/ΣhΣj Ih,j for the intensity (I) of i observations of reflection h.
c r.m.s., root mean square.
FIGURE 2.
Overall structure of the Skp1-Fbx4core complex. A, ribbon diagram of the Skp1-Fbx4core dimer. Fbx4core and Skp1 are shown in red and blue in one molecule and in pink and cyan in the other one. B, ribbon diagram of the Skp1-Fbx4core monomer. The secondary structure elements are labeled. Dotted lines represent disordered regions. The strands in Fbx4core involved in dimerization are colored pink. Deletions of the NTD and finger are indicated by arrows. C, the topology diagram of the Fbx4core dimer. α helices and β strands are shown in red and maroon in one molecule and violet and pink in the other one. Dotted lines represent disordered regions.
The Fbx4 linker domain forms a stem-like structure, thereby positioning the C-terminal G domain well away from the F-box and Skp1 (Fig. 2B). The three helices (H4–H6) of the linker domain pack into a platform that is anchored tightly on the F-box, whereas the extended amphipathic H6–S2 loop projects from H6 to bridge the linker domain to the G domain. Strand S1 in the linker domain forms an extended antiparallel β sheet with strands from the G domain in the other protomer, making up the majority of the dimerization interface (Fig. 2, A and C). The relative orientation of the F-box domain and G domain in the Fbx4 protomer is imposed almost entirely through the conformation of the H6–S2 loop and thus appears less rigid than the corresponding regions in other F-box proteins that mostly employ a tightly anchored helix to bridge the domains (supplemental Fig. S1). We should point out that the truncated finger is located in the beginning of the H6–S2 loop, which is presumably in close proximity to the S1 strand (Fig. 2B). It is thus conceivable that the finger may affect the orientation of the G domain as well as the dimerization of Fbx4 (below).
The overall spatial arrangement of Skp1 and Fbx4 is highly analogous to other Skp1-F-box protein complexes (supplemental Fig. S1). However, the substrate-binding domains of the F-box proteins differ in their relative orientations by up to ∼90°; β-TrCP1 folds into the most curved conformation relative to Skp1, and Fbx4 assumes the most extended fold. Together, the structure of the Skp1-Fbx4core complex is consistent with the notion that plasticity of the linkage between the F-box and the substrate-binding domain is important for SCF function and specificity and that the F-box protein positions and orients the substrate optimally for ubiquitination by the SCF-bound E2 (9).
Structure of the Substrate-binding Domain in Fbx4
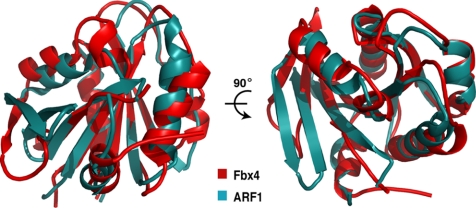
The C-terminal domain of Fbx4 consists of a central seven-stranded β sheet surrounded by seven α helices (Fig. 2, B and C). The mixed β sheet is composed of two antiparallel (S3 and S4) and five parallel strands (S2, S5–S8), which pack around the central S2 strand to form a curved plane. Strand S2 extends to connect the H6 helix in the linker domain. Helices H7, H8, and H13 lie above the plane of the β sheet, whereas H9, H10, and H12 lie below the plane. S3 and H11 on the plane of the β sheet form its outer edges. Unexpectedly, a structure homology search of the Protein Data Bank using the Dali server (42) revealed that the Fbx4 C-terminal domain has a similar fold to many small GTP-binding proteins (G proteins); G proteins are involved in molecular switching by mediating cycling between active GTP-bound and inactive GDP-bound conformational states (43, 44). Indeed, all GDP/GTP-binding domains (called G domain) in G proteins have a common topology consisting of a mixed five- to seven-stranded β sheet and 5–7 α helices located on both sides (43, 44). This overall fold resembles the C-terminal domain of Fbx4. For instance, the Fbx4 C-terminal domain can be superimposed on ADP-ribosylation factor 1 (ARF1) (45) with a root mean square deviation of 2.4 Å for the 180 Cα atoms (out of 210 C-terminal residues in Fbx4) used in the superposition (Fig. 3), although the two proteins have only ∼20% sequence identity. We therefore name the C-terminal domain of Fbx4 as the G domain. Little is presently known about how the G domain of Fbx4 recognizes its substrates.
FIGURE 3.
Structure of the Fbx4 C-terminal domain is similar to that of small GTP-binding proteins. Superimposition of the Fbx4 C-terminal domain (red) and ARF1 (cyan; Protein Data Bank (PDB) code 1HUR) is shown. The unique N-terminal α helix of ARF1 is omitted for clarity.
Dimer Interface
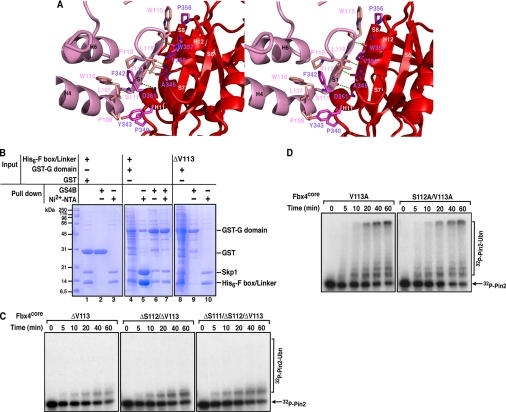
The Fbx4 homodimer is formed in a head-to-tail manner whereby the linker domain of one monomer interacts with the G domain of the other (Fig. 2B). The dimer interface involves the S1 strand, the H4 helix, and the S1–H5 loop from the link domain and the S8 strand and the H11 helix from the G domain; ∼1300 Å2 of solvent-accessible surface area in each monomer is buried in the interfaces. It is likely that this tightly associated antiparallel dimer helps stabilize the relative orientation of the F-box and G domain in each protomer (Fig. 2A). The intermolecular interactions are mediated by backbone hydrogen bonds as well as van der Waals contacts. The most evident feature of the dimer interface is that the S1 strand in the linker domain extends the seven-stranded β sheet in the G domain of another molecule by antiparallel β augmentation (46) (Fig. 2C). The S1 strand (Ser-111, Val-113, and Trp-115) binds the S8 strand (Asp-361, Val-359, and Trp-357) through four β sheet backbone hydrogen bonds (Fig. 4A). In addition, the indole ring of Trp-115 stacks with the pyrrolidine group of Pro-356 to stabilize the edge of the β sheet. Pro-356 is invariant in those Fbx4 orthologs, and Trp-115 is identical in all but the zebrafish Fbx4 homologs, where it is a His (supplemental Fig. S2). It thus appears that the intermolecular interactions between strands S1 and S8 are crucial for dimer formation as well as Fbx4 activity. van der Waals contacts occur on the back of the β sheet leaning toward the linker domain (Fig. 4A). The side chain of Phe-342 from the H11 helix of the G domain dips into a hydrophobic pocket flanked by the S1 strand (Val-113), the H4 (Leu-107, Pro-108, and Trp-110) helix, and the S1–H5 loop (Leu-118 and Pro-119) of the linker domain on one side and the H11 helix (Phe-342 and Ala-345) as well as the S8 strand (Trp-357 and Val-359) of the G domain on the other side (Fig. 4A). In addition, the side chains of Pro-108 (H4), Pro-340 (the S7–H11 loop), and Tyr-343 (H11) act as a lid to stabilize the hydrophobic pocket. Importantly, these Fbx4 residues involved in intermolecular van der Waals interactions are conserved or conservatively substituted in Fbx4 orthologs (supplemental Fig. S2).
FIGURE 4.
Dimer interface in the Skp1-Fbx4core structure. A, a stereo view of the dimer interface of Fbx4core. The two protomers are colored red and pink, respectively. Hydrogen bonds are represented by green dashed lines. The H5 and H13 helices are omitted for clarity. B, reciprocal affinity pulldown assays with the co-expressed His6-F-box-linker-Skp1 complex and GST-G domain. Cell lysates were incubated with Ni2+-NTA and glutathione-Sepharose 4B (GS4B) beads, eluted with imidazole and/or glutathione, respectively, and analyzed with SDS-PAGE and Coomassie Blue staining. Purified GST protein was incubated with purified His6-F-box-linker-Skp1 complex and used as the negative control. C, time course of ubiquitination of the 32P-labeled Pin2FL protein by three Fbx4core deletion mutants. 32P-labeled Pin2 and Pin2-ubiquitin conjugates were analyzed by SDS-PAGE followed by autoradiography. Ubn, more than two Ub molecules attached. D, time course of ubiquitination of the 32P-labeled Pin2FL protein by two Fbx4core Ala mutants. 32P-labeled Pin2 and Pin2-ubiquitin conjugates were analyzed by SDS-PAGE followed by autoradiography.
Importance of the Fbx4 Dimer Interface
Given the dimeric structure of Fbx4core, we next sought to test whether the linker-G domain interactions observed in the crystal structure are indeed important for Fbx4 dimerization in solution. The structure of the Skp1-Fbx4core indicated that in the case of monomer, there is no direct interaction between the G domain and the linker domain from the same molecule (Fig. 2B). In the dimer, however, the G domain of one monomer interacts directly with the linker domain of the other monomer. If this is also the case in solution, we should be able to reconstitute a mixed monomer using the two individual domains. We assessed the interaction between the F-box-linker domain and the G domain using a reciprocal affinity pulldown assay. As shown in Fig. 4B, the His6-tagged F-box-linker-Skp1 complex could pull down the GST-tagged G domain and vice versa (lanes 5 and 6). Importantly, the subsequent glutathione-Sepharose 4B (GS4B) pulldown experiment using the Ni2+-NTA eluates showed that excess His6-tagged F-box-linker-Skp1 could be washed off the glutathione-Sepharose 4B resin so that the GST-G domain and His6-F-box-linker-Skp1 could be eluted in an ∼1:1 molar ratio (Fig. 4B, lane 7). Moreover, deleting even a single residue (i.e. Val-113) in the S1 strand of the His6-tagged F-box-linker domain completely disrupted the interaction between the linker and G domains (Fig. 4B, lanes 9 and 10). These results support our structural observation that the Fbx4 dimer is formed by association of the G domain and the F-box-linker domain.
The Skp1-Fbx4core structure indicates that the backbone hydrogen bond contacts between S1 and S8 strands are important for Fbx4 dimerization and activity. To confirm the importance of the S1–S8 interaction seen in the crystal structure, we investigated the effects of deleting residues from the S1 strand on Fbx4 ubiquitination activity. Removal of Val-113 in the S1 strand abolished ubiquitination activity of Fbx4core (Fig. 4C), and the protein eluted as a mixture of monomers and dimers on a gel filtration column (data not shown). Similarly, the double (Ser-112/Val-113) and triple (Ser-111/Ser-112/Val-113) deletions in the S1 strand substantially impaired Pin2 ubiquitination (Fig. 4C). In contrast, alanine substitutions for Val-113 alone or both Ser-112 and Val-113 had essentially no effect on dimer formation as well as the ubiquitination activity of Fbx4core (Fig. 4D). Therefore, the main-chain interactions between S1 and S8 strands likely make important contributions to Fbx4 activity. More importantly, our data support the notion that the interactions that mediate Fbx4 dimerization are also critical for its activity.
Role of the NTD and Finger in Fbx4 Dimerization and Function
Given that monomeric full-length Fbx4 is inactive, whereas the truncated Fbx4core dimer is active, we sought to examine whether the activity of Fbx4core is due to truncation or dimerization or both. When compared with the full-length protein, Fbx4core lacks the NTD and the finger regions. We thus tested the effects of individually deleting the NTD and the finger on protein dimerization, substrate and Cul1-Rbx1 binding, and ubiquitination activity. Gel filtration chromatography showed that the construct lacking the NTD (Fbx4ΔNTD) eluted as a monomer, whereas the construct lacking the finger (Fbx4Δfinger) eluted as a dimer (Fig. 5A), suggesting that the finger represses Fbx4 dimerization. We next assessed the binding of the His6-tagged Pin2 N-terminal domain (residues 48–268; Pin2NTD) to the Skp1-Fbx4 complexes using the Ni2+-NTA affinity pulldown assay. Mutagenesis studies have suggested that Pin2NTD is sufficient to interact with Skp1-Fbx4 (25). As shown in Fig. 5B, His6-Pin2NTD pulled down both Fbx4core and Fbx4Δfinger (lanes 10 and 12) but did not bind Fbx4FL and Fbx4ΔNTD (lanes 6 and 8 the Skp1-Skp2 complex was used as a negative control). Additionally, His6-Pin2NTD also pulled down the Fbx4 G domain, which is a monomer (Fig. 5B). These data suggest that Fbx4 dimerization is essential for substrate binding, and dimerization likely functions in this regard by exposing the substrate-binding site in the G domain.
Association of the wild-type and mutant Skp1-Fbx4 complexes with Cul1-Rbx1 was also characterized on a gel filtration column. Both Skp1-Fbx4FL and Skp1-Fbx4ΔFinger eluted with Cul1-Rbx1 as two separate peaks, whereas Skp1-Fbx4core and Skp1-Fbx4ΔNTD coeluted with Cul1-Rbx1 in a single peak with a retention volume distinct from those of the subcomplexes (Fig. 5C and supplemental Fig. S3). These data suggest that the N-terminal domain of Fbx4 blocks the formation of a stable SCF complex, consistent with the SCFFbx4-E2 docking model in which the N terminus of Fbx4core is in close proximity to the Cul1-F-box interface (see supplemental Fig. S4 for discussion). In keeping with the substrate and Cul1-Rbx1 binding results, the Pin2 ubiquitination assays showed that both deletion mutants, Fbx4ΔNTD and Fbx4Δfinger, are essentially inactive when compared with Fbx4core, suggesting that both the NTD and the finger are responsible for regulating Fbx4 activity and that the dimerization alone is essential but not sufficient to promote Fbx4-mediated ubiquitination (Fig. 5D). We have also identified the minimal regions in the NTD and finger that can repress Fbx4 activity and dimerization (see supplemental Fig. S5 for discussion). As both the NTD and the finger repress Fbx4 activity in vitro, the underlying mechanism by which Fbx4 dimerization and activation are regulated in vivo becomes a subject of great interest for future studies.
DISCUSSION
Our structural and biochemical data address two specific issues concerning the architecture, substrate binding, and regulation of the SCFFbx4 ubiquitin ligase. First, the unique fold of the C-terminal domain of Fbx4 reinforces the notion that Fbx4 represents a new class of substrate recognition proteins in SCF E3s. Second, the dimer configuration of Fbx4 provides a molecular basis for interpreting the biochemical requirements for SCFFbx4 dimerization in promoting substrate binding and ubiquitin transfer. Moreover, the structural and biochemical studies described here provide a framework for the mechanistic description of how the activity and dimerization of Fbx4 are regulated in the ubiquitination process in the cell.
Fbx4 belongs to the Fbxo class of F-box proteins that includes at least 46 members in humans (16). However, the G domain of Fbx4 shares no significant sequence homology with other Fbxo proteins (data not shown), although this fold is likely conserved among Fbx4 orthologs from human, mouse, rat, cow, dog, chicken, chimp, frog, opossum, and zebrafish (supplemental Fig. S2). Therefore, Fbx4 likely recognizes its two substrates, Pin2 and cyclin D1, in a unique manner, and two distinct substrate recognition modes by the same F-box protein seem to reflect the complexity of SCF E3 actions. Further studies will be required to gain detailed insights into the substrate recognition mechanisms of Fbx4.
Our biochemical analyses demonstrate that Fbx4 dimerization is absolutely required for SCFFbx4 activity as all the monomeric constructs of Fbx4 were essentially inactive and deletion of even a single residue in the Fbx4 dimer interface profoundly impaired the ubiquitination of Pin2 (Figs. 1D, 4C, and 5D). The affinity pulldown assays further showed that substrate binding critically depends on Fbx4 dimerization as only the dimeric constructs (as in Fbx4core and Fbx4ΔFinger) bind to substrate (Fig. 5B). Consistent with the biochemical analyses, our structural data suggest that the formation of the antiparallel Fbx4 dimer can stabilize optimal orientation of the F-box domain and G domain in the protomer and thus may expose the substrate-binding site in the G domain (Figs. 2A and 6A). This is unique because previous studies have shown that dimerization of the F-box protein Cdc4 increases both the initial rate of ubiquitination and the processivity of polyubiquitination for Sic1 but has no effect on substrate recruitment (9, 22, 24). Moreover, the dimer configuration in the SCFFbx4-E2 docking model suggests that the Fbx4 dimer positions the substrate-binding domain on one SCF monomer in closer proximity to the E2 catalytic center of the neighboring molecule than to the E2 on its own SCF, although the exact substrate-binding site in the G domain is not known (supplemental Fig. S4). This configuration supports the model that dimeric SCFFbx4 promotes optimal pairing between E2 and substrate in trans, which is not possible for the monomeric enzyme, thus providing a simple solution to partially overcome the large gap between the substrate-binding site and the bound E2 that is observed in the extant monomeric SCF structures (6, 8, 9, 23).
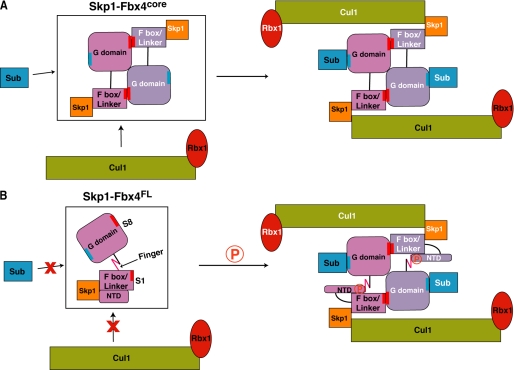
FIGURE 6.
Models for dimerization-dependent ubiquitination by Fbx4core and Fbx4FL. A, the dimeric Skp1-Fbx4core complex can directly interact with both the substrate (Sub) and Cul1-Rbx1 to promote the ubiquitination of the substrate. B, the monomeric Skp1-Fbx4FL complex cannot bind substrate and Cul-Rbx1. It is possible that phosphorylation (circled P) on the Fbx4 NTD induces the conformational changes on both the NTD and the finger, thus enabling Fbx4 to dimerize and to interact with the substrate and Cul1-Rbx1.
Interestingly, the dimer configuration observed in the Fbx4 structure is different from that of the Skp1-Cdc4 complex determined by small angle x-ray scattering (22). In the Skp1-Cdc4 complex, Cdc4 dimerizes in a side-by-side configuration as two substrate-binding sites and two predicated E2 active sites lie in the same plane. These differences likely result from the different modules employed by Cdc4 and Fbx4 to mediate their dimerization. Although Fbx4 dimerizes through the linker-G domain interaction, Cdc4 forms a homotypic dimer through a domain preceding the F-box domain. The two different dimerization configurations are likely due to the conformational polymorphism of F-box proteins and point to a possible structural flexibility in SCF during substrate recruitment and catalysis. In addition, recent studies show that covalent modification of the cullin subunit of SCF by Nedd8 influences SCF-E2 interaction and induces large scale conformational rearrangements in the cullin C-terminal domain and Rbx1, and these latter changes likely reduce the gap between the bound E2 and substrate (47, 48). Thus, complementary to neddylation, SCF dimerization may represent a new key regulatory mechanism by which these complexes acquire conformational variability to accommodate various substrates.
How are Fbx4 dimerization and activation regulated in vivo as the full-length Fbx4 is a monomer and essentially inactive in our in vitro setting using Pin2 as the substrate? By truncating the NTD and the finger in the linker domain, we engineered and produced an active dimeric Fbx4 (Fig. 6A). We demonstrated that the presence of the NTD (as in Fbx4FL and Fbx4ΔFinger) blocks SCF complex formation, whereas the presence of the finger (as in Fbx4FL and Fbx4ΔNTD) represses dimerization and substrate binding. We posit that inducible conformational changes in the NTD and finger are required in vivo to release their autoinhibitory effect on the dimerization-induced activation of Fbx4 (Fig. 6B). Indeed, phosphorylation of a serine residue (Ser-12) in the NTD by glycogen synthase kinase-3-β (GSK3β) can contribute to Fbx4 dimerization and ubiquitination activity (28). Alanine substitution at this position attenuates homodimerization and significantly impairs cyclin D1 ubiquitination and destruction, whereas a phosphomimetic mutation (S12E) results in a reduction of cyclin D1 to levels comparable with that of the wild-type Fbx4 in vivo (28). It is thus possible that single or multiple phosphorylations in the NTD and/or the finger may result in a conformational rearrangement to allow the association of Cul1-Rbx1 to the Skp1-F-box as well as Fbx4 dimerization and substrate binding (Fig. 6B). Alternatively, the activation of Fbx4 in vivo can be achieved by binding of an unidentified partner protein to the NTD and finger of Fbx4. For example, Nedd4 family-interacting protein (NDFIP) and Smad7 stimulate the catalytic activity of Itch-encoded ligase (ITCH) and Smad ubiquitination regulatory factor-2 (Smurf2) E3s, respectively, by binding to E3s to release them from the autoinhibitory intramolecular interactions (49, 50). However, additional studies will be required to determine the molecular details of the activation and regulation of the SCFFbx4 E3 in vivo.
Acknowledgments
We thank David King of the Howard Hughes Medical Institute mass spectrometry laboratory for limited proteolysis analysis, Xuedong Liu of the University of Colorado-Boulder for providing the Cdk1-cyclin B complex, Wuxian Shi and Babu A. Manjasetty of NSLS X3A and X29 beamlines for help with data collection, and Peter Setlow, Barbara Setlow, and Jeff Hoch of University of Connecticut Health Center for critical reading of the manuscript.
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (code 3L2O) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- SCF
- Skp1-Cul1-Rbx1-F-box protein
- NTD
- N-terminal domain
- FL
- full length
- GST
- glutathione S-transferase
- Ni2+-NTA
- Ni2+-nitrilotriacetic acid
- Ub
- ubiquitin.
REFERENCES
- 1.Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2.Pickart C. M. (2001) Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 3.Bosu D. R., Kipreos E. T. (2008) Cell Div. 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardozo T., Pagano M. (2004) Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 5.Deshaies R. J., Joazeiro C. A. (2009) Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 6.Petroski M. D., Deshaies R. J. (2005) Nat. Rev. Mol. Cell Biol 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 7.Willems A. R., Schwab M., Tyers M. (2004) Biochim. Biophys. Acta 1695, 133–170 [DOI] [PubMed] [Google Scholar]
- 8.Dye B. T., Schulman B. A. (2007) Annu. Rev. Biophys. Biomol. Struct. 36, 131–150 [DOI] [PubMed] [Google Scholar]
- 9.Hao B., Oehlmann S., Sowa M. E., Harper J. W., Pavletich N. P. (2007) Mol. Cell 26, 131–143 [DOI] [PubMed] [Google Scholar]
- 10.Mizushima T., Yoshida Y., Kumanomidou T., Hasegawa Y., Suzuki A., Yamane T., Tanaka K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5777–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orlicky S., Tang X., Willems A., Tyers M., Sicheri F. (2003) Cell 112, 243–256 [DOI] [PubMed] [Google Scholar]
- 12.Schulman B. A., Carrano A. C., Jeffrey P. D., Bowen Z., Kinnucan E. R., Finnin M. S., Elledge S. J., Harper J. W., Pagano M., Pavletich N. P. (2000) Nature 408, 381–386 [DOI] [PubMed] [Google Scholar]
- 13.Wu G., Xu G., Schulman B. A., Jeffrey P. D., Harper J. W., Pavletich N. P. (2003) Mol. Cell 11, 1445–1456 [DOI] [PubMed] [Google Scholar]
- 14.Zheng N., Schulman B. A., Song L., Miller J. J., Jeffrey P. D., Wang P., Chu C., Koepp D. M., Elledge S. J., Pagano M., Conaway R. C., Conaway J. W., Harper J. W., Pavletich N. P. (2002) Nature 416, 703–709 [DOI] [PubMed] [Google Scholar]
- 15.Ho M. S., Ou C., Chan Y. R., Chien C. T., Pi H. (2008) Cell Mol. Life Sci. 65, 1977–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin J., Cardozo T., Lovering R. C., Elledge S. J., Pagano M., Harper J. W. (2004) Genes Dev. 18, 2573–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizushima T., Hirao T., Yoshida Y., Lee S. J., Chiba T., Iwai K., Yamaguchi Y., Kato K., Tsukihara T., Tanaka K. (2004) Nat. Struct. Mol. Biol. 11, 365–370 [DOI] [PubMed] [Google Scholar]
- 18.Chew E. H., Poobalasingam T., Hawkey C. J., Hagen T. (2007) Cell. Signal. 19, 1071–1080 [DOI] [PubMed] [Google Scholar]
- 19.Dixon C., Brunson L. E., Roy M. M., Smothers D., Sehorn M. G., Mathias N. (2003) Eukaryot. Cell 2, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kominami K., Ochotorena I., Toda T. (1998) Genes Cells 3, 721–735 [DOI] [PubMed] [Google Scholar]
- 21.Suzuki H., Chiba T., Suzuki T., Fujita T., Ikenoue T., Omata M., Furuichi K., Shikama H., Tanaka K. (2000) J. Biol. Chem. 275, 2877–2884 [DOI] [PubMed] [Google Scholar]
- 22.Tang X., Orlicky S., Lin Z., Willems A., Neculai D., Ceccarelli D., Mercurio F., Shilton B. H., Sicheri F., Tyers M. (2007) Cell 129, 1165–1176 [DOI] [PubMed] [Google Scholar]
- 23.Welcker M., Clurman B. E. (2007) Cell Div. 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolf D. A., McKeon F., Jackson P. K. (1999) Curr. Biol. 9, 373–376 [DOI] [PubMed] [Google Scholar]
- 25.Lee T. H., Perrem K., Harper J. W., Lu K. P., Zhou X. Z. (2006) J. Biol. Chem. 281, 759–768 [DOI] [PubMed] [Google Scholar]
- 26.Lin D. I., Barbash O., Kumar K. G., Weber J. D., Harper J. W., Klein-Szanto A. J., Rustgi A., Fuchs S. Y., Diehl J. A. (2006) Mol. Cell 24, 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbash O., Diehl J. A. (2008) Cell Cycle 7, 2983–2986 [DOI] [PubMed] [Google Scholar]
- 28.Barbash O., Zamfirova P., Lin D. I., Chen X., Yang K., Nakagawa H., Lu F., Rustgi A. K., Diehl J. A. (2008) Cancer Cell 14, 68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ganoth D., Bornstein G., Ko T. K., Larsen B., Tyers M., Pagano M., Hershko A. (2001) Nat. Cell Biol. 3, 321–324 [DOI] [PubMed] [Google Scholar]
- 30.Spruck C., Strohmaier H., Watson M., Smith A. P., Ryan A., Krek T. W., Reed S. I. (2001) Mol. Cell 7, 639–650 [DOI] [PubMed] [Google Scholar]
- 31.Stebbins C. E., Kaelin W. G., Jr., Pavletich N. P. (1999) Science 284, 455–461 [DOI] [PubMed] [Google Scholar]
- 32.Hao B., Zheng N., Schulman B. A., Wu G., Miller J. J., Pagano M., Pavletich N. P. (2005) Mol. Cell 20, 9–19 [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 34.Sheldrick G. M. (2008) Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 35.La Fortelle E. D., Bricogne G. (1997) Methods Enzymol. 276, 472–494 [DOI] [PubMed] [Google Scholar]
- 36.Collaborative Computational Project, Number 4. (1994) Acta Crystallogr. D. Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 37.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D. Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 39.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D. Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 40.Petroski M. D., Kleiger G., Deshaies R. J. (2006) Mol. Cell 24, 523–534 [DOI] [PubMed] [Google Scholar]
- 41.Tan X., Calderon-Villalobos L. I., Sharon M., Zheng C., Robinson C. V., Estelle M., Zheng N. (2007) Nature 446, 640–645 [DOI] [PubMed] [Google Scholar]
- 42.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takai Y., Sasaki T., Matozaki T. (2001) Physiol. Rev. 81, 153–208 [DOI] [PubMed] [Google Scholar]
- 44.Vetter I. R., Wittinghofer A. (2001) Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 45.Amor J. C., Harrison D. H., Kahn R. A., Ringe D. (1994) Nature 372, 704–708 [DOI] [PubMed] [Google Scholar]
- 46.Harrison S. C. (1996) Cell 86, 341–343 [DOI] [PubMed] [Google Scholar]
- 47.Duda D. M., Borg L. A., Scott D. C., Hunt H. W., Hammel M., Schulman B. A. (2008) Cell 134, 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saha A., Deshaies R. J. (2008) Mol. Cell 32, 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., Wrana J. L. (2000) Mol. Cell 6, 1365–1375 [DOI] [PubMed] [Google Scholar]
- 50.Mund T., Pelham H. R. (2009) EMBO Rep. 10, 501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]