Abstract
The metzincins are a clan of metallopeptidases consisting of families that share a series of structural elements. Among them is the Met-turn, a tight 1,4-turn found directly below the zinc-binding site, which is structurally and spatially conserved and invariantly shows a methionine at position 3 in all metzincins identified. The reason for this conservation has been a matter of debate since its discovery. We have studied this structural element in Methanosarcina acetivorans ulilysin, the structural prototype of the pappalysin family, by generating 10 mutants that replaced methionine with proteogenic amino acids. We compared recombinant overexpression yields, autolytic and tryptic activation, proteolytic activity, thermal stability, and three-dimensional structure with those of the wild type. All forms were soluble and could be purified, although with varying yields, and three variants underwent autolysis, could be activated by trypsin, and displayed significant proteolytic activity. All variants were analyzed for the thermal stability of their zymogens. None of the mutants analyzed proved more stable or active than the wild type. Both bulky and small side chains, as well as hydrophilic ones, showed diminished thermal stability. Two mutants, leucine and cysteine, crystallized and showed three-dimensional structures that were indistinguishable from the wild type. These studies reveal that the Met-turn acts as a plug that snugly inserts laterally into a core structure created by the protein segment engaged in zinc binding and thus contributes to its structural integrity, which is indispensable for function. Replacement of the methionine with residues that deviate in size, side-chain conformation, and chemical properties impairs the plug-core interaction and prejudices molecular stability and activity.
Keywords: Enzymes/Mechanisms, Metals/Zinc, Methods/X-ray Crystallography, Protease, Proteases/Metalloprotease, Mutation, Structure-Function Correlation
Introduction
Evolutionary conservation of structural elements in proteins usually results from stringent steric requirements for function. Specific structures enable particular chemical groups to be placed appropriately in three-dimensional space for reaction, transport, regulation, and scaffolding (1). One such case of conserved structural elements is the Met-turn, found in the catalytic domains of all structurally characterized families of the metzincin clan of zinc-dependent metallopeptidases. These include astacins, ADAMs4/adamalysins, serralysins, matrix metalloproteinases (MMPs), leishmanolysins, snapalysins, and pappalysins (2–5). The catalytic domains span between ∼130 and ∼260 residues and fold into globular moieties, which are divided by an active site cleft into an upper N-terminal and a lower C-terminal subdomain when viewed in the standard orientation (Fig. 1A) (5). The N-terminal subdomain comprises a β-sheet and two helices, the backing helix and the active-site helix, as the minimum common core of repetitive secondary structure elements (5). The latter helix includes the first stretch of a zinc-binding consensus sequence, HEXXHXXG/NXXH/D (amino acid one-letter code), which includes three protein ligands of the catalytic zinc and the general base/acid glutamate for catalysis. Features of the C-terminal subdomain common to all metzincins include a C-terminal α-helix and the Met-turn (Fig. 1B). In the course of evolution, distinct structural elements have been introduced into this consensus minimal scaffold for each family, such as helices, strands, structural zinc- and calcium-binding sites, and additional domains (2–5). For example, in the smallest metzincin representative reported for its structure, 132-residue Streptomyces caespitosus snapalysin, only one extra calcium-binding site and a short helix in the N-terminal subdomain are found (Fig. 1A). The result of such insertions is that, overall, the families have evolved along separate pathways. This is reflected by sequence identities well below twilight values to discriminate between similar and dissimilar structures (20–35%) (6), and most sequence alignment protocols only identify the extended zinc-binding sequence and flanking sequence stretches (7). However, the existence of the aforementioned common structural core elements confirms that all of these families are homologous, thus underlining that structural relatedness is more conserved than sequence identity (8).
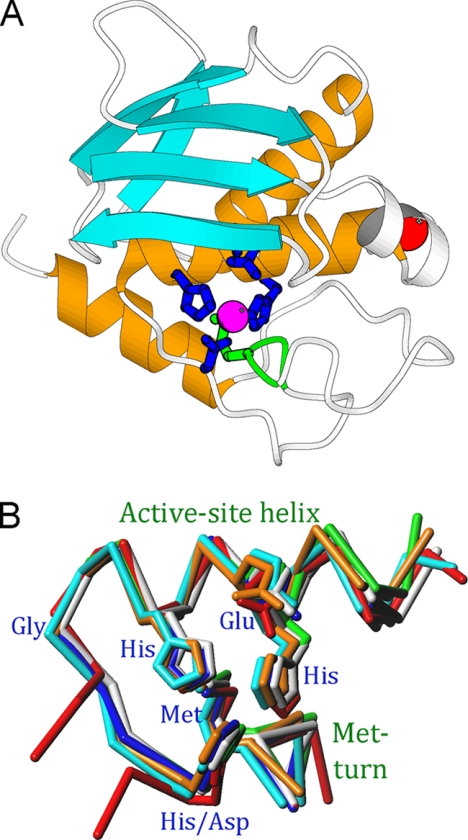
FIGURE 1.
Common structural elements in metzincins. A, Richardson plot depicting the smallest metzincin catalytic domain structurally characterized, snapalysin from S. caespitosus (PDB 1C7K) (33), in standard orientation. The repetitive secondary structure elements common to all metzincins are shown in orange (helices) and cyan (β-strands), and the Met-turn is displayed in green. B, overlay of the seven metzincin prototypes structurally characterized to date as Cα plots showing the active-site helix (including the side chains of the zinc-liganding histidines and aspartate, and the general-base/acid glutamate) and the Met-turn (with the methionine side chain). The catalytic zinc has been omitted for clarity. Color coding: astacin, blue; adamalysin II, cyan; leishmanolysin, red; MMP-8, green; aeruginolysin, white; snapalysin, yellow; and ulilysin, orange (see 3, 5).
The Met-turn is a conserved 1,4-β-turn of type I that contains a methionine at position 3, which is separated from the third zinc-binding histidine/aspartate by connecting segments of 6–53 amino acids in the different metzincin structures (3, 5). This means that the chain traces of the distinct prototypes strongly diverge after the third zinc ligand, but they all converge at the Met-turn. In all structures analyzed, this type I turn is superimposable in space, with the same conformation of the methionine side chain, with dihedral angles χ1, χ2, and χ3 adopting values in the ranges 282–309°, 276–317°, and 286–328°, respectively (Fig. 1B). This residue lies underneath the catalytic zinc. However, the distance from the side-chain atoms to the metal is far too great, and the orientation of the lone electron pairs of the methionine sulfur is incompatible with a metal interaction. This strict conservation of a residue imbedded in a small structural element spanning four residues would suggest a pivotal role in structural and functional integrity because methionine is not a residue particularly conserved during evolution (9). However, mutation studies performed on several metzincin families revealed disparate results.
Replacement of the Met-turn methionine by selenomethionine in matrix metalloproteinase 8 (MMP-8) gave rise to a 50% decrease in catalytic efficiency and diminished conformational stability, but did not affect the crystal structure (10). Similar studies on MMP-2 showed unaltered activity for the serine and leucine mutants but complete loss of activity and enhanced susceptibility to proteolytic degradation for the cysteine mutant (11). In serralysins, substitution of difluoromethionine for methionine revealed no significant differences in activity or in the result of differential scanning calorimetry in Pseudomonas aeruginosa alkaline protease (12). In contrast, studies on Erwinia chrysanthemi PrtC revealed lower levels of protein expression and diminished catalytic efficiency toward resorufin-casein for the leucine (85% of the wild type), isoleucine (50%), and alanine (23%) mutants and subtle changes in the crystal structure at the active site of the first two mutants and a cysteine mutant. However, major structural rearrangement and destabilization of the zinc site and an adjacent protein segment spanning ∼80 residues were observed in the alanine mutant. In the histidine mutant crystal structure, an alternative zinc-binding site was found below the functional site (7, 13). With respect to ADAMs/adamalysins, mutation of methionine in tissue necrosis factor α-converting enzyme to isoleucine, leucine, or serine impaired the ectodomain shedding of physiological substrates (14). In the case of the astacin family, an alanine mutant of crayfish astacin showed significantly lower activity in gelatin zymography but similar thermal stability in CD spectroscopy.5 Finally, a leucine mutant of pregnancy-associated plasma protein A (PAPP-A) from the pappalysin family rendered expression levels similar to wild type, but only ∼5% of its capacity to cleave a physiological protein substrate (15).
To examine further the role of the Met-turn methionine in metzincins, we studied a distinct representative, Methanosarcina acetivoras ulilysin, and mutated its Met290 to 10 different residues. We analyzed recombinant expression levels of these proulilysin variants, (autolytic) activation through limited proteolysis, and activity of the mature ulilysin forms. We further performed thermal shift assays of zymogens and mature enzymes to study structural stability, and we managed to crystallize and solve the structure of the mature forms of two of these mutants, ulilysin M290L and M290C.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis
Plasmids based on pET28a, which were modified to encode for both M. acetivorans proulilysin wild-type and variant C269A (16, 17), were used as a template to mutate Met290 to alanine, leucine, valine, phenylalanine, glycine, serine, cysteine, aspartate, lysine, and histidine (M290X mutants). The vector employed attached a His6 tag to the protein N terminus, followed by a thrombin protease cleavage site, and conferred resistance to kanamycin. Mutations were introduced with the QuikChange Site-directed Mutagenesis kit from Stratagene by using PfuTurbo DNA polymerase (Stratagene) and the oligonucleotides shown in supplemental Table 1. Each reaction product was treated with DpnI endonuclease (Fermentas) to select for mutated DNA and discard parental templates. The mutant vectors were transfected into Escherichia coli DH5α supercompetent cells, and all constructs were verified by sequencing.
Production and Purification of Ulilysin Mutants
All 10 M290X mutant zymogens plus wild-type and C269A proulilysin were expressed in E. coli BL21 StarTM cells (Invitrogen) grown in terrific broth (18) by the addition of 0.25 mm isopropyl 1-thio-β-d-galactopyranoside when A600 reached 0.5–0.6. All proteins were and purified at 4 °C through nickel-nitrilotriacetic affinity chromatography (GE Healthcare) by employing a different column for each mutant. In each case, the His6 tag was removed with thrombin (1 unit/μl; Amersham Biosciences) at a ratio of 1:500 (w/w). The cleavage reaction was quenched with 5 mm phenylmethylsulfonyl fluoride (Fluka), and the mixture was subjected to anion-exchange chromatography purification (Mono Q 4.6/100 PE 1.7 ml; Amersham Biosciences) to remove the serine protease.
Activation of Ulilysin Mutants by Limited Proteolysis
All 10 mutant zymogens plus wild-type proulilysin were analyzed for autolytic activity to render mature forms as described for proulilysin C269A (17). Independently, all zymogens were subjected to activation with trypsin (Sigma) for 6 h at 4 °C at a ratio of 1:815 (w/w). The latter reaction was stopped with 2 mm Pefabloc® inhibitor (Fluka). An extra ion-exchange chromatography step (Mono Q 5/50 GL 1.0 ml; Amersham Biosciences) was performed to isolate the ∼29-kDa mature ulilysin variants, which underwent a final size-exclusion fast protein liquid chromatography step (Superdex 75 10/300 GL; Amersham Biosciences). In each case, central peak fractions were pooled, concentrated, and stored at 4 °C for later analysis. Care was taken in all steps to avoid mutant cross-contamination.
Protease Assays
Purified M290X and C269A ulilysin mutants and the wild-type enzyme were tested (at 1 mg/ml in 20 mm Tris-HCl, pH 7.5, 100 mm NaCl, 5 mm CaCl2) for proteolytic activity against the specific quenched fluorescent peptide substrate, DABCYL-Leu-Ala-Arg-Val-Glu-EDANS (Bachem) at 37 °C for 2 h (16, 17). The increase in fluorescence intensity (λex = 355 nm; λem = 460 nm) upon substrate cleavage was monitored on a fluorescence microplate reader (FLx800; BIOTEK), and kinetic parameters (Km and kcat) were derived by using different substrate concentrations (10, 25, 50, 100, 150, and 175 μm).
Thermal Shift Assays
Aliquots prepared by mixing 7.5 μl of 300× Sypro Orange dye (Molecular Probes), 5 μl of protein solution (2.5 mg/ml), and 37.5 μl of buffer (20 mm Tris-HCl, pH 7.5, 100 mm NaCl) were analyzed in an iCycler iQ Real Time PCR Detection system (Bio-Rad) by using 96-well PCR plates sealed with optical tape. Samples were heated from 25 °C to 90 °C at a rate of 0.5 °C/min, and the change in absorbance (λex = 490 nm; λem = 575 nm) was monitored over time. The melting temperature (Tm) was determined for the mutant and wild-type enzymes and zymogens.
Crystallization and X-ray Data Collection
All proulilysin mutants and the respective mature ulilysin forms, obtained from both autolytic activation and trypsin treatment, were subjected to crystallization assays through sitting-drop vapor diffusion. Despite intensive screening, only mature mutants M290L (autolytically activated) and M290C (trypsin-treated) rendered crystals suitable for x-ray analysis. Both variants (at 10 mg/ml and 5.8 mg/ml, respectively, in 20 mm Tris-HCl, pH 7.5, 0.15 m NaCl, 5 mm CaCl2) crystallized at 20 °C in, respectively, 0.2 m CaCl2, 0.1 m Hepes, pH 7, 20% (w/v) PEG 6000; and 0.2 m CaCl2, 0.1 m Hepes, pH 8, 22% (w/v) PEG 6000 as precipitating agent. Crystals were cryoprotected with 0.2 m CaCl2, 0.1 m Hepes, pH 7, 24% (w/v) PEG 6000, 20% glycerol (M290L); and 0.2 m CaCl2, 0.1 m Hepes, pH 8, 26% (w/v) PEG 6000, 15% glycerol (M290C). Prism-shaped crystals belonging to the same orthorhombic space group, C2221, but with different cell constants, were obtained for both mutants after several weeks. M290L crystals contained four monomers/asymmetric unit (solvent content 46%; Vm = 2.3 Å3/Da) and diffraction data to 1.7 Å were collected at 100 K at beam line ID23-1 of the European Synchrotron Radiation Facility (Grenoble, France). M290C crystals harbored two monomers/asymmetric unit (solvent content 49%; Vm = 2.4 Å3/Da), and diffraction data to 1.8 Å were collected at 100 K at ESRF beam line ID29. Diffraction data were integrated, scaled, merged, and reduced with programs XDS (19) and SCALA (20) within the CCP4 suite (21) (see supplemental Table 2).
Structure Solution and Refinement
The structures were solved by applying Patterson search methods with program AMoRe (22), all diffraction data up to 4 Å resolution, and the co-ordinates of ulilysin C269A (Protein Data Bank (PDB) access code 2CKI) (17), with position 290 replaced by alanine, as searching model. Subsequently, manual model building was carried out with program COOT (23), alternating with crystallographic refinement with REFMAC5 (24) until completion of the model. The final model of ulilysin M290L comprised residues 61–322 for each of the four molecules in the asymmetric unit (chains A–D) plus an Arg-Val dipeptide in each active-site cleft. The final model of ulilysin M290C contained residues 63–321 for each of the two molecules (chains A and B) in the asymmetric unit plus an Arg-Val dipeptide in the active-site cleft of molecule B. Supplemental Table 2 provides crystallographic statistics and the final refinement parameters of both models.
Miscellaneous
Figures were prepared with programs TURBO-Frodo (25), PyMOL (DeLano Scientific LLC), and Molscript (26). Models were superimposed with COOT. The final co-ordinates have been deposited with the Protein Data Bank at the RCSB (PDB access code 3LUM for M290L ulilysin and 3LUN for M290C ulilysin).
RESULTS AND DISCUSSION
Protein Production and Autolytic Activation
All 10 M290X mutant plus wild-type zymogens were overexpressed in E. coli and purified. Comparison of the final yields/500 ml of cell culture revealed that only variant M290L, which harbored the most conservative mutation among those assayed, was obtained in amounts similar to the wild type (supplemental Table 3). The protein yield of the other nine mutants was between 6 and 18% of the wild type. The lowest yields were obtained for the mutants with the bulkiest side chains, lysine and phenylalanine, with slightly higher yields for alanine, valine, histidine, aspartate, serine, cysteine, and glycine. These results suggested that, overall, the mutants were sufficiently well folded to pass the quality control check of the host cell, although they were much less abundant than the wild type (except for leucine).
Both 38-kDa proulilysin C269A and its selenomethionine variant undergo calcium-mediated autoactivation at two sites, one upstream and one downstream of the catalytic moiety, to render mature 29-kDa ulilysin quantitatively after 24 h at room temperature (16, 17, 27). The two sites showed the sequence Ile59-Ser60-Arg61-Met62-Glu63 and Leu321-Ala322-Arg323-Val324-Glu325 (for numbering, see Fig. 1 in 27), respectively, and agreed well with the specificity of the enzyme for basic residues in subsite P1′. We observed that the wild type and M290L underwent complete autolytic activation. Only M290F and M290S were partially self-processed. These results indicated that the most conservative mutant behaved similarly to the wild type, whereas all other mutants but two were autolytically inactive.
Tryptic Activation and Activity
Given the results of the autolysis test, all zymogen variants were subjected to proteolytic activation by trypsin to provide comparable active mature forms. Incubation conditions were fine tuned to obtain cleavage sites near those observed during autolysis, taking into account that trypsin cleaves after basic residues, i.e. one residue downstream from ulilysin. From all variants tested, only the wild type, M290L, M290C, and, partially, M290F could be obtained as stable catalytic domain fragments. The remaining forms were completely degraded, possibly due to enhanced flexibility, which correlates with susceptibility to proteolysis (28). To assay proteolytic activity and determine kinetic parameters, a fluorescent peptide derived from the N-terminal cleavage site of wild-type proulilysin was tested with the four aforementioned variants (supplemental Table 3). Wild-type and M290L ulilysin evinced comparable catalytic efficiency, with similar values for both kcat and Km. M290C showed about 25% efficiency and M290F about 0.3%, the latter due to both a large increase in Km and a large decrease in kcat. In addition, autolytically activated M290S also showed residual activity, but the kinetic parameters could not be derived. These results indicated that only the wild-type, M290L, and M290C catalytic moieties were competent.
Thermal Shift Assays
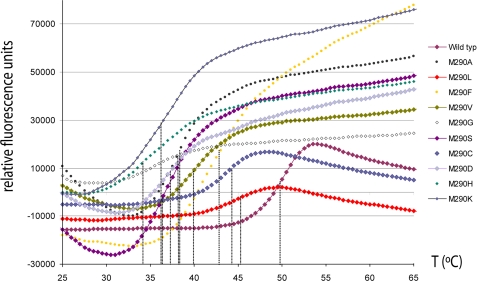
These assays are based on the fluorescence variation of an environmentally sensitive dye, Sypro. The thermal unfolding process exposes hydrophobic regions of the protein to which the dye binds, and this leads to an increase in fluorescence (29). The melting temperature or temperature of midtransition (Tm) is defined as the midpoint in the thermal ramp of a plot depicting relative fluorescence units versus temperature, and it represents the value at which the free energies of native and nonnative forms are equivalent. Higher Tm values indicate higher thermal stability, thus providing a measurement of structural rigidity. The zymogens gave Tm values ranging from 34 °C to 50 °C (supplemental Table 3 and Fig. 2). The lowest value was obtained for M290H, followed by a group consisting of M290K, M290S, M290D, M290G, and M290A (36–38 °C), and, subsequently, by M290V (40°C), M290F (43 °C), M290C (44 °C), M290L (45 °C), peaking with the wild type (50 °C). This test, which was the only biophysical assay that could be performed on all protein variants under comparable conditions, reflected the differences in stability. It also directly correlated with activity, which showed up in the four thermally most stable forms only. In addition, the three top stable variants could also be analyzed for their mature forms. Interestingly, all three forms were much more stable than the zymogens, evincing Tm values around 70 °C (supplemental Table 3). This is consistent with the mature enzyme being a single-domain ellipsoid (see below) rather than a trimodular molecule of 60-, 262-, and 20-residue domains linked by potentially flexible linkers.
FIGURE 2.
Thermal shift assays. Unfolding transition curves showing temperature-dependent change in fluorescence of the M290X variants analyzed. The Tm was obtained from the inflection point of each curve (see supplemental Table 3).
Structural Analysis
Ulilysin variant C269A had been previously crystallized in space groups C2221 (a = ∼119 Å, b = ∼60 Å, c = ∼169 Å; PDB code 2J83) and P21212 (a = ∼50 Å, b = ∼126 Å, c = ∼87 Å; PDB code 2CKI), each with two molecules in the crystallographic asymmetric unit (16, 17, 30). This variant and its selenomethionine derivative rendered crystals that could belong to either space group and were structurally indistinguishable from each other, with root mean square deviation values ranging from 0.22 to 0.30 Å for the different pairwise comparisons of protomers. This compared well with observations for wild-type and selenomethionine-substituted MMP-8, which were likewise structurally indistinguishable (10). In the present study, the C269A mutant was subjected to the same analyses as the wild type and the M290X mutants described above. Except for the melting temperature of the mature enzyme, all values were indistinguishable from the wild type (supplemental Table 3), so the structure of C269A can be considered as a valid model for the wild type.
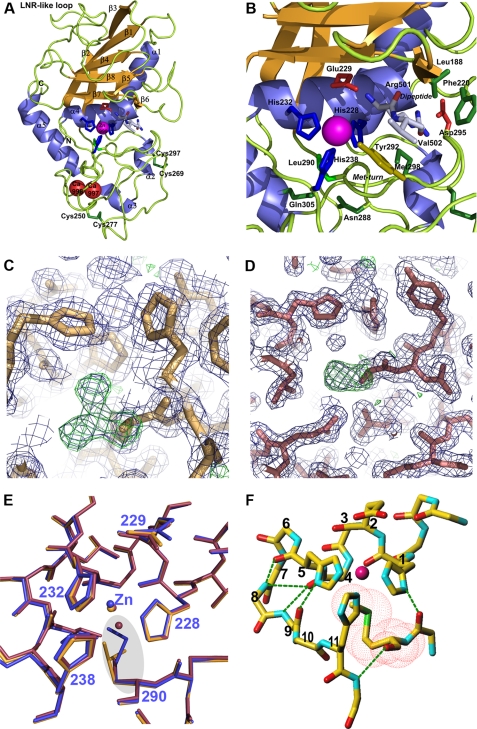
Only the mature forms of the top two methionine mutants in the thermal shift studies, M290L and M290C, could be crystallized and solved for their three-dimensional structures despite extensive trials (Fig. 3, A–D). These two mutants crystallized under slightly different conditions in two crystal forms of orthorhombic space group C2221, with distinct cell parameters (see supplemental Tables 2 and 3) and with four (M290L) and two molecules (M290C)/asymmetric unit. In each case, the identity of the mutants was confirmed by omit electron-density maps (Fig. 3, C and D). Within each structure, the crystallographically independent molecules were equivalent, with root mean square deviation values between chains of 0.10–0.14 Å for M290L and 0.32 Å for M290C. Briefly, mature ulilysin is the largest of all structurally characterized metzincin catalytic domains, with ∼260 residues. Like other metzincins, it is separated into an upper N-terminal and a lower C-terminal subdomain by a central active-site cleft, at whose center-bottom the catalytic zinc ion is bound (Fig. 3, A and B). The former subdomain shows a five-stranded twisted β-sheet at the top, with all strands (β1, β2+β3, β4, β7, and β8) except the fourth parallel to each other and to any substrate that is bound in the cleft. An LNR-like loop bifurcates the second strand into two substrands, β2+β3, and the antiparallel strand, β7, forms the lower edge of this subdomain and creates an upper rim or northern wall of the active-site crevice. In addition, a β-ribbon (β5 plus β6) that protrudes from the molecular surface frames the active site on its primed side. Two long α-helices, the backing helix (α1) and the active-site helix (α4), are found on the concave face of the β-sheet. The segment connecting α1 with β2 is the largest of the metzincins. It covers almost the entire back of the molecule in a cape-like fashion and includes two unique α-helices, α2 and α3. The C-terminal subdomain starts after the active-site helix, which includes the two zinc-binding histidine residues of the zinc-binding consensus sequence and comprises the third zinc-bonding histidine, which contacts the metal from below. This subdomain contains few regular secondary-structure elements, mainly the C-terminal helix α5 at the end of the polypeptide chain and the aforementioned helices within the cape. The C-terminal helix is connected to the active-site helix by a long polypeptide segment that includes the Met-turn (Fig. 3). In addition, the C-terminal subdomain evinces two disulfide bonds, among which Cys269-Cys297 is dispensable for activity (see above), and a unique two-calcium site. This site is a molecular switch for activity, as the proteinase can be reversibly inhibited through calcium chelators (16, 17, 27).
FIGURE 3.
Ulilysin structure. A, Richardson-type plot of ulilysin M290L shown in standard orientation, i.e. with the view into the active-site cleft running left to right. Repetitive secondary structure elements are shown as violet ribbons for helices (labeled α1–α5) and as orange arrows for β-strands (labeled β1–β8). The catalytic zinc ion and the structural calcium ions are shown as magenta and red spheres, respectively. Selected residues engaged in zinc binding or catalysis and a dipeptide bound at the active-site cleft (Arg501-Val502) are shown as stick models. B, close-up view of A around the Met-turn. C, detail of the initial σA-weighted (2mFo − DFc)-type (violet mesh; contoured at 1σ) and (mFo − DFc)-type (green mesh; contoured at 3σ) omit electron-density maps of ulilysin M290L centered on the latter residue. The initial model used for phasing (with an alanine at this position) is superimposed for orientation. D, same as C but for ulilysin M290C. E, superposition of the polypeptide chain traces of ulilysin C269A (blue), M290L (orange), and M290C (mauve). Position 290 is highlighted by a gray ellipse. F, structure of the conserved metzincin core that maintains structural integrity of the zinc-binding site. Hydrogen bonds are shown as green dashed lines, the positions within the zinc-binding consensus sequence (1–11) are labeled, and the van-der-Waals surface of the Met-turn methionine is depicted with red dots.
Detailed inspection of M290L and M290C and their superposition with ulilysin C269A (PDB 2J83) revealed essentially identical structures, with root mean square deviation values for all possible pair-wise comparisons of crystallographically independent molecules of 0.32–0.36 Å and 0.51–0.56 Å, respectively. Replacement of methionine for leucine, which have both apolar and neutral side chains and show similar hydropathy indexes (31), evinced that both side chains occupy essentially the same volume (Fig. 3). The side-chain dihedral angles χ1 (274–276°) and χ2 (294–295°) were comparable with those found for the methionine in wild-type metzincins (see above and Figs. 1B and 3F). Although the first angle is usual for leucine, the second one gives rise to a suboptimal rotamer conformation, found only in 4.5% of cases (32). This angle allows a leucine residue to fit into a space usually occupied by a methionine. With respect to M290C, although cysteine is smaller than methionine, they are both neutral and apolar, and they have a similar hydropathy index (31). The χ1 values (293–296°) were comparable with those of metzincins methionines, and the difference in size was offset by a buried solvent molecule in the space of the methionine Cϵ atom in the wild type (<1 Å away). Finally, no significant deviation was observed in either the position or the orientation of the zinc ion or zinc ligands in either of the mutants (Fig. 3F).
The strict maintenance of the structure around the zinc ion in either structure strongly suggests that what is required for fold and function is an intact core. Indeed, to maintain the architecture of the zinc site and the chain trace of the residues engaged in the zinc-binding consensus sequence, a total of six hydrogen bonds are required in all metzincins, and the Met-turn plays a pivotal role in this bonding pattern (Fig. 3F). In particular, the carbonyl oxygen of the position occupied by the methionine hydrogen bonds the main-chain amide nitrogen of the residue downstream of the third zinc-binding histidine and the carbonyl oxygen preceding the methionine binds the Nδ1 atom of the first zinc-binding histidine, at the first position of the zinc-binding consensus sequence. In addition, the carbonyl oxygens of the residues at positions 4 and 5 bind the amide nitrogens of positions 8 +9 and 9, respectively. Moreover, the carbonyl oxygen of the residue at position 9 hydrogen binds the Nδ1 atom of the zinc-binding histidine at position 5. Finally, the methionine terminal methyl group establishes a hydrophobic interaction with the plane of the latter histidine.
CONCLUSION
The strict conservation of the Met-turn and its methionine among metzincin structures has intrigued structural biologists since its discovery, and it contributed to the name of this metallopeptidase clan (2). Several methionine-replacement studies had shown that this residue is important for fold and function. Our results on 10 ulilysin mutants support its functional relevance. All mutants were expressed and purified, although in varying amounts, thus suggesting at least partial defects in synthesis, secretion, and stability. No mutant proved more stable or active than the wild type. We found an inverse correlation between function/stability and deviation from the optimal size as granted by methionine. In addition, side-chain hydrophobicity was also found to be indispensable because all hydrophilic mutants assayed displayed very low resistance to thermal denaturation. In most cases, they could not be activated but were degraded instead. The next-to-best residues were selenomethionine and leucine, which occupied approximately the same space in the three-dimensional structure as methionine and were similarly hydrophobic and functional. However, the latter was only functional as the wild type under the assumption of an unusual rotamer, thus indicating a suboptimal qualification. The next best option was M290C, which showed lower activity but shared hydrophobicity with the former residues and in which the smaller side chain was offset by the recruitment of a buried solvent molecule. Further ranks were occupied by phenylalanine and valine, with deviating side-chain sizes but similar hydrophobicity. We concluded that there is little margin for variation from a fully competent structure in ulilysin, and activity comparable with the wild type was only observed in structurally equivalent molecules. This was further underlined by detailed inspection of the zinc site in metzincin structures, which revealed a compact core made up by six conserved hydrogen bonds plus a hydrophobic interaction made up by the residues of the zinc-binding consensus sequence and the two central residues of the Met-turn. The latter is a plug that must be snugly inserted into the zinc site to render it functional, and this pinpoints methionine as the “best adapted” residue, which must present a particular combination of conserved side-chain dihedral angles. Mutations entailing significant changes in the size and chemical nature of the side chain would prejudice the functionality of the Met-turn plug, thus giving rise to molecules that were not (auto)activatable, nor crystallizable, or only residually active in best cases. Accordingly, our studies provide an explanation for the absolute conservation of the methionine during evolution, based on steric and chemical requirements for structure and function. Finally, although these results are consistent with most reports on other metzincins, it cannot be ruled out that slight differences in selected structural elements may be observed in certain families, in particular if they do not significantly affect the central zinc-binding core structure.
Supplementary Material
Acknowledgments
We thank Tibisay Guevara for excellent laboratory assistance, Robin Rycroft for helpful discussions on the manuscript, and Isabel Usón for assistance during space group assignment. We further acknowledge the help provided by the European Molecular Biology Laboratory (EMBL) and European Synchrotron Radiation Facility synchrotron local contacts during data collection.
This work was supported by grants and fellowships from European, Catalan, and Spanish public agencies: SAB2002-0102, BIO2009-10336, BIO2008-04080-E, 2009SGR1036, PSE-010000-2009-8, CSD2006-00015, FP7 Strep Project 223101 “AntiPathoGN.” This work was also supported by the Swiss National Science Foundation. Funding for x-ray diffraction data collection at European Synchrotron Radiation Facility (ESRF) synchrotron was provided by ESRF and the European Union.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and additional references.
The atomic coordinates and structure factors (codes 3LUM and 3LUN) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
I. Yiallouros and W. Stöcker, unpublished data.
- ADAM
- a disintegrin and a metalloprotease
- MMP
- matrix metalloprotease
- PEG
- polyethylene glycol
- PDB
- Protein Data Bank.
REFERENCES
- 1.Scheef E. D., Fink J. L. (2003) in Structural Bioinformatics (Bourne P. E., Weissig H. eds) pp. 15–39, Wiley-Liss, Inc., Hoboken, NJ [Google Scholar]
- 2.Bode W., Gomis-Rüth F. X., Stöckler W. (1993) FEBS Lett. 331, 134–140 [DOI] [PubMed] [Google Scholar]
- 3.Gomis-Rüth F. X. (2003) Mol. Biotech. 24, 157–202 [DOI] [PubMed] [Google Scholar]
- 4.Stöcker W., Grams F., Baumann U., Reinemer P., Gomis-Rüth F. X., McKay D. B., Bode W. (1995) Protein Sci. 4, 823–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomis-Rüth F. X. (2009) J. Biol. Chem. 284, 15353–15357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rost B. (1999) Protein Eng. 12, 85–94 [DOI] [PubMed] [Google Scholar]
- 7.Oberholzer A. E., Bumann M., Hege T., Russo S., Baumann U. (2009) Biol. Chem. 390, 875–881 [DOI] [PubMed] [Google Scholar]
- 8.Chothia C., Lesk A. M. (1986) EMBO J. 5, 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tourasse N. J., Li W. H. (2000) Mol. Biol. Evol. 17, 656–664 [DOI] [PubMed] [Google Scholar]
- 10.Pieper M., Betz M., Budiša N., Gomis-Rüth F. X., Bode W., Tschesche H. (1997) J. Protein Chem. 16, 637–650 [DOI] [PubMed] [Google Scholar]
- 11.Butler G. S., Tam E. M., Overall C. M. (2004) J. Biol. Chem. 279, 15615–15620 [DOI] [PubMed] [Google Scholar]
- 12.Walasek P., Honek J. F. (2005) BMC Biochem. 6, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hege T., Baumann U. (2001) J. Mol. Biol. 314, 187–193 [DOI] [PubMed] [Google Scholar]
- 14.Pérez L., Kerrigan J. E., Li X., Fan H. (2007) Biochem. Cell Biol. 85, 141–149 [DOI] [PubMed] [Google Scholar]
- 15.Boldt H. B., Overgaard M. T., Laursen L. S., Weyer K., Sottrup-Jensen L., Oxvig C. (2001) Biochem. J. 358, 359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tallant C., García-Castellanos R., Marrero A., Canals F., Yang Y., Reymond J. L., Solà M., Baumann U., Gomis-Rüth F. X. (2007) Biol. Chem. 388, 1243–1253 [DOI] [PubMed] [Google Scholar]
- 17.Tallant C., García-Castellanos R., Seco J., Baumann U., Gomis-Rüth F. X. (2006) J. Biol. Chem. 281, 17920–17928 [DOI] [PubMed] [Google Scholar]
- 18.Chun Y. J. (1998) Arch. Pharm. Res. 21, 305–309 [DOI] [PubMed] [Google Scholar]
- 19.Kabsch W. (2001) in International Tables for Crystallography. Volume F: Crystallography of Biological Macromolecules (Rossmann M. G., Arnold E. eds), 1st Ed., pp. 730–734, Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 20.Evans P. (1993) in Data Collection and Processing: Proceedings of the CCP4 Study Weekend29–30 January 1993 (Sawyer L., Isaacs N., Bailey S. eds) pp. 114–122, SERC Daresbury Laboratory, Warrington, UK [Google Scholar]
- 21.CCP4 (1994) Acta Crystallogr. Sect. D 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 22.Navaza J. (1994) Acta Crystallogr. Sect. A 50, 157–163 [Google Scholar]
- 23.Emsley P., Cowtan K. (2004) Acta Crystallogr. Sect. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. Sect. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 25.Carranza C., Inisan A.-G., Mouthuy-Knoops E., Cambillau C., Roussel A. (1999) in AFMB Activity Report 1996–1999, pp. 89–90, CNRS-UPR 9039, Marseille, France [Google Scholar]
- 26.Kraulis P. J. (1991) J. Appl. Crystallogr. 24, 946–950 [Google Scholar]
- 27.García-Castellanos R., Tallant C., Marrero A., Solà M., Baumann U., Gomis-Rüth F. X. (2007) Arch. Biochem. Biophys. 457, 57–72 [DOI] [PubMed] [Google Scholar]
- 28.Zappacosta F., Pessi A., Bianchi E., Venturini S., Sollazzo M., Tramontano A., Marino G., Pucci P. (1996) Protein Sci. 5, 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ericsson U. B., Hallberg B. M., Detitta G. T., Dekker N., Nordlund P. (2006) Anal. Biochem. 357, 289–298 [DOI] [PubMed] [Google Scholar]
- 30.García-Castellanos R., Marrero A., Mallorquí-Fernández G., Potempa J., Coll M., Gomis-Rüth F. X. (2003) J. Biol. Chem. 278, 39897–39905 [DOI] [PubMed] [Google Scholar]
- 31.Kyte J., Doolittle R. F. (1982) J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 32.Lovell S. C., Word J. M., Richardson J. S., Richardson D. C. (2000) Proteins 40, 389–408 [PubMed] [Google Scholar]
- 33.Kurisu G., Kinoshita T., Sugimoto A., Nagara A., Kai Y., Kasai N., Harada S. (1997) J. Biochem. 121, 304–308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.