Abstract
Tight junctions are dynamic components of epithelial and endothelial cells that regulate the paracellular transport of ions, solutes, and immune cells. The assembly and permeability of these junctions is dependent on the zonula occludens (ZO) proteins, members of the membrane-associated guanylate kinase homolog (MAGUK) protein family, which are characterized by a core Src homology 3 (SH3)-GUK module that coordinates multiple protein-protein interactions. The structure of the ZO-1 SH3-GUK domain confirms that the interdependent folding of the SH3 and GUK domains is a conserved feature of MAGUKs, but differences in the orientation of the GUK domains in three different MAGUKs reveal interdomain flexibility of the core unit. Using pull-down assays, we show that an effector loop, the U6 region in ZO-1, forms a novel intramolecular interaction with the core module. This interaction is divalent cation-dependent and overlaps with the binding site for the regulatory molecule calmodulin on the GUK domain. These findings provide insight into the previously observed ability of the U6 region to regulate TJ assembly in vivo and the structural basis for the complex protein interactions of the MAGUK family.
Keywords: Calmodulin, Cell Junctions, Cell-Cell Interaction, Crystallography, Protein Structure, MAGUK, Tight Junctions, ZO-1
Introduction
The assembly and functional organization of plasma membrane domains, such as synapses and cell adhesions, depends on the temporally and spatially regulated assembly of multiprotein complexes. In one common paradigm, the trafficking and assembly of the transmembrane receptors and adhesion molecules that form these complexes is organized by a series of cytosolic scaffolding proteins. One of the most common and ubiquitous families of scaffolding molecules is the membrane-associated guanylate kinase homologs (MAGUKs).2 Founding members of this family include the discs large tumor suppressor in Drosophila, Lin2, which is required for RT kinase signaling in Caenorhabditis and PSD-95 and its related synaptic scaffolding proteins, which also control channel activity. This diverse family of proteins is characterized by a core motif of protein-binding domains, including a PSD-95/DLG/ZO-1 (PDZ) domain, an Src homology 3 (SH3) domain, and a region of homology to guanylate kinase referred to as the GUK domain (Fig. 1A) (reviewed in Ref. 1). Most also include additional PDZ domains or other conserved protein-binding domains. These domains are separated by unique regions (U regions), which can also have protein-binding and regulatory roles (1, 2). However, it is the core SH3-GUK module and its adjacent unique domains that often have a dominant role in the regulation of transmembrane ligand binding, protein cross-linking, and localization to the appropriate subcellular domain.
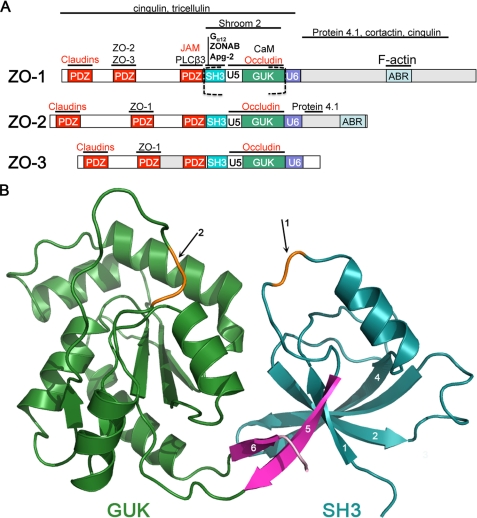
FIGURE 1.
Domain organization of ZO proteins and the structure of the SH3-GUK domains of ZO-1. A, domain organization of the ZO MAGUKs. The canonical MAGUK protein binding motifs (PDZ, SH3, and GUK) are separated by “unique” (U) regions of high sequence diversity. TJ binding partners of ZO-1 are mapped above their known interaction domain with the transmembrane strand components labeled in red (reviewed in Ref. 14). Those interactions that are also conserved in ZO-2 and ZO-3 are indicated, and the section of ZO-1 whose structure is presented here is indicated by dashed square brackets. B, ribbon diagram of the ZO-1 SH3 (cyan) and GUK (green) domains. The fifth strand of the SH3 domain and the strand following the GUK domain form the interdomain linkage (pink). The ZO-1 construct used for the structure determination lacked the U5 region; arrow 1 points to the truncation point (orange highlight). The entire structure could be traced with confidence except for 3 residues located at the NMP-binding region at the GUK domain (arrow 2, orange highlight).
The structural basis for the regulatory properties of the various MAGUKs is poorly understood. To date, the only core motif that has been resolved at an atomic level belongs to PSD-95 (postsynaptic density protein 95) (3, 4), a neuronal protein involved in the regulation of postsynaptic gated ion channel. The structure reveals that the SH3 and GUK domains are not simply arranged like pearls on a string but rather are interconnected. The interdomain interaction is via a β-strand that originates after the GUK domain that returns to become an additional strand of the SH3 β-sheet. The protein binding properties of the core module appear to be dependent upon this interdomain interaction because mutations that disrupt this structure also disrupt interactions of the GUK domain with its ligands (5, 6). Thus, it may be more realistic to consider the SH3-GUK region as a single interdependent structural module. However, although the secondary sequences within the core motifs of MAGUKs are fairly well conserved, there are enough differences to question whether or not the overall structural fold of the SH3-GUK core is conserved.
The U5 region (unique region 5), which is located toward the C terminus of the SH3 domain just prior to the fifth β-strand, plays an important functional role in MAGUKs. In some MAGUKs, it is a protein-binding motif (7–9), whereas in others, it is critical for localization to the appropriate subcellular domain (2, 10, 11). In hDlg/SAP97, this region has multiple alternative splices, and this splicing contributes to diversity in protein ligand binding affinity and function (10). The varied role for this motif is consistent with its sequence being highly diverged between the different MAGUKs. Interestingly, although the U5 region in PSD-95 is rather small compared with other MAGUKs, the structure of this motif was not completely resolved in the crystal structure, suggesting some structural flexibility in the absence of binding partners (3). Thus, in one sense the U5 motif can be thought of as an effector loop that mediates domain-specific functions for a particular MAGUK. However, the functional and biochemical diversity of the U5 region might also affect the packing and relative orientation of the SH3 and GUK domains.
Tight junctions (TJ) are sites of cell-cell adhesion in epithelial and endothelial cells that form a barrier that regulates the movement of ions, solutes, and immune cells between cells. The actual physical barrier is assembled from at least three families of transmembrane proteins, including the claudins, occludin/tricellulin, and junction adhesion molecules, which assemble into highly organized strands in the plasma membrane (12). The assembly of these transmembrane proteins into strands and the formation of the barrier is dependent on the zonula occludens (ZO) family of MAGUKs; ZO-1, ZO-2, and ZO-3 (Fig. 1A) (13). ZO MAGUKs contain the typical PDZ-SH3-GUK core motif, which is preceded by two additional PDZ domains, but are distinguished by unique C-terminal domains of various lengths (reviewed in Ref. 14). The C-terminal domains mediate interactions with the cortical actin cytoskeleton, whereas the N-terminal MAGUK domains bind to the transmembrane components of tight junction strands and other scaffolding or signaling molecules that regulate TJ assembly. The three ZO MAGUKs clearly share some functional roles, and recent genetic studies indicate that there is a certain amount of functional redundancy between ZO-1 and ZO-2 within the TJ (13, 15, 16). Either ZO-1 or ZO-2 appears to be sufficient for formation of a functional barrier in cultured cells, but the deletion of both ZO-1 and ZO-2 in epithelial cells disrupts the TJ barrier and prevents the assembly of TJ transmembrane proteins described above into strands (13). Thus, ZO MAGUKs are a critical scaffold for TJ assembly.
The SH3-GUK module is a critical element of tight junction assembly. Recent studies in cells deficient for ZO MAGUKs indicate that the SH3-GUK module is necessary for rescue of TJ strand assembly and the formation of the barrier (13, 17). This is perhaps not surprising because the SH3 and GUK domains bind to several structural and signaling components of tight junctions (14), and the U5 motif that links these domains is required for localization of ZO MAGUKs to the TJ (2). In addition, we have recently demonstrated that regulated binding to the core module is equally critical. We have found that the U6 region, which is immediately distal to the GUK domain, can regulate occludin binding to the GUK domain in vitro and U5 localization to the TJ in vivo (2). Finally, deletion of the U6 region results in the ectopically located formation of TJ strands throughout the cell. Thus, the U6 region forms a second “effector loop,” which regulates the function of the core module. A clue as to how this regulation may function is provided by the discovery presented here that the U6 region binds to the SH3-GUK (SG) module in a divalent metal-dependent manner and competes with calmodulin for binding to the GUK domain.
To better understand how the SH3-GUK module and its associated effector domains regulate the temporal and spatial assembly of tight junctions, we have solved the crystal structure of the ZO-1 SH3-GUK region to 2.6 Å resolution. The ZO-1 SH3-GUK module has similarities to the homologous module of PSD-95 but also shows several significant differences. Most noticeable is the different interdomain angle, with ZO-1 being more open. A general conclusion concerning MAGUK proteins is that the β-strand following the GUK domain is a component of the SH3 domain. At the same time, the angle between the SH3 and GUK domains is variable among the different MAGUK proteins. Comparison of the ZO-1 SH3-GUK module structure with that of PSD-95 and ZO-3 has allowed us to identify the region within the GUK domain that forms the hinge region between the domains. Furthermore, analysis of the electrostatic surface potential of ZO-1 reveals a highly basic face to the ZO-1 molecule. Binding studies indicate that this basic surface acts to bind calmodulin and the U6 region. Of note, the binding site for calmodulin and U6 has been revealed to be located at the GUK domain, on the same face as the U5 region of the SH3 domain, and both are at opposite ends from the hinge. Furthermore, we present data revealing that the U6 region binds to the SG module in a divalent metal-dependent manner. We propose that yet-to-be-identified ligands of the U5 region and GUK domain are regulated by the binding of calmodulin and/or the U6 region and that these interactions control TJ assembly.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of ZO-1 Fragments
For details on the cloning, expression, and purification of the ZO-1 constructs described here, see the supplemental material.
Crystallization, Data Collection, and Processing
The T709C SGΔU5 (native and selenomethionine) protein crystallized at room temperature by the hanging drop vapor diffusion method in 0.1 m HEPES, pH 7.5, 7% isopropyl alcohol, 15–20% polyethylene glycol 3350 (w/v), and 1–5 mm dithiothreitol. A protein/reservoir volume ratio of 2:1 was used, and the drop was equilibrated against 750 μl of reservoir solution. Crystals appeared within a week and grew up to ∼300 × 80 × 80 μm. To obtain higher resolution diffraction, the crystals were dehydrated by serial transfer to a solution of increasing polyethylene glycol 3350 concentration of up to 45%. The final dehydration solution also acted as the cryoprotectant, and the crystals were flash-frozen in liquid nitrogen. De novo phasing was achieved by soaking the crystals in various mercury-containing compounds at 1 mm concentration. X-ray data were collected at the SERCAT beamlines at the Advanced Photon Source and processed with XDS (18).
Structure Determination and Refinement
Although dehydration of the crystals usually resulted in cell shrinkage, the heavy atom soaks elongated the unit cell despite back-soaks into the dehydration/cryoprotectant solution (supplemental Table 1). The selenomethionine crystals were especially radiation-sensitive; thus, data sets at the peak and edge wavelengths were collected on separate crystals. Only the combination of several data sets from multiple derivatized crystals using multiple isomorphous replacement with anomalous scattering resulted in the program SOLVE (19) finding heavy atom sites with relevant occupancy and a reasonable figure of merit of 0.43. These phases were extended and modified using RESOLVE (20). The electron density maps calculated using the improved phases allowed us to trace both the SH3 and GUK domains. Iterative refinement and model building using O (21) yielded the final model that spans residues 516–588 and 626–803 (residues 589–625 are missing from the SGΔU5 construct) with loop residues 684–686 lacking unambiguous electron density. TLS refinement was applied during the final stages of refinement using two groups, one comprising the SH3 domain from 516 to 641 and the other of the GUK domain from 642 to 803. The stereochemical properties of the final model (Protein Data Bank code 3LH5) were analyzed using PROCHECK. For data collection and refinement statistics, see supplemental Table 1. To compare the overall structure and domain orientations of SGΔU5 relative to those of SG, molecular replacement using PHASER was performed using the SH3 and GUK domains of SGΔU5 as separate search models on a ∼3.7 Å native SG data set. Rigid body refinement with the SH3 and GUK as independent domains was performed on the molecular replacement solution followed by restrained refinement. Figures were made with PyMOL.
Pull-down Assays
The binding assays were done as previously described with minor modifications (22). Prior to performing the binding experiment, the SG and mutant proteins were dialyzed against 50 mm Tris-HCl, pH 7.5, 100 mm sodium citrate, pH 7.5, 100 mm NaCl, and 1 mm dithiothreitol. A 1:3 ratio (10 μm, 30 μm) of GST-CaM/GST-U6 to SG/mutant was used. GST-CaM/GST-U6 was first immobilized onto glutathione-Sepharose beads (GE Healthcare) and washed with a binding buffer of 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1 mm dithiothreitol, and 1 mm CaCl2. SG/mutant protein was then added, incubated for 30 min at room temperature, and washed with binding buffer to remove unbound ZO-1. To test the metal dependence of U6 binding to ZO-1, SG/SGΔU5 was added to immobilized GST-U6 previously equilibrated with 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1 mm dithiothreitol, and either 1 mm MgCl2 or CaCl2 or 20 mm EDTA. After incubation, the samples were washed with their respective binding buffers and analyzed using SDS-PAGE. Control samples of ZO-1 incubated with GST as well as with the glutathione-Sepharose beads were done in parallel with the experimental samples.
RESULTS AND DISCUSSION
Structure Determination of the ZO-1 SH3-GUK Module
The initial crystals of the ZO-1 SG region diffracted poorly. We surmised that flexibility in the U5 region is a causing crystal disorder that limits the diffraction (for a schematic of ZO domain organization, see Fig. 1A). This hypothesis was based on the observation that the homologous region of the MAGUK PSD-95 was only partially observed in the electron density (3, 4). A ZO-1 construct lacking the U5 region, referred to as SGΔU5, crystallized in conditions comparable with those used with SG and produced similar looking crystals. Diffraction data on SGΔU5 crystals extended to ∼3.2 Å in the identical space group and unit cell dimensions comparable with those of SG crystals. We further improved the resolution limit to 2.6 Å by the process of crystal dehydration. Molecular replacement using the known PSD-95 structures of the SH3-GUK domains failed. Thus, the structure was solved using experimentally derived phases from selenomethionine-containing protein and mercury-soaked cysteine mutant of SGΔU5 (see “Experimental Procedures”). Data collection and refinement statistics are presented in supplemental Table 1.
The Intramolecular Interaction between SH3 and GUK Domains Is Conserved in ZO-1
A ribbon diagram of the ZO-1 SH3-GUK region is shown in Fig. 1B. As in PSD-95, we observe that the SH3 and GUK domains form an interdependent structural unit. The SH3 domain (cyan) is linked to the GUK domain (green) by two anti-parallel strands (pink). The first strand of this interdomain linkage is the fifth strand of the SH3, whereas the second strand is an extension from the C terminus of the GUK domain that returns to contribute to the β-sheet in the SH3 domain. In fact, this strand that follows the GUK sequence can be viewed as an integral part of the ZO-1 SH3 domain, and it has been labeled strand 6 in Fig. 1B to highlight this role (in contrast, strands in the GUK domain are indicated by letters in Fig. 2). Thus, as observed in the structure of PSD-95 SH3-GUK, the SH3 domain builds a six-stranded β-sheet, which is different from the canonical five-stranded SH3 domain of Src itself (for a discussion, see McGee et al. (3)). Apart from this interdomain linkage, the SH3 and GUK domains of ZO-1 lack any structurally significant interactions.
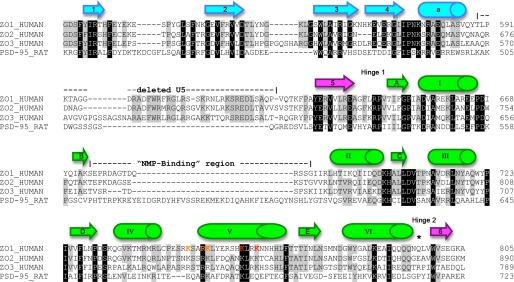
FIGURE 2.
Sequence alignment of SH3-GUK modules from human ZO proteins and rat PSD-95. The initial alignment was accomplished using ClustalW and later improved manually. Residues conserved in all four sequences are highlighted in black, and those present in three sequences are highlighted in gray. The secondary structure elements shown above the sequences are based on ZO-1 and maintain the color-coding of Fig. 1B. The boundaries for the residues deleted from the SGΔU5 construct used for the structure determination are indicated. *, the cis-proline residue in PSD-95 (Pro714) that is responsible for the acute angle between its SH3 and GUK domains. Residues in ZO-1 helix V that were mutated are indicated in orange letters for the triple mutant, with the two additional residues for the pentamutant shown in red.
The U5 region, absent in our structure, would connect the sole helix of the SH3 domain and the loop that precedes the fifth β-strand. The location of the 36-residue U5 deletion is highlighted in orange and indicated by arrow 1 in Fig. 1B. The length and sequence homology of U5 is quite variable among MAGUKs, suggesting that it might be a structurally and functionally distinct binding region in the different proteins. The observation that the U5 domain was not seen in two earlier PSD-95 crystal structures of the SH3-GUK region despite being a part of the construct and the fact that in the recent structure of the ZO-3 SH3-GUK domain the U5 could also not be traced (see below) suggest intrinsic high flexibility for the U5 in the absence of a binding partner. It is unlikely that removal of the U5 from ZO-1 had a significant effect on the overall structure reported here because both SG and SGΔU5 crystallized in the same space group with similar unit cells. This implies that the truncation of U5 did not change the conformation of SG and was confirmed later by our analysis of the 3.7 Å SG data set (see below).
We could trace the entire SH3-GUK module except for three residues in a loop in the GUK domain that were unresolved. The approximate location of the missing three residues is shown in orange and designated by arrow 2 in Fig. 1B. This loop is much shorter in ZO-1 than in PSD-95 (see sequence alignment in Fig. 2). Interestingly, this region is close to and on the same face as the U5 region. Like U5, this loop is highly variable in length and sequence homology among MAGUKs. It is possible that the presence of the U5 region would stabilize this GUK loop. Alternatively, this loop may become more ordered upon binding of a protein-ligand to the U5 region.
The SH3-GUK Regions of ZO-1 Adopt a More Open Conformation than PSD-95
Sequence alignment of the SH3-GUK regions of human ZO-1 and rat PSD-95 reveal a low 25% identity/40% similarity (Fig. 2), and superposition of ZO-1 and PSD-95 SH3-GUK regions reveals major differences in the relative position of the two domains. When the SH3 domain is used as the base for the alignment (Fig. 3A), it is immediately apparent that the domains in PSD-95 adopt a considerably more closed conformation than that adopted by the domains in ZO-1. To understand this difference in domain packing, we analyzed the interactions between the SH3 and GUK domains in the two MAGUKs. Surprisingly, even in the more compact domain arrangement in PSD-95, there are very few interdomain interactions other than those that involve the terminal β-strand complementing the SH3 β-sheet (colored pink for ZO-1 and red for PSD-95). The sole additional interdomain interaction in PSD-95 occurs between the side chain of Arg434 and the main chain carbonyl group of Phe688. Because both of these residues are also present but do not interact in the open conformation in ZO-1, the more closed interdomain conformation in PSD-95 cannot be accounted for by this interaction.
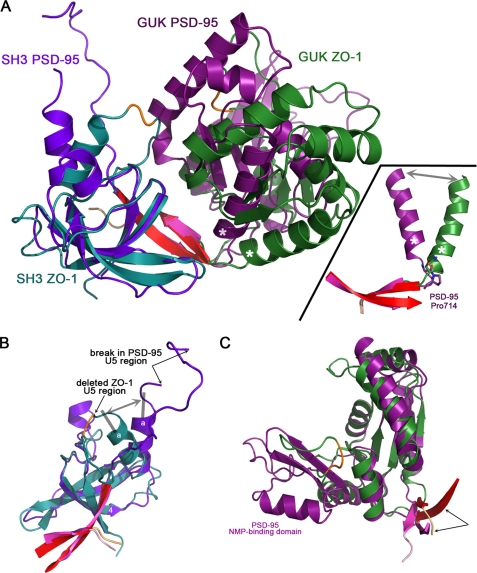
FIGURE 3.
ZO-1 adopts a more open interdomain conformation relative to PSD-95. A, overlay of the SH3-GUK modules of ZO-1 (cyan and green) and PSD-95 (purple and magenta). The superposition was generated by aligning residues of the SH3 domains. The two strands forming the interdomain linkage are colored pink for ZO-1 and red for PSD-95. When the SH3 domains are aligned, the GUK domains occupy significantly different positions, with the PSD-95 adopting a more closed state. This difference in interdomain angle is attributed to the nature of the residue located between the terminal GUK helix VI (labeled with an asterisk) and β-strand 6. In PSD-95, a cis-proline (Pro714) forces an acute angle between these secondary structure elements (see inset). ZO-1 has a glutamine residue at this position, allowing for a more open conformation. B, superposition of the SH3 domains. The deleted U5 region (orange trace) in ZO-1 is indicated by an arrow. In PSD-95, despite the presence of the U5 residues, only a section could be traced, with two arrows indicating the break. Note that the sole helix located within the SH3 domain adopts a different angle in ZO-1 and PSD-95 (double-headed gray arrow) relative to the core of the SH3 domain. C, superposition of the GUK domains. PSD-95 has a significantly longer insert at a region called the NMP-binding domain (see Fig. 2), with ZO-1 having only a short loop (the three residues that could not be modeled in this loop are shown in orange). When using the GUK residues for calculating the superposition, the interdomain strands (pink and red for ZO-1 and PSD-95, respectively) do not overlay well (indicated by two arrows). The converse is true when calculating the overlay with SH3 residues (B). Thus, the interdomain strands are an integral part of the SH3 domain.
Why then do the domains in PSD-95 adopt a more closed arrangement relative to the domain arrangement in ZO-1? The comparison of our ZO-1 structure with that of PSD-95 allowed us to identify the residues that act as hinges between the SH3 and GUK domains. The SH3 and GUK domains are connected via two hinges, which are formed by loop residues placed between secondary structure elements. Hinge 1 is located between strand 5 of the SH3 domain and strand A of the GUK domain, and Hinge 2 is located between helix VI of the GUK domain and strand 6 (Fig. 2). The reason for the acute angle between the SH3 and GUK domains of PSD-95 seems to lie in the presence of a cis-proline (Pro714) at Hinge 2 (Fig. 3A, inset). In contrast, ZO-1 has a glutamine residue at the equivalent position, allowing for an obtuse angle between the domains. Thus, the more compact arrangement of the PSD-95 SH3-GUK region relative to that in ZO-1 is not due to unique interdomain interactions that stabilize the closed state. Rather, this closed state is a result of a hinge region that is limited in the angle it can support between the domains. The analogous hinge region in ZO-1 does not contain the constraining proline residue and can therefore adopt a more open state.
Comparison of the SH3 Domains and Different Angles between the Helical Stem of the U5 Region and the SH3 β-Sheet
Although the aggregate SH3-GUK region of ZO-1 overlays poorly on the homologous region from PSD-95, the individual domains overlay much better. The strands of the SH3 domain overlay nearly perfectly, including strand 6, which originated after the end of the GUK domain (Fig. 3B). In our structure of SGΔU5, the missing U5 domain would be located between helix a and strand 5 (Fig. 1B). PSD-95 has a much shorter U5 region, and in its crystal structure, not all of it could be traced. Interestingly, the angle between strand 4 and helix a of the SH3 domain is significantly different between PSD-95 and ZO-1 (Fig. 3B, gray double-headed arrow). The recently deposited structure of ZO-3, which was solved using a construct that contained the intact U5 region, has a strand-to-helix angle identical to that in our U5-deleted structure of ZO-1. This demonstrates conclusively that this difference in angles (between the SH3 β-sheet and the helix that acts as a stem of the U5 region) between ZO-1 and PSD-95 is not due to the use of the ΔU5 construct but rather to an intrinsic property of ZO proteins. Thus, the helix stem of the U5 region in ZO-1 presents the ligand-binding residues of U5 at a different angle to potential binding partners in comparison with PSD-95. This difference suggests the possibility that upon binding of ligands to U5, the helix responds by changing its relative angle to the SH3 β-sheet.
Comparison of the GUK Domains, ZO-1 Lacks the NMP-binding Region
The name for the GUK domain originates from sequence similarity to guanylate kinase, an enzyme that catalyzes the conversion of GMP to GDP. Of note, no appreciable guanylate kinase activity has been measured for any MAGUK protein (23, 24), and even the question of whether nucleotides bind to MAGUKs has been controversial. McGee et al. (3) could not detect binding of GMP to PSD-95 using equilibrium dialysis experiments. In contrast, Tavares et al. (4) observed GMP in their structure of the PSD-95 SH3-GUK region, but that could have been due to stabilization supplied by a guanidine molecule that was present in the crystallization solutions. Nevertheless, based on homology to the enzyme guanylate kinase and the fact that 10 of 11 residues involved in the binding of GMP are also present in PSD-95, a region of the GUK domain was named as the NMP-binding region (Fig. 2).
However, it is notable that the so-called NMP-binding region is totally missing in ZO-1. Instead, a short flexible loop of ∼14 residues (compared with ∼46 residues in PSD-95) substitutes for this domain. This is the one loop in the GUK domain that we could not build in its entirety (the three missing loop residues are highlighted in orange in Fig. 3C). Since this loop is on the same face as the U5 region that originates from the SH3 domain, it is possible that upon ligand binding to the U5 region, this loop becomes stabilized.
The Interdomain Strands Form a Rigid Link between SH3 and GUK
The overlay of the GUK domains presented in Fig. 3C was carried out by aligning only residues that belong to the GUK domain. Also shown are the residues that build the interdomain linkage of anti-parallel β-strands (pink for ZO-1, red for PSD-95). Although the GUK domains align well between the MAGUKs, the interdomain strands do not (arrows in Fig. 3C). In contrast, the analogous superposition using the SH3 residues results in a good fit for the core SH3 and for the interdomain strands (Fig. 3B). This fixed orientation relative to the SH3 arises from a strictly conserved tryptophan residue (Trp799) located in strand-6 that orients its side chain deep into a hydrophobic depression in the SH3 domain (supplemental Fig. S1). This side chain buttresses the interdomain interaction made via the main chain atoms of strands 5 and 6. Thus, the interdomain strands behave as a rigid body that belongs to the SH3 domain and not to the GUK domain. The hinges between the domains are at the GUK end of these two strands.
Electrostatic Analysis of the ZO-1 SH3-GUK Region and Implications for Ligand Binding
The function of the different modules in MAGUKs is to bind various protein ligands in order to assemble multiprotein complexes. Regulation of these interactions has significant implications for complex assembly and function (2, 10, 25, 26). In the case of ZO-1, we have shown that the GUK domain is sufficient to bind calmodulin and that this interaction is calcium-dependent (22). No canonical calmodulin-binding sequences were identified in the GUK domain, suggesting a novel mode of interaction. Calmodulin is a highly acidic protein. Therefore, we examined the surface charge distribution of SH3-GUK, making the prediction that a basic surface would facilitate the interaction with calmodulin. The GUK domain of ZO-1 is indeed highly basic with 13 arginine and 12 lysine residues (pI of 10). The electrostatic surface potential of ZO-1 SH3-GUK in two orthogonal views is presented in Fig. 4.
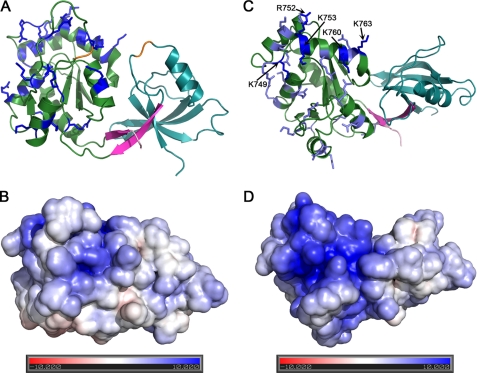
FIGURE 4.
The GUK domain of ZO-1 is characterized by a positively charged surface. A, a ribbon diagram of ZO-1 with all GUK domain lysine and arginine residues shown. B, electrostatic surface potential at the same orientation as in A. C, view rotated 90° relative to A. D, electrostatic surface potential at the same orientation as in C. A congregation of basic residues occurs at and near helix V of the GUK domain. In C, the basic residues probed by mutagenesis are colored dark blue, and all others are shown in light blue.
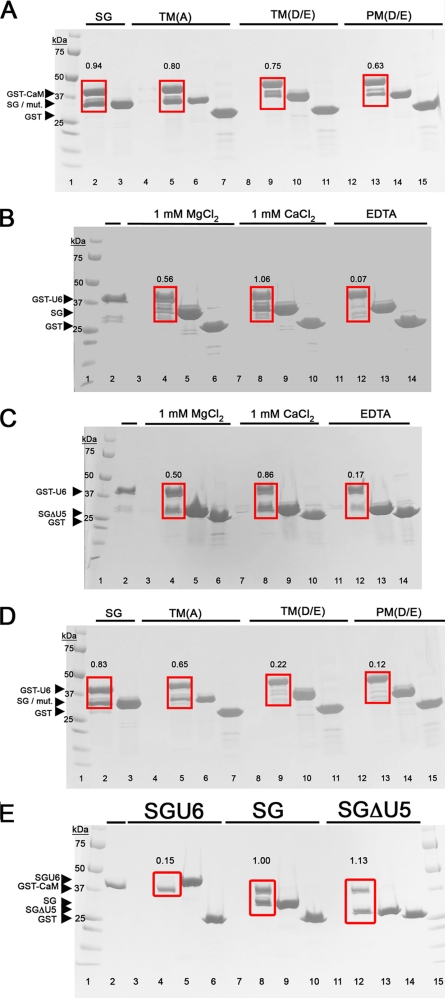
Most pronounced is the concentration of positive charges in and around the vicinity of helix V of the GUK domain (Fig. 2). To test if the basic residues in this helix participate in binding to calmodulin, we generated charge elimination and charge reversal mutations and used a pull-down assay to measure binding. The charge elimination mutants had alanine residues substituting for Lys749, Arg752, and Lys753 (for the location of these residues in the sequence see Fig. 2; for location in the structure, see Fig. 4C). Pull-down assays showed that SG binding to calmodulin was only mildly weaker to this SG variant (Fig. 5A, labeled TM(A)). Similarly weakened binding was observed with a variant in which these basic residues were substituted with oppositely charged carboxylic acid residues (K749D/R752D/K753E; Fig. 5A, TM(D/E)). Mutating additional two basic residues in this helix (K760E/K763E) further weakened the interaction between the SH3-GUK module and calmodulin (Fig. 5A, PM(D/E)). We recognize that the decrease in the intensity of the pull-down band of the SG mutants versus that of the wild type is moderate (range 15–40%, depending on the mutant tested). However, this effect is significant because it was recapitulated in three independent experiments (supplemental Table 2). These pull-down results suggest that the positively charged surface centered about helix V of the GUK domain is a component of the calmodulin binding site but that other elements of SG participate in calmodulin binding. Experiments are under way to identify more precisely the residues that are essential for calmodulin binding.
FIGURE 5.
Helix V of the GUK domain is important for binding to calmodulin and the U6 motif. A, GST-CaM was immobilized on glutathione beads and incubated with various SG constructs in the presence of 1 mm CaCl2 and then washed to remove unbound protein. GST-CaM and bound proteins were resolved by SDS-PAGE and stained with Coomassie Brilliant Blue. Lane 1, molecular weight markers; lanes 2, 5, 9, and 13, the fraction “bound” to GST-CaM after incubation with SG, the triple charge elimination mutant (TM(A)), the triple charge reversal mutant (TM(D/E)), or the penta-charge reversal mutant (PM(D/E)), respectively. Calmodulin binding to PM(D/E) and, to a lesser extent TM(D/E), is attenuated relative to SG. Lanes 3, 6, 10, and 14, supernatant (unbound fraction) after spinning down the beads. Lanes 4, 8, and 12, controls, in which the proteins were simply incubated with the glutathione beads. The lack of a band shows that there is no interaction between the SG constructs and the beads. Lanes 7, 11, and 15, controls in which GST alone on beads (instead of GST-CaM) was incubated with the SG constructs. This shows that the SG constructs do not interact with GST. The ratio of SG or SG mutants relative to GST-CaM, calculated from the intensity of the bands within the red box, is presented for each condition above the box. B, the U6 region binds SG in cis. Lanes 4, 8, and 12, proteins bound to GST-U6 after incubation with SG in the presence of 1 mm MgCl2, 1 mm CaCl2, or EDTA, respectively. The binding of U6 to SG is dependent on divalent metal. Lane 1, molecular weight markers. Lane 2, GST-U6 by itself on the beads. Lanes 3, 7, and 11, controls where the SG was incubated with glutathione beads, showing no nonspecific interactions. Lanes 5, 9, and 13, supernatant (unbound) fraction after the pull-down. About 50% of GST-U6 binds SG in the presence of magnesium and 100% in calcium, but little or none binds in the EDTA-containing condition. Lanes 6, 10, and 14, controls with GST instead of GST-U6. Quantified band ratios are shown above the red box. C, experiment analogous to that in B but with SGΔU5 instead of SG. This demonstrates that the divalent cation-dependent binding of U6 to SG is independent of the U5 region. D, experiment analogous to that in A but with GST-U6 instead of GST-CaM. We see that the charge reversal mutations (lanes 9 and 13) largely abolish the interaction between U6 and SG. The charge elimination mutations (lane 5) only moderately decrease the interaction. E, GST-CaM was immobilized on glutathione beads and incubated with the ZO-1 construct SH3-GUK-U6 (SGU6), SH3-GUK (SG), or SH3-GUK that lacks U5 (SGΔU5). Lanes 1 and 15, marker lanes. Lane 2, GST-CaM bound to the beads. Lanes 3, 7, and 11, control conditions in which the protein, SGU6, SG, or SGΔU5, respectively, was incubated with the glutathione beads; no nonspecific binding was observed. Lanes 4, 8, and 12, the fraction “bound” to GST-CaM after incubation with SGU6, SG, or SGΔU5, respectively. GST-CaM does not pull down SGU6, but it does pull down SG and SGΔU5. Quantified band ratios are shown above the red box. This demonstrates that the binding of calmodulin or the U6 region to the GUK domain of ZO-1 is mutually exclusive. Lanes 5, 9, and 13, supernatant (unbound fraction) after spinning down the beads. Lanes 6, 10, and 14, controls in which GST alone on beads (instead of GST-CaM) was incubated with the SG constructs. This shows that the SG constructs do not interact with GST.
Similar to calmodulin, the U6 region of ZO-1 contains a high density of carboxylic acid residues. Although U6 affects the function of the GUK domain in vivo (2), there are no data addressing whether it does this by binding the GUK domain. Thus, we asked whether the U6 region might bind to the GUK domain and whether the interaction requires helix V. Using a GST-linked U6 construct, we discovered that the U6 region can pull down the SG construct in cis (Fig. 5B). This result was recapitulated using the SGΔU5 construct (Fig. 5C), indicating that the interaction between U6 and SG is independent of the U5 region. Intriguingly, the interaction between U6 and SG was revealed to be divalent cation-dependent. With magnesium, the interaction between U6 and SG or SGΔU5 was weaker, as indicated by about 50% pull-down efficiency. In contrast, calcium promoted a complete 1:1 pull-down. The absence of metal achieved by adding the chelator EDTA abolished binding.
What could be the possible role of the divalent metal in promoting the interaction between SG and U6? One option is that the metal acts as a bridge linking carboxylic residues of the two domains. Alternatively, the metal can be acting to stabilize a binding-compatible structure of U6. Using the charge elimination and charge reversal mutants of the SG helix V, we observed that the basic residues of the helix are required for the interaction with the U6 region (Fig. 5D). The triple charge elimination mutant resulted in diminished binding, whereas both charge reversal mutants totally abrogated binding. The implications of these results are 2-fold. First, helix V of the GUK domain is identified as a necessary component of the U6 binding surface; second, the results are consistent with the divalent metal playing a structural role in the SG-U6 interaction by stabilizing the structure of the U6 region (in the alternative scenario, it is acidic residues that are important for binding, not basic residues).
Because both the U6 region and calmodulin interact with helix V of the GUK domain, we asked whether this binding is mutually exclusive. To address this, we tested the ability of calmodulin to bind to a construct that has the SH3, GUK, and U6 within the same polypeptide (called SGU6). In our pull-down assay, SGU6 failed to bind to calmodulin (Fig. 5E). Our interpretation of this observation is that the U6 region can act to regulate the ability of calmodulin to bind to ZO-1.
These structure-guided ligand-binding studies between the ZO-1 SG module and calmodulin or the U6 region have identified the basic surface on the GUK domain as important for the interaction. Interestingly, both the interaction with calmodulin and that with the U6 region is dependent on the presence of a divalent metal. Also, in both cases, the metal seems to play a structural role that converts either calmodulin or the U6 region into a conformation that is compatible with binding to the GUK domain. However, this interaction with the GUK domain is mutually exclusive between calmodulin and the U6 region (Fig. 5E). Last, the studies with the U5-truncated construct of SG show that these interactions are independent of U5 region (Fig. 5, C and E). This is noteworthy because the U5 region would be on the same face of the molecule as the important helix V of the GUK domain. To date, no proteins that bind solely to the U5 region have been identified. However, the U5 region is required for localization of ZO-1 in vivo, and we have previously demonstrated that the U6 domain can inhibit the localization of GFP-tagged SG to the tight junction. Thus, we propose that calmodulin or U6 binding to the SG domain may act to regulate the binding of ligands to the U5 region.
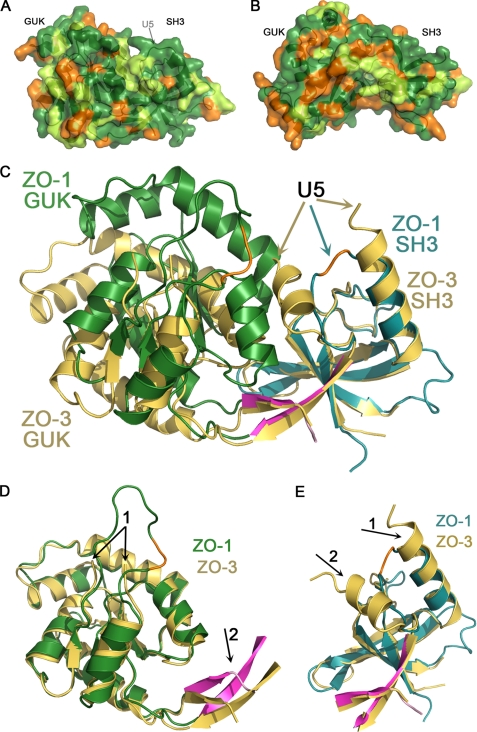
The SH3-GUK Region of ZO-3 Has a More Open Conformation than ZO-1
As we were preparing this paper, the Structural Genomic Consortia deposited the structure of the ZO-3 SH3-GUK region (Protein Data Bank code 3KFV), allowing us to contrast it with our ZO-1 structure. On the level of primary sequence, human ZO-1 and ZO-3 are 51% identical/70% similar (see Figs. 2 and 6, A and B). Interestingly, when the SH3-GUK domains of ZO-1 (cyan and green) and ZO-3 (gold) were overlaid based on superpositioning the SH3 domains, we found that the relative conformation of the SH3 and GUK domains in the ZO-3 structure is even more open than what we observe with ZO-1 (Fig. 6C). The basis for the difference in relative orientation is best illustrated by comparing the orientation of the interdomain strands relative to each domain. For example, when the GUK domains of ZO-1 and ZO-3 are overlaid, the interdomain strands are poorly aligned (Fig. 6D, arrow 2). In contrast, these interdomain strands align perfectly with the rest of the SH3 domain (Fig. 6E) when the superposition is calculated using only residues from the SH3 domain. This recapitulates the previous analysis comparing ZO-1 to PSD-95 and solidifies our conclusion that the hinge region that connects the SH3 to the GUK domain is located between the interdomain strands and the GUK domain (and not, as previously predicted, within U5).
FIGURE 6.
Human ZO-1 adopts a different SH3-GUK interdomain conformation compared with ZO-3. A, surface representation of ZO-1 in which the sequence conservation to ZO-3 is indicated by different colors: green for identical residues, lime for similar residues, and orange for non-conserved residues. An arrow indicates the location of the U5 region. B, an analogous representation rotated 180 degrees. C, superposition of ZO-1 (same color scheme as before) and ZO-3 (gold) based on aligning the SH3 domains. The SH3-GUK interdomain angle is different, with ZO-3 adopting a more open conformation. The U5 region of ZO-3 could not be traced, with the break indicated by two gold arrows. D, superposition of the GUK domains reveals a very similar fold. The NMP-binding loop in ZO-3 could not be traced (arrow 1 points at the break), whereas we could model most of this region in ZO-1 except the orange highlighted stretch. Because the proteins adopt a different interdomain angle, the strands linking the domains do not align well (arrow 2). E, superposition of the SH3 domains shows a nearly perfect overlay. This time the interdomain strands overlay well, indicating that they move as a rigid body with the SH3 domain. Two arrows indicate the break in the U5 region of ZO-3 relative to the break in ZO-1 (orange trace). Note the extra helix in ZO-3 (arrow 2) relative to that of ZO-1.
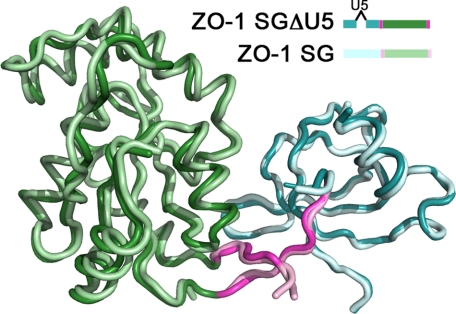
Although the interdomain conformations are significantly different between ZO-1 and ZO-3, the individual domains align well, with a root mean square deviation of 0.95 Å (over 147 atoms) and 0.73 Å (over 73 atoms) for the GUK and SH3 domains, respectively (Fig. 6, D and E). The most pronounced difference in the GUK domains of these proteins is that we could model a larger part of the “NMP-binding loop” in ZO-1 (Fig. 6D, arrow 1). The most significant difference between the SH3 domains of ZO-1 and ZO-3 is in the conformation of the residues flanking the U5 region. It is notable that although the U5 region was present in the ZO-3 construct, the majority of this domain was not modeled (absent from the ZO-3 model are residues Val572–Arg606). This is consistent with our prediction that absent a ligand, the U5 region remains flexible. It is also notable that in the ZO-3 structure, the helix preceding the U5 region is one turn longer than the homologous helix in our U5-deleted ZO-1 structure (Fig. 6E, arrow 1). Moreover, the region following U5 in ZO-3 is a helix in that is replaced by a loop in ZO-1 (Fig. 6E, arrow 2). The presence of this additional helix after the U5 region and the more open interdomain confirmation are likely to be linked because the helix precludes the more closed state we observe in ZO-1. The question remains, are these inherent differences between ZO-1 and ZO-3, or are the differences a result of the U5-truncation in our construct of ZO-1? To address this issue, we took advantage of the x-ray data set that we collected from a crystal of the intact ZO-1 SH3-GUK region (albeit to 3.7 Å resolution). We performed molecular replacement using the model we have built based on the SGΔU5 data. Importantly, we performed this calculation using two independent models, one for the SH3 domain and one for the GUK domain. By doing so, we allow unbiased placement of each domain relative to the other. If the act of truncating the U5 region caused the more closed state, then the structure from the crystal with the U5 would display the more open conformation, as observed with ZO-3. The result is shown in Fig. 7. The domains of the intact region (SG) overlay nearly perfectly on the domains of the truncated protein (SGΔU5).
FIGURE 7.
Presence of the U5 region in ZO-1 does not change the interdomain conformation. A 3.7 Å data set was collected on an SG construct containing the U5 region (SG), and the structure was solved by molecular replacement using the SH3 and GUK as two independent domains. This structure maintains the interdomain positioning as observed in SGΔU5. The color scheme of SG follows that of SGΔU5 but with a lighter shade.
The above analysis suggests that the different conformation we observe between ZO-1 and ZO-3 is a true difference that is not due to the truncation of U5 in our ZO-1 construct. Consistent with this interpretation, examination of the primary sequences reveals that the U5 region of ZO-3 is in fact eight residues longer than that of ZO-1, suggesting that the U5 region confers different functions in the two proteins (Fig. 2). What is the functional relevance of this difference? In ZO-1 the U5 region is required for localization to the tight junction (2). A previous study (13) has shown that ZO-3, although it contains a U5 region, cannot localize to the tight junctions in the absence of either ZO-1 or ZO-2. Thus, we propose that the variation in conformation between ZO-1 and ZO-3 (i.e. ZO-3 being more open), which is directly correlated to the difference in the length of the U5 region, is the reason that ZO-3 lacks this localizing function.
Conclusions
Comparing and contrasting the structure of the ZO-1 SH3-GUK module with that of PSD-95 and ZO-3, in combination with our ligand binding studies, provide us with an important insight into the general characteristics of MAGUK proteins and of ZO-1 specifically. We reveal that although the interdependent folding of the SH3 and GUK domains is conserved in at least three MAGUKs, the angle between the SH3 and GUK domains is variable, and the hinge point between these domains is at the interface between the GUK domain and strands 5 and 6 of the SH3 domain. The U5 region, which is variable in length and sequence among MAGUKs, should be considered a variable surface loop on the SH3 domain rather than a sequence connecting SH3 and GUK. Additionally, this region is quite flexible in all MAGUKs in the absence of a ligand.
The role of the U5 effector loop in ZO-1 is poorly understood. There are currently no well established binding partners, as there are for the U5 in several other MAGUKs (7–9). Nor is there any evidence that the U5 region of ZO-1 regulates binding to the SH3-GUK, as described for SAP97 (25). However, as in several other MAGUKS (10, 11), the U5 effector loop is required for localization of ZO-1 to the tight junction in vivo (2). This observation indicates that the U5 domain of ZO-1, like other MAGUKs, is an important functional domain of the ZO-1 core complex. Additionally, the difference in length of the U5 region between ZO-3 and ZO-1/ZO-2, which results in the SH3-GUK module adopting a different interdomain angle, may explain the inability of ZO-3 to localize to tight junctions (13).
The U6 region is also a critical regulator of ZO-1 function. In vivo, it regulates the temporal and spatial organization of transmembrane proteins like occludin and claudins into functional barrier strands (2). ZO-1 constructs lacking U6 form ectopic TJ strands composed of the barrier-forming claudins, occludin, and junction adhesion molecule proteins. How U6 regulates strand assembly is unclear, but one suggestion from our data is that U6 might regulate binding of various TJ ligands or regulatory proteins to the GUK domain. This hypothesis is supported by our observations that a basic surface on the GUK domain that includes helix V is all or part of the binding surface for calmodulin and the U6 domain and that the U6 domain can compete with calmodulin for binding to this helix. It is also supported by our previously reported studies that demonstrated that the U6 domain inhibits in vitro binding of occludin to the SG (2). Together these observations allow us to speculate that U6 inhibits occludin binding to GUK in vivo, preventing ectopic strand formation. We speculate that there normally exists within the cell a temporal or spatially regulated signal that displaces U6 and permits binding of occludin only within the tight junction. However, we cannot yet rule out the possibility that it is the U6-regulated binding of alternative GUK ligands that contributes to strand assembly. Possible candidates include calmodulin and the actin-binding proteins Shroom and cingulin (Fig. 1). Thus, we conclude that the U6 region is also an important effector loop, and how this loop regulates strand assembly is currently under investigation.
We currently find it difficult rationalize from the SH3-GUK structure how occludin binds to the GUK domain. Peptide binding studies suggest that the occludin binding site in SG includes a combination of residues within U5 and GUK domain helix IV and V (27). This is not inconsistent with our structure and would strongly support our hypothesis that binding of the U6 region to helix V would regulate occludin binding. However, our previous structural and biochemical studies (28), confirmed by our colleagues (29), indicate that the ZO-1-binding domain in occludin is a positive charge face of a coiled-coil domain (28). The addition of negatively charged residues within this region of occludin interfered with ZO-1 binding (28). Thus, it is hard to predict how occludin would bind directly to this region of the GUK domain, whose surface residues are also overwhelmingly positive. We can only hypothesize that some modification, such as phosphorylation of SG, or some intermediary protein may be critical for occludin binding.
Finally, it seems very likely that the activity of the U5 and U6 regions is coordinated in vivo. Our structure suggests that both of these effector domains and the ligand binding sites surrounding helix V of the GUK domain are present on the same surface of the SH3-GUK module, distal from the flexible hinge domain. This implies that any change in the hinge angle would have a significant effect on the distance between the ligand binding sites on the two surfaces. Interestingly, although the U5 is necessary for TJ localization in vivo, this localization is disrupted by the U6 motif, implying that there must be a trigger to move U6 out of the way (2). This observation suggest that these two effector loops functionally interact, and the SG structure should allow further elaboration of how these effector loops regulate TJ assembly in vivo.
Supplementary Material
Acknowledgments
We thank Bernard Santarsiero for help in solving the structure of ZO-1 and the staff of SERCAT for assistance.
This work was supported, in whole or in part, by National Institutes of Health (NIH) Grant R01DK061397 and NIH, NIDDK, Institutional Training Grant T32DK07739, “Training Program in Signal Transduction and Cellular Endocrinology.”

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Fig. S1.
The atomic coordinates and structure factors (code 3LH5) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- MAGUK
- membrane-associated guanylate kinase homolog
- SH3
- Src homology 3
- GUK
- guanylate kinase-like domain
- SG
- SH3-GUK module
- SGΔU5
- SG with a deleted U5 region
- ZO
- zonula occludens
- TJ
- tight junction(s)
- GST
- glutathione S-transferase
- CaM
- calmodulin.
REFERENCES
- 1.Funke L., Dakoji S., Bredt D. S. (2005) Annu. Rev. Biochem. 74, 219–245 [DOI] [PubMed] [Google Scholar]
- 2.Fanning A. S., Little B. P., Rahner C., Utepbergenov D., Walther Z., Anderson J. M. (2007) Mol. Biol. Cell 18, 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGee A. W., Dakoji S. R., Olsen O., Bredt D. S., Lim W. A., Prehoda K. E. (2001) Mol. Cell 8, 1291–1301 [DOI] [PubMed] [Google Scholar]
- 4.Tavares G. A., Panepucci E. H., Brunger A. T. (2001) Mol. Cell 8, 1313–1325 [DOI] [PubMed] [Google Scholar]
- 5.Woods D. F., Hough C., Peel D., Callaini G., Bryant P. J. (1996) J. Cell Biol. 134, 1469–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin H., Hsueh Y. P., Yang F. C., Kim E., Sheng M. (2000) J. Neurosci. 20, 3580–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marfatia S. M., Leu R. A., Branton D., Chishti A. H. (1995) J. Biol. Chem. 270, 715–719 [DOI] [PubMed] [Google Scholar]
- 8.Cohen A. R., Woods D. F., Marfatia S. M., Walther Z., Chishti A. H., Anderson J. M., Wood D. F. (1998) J. Cell Biol. 142, 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuko N., Makino K., Kuwahara H., Fukunaga K., Sudo T., Araki N., Yamamoto H., Yamada Y., Miyamoto E., Saya H. (1999) J. Biol. Chem. 274, 5782–5790 [DOI] [PubMed] [Google Scholar]
- 10.Hanada T., Takeuchi A., Sondarva G., Chishti A. H. (2003) J. Biol. Chem. 278, 34445–34450 [DOI] [PubMed] [Google Scholar]
- 11.Hough C. D., Woods D. F., Park S., Bryant P. J. (1997) Genes Dev. 11, 3242–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Itallie C. M., Anderson J. M. (2006) Annu. Rev. Physiol. 68, 403–429 [DOI] [PubMed] [Google Scholar]
- 13.Umeda K., Ikenouchi J., Katahira-Tayama S., Furuse K., Sasaki H., Nakayama M., Matsui T., Tsukita S., Furuse M., Tsukita S. (2006) Cell 126, 741–754 [DOI] [PubMed] [Google Scholar]
- 14.Fanning A. S., Anderson J. M. (2009) Ann. N.Y. Acad. Sci. 1165, 113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Itallie C. M., Fanning A. S., Bridges A., Anderson J. M. (2009) Mol. Biol. Cell 20, 3930–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNeil E., Capaldo C. T., Macara I. G. (2006) Mol. Biol. Cell 17, 1922–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikenouchi J., Umeda K., Tsukita S., Furuse M., Tsukita S. (2007) J. Cell Biol. 176, 779–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabsch W., Gast W. H., Schulz G. E., Leberman R. (1977) J. Mol. Biol. 117, 999–1012 [DOI] [PubMed] [Google Scholar]
- 19.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terwilliger T. C. (1999) Acta Crystallogr. D 55, 1863–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones T. A., Kjeldgaard M. (1997) Methods Enzymol. 277, 173–208 [DOI] [PubMed] [Google Scholar]
- 22.Paarmann I., Lye M. F., Lavie A., Konrad M. (2008) Protein Sci. 17, 1946–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kistner U., Garner C. C., Linial M. (1995) FEBS Lett. 359, 159–163 [DOI] [PubMed] [Google Scholar]
- 24.Kuhlendahl S., Spangenberg O., Konrad M., Kim E., Garner C. C. (1998) Eur. J. Biochem. 252, 305–313 [DOI] [PubMed] [Google Scholar]
- 25.Wu H., Reissner C., Kuhlendahl S., Coblentz B., Reuver S., Kindler S., Gundelfinger E. D., Garner C. C. (2000) EMBO J. 19, 5740–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabio G., Arthur J. S., Kuma Y., Peggie M., Carr J., Murray-Tait V., Centeno F., Goedert M., Morrice N. A., Cuenda A. (2005) EMBO J. 24, 1134–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Müller S. L., Portwich M., Schmidt A., Utepbergenov D. I., Huber O., Blasig I. E., Krause G. (2005) J. Biol. Chem. 280, 3747–3756 [DOI] [PubMed] [Google Scholar]
- 28.Li Y., Fanning A. S., Anderson J. M., Lavie A. (2005) J. Mol. Biol. 352, 151–164 [DOI] [PubMed] [Google Scholar]
- 29.Murakami T., Felinski E. A., Antonetti D. A. (2009) J. Biol. Chem. 284, 21036–21046 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.