Abstract
Schwanniomyces occidentalis invertase is an extracellular enzyme that hydrolizes sucrose and releases β-fructose from various oligosaccharides and essential storage fructan polymers such as inulin. We report here the three-dimensional structure of Sw. occidentalis invertase at 2.9 Å resolution and its complex with fructose at 1.9 Å resolution. The monomer presents a bimodular arrangement common to other GH32 enzymes, with an N-terminal 5-fold β-propeller catalytic domain and a C-terminal β-sandwich domain for which the function has been unknown until now. However, the dimeric nature of Sw. occidentalis invertase reveals a unique active site cleft shaped by both subunits that may be representative of other yeast enzymes reported to be multimeric. Binding of the tetrasaccharide nystose and the polymer inulin was explored by docking analysis, which suggested that medium size and long substrates are recognized by residues from both subunits. The identified residues were mutated, and the enzymatic activity of the mutants against sucrose, nystose, and inulin were investigated by kinetic analysis. The replacements that showed the largest effect on catalytic efficiency were Q228V, a residue putatively involved in nystose and inulin binding, and S281I, involved in a polar link at the dimer interface. Moreover, a significant decrease in catalytic efficiency against inulin was observed in the mutants Q435A and Y462A, both located in the β-sandwich domain of the second monomer. This highlights the essential function that oligomerization plays in substrate specificity and assigns, for the first time, a direct catalytic role to the supplementary domain of a GH32 enzyme.
Keywords: Carbohydrate-binding Protein, Crystal Structure, Enzyme Kinetics, Enzyme Structure, Protein Domains, Protein Structure, Fructofuranosidase, GH32 Family, Glycosylhydrolase, Invertase
Introduction
Fructans, the fructose-rich polymers derived biosynthetically from sucrose, are important storage oligosaccharides and polysaccharides in many bacteria and fungi and numerous plant species. Furthermore, sucrose is one of the most widespread disaccharides in nature and is especially ubiquitous in higher plants as the first free sugar resulting from photosynthesis. It is the major transport compound to bring energy and carbon skeletons from source to sink tissues. Carbohydrate partitioning and sugar sensing are intimately connected to sucrose metabolism; these processes are vital throughout plant development. Therefore, the enzymes involved in fructans and sucrose processing are essential to plant cell metabolism.
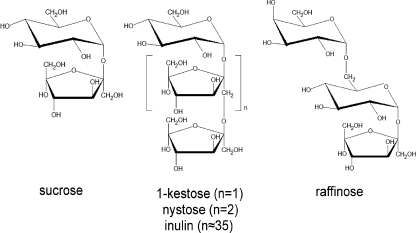
The enzymes that hydrolyze sucrose are referred to collectively as invertases or β-fructofuranosidases (EC 3.2.1.26) and catalyze the release of β-fructose from the nonreducing end of various β-d-fructofuranoside substrates (Fig. 1). The cleavage of the β-glycosidic bond is carried out by a double displacement catalytic mechanism that retains the configuration of the fructose anomeric carbon, two conserved residues, an aspartic and a glutamic acid, being the nucleophile and the general acid-base catalyst, respectively. On the basis of the amino acid sequences (1) they are classified into family 32 of the glycosylhydrolases (GH32),3 which are included into the GHJ clan together with the GH68 (inulosucrase family).
FIGURE 1.
Chemical structure of the general substrates hydrolized by fructofuranosidases from the GH32 family. The average polymerization degree of the inulin used in this study was n ≃ 35.
To date, the three-dimensional structures of exoinulinase from Aspergillus niger (var. awamori; AaEI) (2), a β-fructofuranosidase from Thermotoga maritima (TmInv) (3), a plant FEH from Cichorium intybus (CiFEH) (4), and a cell wall invertase from Arabidopsis thaliana (AtInv) (5), all included in the GH32 family, have been solved by x-ray crystallography. These studies show a common bimodular arrangement for this family, being folded into a catalytic and a β-sandwich domain with unknown function. In addition, the complexes of these enzymes with short substrates, i.e. fructose (2), sucrose (6), and the trisaccharides 1-kestose (7) and raffinose (8), have also been reported. These studies provide insights into the fructose-binding pocket, i.e. the −1 subsite following the nomenclature used for sugar-binding substrates (9), which identifies the cleavage as occurring between subsites −1 and +1. Some general features of the +1 and +2 subsites are also described, and hence, whereas the +1 subsite is formed by several residues making hydrophobic and polar interactions with the substrates, the sugar unit bound at subsite +2 seems to be more loosely recognized, mostly through a few polar interactions mediated by water molecules. Moreover, additional binding subsites for longer substrates have been discarded (8).
It is worth noting that a GH32 yeast invertase structure has not been reported until now, a remarkable fact when taking into account that yeast invertases have been described as multimeric. As shown by electron microscopy (10) the basic structural unit of external and internal yeast invertase is a dimer, but that seems able to associate into larger oligomers upon mannose binding (11). These studies suggest that the extent of glycosylation of invertase determines its oligomeric state, but the molecular mechanisms involved in higher aggregation are uncertain. It remains to be elucidated whether the supplementary β-sandwich domain of the enzyme plays the role of a carbohydrate recognition domain and, therefore, is involved in oligomerization, at least in yeast enzymes.
The nonconventional yeast Schwanniomyces occidentalis (also Debaryomyces occidentalis) has been regarded for years as a biotechnologically interesting organism because of its ability to grow in a broad range of inexpensive carbon sources, such as starch and inulin, using a number of activities (12–15), as well as for its efficient extracellular secretion of high molecular mass proteins (16, 17) and its low attached glycosylation (18). An extracellular invertase/fructofuranosidase activity has also been reported in this yeast when lactose was used as the carbon source (14). This secreted protein showed broad substrate specificity, hydrolyzing sucrose, 1-kestose, nystose, and raffinose (Fig. 1). In addition, the enzyme is able to catalyze the synthesis of fructo-oligosaccharides (FOS), mainly the prebiotic 6-kestose (a β-(2–6)-linked FOS), with 1-kestose (a β-(2–1)-linked FOS) being a side product of the reaction (19). Currently the main industrial FOS producer is the fructosyltransferase from Aspergillus (20, 21), which provides a mixture of FOS of the inulin-type structure β-(2–1) without producing 6-kestose. The protein amino acid sequence from Sw. occidentalis shares considerable sequence similarity with other yeast invertases, inulinases, levanases, and fructosyltransferases, included in the GH32 family (22). In this article we report the crystal structure of the invertase produced by Sw. occidentalis (SoInv), which serves as a model for other yeast GH32 enzymes. Computational docking studies were done to identify the putative position for long substrates binding into the active site. Potential oligosaccharide-binding residues were mutated, and the activities of the mutants were characterized. Our results, illustrating the dimeric structure of yeast invertase, led to the conclusion that this oligomerization is involved directly in substrate recognition and specificity.
EXPERIMENTAL PROCEDURES
Protein Purification, Crystallization, and Data Collection
Extracellular SoInv was purified from Sw. occidentalis ATCC26077 and crystallized in an orthorhombic crystal form as described previously (23). Equal amounts of protein solution (7 mg ml−1 SoInv in 20 mm Tris-HCl, pH 7.5, and 150 mm NaCl) and precipitant buffer (15% (w/v) PEG 3350 in 100 mm Hepes, pH 7) were mixed and vapor-equilibrated against a reservoir containing the latter solution in a sitting drop setup. These rod-shaped crystals belong to P212121 space group, with two molecules/asymmetric unit and 67% solvent content in the cell. For data collection, native crystals were transferred to cryoprotectant solutions containing 18% (w/v) PEG 3350 and 30% (w/v) glycerol before being cooled to 100 K in a stream of nitrogen. A full data set was collected at the European Synchrotron Facility (ESRF, Grenoble, France), using ID14-1 beamline, up to 2.9 Å. Additional data sets collected at beamlines ID23-1 and ID23-2 yielded lower resolution data. In an attempt to increase the diffraction power of the crystals, the sample was deglycosylated by treatment with endoglycosidase H (New England Biolabs), which reduced the molecular mass from 73 kDa, as determined by mass spectrometry, to an apparent molecular mass below 66 kDa, as deduced from the SDS-PAGE analysis. The sample was concentrated to 10 mg ml−1, and the screening of new crystallization conditions was performed with the PACT Suite from Qiagen. Small cubes and thick plates grew in drops containing 20% PEG 6000, 0.2 m MgCl2, with the pH ranging from 6 to 8. These crystals belong to the P21 space group with two molecules in the asymmetric unit and 42% solvent content. For data collection, a plate grown at pH 7 was soaked in a solution containing mother liquor and 50 mm nystose for 3 min before being transferred to cryoprotectant solution consisting of 25% PEG 6000, 0.2 m MgCl2, and 15% PEG 400 and cooled to 100 K in a nitrogen stream. A full data set was collected to 1.9 Å at the ESRF ID14-4 beamline. Analysis of the diffraction patterns was performed using MOSFLM (24), and data were merged with the CCP4 suite (25). A summary of data collection and data reduction statistics is shown in Table 1.
TABLE 1.
X-ray data collection and structure refinement statistics
| Crystal | Native SoInv | Deglycosylated SoInv-Fru |
|---|---|---|
| Data collection | ||
| Unit cell dimensions (Å) | 105.78, 19.49, 137,68 | 60.87, 92.27, 116.28 (β = 104.8°) |
| Space group | P212121 | P21 |
| Molecules in asymmetric unit | 2 | 2 |
| Solvent content (%) | 67 | 42 |
| Limiting resolution (Å) (Outer shell) | 2.9 (2.90-2.95) | 1.9 (2.01-1.90) |
| Unique reflections | 36,939 | 98,012 |
| Rrima | 0.19 (0.54) | 0.13 (0.50) |
| Rpimb | 0.08 (0.30) | 0.05 (0.22) |
| Completeness (%) | 99.7 (99.8) | 99.7 (99.7) |
| Mean multiplicity | 6.9 (6.7) | 7.1 (7.0) |
| Mean I/σI | 9.7 (2.9) | 4.3 (1.9) |
| Final refinement parameters | ||
| Protein atoms (non-H) | 8,100 | 8,100 |
| Solvent molecules | 326 | 823 |
| Ligand molecules | 12 Glycerol | 2 Fructose |
| Glycosylation molecules | 16 NAGc | 8 NAGc |
| R-factord | 0.21 | 0.21 |
| Rfree | 0.27 | 0.25 |
| r.m.s. bonds (Å)/angles (°) | 0.013/1.8 | 0.007/1.24 |
| Averaged B-factors (Å2) | ||
| Main chain | 34.6 | 16.4 |
| Side chain | 34.7 | 17.5 |
| Ligand | 58.9 | 27.6 |
| Solvent | 31.5 | 33.2 |
| Ramachandran plot (Ref. 30) | ||
| Favored (%) | 97.1 | 98.3 |
| Outliers (%) | 1.5 | 0.5 |
| Protein Data Bank code | 3KF5 | 3KF3 |
a Rrim = Σ[n/(n − 1)]1/2Σ|I(h)i − 〈I(h)〉|/ΣΣ〈I(h)〉.
b Rpim = Σ[1/(n − 1)]1/2Σ|I(h)i − 〈I(h)〉|/ΣΣ〈I(h)〉.
c NAG, N-acetylglucosamine.
d R-factor = Σ(|Fobs − Fcalc|)/Σ|Fobs| (Rfree is equivalent to R-factor for a randomly selected 5% subset of reflections not used in structure refinement).
Structure Solution and Refinement
The structure of SoInv was solved using the molecular replacement method with data to 2.9 Å collected from the orthorhombic crystal form. Initial phases were obtained with the MOLREP program (26) using the atomic coordinates of AaEI as the search model (Protein Data Bank code: 1Y4W). The best solution was obtained by using the sequence alignment of both enzymes to correct the model, which after rigid body fitting produced an R-factor of 0.49 with a correlation coefficient of 0.49. Crystallographic refinement was performed by using Refmac (27) in the CCP4 suite (25) with flat bulk solvent correction, including low resolution data to 50 Å, and using maximum likelihood target features. A subset of 5% randomly selected structure factor amplitudes was excluded from automated refinement to compute a free R-factor throughout the refinement. Large regions of the β-sandwich domain were removed from the model, as no electron density was observed at the polypeptide chain. These regions were mainly the loops of the β-sandwich but also some of the β-strands. Several rounds of refinement applying tight NCS (noncrystallographic symmetry) restraints between both molecules in the asymmetric unit were alternated with major rebuilding of these regions using the program O (28), after which the final 2Fo − Fc maps showed continuous density for the whole polypeptide chain. At the final stage, water molecules and several N-acetylglucosamine and glycerol molecules were included, combined with more rounds of positional and individual restrained B-factor refinement and translation/libration/screw-rotation correction. When the monoclinic high resolution crystal form became available, additional rounds of refinement using the same strategy but applying loose NCS restraints between both molecules in the asymmetric unit were carried out, and minor corrections were introduced into the model, mainly concerning the orientation of the peptide bond. This led to a final R-factor of 0.21 (Rfree = 0.25) for all data to 1.9 Å resolution. The final refinement parameters are reported in Table 1. The stereochemistry of the model was checked by using PROCHECK (29) and MOLPROBITY (30), and Figs. 3, 4, and 6–8 were generated with PyMOL (31).
FIGURE 3.
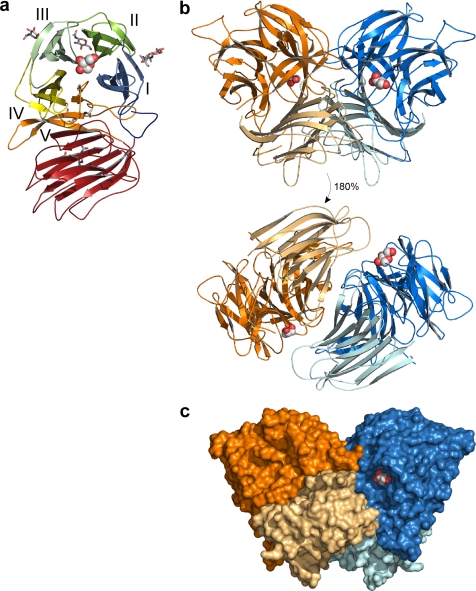
The structure of SoInv. a, ribbon diagram of the monomeric unit of SoInv showing the bimodular arrangement, the cataltytic β-propeller domain colored according to its five blades (I–V), and the C-terminal β-sandwich domain (colored red). A fructose bound at the active site of the deglycosylated form crystals is shown in a spherical representation, and the glycosylation moieties are represented as sticks. b, two SoInv units (blue and orange) associate to form a tight dimer by many polar and hydrophobic interactions between the four domains within the dimer (shaded differently). c, molecular surface of SoInv dimer showing one of the two active site pockets, which is surrounded by the β-sandwich domain from the other monomer.
FIGURE 4.
The active site shape of the GH32 family. a, superimposition of the structures of SoInv (blue), AaEI (violet), TmInv (brown), and CiFEH (green) showing that the major differences in the catalytic domain are concentrated in the highly variable loops connecting the different blades of the propeller (L1–L4) and the turns linking strand B to C within each blade (TI–TV). A long insertion in a SoInv loop located in blade V, involved in dimer formation, is indicated by an arrow. b, molecular surface of the SoInv-fructose (blue), AaEI-fructose (violet), TmInv-raffinose (brown), and CiEI-1-kestose (green) complexes highlighting the above mentioned loops that shape the active site. Loops TIV and TV, defined in Fig. 5, are marked on the SoInv surface. The bound ligands are represented as spherical models.
FIGURE 6.
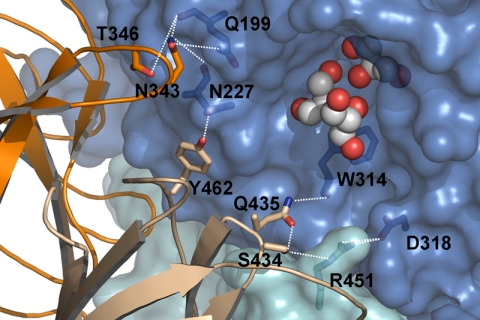
Close-up view of the interface at the active site. A detail of the SoInv active site pocket showing three of the six glycerol molecules bound in the native crystals; these molecules occupy subsites −1, +1, and +2. Gln-199 (TIII), Asn-227 (L3), and Trp-314 and Asp-318 (TV) from one molecule form a network of polar interactions with the Tyr-339—Leu-349 loop and the β-sandwich domain from the other subunit at the dimer interface, which build up the active site pocket. The molecular surface of one subunit is shown, whereas the other is in a ribbon representation for clarity (color-coded as described in the legend for Fig. 3). The glycerol molecules are shown as spheres.
FIGURE 7.
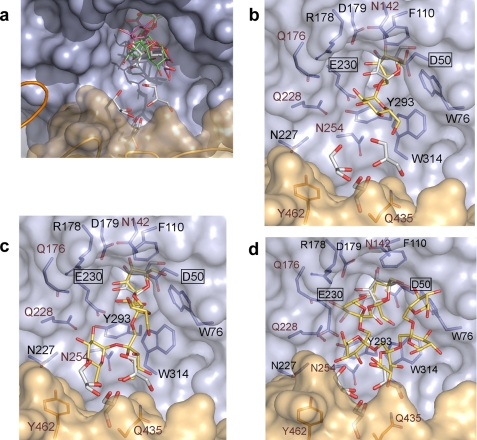
Substrate specificity of SoInv. The active site of SoInv is shown with both subunits in cyan and beige. a, the six glycerol molecules bound at the active site in the native crystals of SoInv and the putative position of 1-kestose (pink) and raffinose (green) inferred from a structural superimposition of SoInv coordinates onto CiEI-1-kestose (7) and TmInv-raffinose (8) complexes. The putative location of the substrate sucrose (b) inferred from the structural superposition of SoInv onto the AtInv-sucrose complex (6) and the positions of nystose (c) and inulin (d), as obtained from docking analysis, are shown as yellow and red sticks. Only six fructose units are shown in the inulin chain. The fructose found in the deglycosylated protein crystals and three of the glycerol molecules found in the native crystals are shown as white and red sticks. The residues suggested to interact with the substrates are shown, with the catalytic residues boxed. The residues that are variable among the GH32 family members and that have been mutated are marked with a brown label.
FIGURE 8.
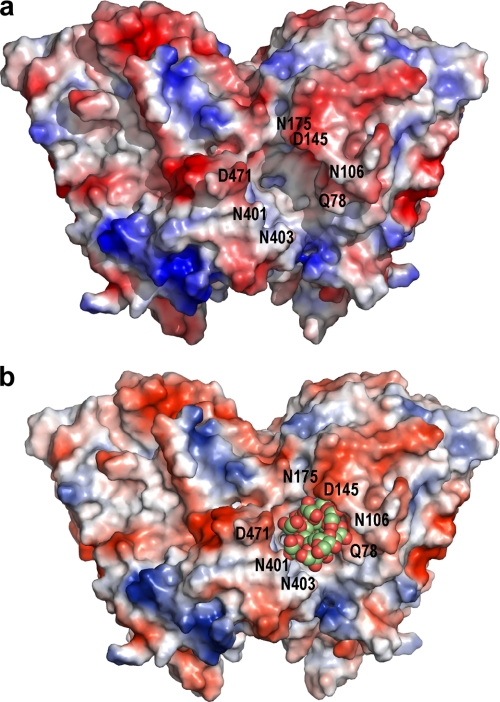
Binding of the polymeric substrate. The molecular surface of the free SoInv dimer shows the electrostatic distribution, with acidic regions in red and basic regions in blue (a) and inulin positioned in the binding cleft by automated docking (b). A chain containing 12 fructose units has been used as a ligand. Several polar residues surround the entrance of the cavity, which is 25 Å deep and is accessible from the solvent by a slot at a maximum 18 Å in width.
Automated Docking of Ligands into SoInv Structure
The nystose coordinates were retrieved from the crystal structure (32) deposited in the Cambridge Structural Database (CSD). All hydrogen atoms were added to the ligand, and charges were assigned by the Gasteiger method using the AutoDockTools program (33). All bonds of hydroxyl groups and glycoside linkages, but not those of the sugar rings, were defined as rotatable bonds. The inulin chain coordinates were also retrieved from the crystal structure determined from electron diffraction data (34) and deposited in the Cambridge Structural Database. In this case, only hydroxyl groups were allowed to rotate, the glycoside linkages being maintained in a fixed helical conformation. The protein model contains the coordinates of the crystals structure of dimeric SoInv presented here after the removal of water molecules and all heteroatoms. Polar hydrogen atoms were then added using AutoDockTools.
AutoDock 4.0 (33) was executed 50 times with the Lamarckian genetic algorithm, a population size of 150, elitism set at 1, mutation rate at 0.02, and crossover rate of 0.80. Simulations were performed with a maximum of 2,500,000 energy evaluations and a maximum of 27,000 generations. Docking results were clustered using a cut-off of 2 Å r.m.s. deviations. In the case of inulin, additional manual docking was followed by optimization of inulin using the local search (LS) algorithm (AutoDock 4.0) executed 30 times with 300 iterations and with the size of the local search space set at 1. In these calculations, the 11 C1–C2 angles linking two fructose units were allowed to rotate, as this has been suggested as the most flexible of the three glycoside torsions within each fructose-fructose pair (34).
Mutagenesis and Protein Purifications
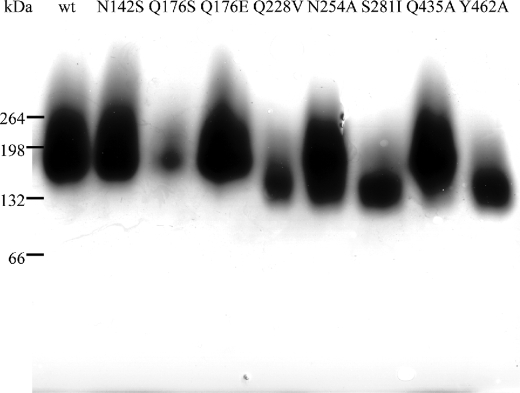
The sequence SoINV (GenBankTM accession number CQ890277) was amplified using the forward primer 5′-TAGGATCCAACATGGTACAAGTTTTAAGTGTATTAG-3′ and the reverse primer 5′-CATCTCTAGACTAGCCCTACTTATTTAGTTCTCT-3′. Restriction sites for BamHI and XbaI (shown in bold) were included in these primers for cloning the PCR product into the pYES2.0 shuttle vector (Invitrogen) under the control of the yeast GAL1 promoter. The generated construct was used as the template for site-directed mutagenesis using the method described previously (35) and the primers listed in Table 2. Saccharomyces cerevisiae EUROSCARF Y02321 (BY4741; Mata; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YIL162w::kanMX4) was used as the expression host, and it was transformed by the lithium acetate method (36). Yeasts were growth on YPGal medium (yeast extract 1%, peptone 1%, and galactose 2%) to the beginning of the stationary phase (OD660 = 6–7). Culture filtrates (1 liter) containing invertase activity were concentrated through a HY200 filter (30,000 molecular weight cut-off) using a VivaFlow 50 system (Vivascience). The concentrated fractions (50 ml) were applied to a DEAE-Sephacel chromatography column (10 ml) equilibrated with 20 mm HCl-Tris, pH 7 (buffer A). The proteins were eluted with a 0–0.5 m NaCl gradient at 1 ml min−1. The 0.1 m fractions (2 ml) were pooled, dialyzed in buffer A, concentrated to the desired volume by using the Microcon YM-10 (Amicon) system, and stored at −70 °C. SDS-PAGE (8%) of the samples confirmed the proteins purification, and protein concentration was estimated photometrically at 280 nm (NanoDrop spectrophotometer ND-1000). The enzyme activities of all mutants were also detected by zymogram analysis (Fig. 2) using nondenaturing gradient gels (4–15%) from Bio-Rad and subsequently were stained with 1% (w/v) 2,3,5-triphenyltetrazolium chloride (19).
TABLE 2.
Oligonucleotides employed for mutagenesis
Forward (F) and reverse (R) sequences are shown with the mutations in bold letters.
| Mutant | Orientation | Sequence |
|---|---|---|
| D50A | F | TGGATGAATGCTCCGAATGGTCTATTCTACGATAAA |
| R | ATTCGGAGCATTCATCCATCCTTTTTCCGGAGTAAA | |
| N142Y | F | CCAACTATATACCAGATTTACAAACCCAAGACATTGC |
| R | GGTATATAGTTGGTATAAATGGCAAGAATTCTTTGGTTTGG | |
| Q176S | F | CTCAAACTCTTTCCGTGATCCAAAAGTTTTCTGGC |
| R | CACGGAAAGAGTTTGAGGAGACATCAATCACAGG | |
| Q176E | F | CTCAAACGAATTCCGTGATCCAAAAGTTTTCTGGC |
| R | CACGGAATTCGTTTGAGGAGACATCAATCACAGG | |
| Q228V | F | TACGGAAATGTCTATGAATGTCCAGGTTTAATTGAA |
| R | ACATTCATAGACATTTCCGTAATAACCAGAAGAAAA | |
| E230A | F | AATCAGTATGCTTGTCCAGGTTTAATTGAAGTTCCTATTGAGAAT |
| R | ACCTGGACAAGCATACTGATTTCCGTAATAACCAGAAGAAAAATT | |
| N254A | F | CAATTGCTCCTTTATCGCCTTTGGGTGGTTCG |
| R | CCCGGAGCAATTGCTAAAAACATAACCCACTTTG | |
| Q435A | F | CATCCGCTTCATCGTTTTATATCGATCGTCATATTCC |
| R | CGATGAAGCGGATGAATCAAAACCAATTTTAATGGAATC | |
| Y462A | F | GCTGCAGCTCTTGAACCTTTAGACTACGATCAAGAC |
| R | GTTCAAGAGCTGCAGCCAACTTATCAGTAAAGAATTGC | |
| S281I | F | AGATGATATCCAAACTAGATTTGTTTGTTGATATTGGAAAAG |
| R | CTAGTTTGGATATGATCTGGAACAAACTGGAAGCCG |
FIGURE 2.
Gel analysis of the SoInv mutants. Culture filtrates (500 ml) of the S. cerevisiae transformants that expressed the wild-type or mutant SoInv proteins (indicated at the top of the image) were processed for protein purification as described under “Experimental Procedures.” Purified proteins (5–20 μg) were subjected to PAGE analysis, and the enzyme activities were revealed in situ. Numbers at the left indicate the positions of the bovine serum albumin molecular masses used as a control (in kDa).
Enzyme and Kinetic Analysis
All of the enzymatic assays were done by the dinitrosalicylic acid method adapted to a 96-well microplate scale as described previously (19). One unit of activity was defined as that catalyzing the formation of 1 μmol of reducing sugar min−1 ml−1. For kinetic analysis, velocity measurements were performed with 0.3–5.3 mg ml−1 enzyme in triplicate at 2.5–100 mm sucrose or nystose and 1–10 mm inulin. Reactions were performed for 20 min in 50 mm NaAc, pH 5. Curve plotting and analysis of the curves were carried out using the SigmaPlot program (version 7.101). Kinetic parameters were calculated by fitting the initial rate values to the Michaelis-Menten equation.
Analytic Ultracentrifuge Analysis
Sedimentation equilibrium experiments were performed in a Beckman Optima XL-A Ultracentrifuge using a Ti50 rotor and six channel centerpieces of Epon-charcoal (optical path length, 12 mm). Samples of purified SoInv in the concentration range of 0.2 to 0.5 mg ml−1 were equilibrated against 20 mm citrate, pH 4.5, 20 mm Tris, pH 7, 20 mm Hepes, pH 7, and 20 mm Tris, pH 9, and centrifuged at 8,000, 10,000, and 14,000 rpm at 293 K. Radial scans at 280 nm were taken at 12, 14, and 16 h. The three scans were identical (equilibrium conditions were reached). The weight average molecular mass (Mw) of SoInv was determined by using the program EQASSOC with the partial specific volume of SoInv set to 0.71 at 293 K as calculated from its amino acid composition and referred to that reported for the monomer (72 kDa) (23).
RESULTS AND DISCUSSION
Crystal Structure of SoInv Shows a Bimodular Folding
Previous attempts to crystallize the native form of SoInv expressed in S. cerevisiae failed to obtain untwined, good quality diffracting crystals as reported; therefore, the extracellular enzyme was isolated directly and purified from Sw. occidentalis (23). This sample gives crystals belonging to the orthorhombic P212121 space group, producing a diffraction pattern up to a 2.9 Å resolution. The three-dimensional structure of the enzyme was solved by molecular replacement and refined. To improve the crystal quality, the sample was partially deglycosylated with endoglycosidase H, a treatment that yielded a monoclinic crystal form giving data to 1.9 Å resolution. Experimental details and structure determination procedures are given under “Experimental Procedures.” The model of the native crystals contains six glycerol molecules from the cryoprotectant solution bound at the active site, whereas the deglycosylated protein crystal was soaked in the substrate nystose prior to data collection and contains the product fructose at the active site (supplemental Fig. S1). The SoInv structure obtained from both crystals is essentially the same, and therefore all of the following structural descriptions will be based on the high resolution deglycosylated form.
As in the other GH32 family enzymes that are known structurally, the invertase molecule is bimodular and folds into a catalytic five-bladed β-propeller (residues 24–356) and a C-terminal β-sandwich (residues 365–535) linked by a short 10-residue segment (Fig. 3). The propeller is assembled from five blades (I–V), each composed of four antiparallel β-strands (A–D from the axis toward the outside of the propeller) connected by turns in a classical “W” pattern. This domain belongs to the β-furanosidase superfamily (37), which contains four other members: GH43 arabinase, GH62 arabinofuranosidase (with no three-dimensional structure known up to now), GH68 levansucrase, and a group of hypothetical proteins, some of which are also predicted to be glycosylhydrolases. Interestingly, this was the first glycosylhydrolase superfamily shown to include enzymes with both inversion (GH43) and retention (GH32 and GH68) mechanisms of action (37). Furthermore, this clan shares the five-bladed β-propeller fold with tachylectin II, which was the first protein described as showing this folding (38), and human apyrase (39).
The second domain of invertase is composed of two antiparallel six-stranded β-sheets folded into a β-sandwich topology. This domain is the region that is most variable in sequence among GH32 family members (sequence identity of SoInv as low as 10–12% with enzymes from bacteria or plants); it belongs to the wide concanavalin A-like lectins/glucanases superfamily with which it shares similar levels of identity.
Five of the eight predicted glycosylation sites were visible in the electron density maps and were assigned to Asn-72, Asn-119, Asn-219, Asn-334, and Asn-394. Four of them are located in the catalytic domain linked to blades I, II, III, and V, and one is in the β-sandwich domain. They are all at the surface of SoInv, far from the active site. A different glycosylation pattern is possibly found in the enzyme expressed in S. cerevisiae, and as reported previously, this likely prevents proper crystal growth (23). In this regard, a single extracellular protein band of about 95 kDa was detected in SDS-PAGE analysis when SoInv was expressed and purified from S. cerevisiae cells, which is 20 kDa higher than the wild-type protein expressed in Sw. occidentalis.
The Loops That Confer Specificity to Enzymes within the GH32 Family
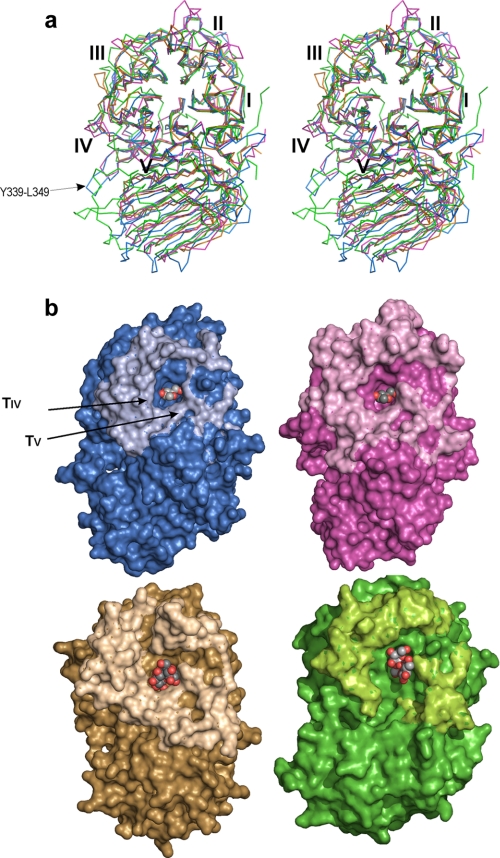
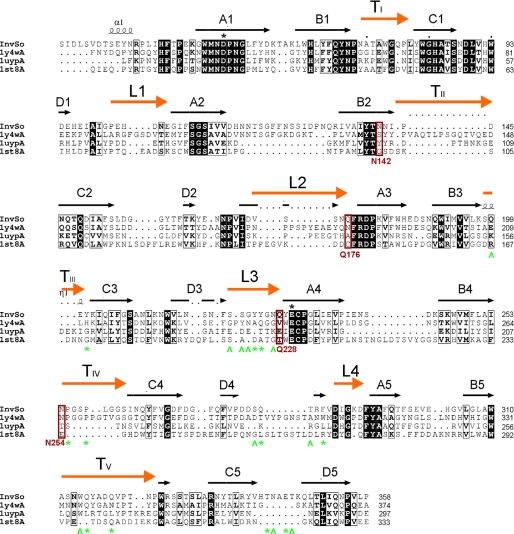
As explained previously, the three-dimensional structure has been reported for four other members of the GH32 family: AaEI (2), TmInv (3), CiFEH (4), and AtInv (5). These structures are good representatives of several of the diverse enzymatic functionalities found among the GH32 family and also correspond to different organisms. SoInv shares the highest identity (35%) with the fungal enzyme, whereas only 25 and 23% identity are found with respect to enzymes from bacteria and plant. However, the sequence conservation within the family is not equally distributed along the polypeptidic chain, the identity being higher in the catalytic domain (39 and 24% in the catalytic and the β-sandwich domains, in the case of SoInv as compared with AaEI). The Cα atoms of the AaEI, TmInv, and CiFEH structures superimposed onto SoInv with r.m.s. deviations of 1.9, 2.1, and 2.6 Å, respectively (based on the overlap of 468, 408, and 449 atoms).
Structural superimposition of these enzymes onto SoInv is shown in Fig. 4a (AtInv is highly similar to CiFEH and has been omitted for clarity). Despite the moderate sequence identity among the family, the core of the propeller is very similar in shape, with blade I the most conserved and blade IV the least conserved region. Moreover, an insertion is observed in strand D4 (Figs. 4 and 5) creating a significant disruption of blade IV in AaEI and CiFEH that is not observed in the enzymes from bacteria and yeast, although this is the least regular sheet in the propeller. It is worth noting that this region is involved directly in the oligomerization of SoInv, which will be explained below.
FIGURE 5.
Structural alignment of known GH32 catalytic domain. SoInv superimposed to AaEI, TmInv, and CiFEH (Protein Data Bank codes 1Y4W, 1UYP, and 1ST8, respectively) as given by the DALI server (44). The black squares indicate amino acid similarity as calculated by the program ALSCRIPT (45). Secondary structure elements for SoInv as suggested by PROCHECK (29) are shown above the amino acid sequences. Letters A–D indicate the β-strands, from the axis toward the outside of the propeller, within each blade. L1–L4, connecting the different blades of the propeller, and TI–TV, linking strand B to C within each blade, are marked with an orange bar. Catalytic residues are highlighted with a black asterisk. Residues of SoInv involved in hydrophobic (caret) and polar (asterisk) interactions within the dimer interface are marked in green. Residues of SoInv involved in substrate recognition and mutated in this work are highlighted in brown.
Furthermore, the analysis of the largest differences observed in the surroundings of the β-propeller center enabled us to identify the substrate binding specificity determinants. As shown in Fig. 4, the topology of this folding leads to the catalytic active site being shaped by the loops connecting the different blades of the propeller (L1–L4) and the turns linking strand B to C within each blade (TI–TV). The structural alignment of the sequences with known structure helps greatly in delimiting the segments involved in shaping these loops (Fig. 5). As seen in Fig. 4, with the exception of TI and L4, these loops are highly variable in length and sequence, with those in AaEI being the longest and those in SoInv the shortest, and they contour a different environment to the catalytic pocket (Fig. 4). Interestingly, loop TV shapes the interface with the β-sandwich domain, and the different conformation presented by this loop in the plant enzymes produces a wider cleft that has been postulated for the inulin-binding site (4). On the contrary, this loop protrudes into the active site in the other enzymes and contains a tryptophan (Trp-314 in SoInv), which is likely involved in substrate recognition. Moreover, loop TIV contains a long insertion in SoInv and AaEI that interacts with the β-sandwich domain to a greater extent, consequently making a deeper pocket in the yeast and fungal enzymes. All of the relevant residues found at the active site of SoInv are located in these loops, as will be discussed later. A final, remarkable structural feature of SoInv is an extended 10-residue loop linking strands C5 to D5 in blade V of the propeller (Tyr-339—Leu-349), located at the opposite side of the active site, which is longer than in the other enzymes (Fig. 4a) and is involved in the oligomerization of the enzyme, as described below.
SoInv Is a Dimeric Enzyme
Two units associate in a “butterfly” shape to form a tight dimer related by noncrystallographic 2-fold symmetry, as shown in Fig. 3. The total surface area for this dimer is 36,680 Å2, with a buried surface of 3,990 Å2 (Pisa server (40)). 60% of the contacts shorter than 4 Å in this interface are produced by the interaction of the two catalytic domains, mainly through blade IV of each unit. This region contains the only two hydrogen bonds that link the main chains of both subunits within the dimer (Table 3): Gly-223-O,N from L3 in one unit is linked to Ser-281-O and Thr-283-N from strand D4 of the other. In addition, the above mentioned extended loop, Tyr-339—Leu-349, from one subunit (significantly longer in SoInv, as noted previously) makes a large number of contacts with L3 and also with TIII from the other subunit. Interestingly, all of these contacts arise at the active site surface, as shown in Fig. 6. Furthermore, the catalytic pocket of one monomer is surrounded by loops from the β-sandwich domain of the other, and thus 16% of the total contacts within the dimer interface arise from the interaction between these two domains, which occurs essentially through TV. There are only two hydrogen bonds in this region, one in this loop TV through the Trp-314 main chain and the other from Asn-227 located in loop L3. However, it is interesting to note that these interactions participate in a net of polar interactions linking residues from the different domains in a fixed conformation that stabilizes the interface and makes the ground of the active site cavity within the dimer (Fig. 6). As Fig. 5 shows, all of the residues of the catalytic domain that participate in polar interactions or short contacts to the other subunit within the dimer are variable among GH32 from different sources.
TABLE 3.
Hydrogen bonds in the dimer interface
C-D, SoInv catalytic domain; β-D, SoInv β-sandwich domain.
| Molecule A | Molecule B | Distance |
|---|---|---|
| Å | ||
| C-D | C-D | |
| Gln-199 (NE2) | Asn-343 (OD1) | 3.20 |
| Gln-199 (O) | Asn-343 (ND2) | 2.99 |
| Gln-199 (O) | Thr-346 (OG1) | 2.51 |
| Ser-221 (O) | Ser-281 (OG) | 3.25 |
| Ser-222 (OG) | Ser-281 (O) | 3.11 |
| Ser-222 (OG) | Ser-281 (OG) | 2.50 |
| Gly-223 (N) | Ser-281 (O) | 3.22 |
| Asn-227 (O) | Asn-343 (ND2) | 3.29 |
| Thr-283 (N) | Gly-223 (O) | 3.11 |
| Thr-283 (OG1) | Thr-283 (OG1) | 2.36 |
| C-D | β-D | |
| Asn-227 (OD1) | Tyr-462 (OH) | 3.26 |
| Trp-314 (O) | Gln-435 (NE2) | 3.82 |
| β-D | β-D | |
| Arg-451 (NE) | Ser-434 (OG) | 3.18 |
| Lys-452 (O) | Lys-458 (NZ) | 3.45 |
| Gln-453 (NE2) | Tyr-439 (OH) | 3.30 |
| Gln-453 (NE2) | Thr-409 (OG1) | 2.37 |
| Phe-455 (O) | Lys-458 (NZ) | 3.86 |
| Thr-456 (OG1) | Lys-458 (O) | 3.63 |
The rest of the interface is made up of direct contacts between both β-sandwich domains, as shown in Fig. 3b. It has been proposed previously that this supplementary domain of yeast invertase might play the role of a carbohydrate recognition domain involved in higher oligomerization formation (3). As seen previously, the SoInv structure reveals additional functions for this domain, even though the arrangement in which both domains are found in the SoInv dimer suggests that such a role could be performed cooperatively by them.
Previous work on S. cerevisiae invertase (ScInv) has reported different oligomerization states of the internal isoform at different pH values, with the enzyme being a monomer at pH 9.4, a dimer at pH 8.3, and an apparent octamer at pH 4.9 (41). It has also been reported that external ScInv exists as a dimer that self-associates into higher aggregates in proportions that vary with the pH, time, and concentration of the enzyme (11). As we observed previously (23), SoInv is a dimer at neutral pH, but to further investigate the oligomeric state of SoInv at different conditions, we performed a series of analytical ultracentrifugation experiments using buffers at different pH values, i.e. 4.5, 7, and 9, which always led to a molecular mass corresponding to the dimer (supplemental Fig. S2). These results indicate that oligomerization of SoInv is apparently not pH-dependent. Moreover, higher aggregates have not been detected in SoInv. The polypeptide sequence of SoInv is only two residues longer than that of ScInv, with both sequences being 66% identical. Nevertheless, an analysis of the alignment between both sequences (not shown) points to the conclusion that only half of the hydrogen bonds found in SoInv could be present in a putative ScInv dimer; therefore, this dimer might present some structural differences that would lead to a different, higher aggregation pattern than the one observed in SoInv.
The Active Site Is Made up from Both Subunits
SoInv presents a narrow pocket, 12 Å deep and 11 Å wide, typical for an exo-enzyme and similar to that found in AaEI. In the bacterial and plant enzymes, the shorter TIV loop mentioned above makes the interface between the catalytic and the β-sandwich domains more open to the solvent, leaving a wider gate to the active site (Fig. 4). Furthermore, the previously reported analysis on the TmInv-raffinose complex (8) shows that the terminal α-galactose unit is located in the active site pocket close to the protein surface, suggesting that longer substrates would situate the additional units dangling out into the solvent with no links to the enzyme. However, oligomerization of SoInv situates the β-sandwich domain of the other subunit facing this pocket (see Fig. 3c) and makes a straight cavity 25 Å deep, which is accessible to the solvent by a slot placed between both subunits with a maximum width of 18 Å. Therefore, additional subsites may be created in this groove, and SoInv seems able to recognize oligosaccharide units distal from the scissile bond, as shown below.
Fig. 7a is a close-up view of the active site pocket of SoInv showing the six glycerol molecules found in the native crystals; the position of the substrates is inferred from the structural superimposition of the TmInv-raffinose (8) and CiEI-1-kestose complexes (7) onto the SoInv coordinates. As observed, the substrates are located in putative subsites close to three of the glycerol moieties that, as generally accepted, are considered a good indicator of sugar-binding sites. This suggests that the other glycerol molecules could reveal additional binding subsites that are able to recognize longer substrates. It is remarkable to find that these glycerol molecules interact with the other subunit of the dimer.
Failure to crystallize the recombinant form of SoInv prevented the genetic replacement of the catalytic residue in order to obtain crystals from the inactivated enzyme-substrate complexes. To overcome this impediment, we tried an alternative strategy proposed for the crystallographic analysis of glycosidases, the exploitation of sugars as a cryoprotectant substitute for glycerol (42), a method that had yielded successful results in our previous studies (43). Following this procedure, we soaked the deglycosylated crystals for a short time in a cryosolution containing as high as possible concentrations of nystose and fructosylnystose, which saturated the active sites of all of the protein molecules and which were subsequently immobilized by flash cryocooling in a nitrogen stream. As we observed some ice formation, a small amount of PEG 400 was also added to the cryoprotectant solution. However, after trying different soaking times, the substrates were processed, and only the product fructose was bound at the active site.
Automatic Docking of Substrates Suggests Relevant Details of the Active Site
In view of the impossibility of getting crystals from the complexes, the binding of long substrates such as tetrasaccharide nystose and polymeric inulin was investigated by docking analysis. The first trials of automatic docking of the substrates into the SoInv active site, as obtained in the crystal structure, failed to produce productive complexes, as the ligands were positioned with the fructose of the nonreducing end out from the catalytic −1 subsite. An inspection of the active site of SoInv showed a different orientation of the Glu-230 and Tyr-293 side chains than found in the above mentioned reported complexes from the GH32 family. Therefore, these two residues were modeled in the same conformation as observed experimentally in the complexes, with this change leading to the successful docking of the nystose molecule into the SoInv active site in a biologically significant position. In the case of the polymer inulin, only an additional change in the Arg-178 side chain allowed the substrate to enter into the active site.
Using nystose as a ligand, we obtained the lowest total docking energy (−11.9 kcal/mol) for a position (yellow in Fig. 7c) in the highest ranked cluster, which contained 24 of 50 runs and had a mean docking energy of −9.0 kcal/mol. The second ranked cluster contains only three positions presenting a less extended oligosaccharide chain (not shown) with a mean docking energy of −9.7 kcal/mol. Finally, the third ranked cluster (with mean binding energy of −7.58 kcal/mol) accounted for 20 conformations that situated the nonreducing fructose ring significantly shifted away from the −1 binding site; these were not considered, as they are not biologically productive conformations. In the case of the polymeric inulin, a segment containing 12 fructose units was docked into a position with the lowest docking energy of −6.6 kcal/mol (Fig. 7d) included within the highest ranked cluster, which contained 37 of 50 runs. As this model displayed the terminal fructose of inulin slightly flipped with respect to that observed in the SoInv-fructose complex, this position was further improved by manual docking followed by local optimization (Fig. 7d).
In summary, docking analysis of the substrates nystose and inulin into the SoInv active site located these ligands in energetically favorable positions, situating the nonreducing end in the fructose site observed in the SoInv-fructose complex and the distal oligosaccharide units in the positions occupied by glycerol molecules in the native crystals. Docking of these ligands involved a rearrangement of some of the residues of the SoInv active site, in particular Arg-178, Glu-230, and Tyr-293, which therefore must be directly implicated in substrate binding. The modeled complexes suggest that the nystose and inulin chains interact directly with residues from both subunits, a unique feature of SoInv. Furthermore, the analysis of its molecular surface revealed that the dimer structure delineates an active site channel that is perfectly adapted to accommodate the polymeric chain of inulin, as illustrated in Fig. 8 showing the SoInv-inulin complex obtained by docking.
Substrate Specificity Is Elucidated by Kinetic Analysis of Mutants
An inspection of the SoInv active site in the putative complexes points to the residues that may be involved in recognizing substrates of different length. Fig. 7b shows the position of the substrate sucrose inferred from the structural superposition of the AtInv-Suc complex (6) into the SoInv coordinates, whereas Fig. 7, c and d, shows the nystose and inulin positions obtained by docking. As observed, the pocket is surrounded by aromatic residues, i.e. Trp-76, Phe-110, Tyr-293, and Trp-314, which are strictly conserved among the GH32 family (see Fig. 3), as is the Asp-50/Glu-230 pair assigned as the catalytic residues. The residues Arg-178 and Asp-179 are also conserved and belong to the well known RDP motif involved in substrate binding. All of these residues appear to play a role in substrate recognition that is shared with the other GH32 enzymes. Therefore, we investigated the role of Asn-142, Gln-176, Gln-228, and Asn-254 in the enzymatic activity of SoInv, as these residues are not conserved and consequently must be responsible for the differences in specificity encountered among the family. The structure of the SoInv dimer interface (Fig. 6) points to Gln-435 and Tyr-462 as possible candidates involved in substrate recognition; these residues were also investigated by mutagenesis experiments. In addition, Ser-281, involved in one of the two main-chain hydrogen bonds linking both subunits (Table 3), was also mutated to a bulky residue with the aim of investigating the possible existence of an active monomer or at least how a distortion in the dimer could affect the enzymatic activity.
Table 4 gives the mutants we produced and their corresponding kinetic parameters. The expected absence of activity in mutants D50A and E230A confirmed the catalytic role of these residues. Furthermore, the experimental results allowed grouping the other mutants into four general groups. First, the Q228V and S281I mutants exhibited a strikingly large reduction in the catalytic efficiency (kcat/Km) of sucrose and inulin, which was even larger when measured against nystose (at least a 100-fold decrease). Gln-228 is a residue located at the highly variable L3; the equivalent TmInv Glu-188 was described previously as linking the glucose unit in the TmInv-raffinose complex (8). Indeed, our docking analysis of SoInv shows that Gln-228 may be involved directly in binding the glucose at the reducing end of nystose through hydrogen bonds (Fig. 7c) and also the third fructose unit of inulin (Fig. 7d); therefore a decrease in catalytic efficiency against long substrates could be expected. However, there is also a 5-fold reduction in activity against sucrose that is not explained by the putative SoInv-sucrose complex (Fig. 7b), as Gln-228 would not interact with the substrate in this case. Nevertheless, it is interesting to note that this residue is next to Asn-227, which is involved in a network of polar links to the other molecule; the Q228V substitution (a Val is found in AaEI) might alter the atomic interactions at the interface and consequently at the active site pocket, leading to a less efficient enzyme. The key role of the dimer in the enzymatic activity is further supported by the fact that the S282I substitution at the dimer interface and far from the active site, yields similar largely reduced kinetic parameters, most probably by producing a somehow distorted dimer (this mutant still migrates as a dimer in a native electrophoresis analysis; see “Experimental Procedures” and Fig. 2).
TABLE 4.
Kinetic analysis of SoInv mutants
The kcat values were calculated assuming a protein molecular mass of 180 kDa. The ± sign refers to standard errors based on curve fitting using SigmaPlot. The mutants D50A and E230A gave no activity against any of the substrates. WT, wild type.
| SoINV mutant |
kcat |
Km |
kcat/Km |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Sucrose | Nystose | Inulin | Sucrose | Nystose | Inulin | Sucrose | Nystose | Inulin | |
| s−1 | mm | s−1·mm−1 | |||||||
| WT | 106 ± 7 | 84 ± 14 | 25 ± 4 | 6.4 ± 0.4 | 2.4 ± 0.4 | 12 ± 2 | 16.6 | 35 | 2.1 |
| N142Y | 262 ± 30 | 87 ± 17 | 68 ± 17 | 29 ± 3 | 8 ± 1 | 60 ± 15 | 9 | 11 | 1.1 |
| Q176E | 88 ± 6 | 92 ± 11 | 39 ± 5 | 7 ± 0.5 | 1.8 ± 0.2 | 24 ± 3 | 12.6 | 51 | 1.6 |
| Q176S | 134 ± 14 | 230 ± 29 | 1710 ± 172 | 5 ± 0.5 | 11 ± 1 | 575 ± 58 | 26.8 | 21 | 3 |
| Q228V | 33 ± 2 | 2 ± 0.5 | 3 ± 0.5 | 11 ± 0.6 | 9 ± 3 | 47 ± 7 | 3 | 0.2 | 0.06 |
| N254A | 85 ± 11 | 60 ± 8 | 12 ± 2 | 8 ± 1 | 8 ± 1 | 7 ± 1 | 10.6 | 7.5 | 1.7 |
| S281I | 20 ± 3 | 6 ± 1 | 2 ± 0.4 | 13 ± 2 | 15 ± 3 | 44 ± 8 | 1.5 | 0.4 | 0.04 |
| Q435A | 99 ± 16 | 76 ± 6 | 10 ± 1 | 9 ± 1 | 3.8 ± 0.3 | 14 ± 2 | 11 | 20 | 0.7 |
| Y462A | 43 ± 6 | 22 ± 3 | 5 ± 0.6 | 7 ± 1 | 8 ± 1 | 9 ± 1 | 6 | 2.7 | 0.5 |
A second group is formed by the mutants Q435A and Y462A, leading to both a 3-fold decrease in catalytic efficiency against inulin and a slighter effect against the other two substrates, with the exception found in the Tyr-462 activity versus nystose, which has a lower kcat and reduced affinity leading to a large 12-fold decrease in the overall catalytic efficiency. Both residues are located at the β-sandwich domain of molecule B and participate in the polar interaction network that shapes the interface and the active site pocket (Fig. 6); therefore their substitution must affect the interface and efficient positioning of the oligosaccharides within the catalytic pocket. As deduced from the complexes shown in Fig. 7, c and d, Gln-435 could be hydrogen-bonded to nystose and inulin, whereas Tyr-462 might participate in nystose binding but seems too far from the inulin chain, and none of these residues participate in sucrose binding. This fact reinforces the idea that even a small change in the dimer interface has a deleterious effect on activity, and the concerted action of both subunits determines the catalytic efficiency of SoInv.
The third group is composed of the N142Y and N254A mutants, which have a reduced catalytic efficiency most apparent when the activity is assayed against nystose. Asn-142, located at the end of strand B2, is a Tyr in the fungal and bacterial enzymes and has been described as binding the terminal galactose moiety of raffinose, through a water molecule, in the TmInv-raffinose complex (8). As seen in Fig. 7, direct interaction of Asn-142 to the substrates cannot be deduced from any of the complexes, but interaction through solvent molecules cannot be discarded. Moreover, the SoInv crystal structure shows that the side chain of Asn-142 is hydrogen-bonded to Glu-107 from loop L1 and Gln-147 located at strand C2; its replacement by a Tyr probably rearranges the conformation of the active site residues, lowering the affinity for the substrates (Table 4). Unexpectedly, this lower affinity is compensated by a higher kcat as found when it was measured against sucrose or inulin. On the other hand, Asn-254 from the variable loop TIV is located in the ground of the pocket, and its side chain could be hydrogen-bonded to the fructose unit at subsite +2 in the putative SoInv-nystose complex (Fig. 7c). Accordingly, the substitution N254A decreases the affinity for nystose by 4-fold. No interaction is observed in the other complexes. It is noteworthy that the equivalent TmInv Thr-208 has been described previously as interacting also with the glucose unit of the TmInv-raffinose complex. However, our results showed no significant influence on activity against sucrose or inulin upon mutation, and therefore, this residue possibly plays a role in the binding of medium-size substrates.
Finally, we studied the role of Gln-176, located at the end of L2 and preceding the conserved RDP motif. This residue is linked to the essential Gln-228 and, more significantly, to Arg-178 from this RDP motif, which, as explained previously, was necessarily shifted to succeed in properly docking inulin into the SoInv active site. However, the replacement of Gln-176 by either Glu or Ser does not reveal any clear influence on SoInv catalytic efficiency, as it is slightly increased against nystose but somewhat reduced against the other two substrates in the Q176E mutant, but curiously shows the opposite effect in the Q176S mutant. Although the moderate magnitude of the changes in catalytic efficiency indicates that this residue may not be critical, the deviations observed in the kcat and Km values of the Gln-176 mutant suggest that this position should be investigated through the analysis of further mutants to determine the precise role of this residue in the activity of SoInv.
In summary, docking and mutagenesis analyses of SoInv reveal that substrates longer than sucrose might be recognized by hydrogen links to residues Gln-228 and Asn-254 from one subunit and Gln-435 and Tyr-462 from the other. However, the amino acid replacements that have a deleterious effect on catalytic activity, i.e. Q228V and S281I, seem to be related also to some change in the interface within the dimer or, at least, to the conformation of the active site cavity, supporting the idea that oligomerization is the key to delineating SoInv functionality.
Conclusions
Fructans and related compounds play a key role in metabolism and energy exchange and may be employed as storage polymers in organisms as plants. Several microorganisms use them as regular energy and carbon sources by the expression of hydrolytic enzymes. In this study, we have described the structure of dimeric Sw. occidentalis invertase, which is an example of how these proteins have adapted, in many yeasts, to the hydrolysis of fructans. The crystal structure of the SoInv subunit shows a bimodular arrangement similar to that described previously for other GH32 enzymes. A detailed comparison of the catalytic domain with those of other known structures helped delimitate the regions that are essential in determining substrate specificity among the family members. However, the active site of SoInv is exceptional, as it is built up by both subunits and presents substrate-binding sites additional to those described for known enzymes. This structure is an interesting example of how oligomerization can be a molecular mechanism for fine-tuning the functionality within a family of homologous enzymes. Moreover, the supplementary β-sandwich domain was previously postulated to play a role as an ancillary carbohydrate recognition domain, owing to its lectin-like fold and to what had been observed in other glycosylhydrolases enzymes. However, the SoInv structure reveals that this domain is involved in contouring the active site cleft and also plays a direct role in binding the substrates, both medium-size oligosaccharides and the polymeric chain of inulin. This unique feature of the structure presented here may help in understanding the protein-carbohydrate recognition molecular mechanisms implicated in modularity, a pivotal property within the glycosylhydrolase enzymes that is crucial to many biologically relevant degradative processes. The specificity of SoInv has been investigated by a docking analysis of long substrates into its active site, which points to the key role of residues from both subunits in substrate binding; this has been supported by kinetic analysis of mutants. The replacements that presented the most deleterious effect on activity were Q228V, a residue involved directly in substrate recognition, and S281I, a residue responsible for one of the two main-chain hydrogen-bonding links between both subunits and, therefore, significant to proper oligomerization. Mutation of the other identified residues, Asn-142, Gln-176, and Asn-254 from the catalytic domain and Gln-435 and Tyr-462 from the β-sandwich domain of the other subunit, was also shown to have an effect on substrate binding or the catalytic rate, confirming the interactions of these residues with the substrate. Consequently, our results assign a direct catalytic role to the supplementary β-sandwich domain of SoInv, the first time that such a role has been observed within GH32 enzymes.
Supplementary Material
Acknowledgment
We thank the staff of the European Synchrotron Radiation Facility for providing beam time and for technical assistance at beamlines ID14-1, ID14-4, ID23-1, and ID23-2.
This work was supported by Grants BIO2007-67708-C04-04, BIO2007-67708-C04-03, BIO2004-67708-C04-04, and BIO2004-67708-C04-03 from the Ministerio de Educación y Ciencia, Spain, and by an institutional grant from the Fundación Ramón Areces to the Centro de Biología Molecular Severo Ochoa.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- GH
- glycosylhydrolase
- AaEI
- exo-inulinase from Aspergillus awamori
- TmInv
- invertase from Thermotoga maritima
- CiFEH
- fructan 1-exohydrolase IIa from Cichorium intybus
- AtInv
- cell wall invertase 1 from Arabidopsis thaliana
- FOS
- fructo-oligosaccharide
- SoInv
- invertase from Schwanniomyces occidentalis
- ScInv
- invertase from Saccharomyces cerevisiae
- r.m.s.
- root mean square
- PEG
- polyethylene glycol.
REFERENCES
- 1.Coutinho P. M., Henrissat B. (1999) in Recent Advances in Carbohydrate Bioengineering (Gilbert H. J., Davies G., Henrissat B., Svensson B., eds) pp. 3–12, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 2.Nagem R. A., Rojas A. L., Golubev A. M., Korneeva O. S., Eneyskaya E. V., Kulmisnkaya A. A., Neustroev K. N., Polikarpov I. (2004) J. Mol. Biol. 334, 471–480 [DOI] [PubMed] [Google Scholar]
- 3.Alberto F., Bignon C., Sulzenbacher G., Henrissat B., Czjzek M. (2004) J. Biol. Chem. 279, 18903–18910 [DOI] [PubMed] [Google Scholar]
- 4.Verhaest M., Van den Ende W., Le Roy K., De Ranter C. J., Van Laere A., Rabijns A. (2005) Plant J. 41, 400–411 [DOI] [PubMed] [Google Scholar]
- 5.Verhaest M., Lammens W., Le Roy K., De Coninck B., De Ranter C. J., Van Laere A., Van den Ende W., Rabijns A. (2006) Acta Crystallog. D Biol. Crystallogr. 2, 1555–1563 [DOI] [PubMed] [Google Scholar]
- 6.Mátrai J., Lammens W., Jonckheer A., Le Roy K., Rabijns A., Van den Ende W., Maeyer M. (2008) Proteins 71, 552–564 [DOI] [PubMed] [Google Scholar]
- 7.Verhaest M., Lammens W., Le Roy K., De Ranter C. J., Van Laere A., Rabijns A., Van den Ende W. (2006) New Phytol. 174, 90–100 [DOI] [PubMed] [Google Scholar]
- 8.Alberto F., Jordi E., Henrissat B., Czjzek M. (2006) Biochem. J. 395, 457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies G. J., Wilson K. S., Henrissat B. (1997) Biochem. J. 321, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tammi M., Ballou L., Taylor A., Ballou C. E. (1987) J. Biol. Chem. 262, 4395–4401 [PubMed] [Google Scholar]
- 11.Reddy A. V., MacColl R., Maley F. (1990) Biochemistry 29, 2482–2487 [DOI] [PubMed] [Google Scholar]
- 12.Abarca D., Fernández-Lobato M., Claros M. G., Jiménez A. (1989) FEBS Lett. 255, 455–459 [DOI] [PubMed] [Google Scholar]
- 13.Abarca D., Fernández-Lobato M., del Pozo L., Jiménez A. (1991) FEBS Lett. 279, 41–44 [DOI] [PubMed] [Google Scholar]
- 14.Klein R. D., Deibel M. R., Jr., Sarcich J. L., Zurcher-Neely H. A., Reardon I. M., Heinrikson R. L. (1989) Prep. Biochem. 19, 293–319 [DOI] [PubMed] [Google Scholar]
- 15.Dohmen R. J., Strasser A. W., Dahlems U. M., Hollenberg C. P. (1990) Gene 95, 111–121 [DOI] [PubMed] [Google Scholar]
- 16.Deibel M. R., Jr., Hiebsch R. R., Klein R. D. (1988) Prep. Biochem. 18, 77–120 [DOI] [PubMed] [Google Scholar]
- 17.Wang T. T., Lee C. F., Lee B. H. (1999) Crit. Rev. Biotechnol. 19, 113–143 [DOI] [PubMed] [Google Scholar]
- 18.Yáñez E., Carmona T. A., Tiemblo M., Jiménez A., Fernández-Lobato M. (1998) Biochem. J. 329, 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Álvaro-Benito M., Abreu M., Fernández-Arrojo L., Plou F. J., Jiménez-Barbero J., Ballesteros A., Polaina J., Fernández-Lobato M. (2007) J. Biotechnol. 1, 75–81 [DOI] [PubMed] [Google Scholar]
- 20.Sangeetha P. T., Ramesh M. N., Prapulla S. G. (2005) Process Biochem. 40, 1085–1088 [Google Scholar]
- 21.Ghazi I., Fernandez-Arrojo L., Garcia-Arellano H., Ferrer M., Ballesteros A., Plou F. J. (2007) J. Biotechnol. 128, 204–211 [DOI] [PubMed] [Google Scholar]
- 22.Pons T., Naumoff D. G., Martínez-Fleites C., Hernández L. (2004) Proteins 54, 424–432 [DOI] [PubMed] [Google Scholar]
- 23.Polo A., Alvaro-Benito M., Fernández-Lobato M., Sanz-Aparicio J. (2009) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 1162–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leslie A. G. (1992) in Crystallographic Computing 5: From Chemistry to Biology (Moras D., Podjarny A. D., Thierri J. C. eds) pp. 39–50, Oxford University Press, Oxford, UK [Google Scholar]
- 25.Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 26.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 27.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 28.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 29.Laskowsky R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 30.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 32.Okuyama K., Noguchi K., Saitoh M., Ohno S., Fujii S., Tsukada M., Takeda H., Hidano T. (1993) Bull. Chem. Soc. Jpn. 66, 374–379 [Google Scholar]
- 33.Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., Olson A. J. (2009) J. Comput. Chem. 30, 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.André I., Mazeau K., Putaux W., Winter W. T., Taravel F. R., Chanzy H. (1996) Macromolecules 29, 4626–4635 [Google Scholar]
- 35.Zheng. L., Baumann U., Reymond J. L. (2004) Nucleic Acids Res. 32, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burke D., Dawson D., Stearns T. (2000) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37.Naumoff D. G. (2001) Proteins 42, 66–76 [DOI] [PubMed] [Google Scholar]
- 38.Beisel H. G., Kawabata S., Iwanaga S., Huber R., Bode W. (1999) EMBO J. 18, 2313–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai J., Liu J., Deng Y., Smith T. M., Lu M. (2004) Cell 116, 649–659 [DOI] [PubMed] [Google Scholar]
- 40.Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 41.Williams R. S., Trumbly R. J., MacColl R., Trimble R. B., Maley F. (1985) J. Biol. Chem. 260, 13334–13341 [PubMed] [Google Scholar]
- 42.Schmidt A., Gübitz G. M., Kratky C. (1999) Biochemistry 38, 2403–2412 [DOI] [PubMed] [Google Scholar]
- 43.Isorna P., Polaina J., Latorre-García L., Cañada F. J., González B., Sanz-Aparicio J. (2007) J. Mol. Biol. 371, 1204–1218 [DOI] [PubMed] [Google Scholar]
- 44.Holm L. (1998) Curr. Opin. Struct. Biol. 8, 372–379 [DOI] [PubMed] [Google Scholar]
- 45.Barton G. J. (1993) Protein Eng. 6, 37–40 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.