Abstract
The nucleocytoplasmic egress of viral capsids is a rate-limiting step in the replication of the human cytomegalovirus (HCMV). As reported recently, an HCMV-specific nuclear egress complex is composed of viral and cellular proteins, in particular protein kinases with the capacity to induce destabilization of the nuclear lamina. Viral protein kinase pUL97 and cellular protein kinase C (PKC) play important roles by phosphorylating several types of nuclear lamins. Using pUL97 mutants, we show that the lamin-phosphorylating activity of pUL97 is associated with a reorganization of nuclear lamin A/C. Either pUL97 or PKC has the potential to induce distinct punctate lamina-depleted areas at the periphery of the nuclear envelope, which were detectable in transiently transfected and HCMV-infected cells. Using recombinant HCMV, which produces green fluorescent protein-labeled viral capsids, the direct transition of viral capsids through these areas could be visualized. This process was sensitive to an inhibitor of pUL97/PKC activity. The pUL97-mediated phosphorylation of lamin A/C at Ser22 generated a novel binding motif for the peptidyl-prolyl cis/trans-isomerase Pin1. In HCMV-infected fibroblasts, the physiological localization of Pin1 was altered, leading to recruitment of Pin1 to viral replication centers and to the nuclear lamina. The local increase in Pin1 peptidyl-prolyl cis/trans-isomerase activity may promote conformational modulation of lamins. Thus, we postulate a novel phosphorylation-triggered mechanism for the reorganization of the nuclear lamina in HCMV-infected cells.
Keywords: Nucleus, Prolyl Isomerase, Protein Export, Protein Kinase C (PKC), Protein Phosphorylation, Human Cytomegalovirus, Multiprotein Complex, Nuclear Lamina-depleted Areas, Viral Kinase pUL97, Viral Nuclear Egress
Introduction
Human cytomegalovirus (HCMV)2 belongs to the β-herpesvirus subfamily, exhibiting worldwide distribution. When infecting immunocompetent individuals, HCMV possesses low pathogenicity, causing mainly asymptomatic infections. In immunocompromised or immunosuppressed hosts, HCMV infection can cause severe and even life-threatening diseases, including pneumonitis, retinitis, hepatitis, encephalitis, and gastroenteritis (1–3).
HCMV replication is based on a nuclear phase, a characteristic of most DNA viruses. Transition from the nuclear to the cytoplasmic phase is determined by the nuclear exit of DNA-filled capsids budding through the inner nuclear membrane (INM) (4–6). The site-specific budding of viral capsids through distinct locally occurring invaginations in the INM of HCMV-infected cells was clearly illustrated by Buser et al. (7) using electron microscopic analysis. The proteinaceous network of the nuclear lamina, underlying the INM, constitutes a major obstacle for the nuclear egress of capsids. Lamins, belonging to type V intermediate filament proteins, are the main constituents of the nuclear lamina and are grouped into A and B types. A-type lamins (A, C, AΔ10, and C2; collectively lamin A/C) result from alternative splicing of the LMNA gene. B-type lamins are encoded by the LMNB1 (B1) or LMNB2 (B2, B3) gene (8, 9). A major function of the nuclear lamina is to maintain the structure of the nuclear envelope. During mitosis, the nuclear lamina has to be transiently disassembled. This dynamic process is regulated by destabilizing phosphorylation of lamins at specific sites. In particular, it is well established that CDK1 (cyclin-dependent kinase 1; Cdc2) is mainly responsible for phosphorylation of lamins during mitosis (10–13). CDK1-dependent phosphorylation of lamin A/C occurs at Ser22, Ser392, and probably farther sites (10, 14, 15). HCMV blocks the cell cycle through the action of viral regulatory proteins (16, 17); however, it remains unclear whether HCMV is able to utilize the CDK1-based pathway for distortion of the nuclear lamina. A number of studies provided evidence that CMVs (including HCMV as well as murine CMV) and also other herpesviruses recruit another cellular lamin-phosphorylating kinase, namely protein kinase C (PKC). PKC is important for the phosphorylation and dissolution of the nuclear lamina (18, 19). During mitosis of uninfected cells, there is substantial evidence that PKC is important for lamin phosphorylation and mitotic nuclear lamina disassembly (20). In HCMV-infected cells, the lamina-reorganizing activity of PKC appears to be supported by further lamin-phosphorylating protein kinases. The HCMV-encoded protein kinase pUL97 is recruited to the nuclear lamina (21, 22) and is highly important for efficient export of viral capsids to the cytoplasm (23, 24). In particular, pUL97 phosphorylates lamins and has a destabilizing effect on the integrity of the nuclear lamina (22). A recent study by Hamirally et al. (14) confirmed the important regulatory role of pUL97 for nuclear egress by demonstrating a pUL97-dependent phosphorylation of A-type lamins at Ser22. This was causatively linked with morphological alterations of the nuclear lamina. On the basis of these observations, we postulated the formation of an HCMV-specific nuclear egress complex (NEC) composed of viral and cellular components (21). This complex may basically consist of pUL50, pUL53, pUL97, PKC, p32, and the lamin B receptor (21). In particular, the direct and indirect recruitment of PKC and pUL97, respectively, is mediated by viral pUL50. Whereas pUL50 is principally sufficient for PKC recruitment (21), other viral proteins such as pUL53 appear to enhance this function. pUL50 and pUL53 represent an interactor pair of essential nuclear egress proteins that are conserved among herpesviruses (5, 18, 19, 25, 26). Here, we provide novel information about the composition, function, and regulatory complexity of the NEC. In particular, our study describes the formation of distinct punctate lamin A/C-depleted areas at the periphery of the nuclear envelope that can be induced by HCMV infection or by transient overexpression of individual NEC kinases. Searching for the molecular mode of the phosphorylation-triggered process of lamina reorganization, we detected the involvement of the peptidyl-prolyl cis/trans-isomerase (PPIase) Pin1. These findings led us to propose a model that may explain the regulatory key features of the NEC and HCMV nuclear egress.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
Expression plasmids coding for FLAG-tagged versions of pUL97 (i.e. pcDNA-UL97-FLAG, pcDNA-UL97(K355M)-FLAG, pcDNA-UL97-(1–595)-FLAG, and pcDNA-UL97-(181–707)-FLAG)), hemagglutinin-tagged pUL50 (pcDNA-UL50-HA), and PKCα fused to enhanced green fluorescent protein (EGFP; pEGFP-N1-PKCα) were described previously (19, 21, 22, 27). Plasmid pHM990, coding for a fusion protein of IE2p86 and GFP, was kindly provided by Dr. N. Tavalai (Institute for Clinical and Molecular Virology, University of Erlangen-Nuremberg) (28). The plasmids pEGFP-N1-lamin A and pEGFP-N1-lamin C code for lamin A and C, respectively, which are fused to EGFP (kindly provided by Dr. J. Broers, Cardiovascular Research Institute, University of Maastricht, Maastricht, The Netherlands) (29).
Cell Culture, Plasmid Transfections, and Virus Infections
HeLa and 293T cells were cultivated and transfected as described previously (19, 21). Human foreskin fibroblasts (HFFs) were cultivated in minimal essential medium containing 7.5% fetal calf serum. HCMV infection experiments were performed at a multiplicity of infection (m.o.i.) of 1.0 (or lower as indicated for specific experiments) using the laboratory strain AD169, the UL97 deletion mutant BAC213 (22), or the recombinant TB40-UL32-EGFP virus expressing GFP fused to the capsid-associated tegument protein pUL32 (pp150, kindly provided by Prof. C. Sinzger, Institute of Medical Virology, University of Tübingen, Tübingen, Germany) (6).
Protein Kinase Inhibitors
Gö6976 is an inhibitor of serine/threonine protein kinases (particularly pUL97 and PKC) (30). AG490 (tyrphostin) is an inhibitor of tyrosine protein kinases (31). Compounds were purchased from Calbiochem. Stock solutions were prepared in Me2SO, and aliquots were stored at −20 °C.
Indirect Immunofluorescence Double Staining and Confocal Laser Scanning Microscopy (CLSM)
HeLa cells or primary HFFs were grown on coverslips for transient transfection or HCMV infection, respectively. At the indicated time points, cells were fixed and permeabilized following indirect immunofluorescence staining as described (19, 21). The following polyclonal (pAb) and monoclonal (mAb) antibodies were used to detect cellular and viral proteins: anti-lamin A/C mAb (636) and anti-Pin1 pAb (H-123, raised against amino acids 41–163 of human Pin1) (Santa Cruz Biotechnology), anti-FLAG mAb (M2) and anti-FLAG pAb (Sigma), anti-hemagglutinin pAb (HA.11; HiSS Diagnostics GmbH), anti-GFP mAb (clone 7.1/13.1; Roche Applied Science), anti-UL97 pAb (kindly provided by Prof. D. Michel, University of Ulm, Ulm, Germany) (32), and anti-UL44 mAb (BS 510; kindly provided by Prof. B. Plachter, University of Mainz, Mainz, Germany). Secondary antibodies used for double staining were fluorescein isothiocyanate- and Cy3-conjugated (Dianova). Images were acquired using a Leica TCS SP5 confocal laser scanning microscope equipped with a 63× HCX PL APO CS oil immersion objective lens (Leica) and analyzed using LAS AF software (Leica).
Confocal Time-lapse Microscopy of HCMV-infected Cells with Live Cell Staining
HFFs were seeded into 2-well chambered coverglass units with coverslip quality glass bottoms (Lab-Tek, Nunc) at a density of 1 × 105 cells/well. The next day, the cells were infected with recombinant TB40-UL32-EGFP virus at a m.o.i. of 1.0. 62 h post-infection (hpi), the cells were washed with Hanks' balanced salt solution with calcium and magnesium (Invitrogen) and then incubated with prewarmed staining solution for live cell endoplasmic reticulum labeling (ER-Tracker red dye, Invitrogen) at a concentration of 1 μm for 20 min at 37 °C and 5% CO2. After replacing the staining solution with fresh probe-free medium, the cells were examined using a TCS SP5 confocal laser scanning microscope. The intracellular trafficking velocity of viral particles was determined by using LAS AF Version 1.8.2 (build 1465, Leica).
In Vitro Kinase Assay
The kinase activity of FLAG-tagged pUL97 was determined in vitro (2.5 μCi of [γ-33P]ATP) after immunoprecipitation of the kinase and the putative substrate proteins lamin A-GFP and lamin C-GFP from lysates of transfected 293T cells. Immunoprecipitates were subsequently pelleted, washed, and subjected to in vitro kinase assay reaction as described previously (19, 21). Finally, samples were prepared for separation by 12.5% SDS-PAGE, followed by transfer to nitrocellulose membrane (A. Hartenstein) by Western blotting. Autoradiographic membranes were exposed to a phosphorimager plate and measured using a BAS-2000 phosphorimager (Fuji Film. Co., Tokyo, Japan).
Co-immunoprecipitation Assay
HFFs were seeded into cell culture flasks at a density of 3.6 × 106 cells/flask. The next day, cells were infected with HCMV strain AD169 at m.o.i. = 0.1 and 1.0. Three days post-infection, immunoprecipitation was performed as described previously (21) using 10 μl of anti-Pin1 pAb (A302-315A, recognizing epitope 50-100 of human Pin1; BIOMOL) or preimmune rabbit antiserum, respectively. Co-immunoprecipitated samples and expression controls were subjected to standard Western blot analysis using anti-Pin1 pAb (A302-316A, recognizing epitope 113-163 of human Pin1; BIOMOL) and anti-lamin A/C mAb.
Bioinformatic Analysis
Candidates for functional protein interaction motifs in lamin A/C were identified in the ELM Motif Database (33) using the algorithm from Dinkel and Sticht (34). The structure of lamin A in complex with Pin1 was modeled based on the known crystal structure of Pin1 in complex with a peptide from the RNA polymerase II C-terminal domain (Protein Data Bank code 1f8a) (35). For this purpose, the C-terminal domain ligand sequence was replaced with that of lamin A using the lowest energy rotamers for the non-conserved amino acid side chains. The complex was subsequently refined by 100 steps of energy minimization using SYBYL 7.3 software (Tripos, L.P.). Structural analysis and visualization were performed using the program DS ViewerPro (Accelrys Inc.).
RESULTS
Morphological Alteration of the Nuclear Lamina Induced by Protein Kinases pUL97 and PKC
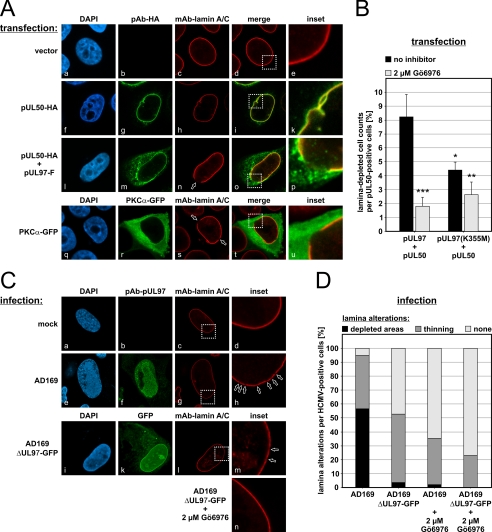
Previous investigations demonstrated that HCMV replication exerts drastic morphological alterations on the nuclear lamina (21, 22). The effects described include a thinning of the lamina layer, loss of the typical rim staining of individual lamina components, internal nuclear speckling of lamins, and modified antigenic detectability (7, 14, 22). We investigated the fine-structured morphological organization of the nuclear lamina by CLSM and visualized effects produced by the expression of the viral protein kinase pUL97 (supplemental Fig. S1A). In vector-transfected cells, the staining for lamin A/C showed a strict rim signal (supplemental Fig. S1A, panels a–e). As a striking result, those cells expressing a catalytically active version of pUL97 (full-length pUL97 or N-terminally truncated pUL97-(181–707)) showed an increase of lamin signals throughout the nuclear plasma (supplemental Fig. S1A, panels f–p). The rim staining changed to a more homogeneous nuclear distribution (supplemental Fig. S1A, panels k and p). No induction of similar effects was observed for catalytically inactive versions of pUL97 (point mutant pUL97(K355M) or C-terminally truncated pUL97-(1–595)) (22) (supplemental Fig. S1A, panels q–z). To ensure comparable levels of pUL97 expression, semiquantitative Western blot analysis was performed (supplemental Fig. S1B). Interestingly, transient expression of pUL97 produced only a rather moderate phenotype of lamin A/C reorganization (as shown in supplemental Fig. S1), whereas coexpression of pUL97 and egress protein pUL50 potentiated the lamina-reorganizing activity (Fig. 1A). Coexpression of pUL50 and pUL97 led to punctate distortions at the periphery of the nuclear envelope, which clearly represented centers of massive lamin A/C reorganization (Fig. 1A, panels l–p). The potentiating effect of pUL50 might be explained by an indirect protein interaction bridged by the pUL97-interacting cellular adapter protein p32 (22). Coexpression of pUL53 and pUL97 did not induce a lamina reorganization as demonstrated for pUL50 and pUL97 (data not shown). Using pUL50 only induced some minor lamina alterations restricted to a small fraction of cells; however, in no case was a local depletion of the nuclear lamina observed (Fig. 1A, panels f–k). Quantification of pUL50/pUL97-coexpressing cells revealed that 8.2% of cells staining positive for pUL50 showed distinct distortions of nuclear lamin A/C (Fig. 1B). Treatment with the kinase inhibitor Gö6976 significantly reduced this effect to 1.8%, confirming the central role of protein kinase activity. The indolocarbazole compound Gö6976 possesses strong inhibitory potential against pUL97 (30, 36) and an additional activity against PKC (37). The use of a kinase-inactive point mutant (K355M) led to a statistically significant difference in measurable lamina distortions compared with wild-type pUL97. However, a substantial portion of cells still showed measurable effects (4.4%) (Fig. 1B), suggesting that endogenous PKC activity, possibly associated with pUL97(K355M)-derived protein complexes, might also contribute to the alterations in lamin A/C. This notion was underscored by the finding that Gö6976 treatment markedly reduced the effect of mutant K355M (2.6%).
FIGURE 1.
Induction of nuclear lamina-depleted areas in HCMV-infected and plasmid-transfected cells. A, HeLa cells were single (panels a–k and q–u) or cotransfected (panels l–p) with plasmids encoding tagged versions of pUL50, pUL97, or PKCα before cells were fixed and immunostained with the indicated antibodies 2 days post-transfection. DAPI, 4′,6-diamidino-2-phenylindole. C, HFFs were mock-infected (panels a–d) or infected with HCMV strain AD169 (panels e–h) or AD169-derived UL97 deletion virus expressing a GFP reporter (panels i–n). A subset of cells was treated with the inhibitor Gö6976 at 2 μm (panel n). At 3 days post-infection (AD169) or 3 weeks post-infection (AD169ΔUL97-GFP), cells were fixed and immunostained. Open arrowheads indicate lamin A/C-depleted areas. Inset images show enlargements of representative areas of the nuclear lamina. B and D, kinase-dependent nuclear lamin A/C alterations were quantitated in transfected (B) and HCMV-infected (D) cells in the presence or absence of Gö6976. Statistical significance compared with pUL97/pUL50 without Gö6976 treatment was calculated by Student's t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
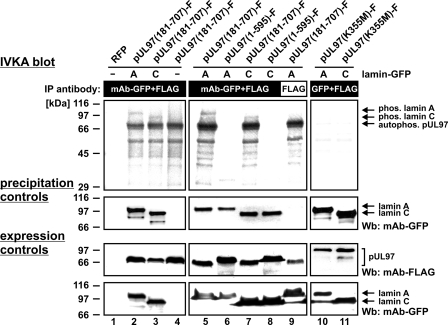
The direct phosphorylation of lamins by pUL97 was analyzed by performing in vitro kinase assay. pUL97 and its putative substrates lamin A-GFP and lamin C-GFP were immunoprecipitated from lysates of transiently transfected cells. In the in vitro phosphorylation reaction, lamin A-GFP and lamin C-GFP were phosphorylated by pUL97 (Fig. 2, lanes 2, 3, 5, and 7). The vector control (Fig. 2, lane 4), red fluorescent protein (lane 1), or GFP (data not shown) did not produce a phosphorylation signal. An additional antibody control confirmed the specificity of the reaction (Fig. 2, lane 9). Efficient precipitation of the substrate proteins was confirmed by precipitation control staining (Fig. 2, middle panels). In accordance with the findings shown in supplemental Fig. S1, lamin phosphorylation was exclusively detectable for catalytically active pUL97 (N-terminally truncated pUL97-(181–707)) (Fig. 2, lanes 5 and 7), whereas an inactive C-terminally truncated version (pUL97-(1–595)) (lanes 6 and 8) or a kinase-inactive point mutant (pUL97(K355M)) (lanes 10 and 11) did not produce a phosphorylation signal.
FIGURE 2.
Phosphorylation of lamins A and C by pUL97 in vitro. 293T cells were cotransfected with catalytically active pUL97 (lanes 2–4, 5, 7, and 9), inactive C-terminally truncated pUL97 (lanes 6 and 8), or kinase-inactive point mutant K355M (lanes 10 and 11) together with constructs encoding lamin A or C fused to GFP. At 2 days post-transfection, cells were lysed and subjected to immunoprecipitation (IP) with the indicated antibodies, followed by in vitro kinase reaction with the precipitates. Labeled phosphorylation products were separated by SDS-PAGE/Western blot (Wb) transfer and visualized by exposure of the blots to autoradiography films (upper panels). Lysate control samples taken prior to immunoprecipitation were used for Western blot analysis with the indicated antibodies to monitor the levels of expressed proteins (lower panels). Control staining of the in vitro kinase assay (IVKA) blot using anti-GFP mAb confirmed the efficient precipitation of the substrate proteins (middle panels). RFP, red fluorescent protein; Phos., phosphorylated.
We addressed the question as to whether additional protein kinases are involved in lamina-modifying processes. Expression experiments with PKCα-GFP were performed to investigate whether PKC can induce morphological alteration of the nuclear lamina similarly to pUL97. In fact, the formation of distinct punctate distortions of nuclear lamin A/C was also detected in PKC-expressing cells (Fig. 1A, panels q–u). The inset magnification of the merged picture (Fig. 1A, panel u) shows PKCα-GFP mainly in a cytoplasmic localization adjacent to the regular nuclear rim shaped by lamin A/C. The nuclear rim is significantly broken so that no lamin signal remains detectable. Similar alterations were not detectable in control cells lacking PKCα-GFP expression (Fig. 1A, panel e).
Induction of Lamina-depleted Areas in HCMV-infected Primary Fibroblasts
We monitored the induction of nuclear lamina alterations (lamin A/C) during HCMV replication and the localization of pUL97. In HCMV-infected primary HFFs, pUL97 was found mostly associated with viral replication centers at late time points of infection (60–120 hpi) as described previously (38). With increasing time, a number of cells also contained pUL97 in a perinuclear localization, which was at least in part associated with the nuclear envelope (Fig. 1C, panel f). Pronounced distortions of nuclear lamin A/C were increasingly induced in the late phase of viral replication (Fig. 1C, panel h). When using an AD169-derived UL97 deletion virus expressing a GFP reporter (AD169ΔUL97-GFP), limited distortions of the nuclear lamina were observed (Fig. 1C, panels i–m). This type of distortion was characterized by a limited thinning of the nuclear lamina (lamin A/C) and by smaller depletions appearing with lower quantity compared with parental HCMV AD169. Quantification of the data revealed that most of the lamina-specific effect could be attributed to the presence of pUL97 (Fig. 1D). The lack of viral pUL97 expression (AD169ΔUL97-GFP) and/or the inhibition of kinase activity (Gö6976) substantially reduced the effect of nuclear lamina depletion. Thus, the measurable inhibitory potency of Gö6976 in cells infected with the UL97 deletion virus pointed to the additional importance of PKC for this phenotype (Fig. 1C, panel n).
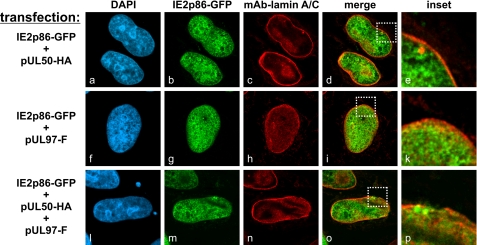
On the basis of the identified lamina-depleted areas, the question was addressed as to whether the physical stress of cells exposed to lamina-reorganizing protein complexes may cause a dysfunction of the nuclear membrane permeability. To monitor the localization of a large nuclear protein, a construct for the expression of IE2p86-GFP (∼115 kDa) was used for cotransfection experiments (Fig. 3). Interestingly, the massive morphological distortion of the nuclear rim did not lead to any detectable nucleocytoplasmic diffusion of this marker protein (Fig. 3, panels l–p). An invariable nuclear localization was noted for IE2p86-GFP, and no difference was found when comparing cells coexpressing IE2p86-GFP with those expressing pUL50 (Fig. 3, panel d), pUL97 (panel i), pUL50 plus pUL97 (panel o), or a vector control (data not shown).
FIGURE 3.
pUL97/pUL50-induced reorganization of the nuclear lamina does not lead to free diffusion of coexpressed IE2p86. HeLa cells were cotransfected with combinations of constructs coding for pUL97, pUL50, and IE2p86 as indicated. Cells were fixed at 12 h post-transfection, immunostained with anti-lamin A/C mAb, and analyzed for IE2p86-GFP fluorescence. DAPI, 4′,6-diamidino-2-phenylindole; HA, hemagglutinin.
Monitoring Nucleocytoplasmic Trafficking of Fluorescent Viral Capsids by Confocal Imaging
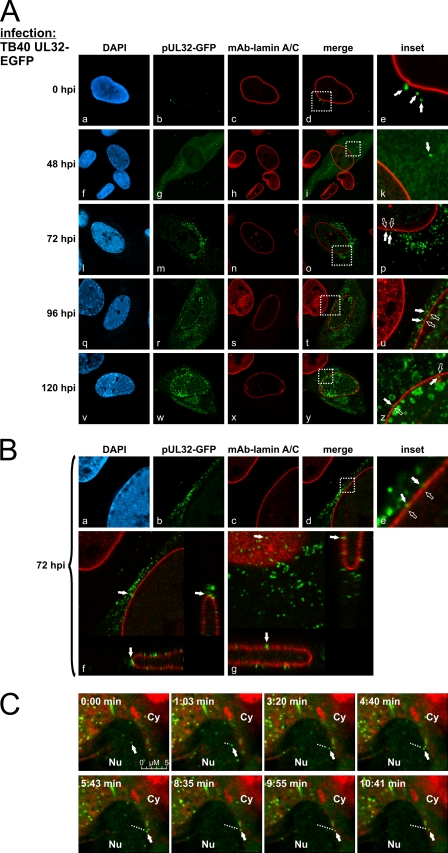
The phenomenon of nuclear lamina reorganization was investigated in more detail using HCMV TB40-UL32-EGFP, recombinantly expressing pUL32-EGFP (pp150-EGFP) as a reporter protein (Fig. 4, A–C). The reporter protein pUL32-EGFP is functional as a tegument protein and associates with viral capsids in the nucleus, so the trafficking of viral capsids from the nucleus to the cytoplasm can be visualized as demonstrated by Sinzger and co-workers (6). Entry of GFP-labeled particles was detectable directly after the experimental adsorption phase (0 hpi) (Fig. 4A, panels a–e). The replication kinetics analyzed from early to late time points indicated that newly synthesized pUL32-EGFP was detectable mostly in the cytoplasm in an even distribution (24–48 hpi) (Fig. 4A, panel g), whereas at later time points (starting at 72 hpi), the reporter protein appeared in a particle-associated, speckled pattern in both the nucleus and the cytoplasm (panels m, r, and w). Notably, lamina-depleted areas were induced starting at 72 hpi (Fig. 4A, panels n, s, and x). The accumulation of GFP-labeled viral particles was detectable close to rearranged sites of nuclear lamin A/C and occurred in a temporally coordinated fashion (Fig. 4A, panels p, u, and z). High magnifications of CLSM imaging revealed viral particles in close proximity to the nuclear lamina (Fig. 4B, panels a–e). Moreover, selected z stacks of those regions demonstrated viral particles passing through lamina-depleted areas (Fig. 4B, panels f and g). This finding was further illustrated by confocal live cell imaging in a time-lapse experiment started 64 h post-infection. Virus-producing cells were monitored over a period of 8 h. The use of direct live cell staining of the endoplasmic reticulum allowed the visualization of the trafficking of individual fluorescent viral particles from the internal area of the nucleus toward the nuclear envelope (Fig. 4C, see trafficking marked by dotted lines). As a striking result, the transition of particles through specific sites of the nuclear envelope could be detected (Fig. 4C, lower panels, representing selected frames of Movie 1). When examining the trafficking velocity of intranuclear viral particles, a mean of 0.3–0.4 μm/s was determined (mean of individual particles measured for 1.3–1.6 s over a distance of ∼500 nm). Slightly different velocities were noted for nuclear compared with cytoplasmic trafficking of particles (0.7–0.8 μm/s). The faster cytoplasmic movement was consistent with data published by others (6). Combined, the data presented in Fig. 4 suggest an important role of lamina-depleted areas during HCMV nuclear egress.
FIGURE 4.
Visualization of HCMV nuclear egress by tracking viral capsids by CLSM. HFFs were infected with HCMV TB40-UL32-EGFP. A and B, cells were fixed at various time points post-infection and immunostained with anti-lamin A/C mAb. A, monitoring of GFP-labeled capsids during the time course of infection. DAPI, 4′,6-diamidino-2-phenylindole. B, shown is the high resolution imaging of the nuclear egress of viral capsids (z stacks of two examples of HCMV-infected cells; panels f and g). Filled arrowheads indicate viral capsids; open arrowheads indicate lamina-depleted areas. C, shown is a time-lapse series of the movement of a GFP-labeled viral capsid at 3 days post-infection. At 62 hpi, live cell staining of the endoplasmic reticulum was performed (red). Filled arrowheads indicate the current position of the viral capsid; the dotted lines indicate the distance to the starting point of the viral capsid. Nu, nucleus; Cy, cytoplasm.
Prediction of Interaction Partners of Phosphorylated Lamins
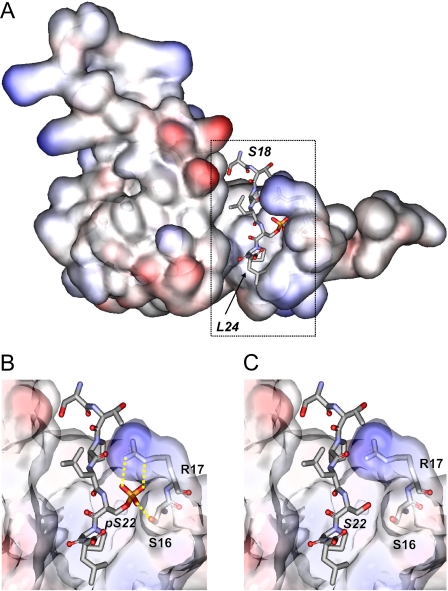
In the search for a possible molecular mechanism responsible for the induction of nuclear lamina reorganization, we performed bioinformatic analyses. A recent report showed that lamin A is phosphorylated at Ser22 by pUL97 (14). Ser22 is located in the N-terminal region of lamin A, which is predicted to adopt a non-globular structure. Such regions represent candidates for protein interactions via linear sequence motifs (34). A search in the ELM Database (33) for known interaction motifs revealed that the phosphorylated Ser22 residue is part of a phosphoserine-proline motif that is known to mediate interaction with a subset of WW domains (i.e. small structurally conserved adapter domains) (39). In general, WW domains differ considerably in their ligand-binding preferences: group I, II, and III WW domains recognize unphosphorylated proline-rich sequence motifs (e.g. PPXY, PPLP, and PPR), whereas group IV WW domains recognize phosphorylated serine and threonine residues (33, 39). In supplemental Fig. S2, the strategy for the identification of those WW domains that represent potential interaction partners of lamins is summarized; because the phosphorylation of lamins occurs in the nucleus, we first aimed to identify those nuclear proteins that contain WW domains and therefore represent putative binding partners of lamins. A keyword search in the UniProt Database yields a total of 4386 human nuclear proteins, of which 14 contain WW domains (supplemental Fig. S3). The alignment of the 25 WW domains present in these 14 proteins is shown in supplemental Fig. S4. In the next step, these WW domains were analyzed with respect to their ligand-binding properties. For four WW domains, experimental complex structures with peptide ligands exist (supplemental Fig. S5). Three of these WW domains (supplemental Fig. S5, panels a, b, and d) are group I or II WW domains, which recognize either unphosphorylated PPYP or PPLP ligand sequences. The crystal structure of the Pin1 WW domain in complex with a phosphorylated ligand reveals a different mode of recognition (supplemental Fig. S5, panel c). A key role for the recognition of the phosphorylated residue comes from an arginine (Arg17) of Pin1. This is consistent with the observation that a basic residue at the respective sequence position is a prerequisite for the recognition of phosphorylated ligands in this class of WW domains. Other types of WW domains with distinct ligand-binding properties frequently contain a sequence gap at the respective position (39). Taking these observations into account, Pin1 is the only nuclear protein that exhibits the sequence properties (supplemental Fig. S4) required for the binding of a phosphorylated serine residue. Pin1 is a nuclear PPIase that is known to be essential for cell cycle progression (40). In addition to Pin1, a closely related protein exists, the Pin1-like protein (PINL_HUMAN). However, this protein most likely represents a pseudogene and therefore was excluded from further analysis. Additional evidence for lamin binding by Pin1 comes from the striking similarity between lamin and known high affinity Pin1 ligands (35). One of these ligands, which originates from the C-terminal domain of RNA polymerase II, shares significant sequence similarities with the N terminus of lamin A (RNA polymerase II, SPTpSPS; and lamin A, TPLpSPT). This observation suggests that the phosphorylation of Ser22 in lamin A creates a Pin1-binding site. Modeling the respective Pin1-lamin A interaction complex (Fig. 5A) further substantiated this hypothesis. The model shows that the phosphoryl group of phospho-Ser22 in lamin A favorably interacts with a conserved serine (Ser16) and arginine (Arg17) of a Pin1 loop (Fig. 5B), whereas none of these interactions can be formed by the unphosphorylated Ser22 (Fig. 5C). Thus, the computational investigations suggested that phosphorylation of lamin A/C at Ser22 generates a Pin1-binding motif.
FIGURE 5.
A, model of the Pin1-lamin A interaction. The WW domain of Pin1 is shown in surface presentation and colored according to the electrostatic potential (red, most negative; blue, most positive). Ser18–Leu24 of lamin A, which comprise the binding site, are shown in stick presentation and colored according to the atom type. The boxed region is shown as an enlargement below. B, detailed view of the interactions formed by phosphorylated Ser22. The phosphoryl group forms tight interactions with a serine (Ser16) and arginine (Arg17) of Pin1 (yellow dotted lines). C, the respective interactions cannot be formed by unphosphorylated Ser22.
Interaction of Pin1 with Lamin A in HCMV-infected Cells
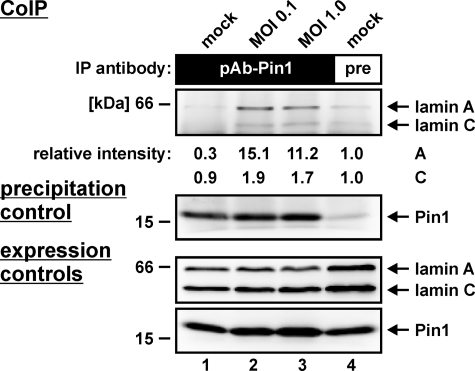
The question of a Pin1-lamin A interaction was addressed by co-immunoprecipitation analysis. HCMV-infected HFFs were harvested during the late phase of viral replication (72 hpi), and total cell lysates were used for co-immunoprecipitation with a Pin1-specific antibody. As demonstrated in Fig. 6 (upper panel, lanes 2 and 3), Pin1 was co-immunoprecipitated with lamin A in HCMV-infected cells at both low and high m.o.i. The signal in mock-infected cells remained below the detection limit (Fig. 6, lane 1). A control with preimmune serum illustrated the specificity of the reaction with only a marginal background (Fig. 6, lane 4). Relative intensities of the co-immunoprecipitation signals were quantified by densitometry, indicating a significant Pin1-specific co-immunoprecipitation of lamin A (15.1-fold/11.2-fold increase versus background), whereas lamin C appeared to associate with Pin1 at a much lower affinity (1.9-fold/1.7-fold). The additional staining patterns on precipitation and expression control Western blots confirmed the Pin1 specificity of the co-immunoprecipitation antibody and the reliable expression of all proteins (Fig. 6, lower panels as indicated). These data provide the first experimental evidence for the binding of Pin1 to lamin A in HCMV-infected fibroblasts.
FIGURE 6.
Interaction of Pin1 with lamin A in HCMV-infected cells. HFFs were infected with HCMV strain AD169 at m.o.i. = 0.1 and 1.0 or were left uninfected (mock). At 3 days post-infection, cells were lysed and used for co-immunoprecipitation (CoIP) analysis with anti-Pin1 pAb (A302-315A; lanes 1–3) or preimmune (pre) rabbit antiserum as a control (lane 4). Detection of co-immunoprecipitates (upper panels) and expression controls (lower panels) was performed on Western blots using anti-lamin A/C mAb or anti-Pin1 pAb (A302-315A), respectively. IP, immunoprecipitate.
Pin1 Is Partly Recruited to the Nuclear Envelope at Late Times of HCMV Replication and Represents a Putative Lamina-reorganizing Effector Molecule
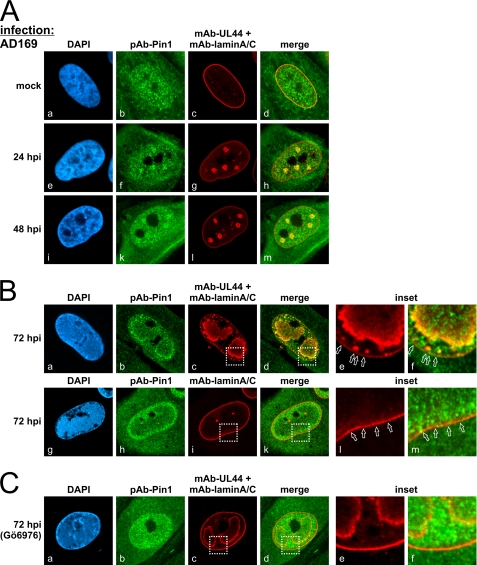
Next, we analyzed the intracellular localization of Pin1 by confocal imaging of HFFs infected with HCMV AD169. Co-staining with antibodies directed to lamin A/C and viral polymerase processivity factor pUL44 was performed to obtain markers for the localization of nuclear lamina and viral replication centers, respectively (Fig. 7). In uninfected cells (mock), Pin1 was predominantly localized diffusely throughout the nucleus, with a slight accumulation at the nuclear rim (Fig. 7A, panels a–d). At early time points of infection, starting with 24 hpi, the Pin1 distribution altered toward a distinct accumulation in viral replication centers (Fig. 7A, panels e–m). This was an unexpected finding, and its functional relevance remains to be elucidated in future studies. Even more striking was the observation that, during the late phase of viral replication (72 hpi), Pin1 was recruited not only to the periphery of viral replication centers (Fig. 7B, panels a–f) but also partly to specific depletion sites of nuclear lamin A/C (panels g–m). The specificity of the observed recruitment of Pin1 was additionally confirmed with a number of single-stained preparations to exclude cross-talk of the dyes used.
FIGURE 7.
Subnuclear relocalization of Pin1 in HCMV-infected cells. HFFs were infected with HCMV AD169 and fixed at early (A) and late (B) time points post-infection for immunostaining with anti-Pin1 pAb (H-123), anti-lamin A/C mAb, and anti-UL44 mAb as indicated. HCMV-infected HFFs were treated with 2 μm Gö6976 (C). Open arrowheads indicate lamina-depleted areas. Inset images show enlargements of representative areas of the nuclear lamina. DAPI, 4′,6-diamidino-2-phenylindole.
To assess whether the observed redistribution of Pin1 depends on pUL97 kinase activity, Pin1 localization was also visualized in infected cells treated with protein kinase inhibitors. Treatment with the pUL97 inhibitor Gö6976 considerably reduced the formation of lamina depletion sites and the accumulation of Pin1 at the nuclear lamina (Fig. 7C). In a control setting with the unrelated tyrosine kinase inhibitor AG490, no reduction of HCMV-induced Pin1 relocalization was noted (data not shown).
Further information about the HCMV-specific Pin1 relocalization was provided by the analysis of a second cell type (human astrocytoma cell line U373) known as a low permissive host for HCMV replication in vitro. Importantly, the phenomenon of Pin1 relocalization to nuclear replication centers and lamina-depleted areas, as described for HCMV-infected primary fibroblasts, could not be similarly detected in U373 cells. Only very poor alteration of intracellular Pin1 localization was observed during early and late phases of HCMV replication (data not shown). This might indicate a fibroblast-specific phenotype of Pin1 and might reflect one of the reasons why U373 cells produce lower amounts of infectious HCMV compared with HFFs. The rate-limiting efficiency of HCMV nuclear egress in U373 cells had been addressed by our earlier investigations (22). As far as the Pin1 relocalization in HFFs is concerned, a confirmation of the phenotype was obtained when different batches of these primary cells (i.e. derived from different recipients) were used for HCMV infection and produced identical results. Combined, these novel findings illustrate the role of Pin1 in HCMV replication and strongly support the prediction of a phosphorylation-triggered generation of a Pin1-binding site in lamin A/C. Thus, the cellular PPIase Pin1 is a candidate for exerting a so-far unrecognized mechanism of lamina reorganization in HCMV-infected fibroblasts.
DISCUSSION
Functionality of the HCMV-specific NEC Is Based on Viral and Cellular Components
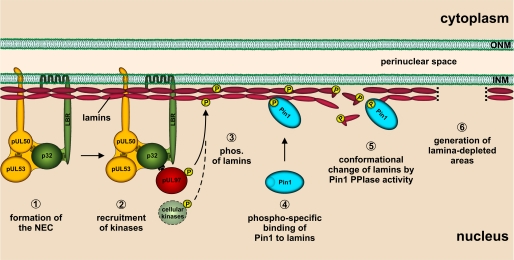
Here, we have provided novel experimental data for the reorganization of the nuclear lamina during the nuclear egress of HCMV. In particular, various forms of distortions of the nuclear lamina (lamin A/C) were detected in HCMV-infected primary fibroblasts, including the occurrence of punctate lamina-depleted areas. Based on data obtained from experiments with protein kinase inhibitors, these distortions of the nuclear lamina could be attributed mainly to the viral protein kinase pUL97 and cellular PKC. Using HCMV TB40-UL32-EGFP, we were able to visualize CMV particles approaching and traversing these lamina-depleted areas. Searching for the underlying effector mechanism, we combined molecular-virological analyses with bioinformatics and present a novel scenario of nuclear lamina reorganization (Fig. 8).
FIGURE 8.
Hypothetical model of the course of events triggering the formation of nuclear lamina-depleted areas in HCMV-infected cells. ONM, outer nuclear membrane; phos., phosphorylation.
NEC-associated Protein Kinases pUL97 and PKC Trigger Regulatory Events by Phosphorylating Nuclear Lamins
In previous work (19, 21), we demonstrated that the multicomponent NEC of HCMV-infected cells contains at least two essential egress proteins, pUL50 and pUL53 (Fig. 8, step 1). In addition, a number of regulatory proteins and protein kinases are also associated, such as pUL97, PKC, p32, and the lamin B receptor (18, 19, 21, 22, 41, 42). The NEC-mediated recruitment of protein kinases (Fig. 8, step 2) appears to be essential for hyperphosphorylation of nuclear lamins and lamina-associated proteins (18, 21). Previously, we described the phosphorylation of endogenous lamins A/C and B by pUL97 (22). Hamirally et al. (14) demonstrated the direct pUL97-mediated phosphorylation of lamin A by the use of purified proteins. In this study, we described the direct phosphorylation of transiently expressed versions of lamins A and C by pUL97. The pUL97-mediated phosphorylation of lamins could be correlated to the lamina-reorganizing potential of pUL97. This was concluded from the finding that catalytically active pUL97 was sufficient to induce morphological alterations of the nuclear lamina, whereas catalytically inactive pUL97 was not. In addition, cellular PKCα was also found to be involved in lamin phosphorylation and in HCMV-induced distortion of the nuclear lamina. Supporting this, in herpes simplex virus-1-infected cells, PKCα and PKCδ are very efficiently recruited to the nuclear rim (43). Notably, pro-apoptotic PKCδ is able to phosphorylate lamin B in vitro, which is a prerequisite for efficient nuclear lamina disassembly during apoptosis (44). This might play an additional role in the context of herpesvirus replication. Thus, several isoforms of PKC appear to be involved in the nuclear egress of herpesviruses.
Site-specific Lamin Phosphorylation Can Generate a Binding Motif for the Putative Downstream Effector Pin1
An important hallmark of lamin phosphorylation in HCMV-infected cells is the site-specific phosphorylation of lamin A/C (Ser22) by pUL97 (Fig. 8, step 3) (14). Notably, Ser22 is also a phosphorylation site of CDK1 (45). The CDK1 phosphorylation sites of the lamin types are part of conserved sequence stretches. For lamin B, the respective phosphorylated residues are Ser23 (lamin B1) and Ser17 (lamin B2) (45). Lamins A and C are subject to alternative splicing of the LMNA gene and share the same N-terminal sequence, including the CDK1/pUL97 phosphorylation site. In this study, we have provided the first evidence that phosphorylation of Ser22 of lamin A/C generates a sequence pattern (TPLpSPT) that fulfills the minimal binding motif for the PPIase Pin1 (pSP) and, moreover, shares similarities with the respective sequence stretches of known ligands of Pin1. It was highly encouraging to detect a Pin1-lamin A interaction and a relocalization of Pin1 in HCMV-infected fibroblasts that appeared to be dependent on protein kinase activity (pUL97 and/or PKC). This conclusion was based on the finding that the protein kinase inhibitor Gö6976 strongly reduced Pin1 relocalization. Thus, we infer that Pin1 may be part of a phosphorylation-triggered regulation pathway in HCMV-infected cells (Fig. 8, step 4).
Pin1 Recruitment May Locally Provide Increased PPIase Activity to Execute a Reorganization of the Nuclear Lamina
Our data describe the early recruitment of Pin1 to viral replication centers and a later partial recruitment to the nuclear envelope, suggesting a novel scenario for the mechanism of nuclear lamina distortion. This mechanism might be explained by a Pin1-mediated modulation of the lamin head domain, possibly resulting in a decreased potency to form head-to-tail polymers (Fig. 8, step 5). PPIases such as Pin1 catalyze the cis/trans-isomerization of peptidyl-prolyl peptide bonds, which mediates conformational changes of proteins leading to altered protein folding. Pin1-catalyzed switches regulate a spectrum of target activities, including decreases and increases in protein stability or transcriptional activity, targeting of the subcellular protein localization, and many more (46–48). An alternative indirect mechanism may involve so far unknown effector proteins that bind lamins as a consequence of Pin1-induced conformational changes. Such effector binding could eventually provide the stimulus for another molecular mechanism leading to disassembly of the nuclear lamina.
Lamina-depleted Areas May Represent Preferred Sites of HCMV Nuclear Capsid Egress
In HCMV-infected fibroblasts, we identified by CLSM analysis very characteristic punctate distortions of nuclear lamin A/C. The pivotal role of viral protein kinase pUL97 in this process was illustrated by the finding that infection with a HCMV UL97 deletion mutant induced only a limited degree of nuclear lamina reorganization. It should be mentioned that previous studies already reported effects of nuclear lamina reorganization during HCMV replication (14, 22, 41), which remained, however, poorly characterized so far. Our present study describes for the first time the HCMV-specific induction of distinct lamina-depleted areas (Fig. 8, step 6). This observation is strongly reminiscent of the infoldings of the INM free of lamina described by Buser et al. (7) by transmission electron microscopy. These infoldings were postulated as preferred sites where HCMV and murine CMV capsids acquire their primary envelope by budding into the perinuclear space. Our CLSM data on the morphological alteration of the nuclear lamina in HCMV-infected cells, including live cell imaging of freely moving viral particles, strongly support this report. In summary, our data suggest that the combined activities of cellular and CMV protein kinases trigger the formation of lamina-depleted areas required for viral nuclear egress on the basis of a novel Pin1-driven mechanism. Further investigation will be necessary to gain a deeper insight into the specific molecular steps regulating this mechanism. Pin1 recruitment might be a conserved mode of phosphorylation-triggered reorganization of the nuclear lamina during the replication of HCMV and other herpesviruses.
Supplementary Material
Acknowledgments
We thank Christian Sinzger for kindly providing the HCMV reporter virus TB40-UL32-EGFP; Thomas Stamminger, Nina Tavalai, and co-workers (Institute for Clinical and Molecular Virology, University of Erlangen-Nuremberg) for providing plasmid pHM990 and for continuous fruitful cooperation; Sabine Rechter for scientific discussion and support; Jos Broers for lamin A/C-GFP-encoding plasmids; and Hendrik Coldenstrodt-Ronge (Merton College, Oxford, UK) for reading the manuscript.
This work was supported by Deutsche Forschungsgemeinschaft Grants SFB 796 (C3 and A2), MA 1289/4-1, and MA 1289/6-1 and by the Argus Foundation (Berlin).

The on-line version of this article (available at http://www.jbc.org) contains Figs. S1–S5 and Movie 1.
- HCMV
- human cytomegalovirus
- INM
- inner nuclear membrane
- PKC
- protein kinase C
- NEC
- nuclear egress complex
- PPIase
- peptidyl-prolyl cis/trans-isomerase
- EGFP
- enhanced green fluorescent protein
- HFFs
- human foreskin fibroblasts
- m.o.i.
- multiplicity of infection
- CLSM
- confocal laser scanning microscopy
- pAb
- polyclonal antibody
- mAb
- monoclonal antibody
- hpi
- hours post-infection.
REFERENCES
- 1.Mocarski E. S., Shenk T., Pass R. F. (2007) in Fields Virology (Knipe D. M., Howley P. M. eds) pp. 2701–2772, Lippincott/Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2.Vancíková Z., Dvorák P. (2001) Curr. Drug Targets Immune Endocr. Metab. Disord. 1, 179–187 [PubMed] [Google Scholar]
- 3.Drew W. L. (1992) Clin. Microbiol. Rev. 5, 204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mettenleiter T. C., Klupp B. G., Granzow H. (2009) Virus Res. 143, 222–234 [DOI] [PubMed] [Google Scholar]
- 5.Sanchez V., Spector D. H. (2002) Science 297, 778–779 [DOI] [PubMed] [Google Scholar]
- 6.Sampaio K. L., Cavignac Y., Stierhof Y. D., Sinzger C. (2005) J. Virol. 79, 2754–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buser C., Walther P., Mertens T., Michel D. (2007) J. Virol. 81, 3042–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldman R. D., Gruenbaum Y., Moir R. D., Shumaker D. K., Spann T. P. (2002) Genes Dev. 16, 533–547 [DOI] [PubMed] [Google Scholar]
- 9.Gruenbaum Y., Margalit A., Goldman R. D., Shumaker D. K., Wilson K. L. (2005) Nat. Rev. Mol. Cell Bio. 6, 21–31 [DOI] [PubMed] [Google Scholar]
- 10.Heald R., McKeon F. (1990) Cell 61, 579–589 [DOI] [PubMed] [Google Scholar]
- 11.Ward G. E., Kirschner M. W. (1990) Cell 61, 561–577 [DOI] [PubMed] [Google Scholar]
- 12.Peter M., Nakagawa J., Dorée M., Labbé J. C., Nigg E. A. (1990) Cell 61, 591–602 [DOI] [PubMed] [Google Scholar]
- 13.Likhacheva E. V., Bogachev S. S. (2001) Membr. Cell Biol. 14, 565–577 [PubMed] [Google Scholar]
- 14.Hamirally S., Kamil J. P., Ndassa-Colday Y. M., Lin A. J., Jahng W. J., Baek M. C., Noton S., Silva L. A., Simpson-Holley M., Knipe D. M., Golan D. E., Marto J. A., Coen D. M. (2009) PLoS Pathog. 5, e1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peter M., Heitlinger E., Häner M., Aebi U., Nigg E. A. (1991) EMBO J. 10, 1535–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maul G. G., Negorev D. (2008) Med. Microbiol. Immunol. 197, 241–249 [DOI] [PubMed] [Google Scholar]
- 17.Bain M., Sinclair J. (2007) Rev. Med. Virol. 17, 423–434 [DOI] [PubMed] [Google Scholar]
- 18.Muranyi W., Haas J., Wagner M., Krohne G., Koszinowski U. H. (2002) Science 297, 854–857 [DOI] [PubMed] [Google Scholar]
- 19.Milbradt J., Auerochs S., Marschall M. (2007) J. Gen. Virol. 88, 2642–2650 [DOI] [PubMed] [Google Scholar]
- 20.Thompson L. J., Fields A. P. (1996) J. Biol. Chem. 271, 15045–15053 [DOI] [PubMed] [Google Scholar]
- 21.Milbradt J., Auerochs S., Sticht H., Marschall M. (2009) J. Gen. Virol. 90, 579–590 [DOI] [PubMed] [Google Scholar]
- 22.Marschall M., Marzi A., aus dem Siepen P., Jochmann R., Kalmer M., Auerochs S., Lischka P., Leis M., Stamminger T. (2005) J. Biol. Chem. 280, 33357–33367 [DOI] [PubMed] [Google Scholar]
- 23.Wolf D. G., Courcelle C. T., Prichard M. N., Mocarski E. S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1895–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krosky P. M., Baek M. C., Coen D. M. (2003) J. Virol. 77, 905–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mettenleiter T. C. (2004) Virus Res. 106, 167–180 [DOI] [PubMed] [Google Scholar]
- 26.Mettenleiter T. C. (2006) Vet. Microbiol. 113, 163–169 [DOI] [PubMed] [Google Scholar]
- 27.Thomas M., Rechter S., Milbradt J., Auerochs S., Müller R., Stamminger T., Marschall M. (2009) J. Gen. Virol. 90, 567–578 [DOI] [PubMed] [Google Scholar]
- 28.Tavalai N., Papior P., Rechter S., Leis M., Stamminger T. (2006) J. Virol. 80, 8006–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broers J. L., Kuijpers H. J., Ostlund C., Worman H. J., Endert J., Ramaekers F. C. (2005) Exp. Cell Res. 304, 582–592 [DOI] [PubMed] [Google Scholar]
- 30.Marschall M., Stein-Gerlach M., Freitag M., Kupfer R., van den Bogaard M., Stamminger T. (2001) J. Gen. Virol. 82, 1439–1450 [DOI] [PubMed] [Google Scholar]
- 31.Meydan N., Grunberger T., Dadi H., Shahar M., Arpaia E., Lapidot Z., Leeder J. S., Freedman M., Cohen A., Gazit A., Levitzki A., Roifman C. M. (1996) Nature 379, 645–648 [DOI] [PubMed] [Google Scholar]
- 32.Michel D., Pavić I., Zimmermann A., Haupt E., Wunderlich K., Heuschmid M., Mertens T. (1996) J. Virol. 70, 6340–6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puntervoll P., Linding R., Gemünd C., Chabanis-Davidson S., Mattingsdal M., Cameron S., Martin D. M., Ausiello G., Brannetti B., Costantini A., Ferrè F., Maselli V., Via A., Cesareni G., Diella F., Superti-Furga G., Wyrwicz L., Ramu C., McGuigan C., Gudavalli R., Letunic I., Bork P., Rychlewski L., Küster B., Helmer-Citterich M., Hunter W. N., Aasland R., Gibson T. J. (2003) Nucleic Acids Res. 31, 3625–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dinkel H., Sticht H. (2007) Bioinformatics 23, 3297–3303 [DOI] [PubMed] [Google Scholar]
- 35.Verdecia M. A., Bowman M. E., Lu K. P., Hunter T., Noel J. P. (2000) Nat. Struct. Biol. 7, 639–643 [DOI] [PubMed] [Google Scholar]
- 36.Marschall M., Stein-Gerlach M., Freitag M., Kupfer R., van den Bogaard M., Stamminger T. (2002) J. Gen. Virol. 83, 1013–1023 [DOI] [PubMed] [Google Scholar]
- 37.Goekjian P. G., Jirousek M. R. (1999) Curr. Med. Chem. 6, 877–903 [PubMed] [Google Scholar]
- 38.Marschall M., Freitag M., Suchy P., Romaker D., Kupfer R., Hanke M., Stamminger T. (2003) Virology 311, 60–71 [DOI] [PubMed] [Google Scholar]
- 39.Kato Y., Ito M., Kawai K., Nagata K., Tanokura M. (2002) J. Biol. Chem. 277, 10173–10177 [DOI] [PubMed] [Google Scholar]
- 40.Sudol M., Sliwa K., Russo T. (2001) FEBS Lett. 490, 190–195 [DOI] [PubMed] [Google Scholar]
- 41.Camozzi D., Pignatelli S., Valvo C., Lattanti G., Capanni C., Dal Monte P., Landini M. P. (2008) J. Gen. Virol. 89, 731–740 [DOI] [PubMed] [Google Scholar]
- 42.Lötzerich M., Ruzsics Z., Koszinowski U. H. (2006) J. Virol. 80, 73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park R., Baines J. D. (2006) J. Virol. 80, 494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cross T., Griffiths G., Deacon E., Sallis R., Gough M., Watters D., Lord J. M. (2000) Oncogene 19, 2331–2337 [DOI] [PubMed] [Google Scholar]
- 45.Isobe K., Gohara R., Ueda T., Takasaki Y., Ando S. (2007) Biosci. Biotechnol. Biochem. 71, 1252–1259 [DOI] [PubMed] [Google Scholar]
- 46.Lippens G., Landrieu I., Smet C. (2007) FEBS J. 274, 5211–5222 [DOI] [PubMed] [Google Scholar]
- 47.Lu K. P., Finn G., Lee T. H., Nicholson L. K. (2007) Nat. Chem. Biol. 3, 619–629 [DOI] [PubMed] [Google Scholar]
- 48.Lu K. P., Zhou X. Z. (2007) Nat. Rev. Mol. Cell Biol. 8, 904–916 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.