Abstract
Synaptic cell adhesion molecules regulate various steps of synapse formation. The trans-synaptic adhesion between postsynaptic NGL-3 (for netrin-G ligand-3) and presynaptic LAR (for leukocyte antigen-related) regulates excitatory synapse formation in a bidirectional manner. However, little is known about the molecular details of the NGL-3-LAR adhesion and whether two additional LAR family proteins, protein-tyrosine phosphatase δ (PTPδ), and PTPσ, also interact with NGL-3 and are involved in synapse formation. We report here that the leucine-rich repeat (LRR) domain of NGL-3, containing nine LRRs, interacts with the first two fibronectin III (FNIII) domains of LAR to induce bidirectional synapse formation. Moreover, Gln-96 in the first LRR motif of NGL-3 is critical for LAR binding and induction of presynaptic differentiation. PTPδ and PTPσ also interact with NGL-3 via their first two FNIII domains. These two interactions promote synapse formation in a different manner; the PTPσ-NGL-3 interaction promotes synapse formation in a bidirectional manner, whereas the PTPδ-NGL-3 interaction instructs only presynaptic differentiation in a unidirectional manner. mRNAs encoding LAR family proteins display overlapping and differential expression patterns in various brain regions. These results suggest that trans-synaptic adhesion between NGL-3 and the three LAR family proteins regulates excitatory synapse formation in shared and distinct neural circuits.
Keywords: Cell/Neuron, Cell/Adhesion, Cell/Surface, Neurobiology/Neuroscience, Receptors/Membrane, Receptors/Phosphatases
Introduction
Synaptic cell adhesion molecules have been implicated in the regulation of the initial contacts of dendrites and axons, early synapse formation and maturation, and maintenance and structural plasticity of established synapses (1–16). Recent studies have identified a large number of adhesion molecules that are capable of inducing pre- and postsynaptic differentiation in contacting axons and dendrites, respectively. Examples of such molecules include neuroligins, neurexins, SynCAMs, NGLs (for netrin-G ligand),3 LAR (for leukocyte antigen-related), LRRTMs, and EphB receptors (17–22).
The NGL family of synaptic adhesion molecules contains three known members: NGL-1, NGL-2, and NGL-3 (16, 20, 23). NGL proteins are mainly detected at the postsynaptic site of excitatory synapses (20). NGLs share a common domain structure, comprising nine LRRs and an immunoglobulin (Ig) domain in the extracellular region, followed by a single transmembrane domain and a cytoplasmic region that ends with a PDZ domain-binding motif. The C-terminal PDZ-binding motifs of NGLs bind to the PDZ domains of PSD-95, an abundant postsynaptic scaffolding protein (20). This interaction is thought to couple NGL-dependent trans-synaptic adhesions with postsynaptic differentiation.
The extracellular regions of NGLs interact with distinct presynaptic ligands (16). NGL-1 and NGL-2 interact with netrin-G1 and netrin-G2 (also known as laminet-1 and laminet-2), respectively (20, 23), which are glycosylphosphatidylinositol-anchored adhesion molecules (24–26). NGL-3 interacts with LAR (21), a receptor tyrosine phosphatase that contains adhesion domains in the extracellular region and two tyrosine phosphatase domains in their cytoplasmic regions, a membrane-proximal (D1) domain and a membrane-distal (D2) domain, of which only the D1 domain is catalytically active (27, 28). The interaction between NGL-3 and LAR is thought to induce excitatory synapse formation in a bidirectional manner (21). Little is known, however, about the molecular determinants of NGL-3 interaction with LAR, and whether LAR and NGL-3 are the main receptors that mediate NGL-3- and LAR-induced pre- and postsynaptic differentiation, respectively. In addition, it is unknown whether protein-tyrosine phosphatase δ (PTPδ) and PTPσ, two additional members of the LAR family, also interact with NGL-3, and if so, whether these interactions contribute to synapse formation.
Here we demonstrate that the LRR domain of NGL-3 interacts with the first two FNIII domains of LAR to mediate bidirectional synapse formation. Gln-96 in the first LRR motif of NGL-3 is important for LAR binding and induction of presynaptic differentiation. PTPδ and PTPσ interact with NGL-3 via their first two FNIII domains, and these interactions promote synaptogenesis in distinct manners. mRNAs encoding the three LAR family members show overlapping and differential distribution patterns in the brain. These results suggest that adhesion between NGL-3 and LAR family proteins contribute to synapse formation in shared and distinct neural circuits.
EXPERIMENTAL PROCEDURES
DNA Constructs and Antibodies
Full-length ectodomain of Rat NGL-3 (NGL-3-Ecto, XM_218615, aa 41–565), NGL-3-LRR (aa 40–377), and NGL-3-Ig (aa 365–458) were subcloned into pDisplay (Invitrogen). NGL-3-LRR (aa 1–373) and NGL-3-LRR-Q96A were subcloned into pEGFP-N1, in which EGFP was replaced with a human Fc domain. Point mutants of C-terminally EGFP-tagged NGL-3 (Q96A, K126A, D244A, H264A, D277A, K279A, and E282A) were subcloned to pEGFP-N1 (Clontech). The following deletion variants of LAR and LAR-related proteins were also subcloned into pDisplay; full-length ectodomain of human LAR (LAR-Ecto, Y00815, aa 17–1163), LAR-Ig1–3 (aa 35–295), LAR-FN1–8 (aa 309–1078), LAR-FN1–4 (aa 309–700), LAR-FN5–8 (aa 697–1078), LAR-FN1–2 (aa 309–504), LAR-FN3–4 (aa 498–700), LAR-FN1 (aa 309–420), LAR-FN2 (aa 399–520), human PTP-δ-Ecto (NM_002839, aa 21–1174), PTP-δ-FN1–2 (aa 323–518), human PTP-σ-Ecto (NM_130854, aa 30–1167), and PTP-σ-FN1–2 (aa 319–514). Constructs for LAR-CFP, NGL-3-EGFP, and LAR-Ecto-Fc have been described previously (21). Guinea pig polyclonal EGFP antibodies (#1431) were raised against H6-EGFP (aa 1–240). Other antibodies were purchased; synapsin I (Chemicon), HA (Santa Cruz Biotechnology), PSD-95 (Affinity BioReagents), FLAG (Sigma), and LAR (BD Biosciences, #610350).
Cell Adhesion Assay
Two groups of L-cells grown in 6-well plates were transfected with either EGFP and NGL-3, or RFP (DsRed) and LAR, using Lipofectamine (Invitrogen) according to the manufacturer's manual. After 48 h, L-cells were trypsinized and resuspended in 1 ml of serum-free Dulbecco's modified essential medium. Approximately half of this cell suspension (500 μl) was transferred to microtubes and rotated at room temperature for 1 h to allow cells to recover from possible damage from trypsin digestion and to prevent the cells from settling. The two groups of transfected L-cells were mixed together and rotated at room temperature for 30 min to allow cells to aggregate. Cell mixtures (100 μl) were added to 400 μl of serum-free Dulbecco's modified essential medium in 4-well culture slides (Falcon), and then imaged by confocal microscopy.
Dot Blot Analysis
Fc fusion proteins of NGL-3 (NGL-3-LRR-Fc, NGL-3-LRR-Q96A-Fc, and Fc alone, 300 ng) were spotted on a nitrocellulose membrane. The filter was then incubated with LAR-Ecto-Fc, followed by immunoblotting with LAR-Ecto antibodies (BD Biosciences) and secondary horseradish peroxidase-conjugated anti-mouse IgG.
Hippocampal Neuron Culture, Transfection, and Immunocytochemistry
Cultured hippocampal neurons were prepared from embryonic day 18 rat brain. The neurons were cultured on coverslips coated with poly-l-lysine and laminin and grown in Neurobasal medium supplemented with B27 (Invitrogen), 2% fetal bovine serum, 0.5 mm glutamine in 10% CO2 incubator. Cultured neurons were transfected by using a CalPhos mammalian transfection kit (Clontech). For immunohistochemistry, neurons were fixed with 4% paraformaldehyde/4% sucrose, permeabilized with 0.2% Triton X-100 in phosphate-buffered saline, and immunostained with primary antibodies, followed by Cy3-, Cy5-, or fluorescein isothiocyanate-conjugated secondary antibodies (Jackson ImmunoResearch).
Mixed-culture Assay
Mixed-culture assays were carried out as previously described (29). Briefly, cultured hippocampal neurons at DIV 10 were cocultured for 3 days with HEK293T cells expressing NGL-3 or LAR in the presence of 2 μm arabinocytidine hydrochloride, included to suppress HEK293T cell proliferation. For mixed-culture assays between neurons exogenously expressing NGL-3-FLAG and HEK293T cells expressing LAR, PTPδ, or PTPσ, neurons at DIV 13 were transfected with NGL-3-FLAG for 6 h, followed by coculture with HEK293T cells for 2 days.
Image Acquisition and Quantification
Z-stacked images were randomly acquired by confocal microscopy (LSM510, Zeiss), followed by image analysis by using the MetaMorph program (Universal Imaging). Images acquired from mixed-culture assays were thresholded, and the integrated intensities of synaptic marker proteins on transfected HEK293T cells were normalized to the cell area. For the quantification of L-cell clustering, Z-stacked images were used for analysis. Cell clusters were defined as cell aggregates containing four or more cells, which include at least one green (EGFP) and one red (red fluorescent protein) cells. However, frames with no detectable cell clusters were counted as zero in the quantification of cell number per cluster. Bar graph data represent mean ± S.E., and their statistical significances were determined by Student's t test or one-way ANOVA (Tukey test).
In Situ Hybridization
In situ hybridization was performed on mouse brain sections (12-μm thick) from 1-, 2-, 3-, and 6-week-old mice. Hybridization probes for NGL-3, LAR, PTPδ, and PTPσ were generated using the following constructs: pGEM7zf containing nt 2040–2557 of NGL-3 (NM_198250.1), nt 5973–6492 of LAR (NM_011213.2), nt 4573–5092 of PTPδ (NM_011211.2), and nt 5892–6424 of PTPσ (NM_011218.2). Antisense Riboprobes were prepared by RNA polymerase transcription using a Riboprobe System (Promega) in the presence of [α-35S]UTP. In situ hybridization histochemistry was carried out as described previously (30). Briefly, fresh-frozen sections of brains were thaw-mounted on 3-aminopropyltriethoxysilane-coated glass slides, fixed in 4% paraformaldehyde, washed with phosphate-buffered saline, acetylated with 0.25% acetic anhydrides in 0.1 m triethanolamine/0.9% NaCl (pH 8.0), dehydrated/defatted in ethanol and chloroform, and air-dried. The sections were hybridized overnight with 35S-labeled probes (1.2 × 106 cpm/slide) at 55 °C, followed by 4 washes in 2× SSC solution at room temperature. After RNase treatment, slides were sequentially rinsed with 2× SSC, 1× SSC, 0.5× SSC, and 0.1× SSC containing 1 mm dithiothreitol for 10 min each at room temperature. Finally, the sections were dehydrated, air-dried, and exposed to x-ray film (Biomax MR, Kodak, Rochester, NY).
RESULTS
The LRR Domain of NGL-3 Is Sufficient for LAR Binding
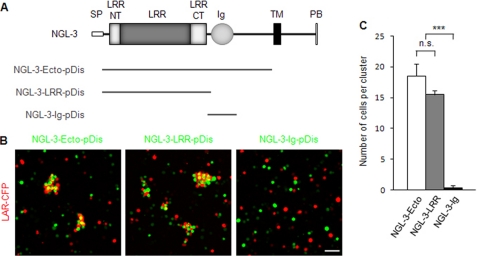
NGL-3 contains two distinct domains, LRR and Ig, in the extracellular region. We previously demonstrated that the LRR domain is required for LAR binding, as evidenced by the failure of a mutant NGL-3 lacking the LRR domain to interact with LAR in cell aggregation assays (21). To determine whether the LRR domain is sufficient for LAR binding, we generated three different pDisplay NGL-3 constructs containing different regions of the ectodomain: NGL-3-Ecto, containing the full-length ectodomain; NGL-3-LRR, containing the LRR domain only; and NGL-3-Ig, containing only the Ig domain (Fig. 1A). In L-cell adhesion assays, two groups of L-cells, one expressing NGL-3-LRR and one expressing LAR, formed cell aggregates when mixed together (Fig. 1, B and C). The extent of cell aggregation in NGL-3-LRR was similar to that observed with the full-length NGL-3-Ecto construct. In contrast, NGL-3-Ig did not mediate cell aggregation with LAR (Fig. 1, B and C). Control experiments showed that this absence of aggregation was not due to a failure of NGL-3-Ig to localize to the plasma membrane; in fact, surface expression levels of NGL-3-Ig were slightly higher than those of NGL-3-Ecto and NGL-3-LRR (supplemental Fig. 1). These results indicate that the LRR domain of NGL-3 is sufficient to mediate LAR binding.
FIGURE 1.
The LRR domain of NGL-3 is sufficient for LAR interaction in cell adhesion assays. A, NGL-3 variants carrying the full-length ectodomain of NGL-3 (NGL-3-Ecto), the LRR domain (NGL-3-LRR), and the Ig domain (NGL-3-Ig). pDis, pDisplay vector; SP, signal peptide; LRRNT, leucine-rich repeat N-terminal domain; LRRCT, leucine-rich repeat C-terminal domain; TM, transmembrane domain; PB, PDZ domain-binding motif. B, the LRR domain, but not the Ig domain, of NGL-3 is sufficient to mediate the interaction with LAR in cell adhesion assays. L-cells doubly expressing EGFP and NGL-3 variants (Ecto, LRR, or Ig) were mixed with another group of L-cells coexpressing DsRed and LAR-CFP for cell aggregation. Scale bar, 20 μm. C, quantification (average number of cells per cell cluster) of results shown in B. Mean ± S.E., n = 10; ***, p < 0.001, ANOVA; n.s., not significant.
The LRR Domain of NGL-3 Is Sufficient for Presynaptic Induction
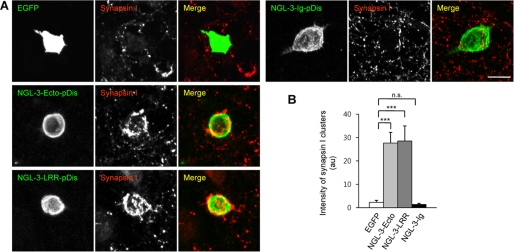
We next tested whether the LRR domain of NGL-3 is sufficient to induce presynaptic differentiation in contacting axons in coculture (or mixed-culture) assays (17, 29). NGL-3-LRR induced the clustering of synapsin I in contacting axons of cocultured neurons, to an extent similar to that of NGL-3-Ecto (Fig. 2, A and B). In contrast, NGL-3-Ig did not induce synapsin I clustering, compared with control cells expressing EGFP alone (Fig. 2, A and B). These results indicate that the LRR domain of NGL-3 is sufficient for inducing presynaptic differentiation.
FIGURE 2.
The LRR domain of NGL-3 is sufficient to induce presynaptic differentiation. A, NGL-3-Ecto is sufficient to induce synapsin I clustering in contacting axons of cocultured neurons. HEK293T cells expressing NGL-3 variants (Ecto, LRR, or Ig), or EGFP alone, were cocultured with hippocampal neurons (10–13 days in vitro or DIV) and stained for synapsin I. Scale bar, 20 μm. B, quantification of the intensity of synapsin I clusters induced by NGL-3 variants. Integrated fluorescence intensity of synapsin I was normalized to the cell area. Mean ± S.E., n = 14 for EGFP, n = 15 for NGL-3-Ecto, n = 15 for NGL-3-LRR, and n = 15 for NGL-3-Ig; ***, p < 0.001, ANOVA; n.s., not significant.
Gln-96 in the First LRR Motif of NGL-3 Is Important for LAR Binding and Adhesion with LAR-expressing Cells
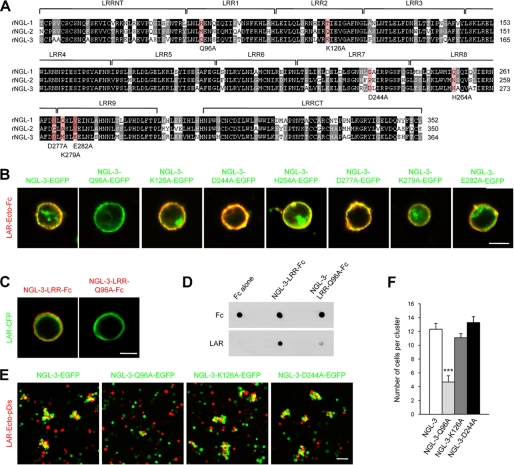
The three NGLs share ∼70–73% amino acid sequence identity in the LRR domain, which contains nine LRRs flanked by cysteine-rich capping structures known as LRRNT and LRRCT. Despite this similarity, the LRR domains of NGLs exhibit distinct ligand specificities: NGL-3 selectively binds LAR (21), whereas NGL-1 and NGL-2 bind netrin-G1 and netrin-G2, respectively (20, 23). To gain molecular insight into these distinct interactions, we first compared the amino acid sequences of the LRR domains of NGL-1, NGL-2, and NGL-3 and selected a total of seven amino acid residues that are uniquely present in NGL-3 (Fig. 3A). We then mutated the seven residues to alanine and assessed the effect of these changes on NGL-3 binding to LAR. Additional residues were unique to NGL-3, but the seven selected residues were chosen, because their properties (e.g. polarity and charge) were significantly different from those of the corresponding residues in NGL-1 and NGL-2. Interestingly, a glutamine-to-alanine mutation in residue 96 (Q96A) of the first LRR motif of NGL-3 markedly reduced LAR binding, as shown by the failure of exogenously expressed mutant NGL-3-Q96A proteins to bind recombinant LAR proteins in HEK293T cells (Fig. 3B). The other six point mutations had no effect on NGL-3 binding to LAR. Surface expression levels of wild-type and mutant (Q96A, K126A, and D244A) NGL-3 proteins were similar (supplemental Fig. 2). Conversely, recombinant NGL-3-Q96A proteins showed significantly reduced binding to LAR expressed in HEK293T cells (Fig. 3C). The effect of the Q96A mutation was further confirmed in a dot-blot assay in which recombinant NGL-3 and LAR proteins were shown to directly bind to each other (Fig. 3D). Consistent with these observations, L-cells expressing NGL-3-Q96A showed significantly reduced co-aggregation with LAR-expressing cells, whereas two other control mutations (K126A and D244A) had no effect on the cell aggregation (Fig. 3, E and F). These results indicate that Gln-96 in the first LRR motif of NGL-3 is important for LAR binding.
FIGURE 3.
Gln-96 in the first LRR motif of NGL-3 is important for LAR binding and for adhesion with LAR-expressing cells. A, comparison of the amino acid sequences of the LRR domain of rat NGLs. Seven residues unique to NGL-3, indicated in red, were mutated to alanine. The residues shown in black and gray backgrounds denote those that are identical in all three sequences and in two sequences, respectively. Boundaries of LRRNT, LRRs, and LRRCT were predicted by the SMART program (available on-line). B, a Q96A point mutation in the first LRR motif of NGL-3, but not other NGL-3 mutations, significantly reduces the binding of recombinant LAR to NGL-3. HEK293T cells expressing full-length NGL-3 proteins (C-terminally EGFP tagged) carrying seven distinct mutations were incubated with recombinant LAR proteins (the ectodomain of LAR fused to Fc; LAR-Ecto-Fc). Scale bar, 10 μm. C, recombinant NGL-3 Q96A mutant proteins show significantly weakened binding to LAR expressed in HEK293T cells. HEK293T cells expressing C-terminally ECFP-tagged, full-length LAR were incubated with purified Fc fusion proteins containing the LRR domain of NGL-3 (WT and Q96A). Scale bar, 10 μm. D, direct binding between recombinant NGL-3 and LAR proteins is demonstrated in a dot-blot assay. NGL-3-LRR fusion proteins (WT and Q96A) and Fc alone were spotted onto a nitrocellulose membrane and incubated with LAR-Ecto-Fc, followed by immunoblotting with anti-LAR-Ecto antibodies. E, a Q96A point mutation in NGL-3 selectively suppresses cell adhesion mediated by NGL-3 and LAR. L-cells expressing NGL-3 proteins (wild type, Q96A, K126A, or D244A), and EGFP were mixed with those expressing LAR (LAR-Ecto-pDis) and DsRed for cell adhesion. Scale bar, 20 μm. F, quantification of the average number of cells per cell cluster in E. Mean ± S.E., n = 10; ***, p < 0.001, ANOVA.
Gln-96 in NGL-3 Is Important for Presynaptic Induction
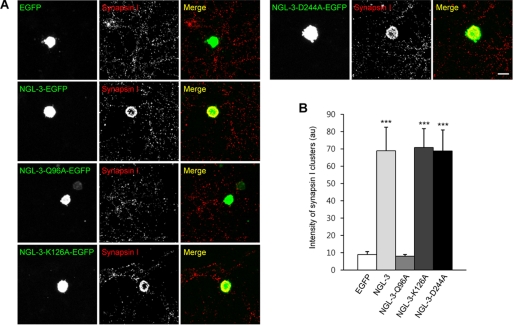
We next tested whether NGL-3-induced presynaptic differentiation was altered by the Q96A mutation in coculture assays. HEK293T cells expressing NGL-3-Q96A did not induce synapsin I clustering in contacting axons, exhibiting a presynaptic induction level similar to that of control cells expressing EGFP alone (Fig. 4, A and B). In contrast, HEK293T cells expressing wild-type NGL-3 proteins or other mutant NGL-3 proteins (K126A and D244A) normally induced presynaptic differentiation, showing significantly higher levels of synapsin I clustering compared with the EGFP control (Fig. 4, A and B). These results indicate that Gln-96 in the first LRR motif of NGL-3 is important for the induction of presynaptic differentiation. In addition, these results suggest that LAR, which shows a weakened interaction with NGL-3-Q96A, is likely the major presynaptic receptor that mediates NGL-3-induced presynaptic differentiation.
FIGURE 4.
Gln-96 of NGL-3 is important for the induction of presynaptic differentiation. A, NGL-3-Q96A, but not other NGL-3 mutants, fails to induce presynaptic differentiation. HEK293T cells expressing wild-type or mutant (Q96A, K126A, and D244A) NGL-3 proteins (C-terminally EGFP tagged), or EGFP alone, were cocultured with hippocampal neurons (10–13 DIV) and stained for synapsin I. Scale bar, 20 μm. B, quantification of the intensity of synapsin I clusters induced by NGL-3 mutants. Mean ± S.E., n = 15 for EGFP, n = 11 for NGL-3, n = 11 for NGL-3-Q96A, n = 11 for NGL-3-K126A, and n = 11 for NGL-3-D244A; ***, p < 0.001, ANOVA.
The First Two FNIII Domains of LAR Are Sufficient for NGL-3 Binding
We next sought to identify key domains of LAR involved in NGL-3 binding. To this end, we generated several LAR variants carrying different regions of the LAR ectodomain (Fig. 5A). Among these constructs, only those that carried the first two FNIII domains (FN1–8, FN1–4, and FN1–2) were able to interact with NGL-3 in cell adhesion assays (Fig. 5, B and C). Smaller FN1–2-containing constructs had a tendency to show greater cell adhesion activities (Fig. 5, B and C). In contrast, constructs that lacked the first two FN domains (Ig1–3, FN5–8, and FN3–4) did not interact with NGL-3. The smallest LAR variants carrying only the first or second FNIII domain did not bind to NGL-3, indicating that both FN1 and FN2 domains are required for NGL-3 binding. In addition to interacting with the full-length ectodomain of NGL-3, FN1–2 of LAR also interacted with the LRR domain of NGL-3 in cell adhesion assays (Fig. 5, D and E). Surface expression levels of the LAR variants were similar, except for a small increase in FN1–2 (supplemental Fig. 3). These results indicate that the first two FNIII domains of LAR are sufficient to mediate the interaction with the LRR domain of NGL-3.
FIGURE 5.
The first two FNIII domains of LAR are sufficient for NGL-3 binding. A, LAR variants carrying different regions of the ectodomain. D1 and D2, tyrosine phosphatase domains. B, all LAR variants carrying the first two FNIII domains (FN1–8, FN1–4, and FN1–2) interact with NGL-3 in cell adhesion assays. L-cells expressing LAR variants and DsRed were mixed with another group of L-cells expressing full-length NGL-3 and EGFP for cell aggregation. Note that smaller FN1–2-containing LAR variants tend to have greater cell-adhesion activities. Scale bar, 20 μm. C, quantification of the average number of cells per cell cluster in B. Mean ± S.E., n = 10. D, FN1–2 of LAR interacts with the LRR domain of NGL-3. L-cells expressing FN1–2 of LAR and dsRed were mixed with those expressing NGL-3-LRR-pDis (or NGL-3-Ecto for comparison) and EGFP. Scale bar, 20 μm. E, quantification of the results in D. Mean ± S.E., n = 10; ***, p < 0.001, Student's t test.
The First Two FNIII Domains of LAR Induce Postsynaptic PSD-95 Clustering
We have recently shown that LAR expressed in HEK293T cells induces clustering of excitatory postsynaptic proteins in contacting dendrites of cocultured neurons (21). We thus tested whether the first two FNIII domains of LAR are sufficient to induce postsynaptic protein clustering in coculture assays. Two LAR variants, FN1–4 and FN1–2, expressed in HEK293T cells induced PSD-95 clustering in contacting dendrites, whereas control cells expressing EGFP alone did not (Fig. 6, A and B). These results indicate that FN1–2 of LAR is sufficient to induce postsynaptic PSD-95 clustering. In addition, these results suggest that NGL-3, which binds FN1–2 of LAR, is likely the major postsynaptic receptor that mediates LAR-induced postsynaptic protein clustering.
FIGURE 6.
The first two FNIII domains of LAR are sufficient to induce postsynaptic PSD-95 clustering. A, FN1–2 and FN1–4 induce PSD-95 clustering in contacting dendrites of cocultured neurons. HEK293T cells expressing LAR variants, or EGFP alone (control), were cocultured with hippocampal neurons (10–13 DIV) and stained for PSD-95. Scale bar, 20 μm. B, quantification of the intensity of PSD-95 clusters normalized to cell area. Mean ± S.E., n = 26 for EGFP, n = 28 for LAR-Ecto, n = 25 for LAR-Ig1–3, n = 26 for LAR-FN1–8, n = 28 for LAR-FN1–4, n = 27 for LAR-FN5–8, n = 25 for LAR-FN1–2, n = 25 for LAR-FN3–4, n = 25 for LAR-FN1, and n = 25 for LAR-FN2. **, p < 0.01; ***, p < 0.001; ANOVA.
Notably, however, neither LAR-Ecto (full-length ectodomain) nor LAR-FN1–8 induced detectable PSD-95 clustering (Fig. 6, A and B), despite the fact that LAR-Ecto and LAR-FN1–8 interacted normally with NGL-3 in cell adhesion assays (Fig. 5). This contrasts with the clear PSD-95 clustering induced by FN1–2 and FN1–4 described above, and the previously reported PSD-95 clustering induced by full-length LAR (C-terminally EGFP-tagged) (21). A possible reason for this discrepancy is that the PSD-95 clustering assay, which likely involves LAR-induced clustering of endogenous NGL-3, may require an affinity of LAR for NGL-3-binding that is much greater than that required for cell adhesion assays. In support of this possibility, FN1–2 and FN1–4 exhibited higher levels of cell aggregation, compared with LAR-Ecto and LAR-FN1–8 (Fig. 5C). Alternatively, it may stem from the fact that these extracellular domains of LAR are displayed on the surface in the context of pDisplay vector; i.e. the ectodomains are surrounded by upstream HA and downstream Myc epitopes.
PTPδ and PTPσ Interact with NGL-3 through Their First Two FNIII Domains
We extended our analysis to two additional members of the LAR family, PTPδ and PTPσ (27, 28), testing their interaction with NGL-3. The full-length ectodomain of PTPδ and PTPσ interacted with NGL-3 in cell adhesion assays, similar to LAR (Fig. 7, A and B). The cell adhesion activities of PTPδ and PTPσ were lower than that of LAR, differences that were partially correlated with the lower surface expression levels of PTPδ and PTPσ (supplemental Fig. 4A). In addition, PTPδ- and PTPσ-expressing HEK293T cells induced the clustering of (exogenously expressed) NGL-3 in contacting dendrites of cocultured neurons (Fig. 7, C and D), further indicating that NGL-3 interacts with PTPδ and PTPσ. In this experiment, endogenous NGL-3 could not be visualized due to the lack of antibodies suitable for immunocytochemistry (21).
FIGURE 7.
PTPδ and PTPσ interact with NGL-3 through their first two FNIII domains. A, PTPδ and PTPσ interact with NGL-3 in cell adhesion assays. L-cells expressing DsRed and the full-length ectodomains of LAR, PTPδ, or PTPσ, were mixed with another group of L-cells expressing EGFP and NGL-3. Scale bar, 20 μm. B, quantification of the average number of cells per clusters in A. Mean ± S.E., n = 10; **, p < 0.01; ***, p < 0.001, ANOVA. C, PTPδ or PTPσ expressed in HEK293T cells induce NGL-3 clustering in dendrites of cocultured neurons. HEK293T cells expressing PTPδ, PTPσ, LAR, or EGFP were cocultured (10–13 DIV) with hippocampal neurons transfected with NGL-3-FLAG (13–15 DIV) and stained for HA (for LAR family proteins) and FLAG. D, quantification of the NGL-3 clustering in C (mean ± S.E., n = 14 for EGFP-pDis, n = 13 for LAR-pDis, n = 16 for PTPδ-pDis, and n = 13 for PTPσ-pDis; ***, p < 0.001, ANOVA). Scale bar, 5 μm. E, the FN1–2 domains of PTPδ and PTPσ interact with NGL-3 in cell adhesion assays. Scale bar, 20 μm. F, quantification of the average number of cells per clusters in E (mean ± S.E., n = 10; *, p < 0.05; ***, p < 0.001, ANOVA). G, the FN1–2 domains of LAR, PTPδ, and PTPσ weakly interact with NGL-3-Q96A, relative to wild-type NGL-3, in cell adhesion assays. Scale bar, 20 μm. H, mean ± S.E., n = 10; ***, p < 0.001, Student t test.
Because the first two FNIII domains of LAR mediated NGL-3 binding, we tested whether the FN1–2 domains of PTPδ and PTPσ are also sufficient for NGL-3 binding. Both PTPδ-FN1–2 and PTPσ-FN1–2 interacted with NGL-3 in cell adhesion assays (Fig. 7, E and F), similar to their full-length ectodomains (Fig. 7, A and B). It should be noted, however, that the cell adhesion activity of PTPδ was weaker than that of LAR or PTPσ (Fig. 7, E and F), despite the fact that PTPδ, LAR, and PTPσ were all expressed at similar levels on the cell surface (supplemental Fig. 4B). Importantly, PTPδ-FN1–2 and PTPσ-FN1–2 showed reduced binding to NGL-3-Q96A relative to wild-type NGL-3 in cell adhesion assays (Fig. 7, G and H), suggesting that Gln-96 in NGL-3 is a common molecular determinant for the interactions with LAR, PTPδ, and PTPσ. Collectively, these results indicate that PTPδ and PTPσ interact with NGL-3 via the FN1–2 domains, and that Gln-96 in the first LRR motif of NGL-3 is a key determinant of NGL-3 interactions with PTPδ and PTPσ.
The First Two FNIII Domains of PTPσ, but Not PTPδ, Induce Postsynaptic PSD-95 Clustering
We next tested whether the FN1–2 domains of PTPδ and PTPσ can induce postsynaptic PSD-95 clustering. FN1–2 of PTPσ did induce PSD-95 clustering in contacting dendrites of cocultured neurons, similar to LAR-FN1–2 (Fig. 8, A and B). In contrast, FN1–2 of PTPδ failed to induce PSD-95 clustering beyond that observed in the EGFP-alone control (Fig. 8, A and B). Despite this, PTPδ-FN-1–2 did interact with NGL-3 in cell adhesion assays, albeit to a lesser extent than LAR and PTPσ (Fig. 7, E and F). This suggests that PTPδ-FN1–2 might have an affinity for NGL-3 that is strong enough to mediate cell adhesion, but not to induce postsynaptic protein clustering. Therefore, PTPσ-FN1–2, but not PTPδ-FN1–2, is capable of inducing postsynaptic PSD-95 clustering, similar to LAR.
FIGURE 8.
The FN1–2 domain of PTPσ, but not PTPδ, induces postsynaptic PSD-95 clustering. A, PTP-σ-FN1–2, but not PTP-δ-FN1–2, induces PSD-95 clustering in contacting dendrites of cocultured neurons. HEK293T cells expressing the FN1–2 domains of LAR, PTP-σ, PTP-δ, or EGFP alone, were cocultured with hippocampal neurons (10–13 DIV) and stained for PSD-95. Scale bar, 20 μm. B, quantification of the intensity of PSD-95 clusters in A (mean ± S.E., n = 20 for EGFP, n = 21 for LAR-FN1–2, n = 21 for PTP-δ-FN1–2, and n = 20 for PTP-σ-FN1–2). ***, p < 0.001, ANOVA; n.s., not significant.
Overlapping and Differential Expression Patterns of mRNAs Encoding LAR Family Proteins
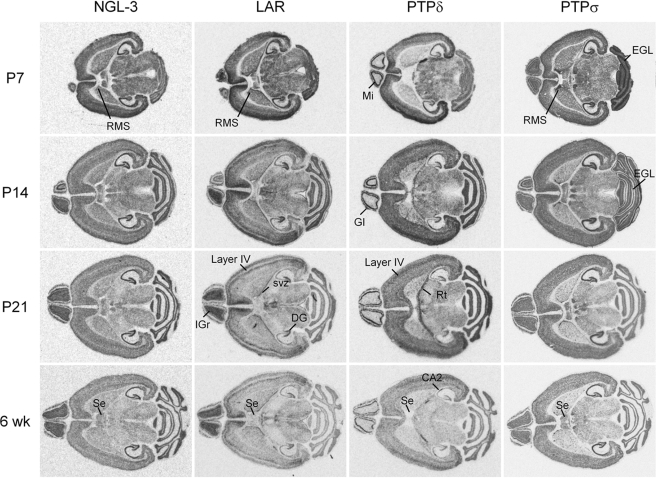
The adhesions between NGL-3 and LAR family proteins may regulate the formation of excitatory synapses in distinct neural circuits of the brain. To this end, we compared expression patterns of mRNAs encoding these proteins in mouse brain regions. Although mRNAs distribution patterns of LAR family members in mice and rat brains have been reported previously (31–35), we attempted here a comprehensive comparison of mRNA distribution patterns of LAR family members, along with NGL-3, in mouse brain sections at several different developmental stages, including P7, P14, and P21, during which active synapse formation occurs. The overall expression levels of LAR family mRNAs gradually diminished toward the adult stage (6 weeks) (Figs. 9 and 10 and supplemental Fig. 5). LAR family mRNAs showed widespread distribution patterns in various mouse brain regions. NGL-3 mRNAs were also widespread, similar to their distribution patterns in rat brains (20).
FIGURE 9.
Overlapping and differential distribution patterns of mRNAs encoding LAR family proteins and NGL-3 in horizontal mouse brain sections at different developmental stages (P7, P14, P21, and 6 weeks) revealed by in situ hybridization analysis. DG, dentate gyrus; EGL, external granular layer of cerebellum; IGr, internal granular layer of olfactory bulb; RMS, rostral migratory stream; Rt, thalamic reticular nucleus; Se, septal areas; and svz, subventricular zone.
FIGURE 10.
Selected regions in Fig. 9 (P21 sections; hippocampus, cortex, and olfactory bulb) enlarged for better comparisons. CA1, CA2, and CA3, subregions of the Ammon's horn in the hippocampus; DG, dentate gyrus; Gl, glomerular layer of olfactory bulb; IGr, internal granular layer of olfactory bulb; and Mi, mitral cell layer of olfactory bulb. The numbers in the cortex region indicate cortical layers.
mRNAs of LAR family proteins showed overlapping as well as distinct distribution patterns (Figs. 9 and 10 and supplemental Fig. 5). For instance, mRNAs of LAR, PTPδ, and PTPσ were found in all hippocampal subregions, similar to the distribution pattern of NGL-3, although LAR mRNAs were more abundant in the dentate gyrus, and PTPδ mRNA signals were stronger in the dentate gyrus and the CA2 region. This suggests the possibility that the synapses linking CA3 Schaffer collaterals with CA1 pyramidal neurons likely contain three LAR family proteins and NGL-3 at pre- and postsynaptic sites, respectively.
Importantly, distinct distribution patterns of LAR, PTPδ, and PTPσ mRNAs were observed in several brain regions (Figs. 9 and 10 and supplemental Fig. 5). PTPδ mRNAs were minimally detected in the internal granule layer of olfactory bulb, contrary to LAR and PTPσ. In contrast, PTPδ mRNAs were abundantly detected in the reticular thalamic area. In addition, PTPδ mRNAs were more abundant in the layer IV of cortex, compared with other layers, whereas LAR mRNAs were less abundant in the layer IV. In the septal area, PTPσ signals were strong, whereas LAR signals were faint, and PTPδ were almost undetectable. Other regions also showed differential expression of LAR family transcripts; LAR mRNAs were abundant in the subventricular zone of caudate putamen, and PTPσ signals were stronger in external granule layers of early stage cerebellum and rostral migratory streams. These results collectively suggest that mRNAs encoding LAR family proteins exhibit both overlapping and differential distribution patterns in the brain.
DISCUSSION
Molecular Determinants of the Adhesion between NGL-3 and LAR
In this study, we identified that the LRR domain of NGL-3 is a minimal LAR-binding region and demonstrated that this domain is sufficient for the induction of presynaptic differentiation in contacting axons. The human genome contains a large number of genes encoding LRR-containing proteins. Of these proteins, 139 are known to contain LRRs in their extracellular regions (36). A single LRR protein contains several to dozens of LRRs, which are tandemly arranged to form horseshoe- or crescent-shaped solenoid structures (36–38). LRR proteins in neurons regulate neurodevelopmental processes, including neurite outgrowth (39) and target-muscle selection by motor neurons (40). Synaptically localized LRR proteins have also been identified (41). These include NGLs (20, 21), SALMs (42, 43), LRRTMs (22), LGI1 (44), densin-180 (45), and erbin (46). Among these, NGLs and LRRTMs are adhesion molecules that have the ability to induce synapse formation (21, 22). This study and our previous report (21) demonstrate that the LRR domain of NGL-3 is necessary and sufficient for the induction of presynaptic differentiation. In addition, the LRR domain of LRRTM2, a recently identified synaptogenic LRR protein, is necessary and sufficient for presynaptic induction (22). These results collectively suggest a critical role for LRRs in synaptogenesis.
Our results indicate that the first two FNIII domains (FN1–2) of LAR are sufficient for NGL-3 binding. Three specific ligands of LAR have been identified (27, 28). Syndecan and dallylike in Drosophila are heparan sulfate proteoglycans that bind to the Ig domains of LAR and regulate presynaptic development at the neuromuscular junction (47, 48), which mimics the LAR-dependent presynaptic regulation in Drosophila and Caenorhabditis elegans (49, 50). nidogen is a basement membrane protein that binds to the fifth FNIII domain of LAR (51). A C. elegans homolog of nidogen has been shown to regulate synaptic localization of PTP-3A, a C. elegans homolog of LAR, and PTP-3A-dependent synaptic morphogenesis (50). Therefore, our study identifies a distinct region of LAR (FN1–2) involved in ligand (NGL-3) binding and excitatory synapse formation in mammals.
Notably, the deletion variants of LAR differentially promote cell aggregation and presynaptic induction in the order of LAR-FN1–2, LAR-FN1–4, and LAR-FN1–8 (Figs. 5 and 6). This may be partly attributable to that LAR-FN1–2 has a higher surface expression level. Another interesting possibility is that the first two FNIII domains of LAR might be inhibited by the following FNIII domains, which remains to be determined in future studies.
The trans-synaptic interaction between NGL-3 and LAR regulates bidirectional excitatory synapse formation (21). An important question would be whether the direct interaction between LAR and NGL-3 mediates NGL-3- and LAR-induced pre- and postsynaptic differentiation, respectively. We found in the present study that NGL-3-Q96A, which has substantially weakened affinities for LAR in cell adhesion assays (Fig. 3, E and F), does not induce presynaptic differentiation in contacting axons (Fig. 4). Similarly, a small region (FN1–2) in the ectodomain of LAR, which interacts with NGL-3 (Fig. 5), is sufficient to induce PSD-95 clustering in contacting dendrites (Fig. 6). These results suggest that NGL-3 and LAR induce pre- and postsynaptic differentiation via LAR and NGL-3, respectively.
Interaction of NGL-3 with PTPδ and PTPσ
Our study reveals novel interactions of NGL-3 with PTPδ and PTPσ. Previous studies have identified several ligands of PTPδ and PTPσ. PTPδ exhibits a homophilic adhesion (52). PTPσ interacts with two heparan sulfate proteoglycans, agrin and collagen XVIII, through the first Ig domain (53) and with α-latrotoxin of black widow spider venom through the FNIII 2–3 domains (54). Nucleolin, a protein detected on the surface of developing myotubes, has been suggested to bind PTPσ (55). More recently, the first Ig domain of PTPσ was shown to bind chondroitin sulfate proteoglycans, which are produced by cell types, including reactive astroglia at sites of neuronal injury for the inhibition of axonal regeneration (56). Therefore, NGL-3 represents the first heterophilic ligand for PTPδ and a novel ligand for PTPσ.
NGL-3-Q96A, which fails to induce presynaptic differentiation, exhibited lowered affinities for PTPδ and PTPσ, in addition to LAR (Fig. 7, G and H), suggesting that PTPδ and PTPσ interact with NGL-3 through mechanisms shared by the LAR-NGL-3 interaction. In addition, this suggests that, when all three LAR family proteins (LAR, PTPδ, and PTPσ) are present in contacting axons, NGL-3 may induce presynaptic differentiation by interacting with all three of them, meaning that they may collectively function as the major presynaptic receptors for NGL-3.
Our data indicate that the first two FNIII domains of PTPσ are sufficient to bind NGL-3 (Fig. 7, E–H) and induce postsynaptic clustering of PSD-95 (Fig. 8). In contrast, FN1–2 of PTPδ lacked PSD-95-clustering activity (Fig. 8), despite its NGL-3-binding activity (Fig. 7, E–H). These results suggest that the adhesion between PTPσ and NGL-3 promotes bidirectional excitatory synapse formation, similar to the LAR-NGL-3 interaction. In contrast, the adhesion between PTPδ and NGL-3 seems to promote only presynaptic differentiation in a unidirectional manner, although it may still contribute to the strength of trans-synaptic adhesion, similar to other NGL-3-based interactions.
How might PTPδ and PTPσ promote presynaptic differentiation? Similar to LAR, the second cytoplasmic phosphatase (D2) domains of PTPδ and PTPσ, which are catalytically inactive, interact with liprin-α (57), a cytoplasmic adaptor protein that is important for presynaptic development (49, 58) and is coupled to other presynaptic active zone proteins, including RIM and ELKS/ERC (59, 60). Therefore, LAR, PTPδ, and PTPσ may converge onto liprin-α to promote synaptic differentiation in a synergistic manner. In addition, we cannot exclude the possibility that the first phosphatase (D1) domains of the LAR family proteins, which are catalytically active, may contribute to NGL-3-induced presynaptic differentiation.
Previous studies have identified several functions of PTPδ and PTPσ. PTPδ regulates cell adhesion, neurite outgrowth, and axon guidance (52, 61). Different functions of PTPδ seem to be mediated by distinct domains of PTPδ; a deletion variant of PTPδ that lacks the FNIII 4–8 domains poorly mediates cell adhesion but normally promotes neurite outgrowth (62). PTPσ regulates neurite outgrowth, axon guidance, axonal target finding, and axon regeneration (63–68).
Mice that lack the expression of PTPδ or PTPσ show relatively severe phenotypes (69–71), relative to those exhibited by LAR-deficient mice (72, 73). PTPδ-deficient mice show postnatal semi-lethality due to limitations in food intake, and mice that survive to adulthood show enhanced long term potentiation and impaired learning and memory (69). PTPσ-deficient mice exhibit perinatal semi-lethality, and show endocrine defects in the hypothalamo-pituitary axis, and various neuroanatomical and behavioral abnormalities (70, 71, 74). It remains to be determined whether the adhesion between NGL-3 and PTPδ, or PTPσ, underlie any of the phenotypic abnormalities in knock-out mice or suggested functions of PTPδ/PTPσ noted above.
NGL-3 and LAR Family Proteins in Shared and Distinct Neural Circuits
Our results indicate that mRNAs encoding the three LAR family proteins show overlapping and differential distribution patterns. This suggests that NGL-3 and LAR family proteins contribute to the formation of excitatory synapses in both shared and distinct neural circuits of the brain. It is conceivable that different axonal populations express different combinations of LAR family proteins, creating collective properties that would likely regulate the strength and specificity of synaptic adhesions involving interactions with NGL-3.
Notably, the promiscuous interaction of NGL-3 with the three LAR family proteins sharply contrasts with the specific interactions of NGL-1 and NGL-2 with netrin-G1 and netrin-G2, respectively (20, 23). The specificity of NGL-netrin-G interactions is strongly supported by the observation that NGL-1 and NGL-2, which normally show lamina-specific distribution patterns in subdendritic segments, are dispersed in contacting dendrites by genetic ablation of netrin-G1 and netrin-G2, respectively, in presynaptic axons (75). Our data, however, indicate that the distribution patterns of mRNA for LAR family proteins overlap in several brain regions. This predicts that the removal of one type of LAR family protein in presynaptic axons is unlikely to abolish NGL-3-dependent synapse formation if the other two remain intact. This might act as a redundancy mechanism, preventing NGL-3-dependent synapse formation from being easily disturbed.
Another potential function of the interaction of NGL-3 with all three LAR family proteins would be to achieve a maximum-strength trans-synaptic adhesion, something that might not be readily accomplished with any single pair of adhesion molecules. Consistent with this idea, the presynapse-inducing activity of NGL-3 in coculture assays is stronger than that of either NGL-1 or NGL-2 (21). Lastly, the interaction of NGL-3 with different LAR family proteins may provide a mechanism by which the strength of trans-synaptic adhesion could be graded at several levels.
Supplementary Material
Acknowledgment
We thank the In Situ Hybridization facility at Korea University supported by the Brain Research Center under the 21st Century Frontier Research Program.
This work was supported by the National Creative Research Initiative Program of the Korean Ministry of Education, Science and Technology (to E. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- NGL
- netrin-G ligand
- LAR
- leukocyte antigen-related
- PTP
- protein-tyrosine phosphatase
- FNIII
- fibronectin III
- aa
- amino acid(s)
- EGFP
- enhanced growth factor protein
- DIV
- days in vitro
- ANOVA
- analysis of variance
- nt
- nucleotide(s).
REFERENCES
- 1.Akins M. R., Biederer T. (2006) Curr. Opin. Neurobiol. 16, 83–89 [DOI] [PubMed] [Google Scholar]
- 2.Biederer T., Stagi M. (2008) Curr. Opin. Neurobiol. 18, 261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brose N. (2009) Neuron 61, 650–652 [DOI] [PubMed] [Google Scholar]
- 4.Craig A. M., Kang Y. (2007) Curr. Opin. Neurobiol. 17, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalva M. B., McClelland A. C., Kayser M. S. (2007) Nat. Rev. Neurosci. 8, 206–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerrow K., El-Husseini A. (2006) Front. Biosci. 11, 2400–2419 [DOI] [PubMed] [Google Scholar]
- 7.Han K., Kim E. (2008) Prog. Neurobiol. 84, 263–283 [DOI] [PubMed] [Google Scholar]
- 8.Huang Z. J., Scheiffele P. (2008) Curr. Opin. Neurobiol. 18, 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McAllister A. K. (2007) Annu. Rev. Neurosci. 30, 425–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheiffele P. (2003) Annu. Rev. Neurosci. 26, 485–508 [DOI] [PubMed] [Google Scholar]
- 11.Südhof T. C. (2008) Nature 455, 903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tai C. Y., Kim S. A., Schuman E. M. (2008) Curr. Opin. Cell Biol. 20, 567–575 [DOI] [PubMed] [Google Scholar]
- 13.Washbourne P., Dityatev A., Scheiffele P., Biederer T., Weiner J. A., Christopherson K. S., El-Husseini A. (2004) J. Neurosci. 24, 9244–9249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamagata M., Sanes J. R., Weiner J. A. (2003) Curr. Opin. Cell Biol. 15, 621–632 [DOI] [PubMed] [Google Scholar]
- 15.Aoto J., Chen L. (2007) Brain Res. 1184, 72–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo J., Kwon S. K., Kim E. (2009) Mol. Cell Neurosci. 42, 1–10 [DOI] [PubMed] [Google Scholar]
- 17.Scheiffele P., Fan J., Choih J., Fetter R., Serafini T. (2000) Cell 101, 657–669 [DOI] [PubMed] [Google Scholar]
- 18.Ichtchenko K., Hata Y., Nguyen T., Ullrich B., Missler M., Moomaw C., Südhof T. C. (1995) Cell 81, 435–443 [DOI] [PubMed] [Google Scholar]
- 19.Biederer T., Sara Y., Mozhayeva M., Atasoy D., Liu X., Kavalali E. T., Süudhof T. C. (2002) Science 297, 1525–1531 [DOI] [PubMed] [Google Scholar]
- 20.Kim S., Burette A., Chung H. S., Kwon S. K., Woo J., Lee H. W., Kim K., Kim H., Weinberg R. J., Kim E. (2006) Nat. Neurosci. 9, 1294–1301 [DOI] [PubMed] [Google Scholar]
- 21.Woo J., Kwon S. K., Choi S., Kim S., Lee J. R., Dunah A. W., Sheng M., Kim E. (2009) Nat. Neurosci. 12, 428–437 [DOI] [PubMed] [Google Scholar]
- 22.Linhoff M. W., Laurén J., Cassidy R. M., Dobie F. A., Takahashi H., Nygaard H. B., Airaksinen M. S., Strittmatter S. M., Craig A. M. (2009) Neuron 61, 734–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin J. C., Ho W. H., Gurney A., Rosenthal A. (2003) Nat. Neurosci. 6, 1270–1276 [DOI] [PubMed] [Google Scholar]
- 24.Nakashiba T., Ikeda T., Nishimura S., Tashiro K., Honjo T., Culotti J. G., Itohara S. (2000) J. Neurosci. 20, 6540–6550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakashiba T., Nishimura S., Ikeda T., Itohara S. (2002) Mech. Dev. 111, 47–60 [DOI] [PubMed] [Google Scholar]
- 26.Yin Y., Miner J. H., Sanes J. R. (2002) Mol Cell Neurosci. 19, 344–358 [DOI] [PubMed] [Google Scholar]
- 27.Johnson K. G., Van Vactor D. (2003) Physiol. Rev. 83, 1–24 [DOI] [PubMed] [Google Scholar]
- 28.Stryker E., Johnson K. G. (2007) J. Cell Sci. 120, 3723–3728 [DOI] [PubMed] [Google Scholar]
- 29.Biederer T., Scheiffele P. (2007) Nat. Protoc. 2, 670–676 [DOI] [PubMed] [Google Scholar]
- 30.Kim S. Y., Chung H. S., Sun W., Kim H. (2007) Neuroscience 147, 996–1021 [DOI] [PubMed] [Google Scholar]
- 31.Mizuno K., Hasegawa K., Katagiri T., Ogimoto M., Ichikawa T., Yakura H. (1993) Mol. Cell. Biol. 13, 5513–5523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaapveld R. Q., Schepens J. T., Bächner D., Attema J., Wieringa B., Jap P. H., Hendriks W. J. (1998) Mech. Dev. 77, 59–62 [DOI] [PubMed] [Google Scholar]
- 33.Sommer L., Rao M., Anderson D. J. (1997) Dev. Dyn. 208, 48–61 [DOI] [PubMed] [Google Scholar]
- 34.Yan H., Grossman A., Wang H., D'Eustachio P., Mossie K., Musacchio J. M., Silvennoinen O., Schlessinger J. (1993) J. Biol. Chem. 268, 24880–24886 [PubMed] [Google Scholar]
- 35.Zhang J. S., Honkaniemi J., Yang T., Yeo T. T., Longo F. M. (1998) Mol. Cell Neurosci. 10, 271–286 [DOI] [PubMed] [Google Scholar]
- 36.Dolan J., Walshe K., Alsbury S., Hokamp K., O'Keeffe S., Okafuji T., Miller S. F., Tear G., Mitchell K. J. (2007) BMC Genomics 8, 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobe B., Kajava A. V. (2001) Curr. Opin. Struct. Biol. 11, 725–732 [DOI] [PubMed] [Google Scholar]
- 38.Bella J., Hindle K. L., McEwan P. A., Lovell S. C. (2008) Cell Mol. Life Sci. 65, 2307–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y., Aulia S., Li L., Tang B. L. (2006) Brain Res. Brain Res. Rev. 51, 265–274 [DOI] [PubMed] [Google Scholar]
- 40.Kurusu M., Cording A., Taniguchi M., Menon K., Suzuki E., Zinn K. (2008) Neuron 59, 972–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ko J., Kim E. (2007) J. Neurosci. Res. 85, 2824–2832 [DOI] [PubMed] [Google Scholar]
- 42.Wang C. Y., Chang K., Petralia R. S., Wang Y. X., Seabold G. K., Wenthold R. J. (2006) J. Neurosci. 26, 2174–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ko J., Kim S., Chung H. S., Kim K., Han K., Kim H., Jun H., Kaang B. K., Kim E. (2006) Neuron 50, 233–245 [DOI] [PubMed] [Google Scholar]
- 44.Fukata Y., Adesnik H., Iwanaga T., Bredt D. S., Nicoll R. A., Fukata M. (2006) Science 313, 1792–1795 [DOI] [PubMed] [Google Scholar]
- 45.Apperson M. L., Moon I. S., Kennedy M. B. (1996) J. Neurosci. 16, 6839–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Y. Z., Wang Q., Xiong W. C., Mei L. (2001) J. Biol. Chem. 276, 19318–19326 [DOI] [PubMed] [Google Scholar]
- 47.Fox A. N., Zinn K. (2005) Curr. Biol. 15, 1701–1711 [DOI] [PubMed] [Google Scholar]
- 48.Johnson K. G., Tenney A. P., Ghose A., Duckworth A. M., Higashi M. E., Parfitt K., Marcu O., Heslip T. R., Marsh J. L., Schwarz T. L., Flanagan J. G., Van Vactor D. (2006) Neuron 49, 517–531 [DOI] [PubMed] [Google Scholar]
- 49.Kaufmann N., DeProto J., Ranjan R., Wan H., Van Vactor D. (2002) Neuron 34, 27–38 [DOI] [PubMed] [Google Scholar]
- 50.Ackley B. D., Harrington R. J., Hudson M. L., Williams L., Kenyon C. J., Chisholm A. D., Jin Y. (2005) J. Neurosci. 25, 7517–7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Grady P., Thai T. C., Saito H. (1998) J. Cell Biol. 141, 1675–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J., Bixby J. L. (1999) Mol. Cell Neurosci. 14, 370–384 [DOI] [PubMed] [Google Scholar]
- 53.Aricescu A. R., McKinnell I. W., Halfter W., Stoker A. W. (2002) Mol. Cell. Biol. 22, 1881–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krasnoperov V., Bittner M. A., Mo W., Buryanovsky L., Neubert T. A., Holz R. W., Ichtchenko K., Petrenko A. G. (2002) J. Biol. Chem. 277, 35887–35895 [DOI] [PubMed] [Google Scholar]
- 55.Alete D. E., Weeks M. E., Hovanession A. G., Hawadle M., Stoker A. W. (2006) FEBS J. 273, 4668–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen Y., Tenney A. P., Busch S. A., Horn K. P., Cuascut F. X., Liu K., He Z., Silver J., Flanagan J. G. (2009) Science 326, 592–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pulido R., Serra-Pagès C., Tang M., Streuli M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 11686–11690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhen M., Jin Y. (1999) Nature 401, 371–375 [DOI] [PubMed] [Google Scholar]
- 59.Schoch S., Castillo P. E., Jo T., Mukherjee K., Geppert M., Wang Y., Schmitz F., Malenka R. C., Südhof T. C. (2002) Nature 415, 321–326 [DOI] [PubMed] [Google Scholar]
- 60.Ko J., Na M., Kim S., Lee J. R., Kim E. (2003) J. Biol. Chem. 278, 42377–42385 [DOI] [PubMed] [Google Scholar]
- 61.Sun Q. L., Wang J., Bookman R. J., Bixby J. L. (2000) Mol. Cell Neurosci. 16, 686–695 [DOI] [PubMed] [Google Scholar]
- 62.Gonzalez-Brito M. R., Bixby J. L. (2006) Int. J. Dev. Neurosci. 24, 425–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rashid-Doubell F., McKinnell I., Aricescu A. R., Sajnani G., Stoker A. (2002) J. Neurosci. 22, 5024–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stepanek L., Stoker A. W., Stoeckli E., Bixby J. L. (2005) J. Neurosci. 25, 3813–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thompson K. M., Uetani N., Manitt C., Elchebly M., Tremblay M. L., Kennedy T. E. (2003) Mol. Cell Neurosci. 23, 681–692 [DOI] [PubMed] [Google Scholar]
- 66.Sapieha P. S., Duplan L., Uetani N., Joly S., Tremblay M. L., Kennedy T. E., Di Polo A. (2005) Mol. Cell Neurosci. 28, 625–635 [DOI] [PubMed] [Google Scholar]
- 67.Sajnani G., Aricescu A. R., Jones E. Y., Gallagher J., Alete D., Stoker A. (2005) J. Neurobiol. 65, 59–71 [DOI] [PubMed] [Google Scholar]
- 68.McLean J., Batt J., Doering L. C., Rotin D., Bain J. R. (2002) J. Neurosci. 22, 5481–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Uetani N., Kato K., Ogura H., Mizuno K., Kawano K., Mikoshiba K., Yakura H., Asano M., Iwakura Y. (2000) EMBO J. 19, 2775–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Elchebly M., Wagner J., Kennedy T. E., Lanctôt C., Michaliszyn E., Itié A., Drouin J., Tremblay M. L. (1999) Nat. Genet. 21, 330–333 [DOI] [PubMed] [Google Scholar]
- 71.Wallace M. J., Batt J., Fladd C. A., Henderson J. T., Skarnes W., Rotin D. (1999) Nat. Genet. 21, 334–338 [DOI] [PubMed] [Google Scholar]
- 72.Yeo T. T., Yang T., Massa S. M., Zhang J. S., Honkaniemi J., Butcher L. L., Longo F. M. (1997) J. Neurosci. Res. 47, 348–360 [DOI] [PubMed] [Google Scholar]
- 73.Van Lieshout E. M., Van der Heijden I., Hendriks W. J., Van der Zee C. E. (2001) Neuroscience 102, 833–841 [DOI] [PubMed] [Google Scholar]
- 74.Meathrel K., Adamek T., Batt J., Rotin D., Doering L. C. (2002) J. Neurosci. Res. 70, 24–35 [DOI] [PubMed] [Google Scholar]
- 75.Nishimura-Akiyoshi S., Niimi K., Nakashiba T., Itohara S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14801–14806 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.