Abstract
Reversible ubiquitination is essential for the endocytic sorting and down-regulation of G protein-coupled receptors, such as the chemokine receptor CXCR4. The deubiquitinating enzyme AMSH has been implicated in the endocytic sorting of both G protein-coupled receptors and receptor-tyrosine kinases. Herein, we examine the role of AMSH in the regulation of CXCR4 stability and trafficking and characterize protein-protein interactions critical for this function. Loss of AMSH catalytic activity or depletion by RNAi results in increased steady-state levels of CXCR4 under basal conditions. Analysis of truncation and point mutation of AMSH reveal the importance of an RXXK motif for CXCR4 degradation. The RXXK motif of AMSH interacts with the SH3 domains of the STAM and Grb2 families of adaptor proteins with high affinity. Cells expressing a catalytically inactive mutant of AMSH show basal hyperubiquitination, but not increased degradation, of the ESCRT-0 components STAM1 and Hrs. This is dependent on the RXXK motif of AMSH. Ubiquitination of endocytic machinery modulates their activity, suggesting that AMSH may directly regulate endocytic adaptor protein function. This is reflected in CXCR4 trafficking and provides a mechanism by which AMSH specifies the fate of endocytosed receptors. Taken together, these studies implicate AMSH as a key modulator of receptor fate determination through its action on components of the endocytic machinery.
Keywords: Proteases/Ubiquitin, Protein/Sorting, Receptors/Chemokine, Receptors/Endocytosis, Stem Cells, Subcellular Organelles/Endosomes, Trafficking, Deubiquitination
Introduction
The sorting and trafficking of cell surface receptors through endosomal compartments is a highly regulated process that is essential for maintaining cellular homeostasis and generating adaptive and coordinated responses to external stimuli. To avoid prolonged receptor activation and signaling, receptor-ligand complexes are endocytosed and either recycled back to the plasma membrane or sorted to lysosomes for degradation (1–4). This process is mediated by reversible ubiquitination. The modification of a target protein with ubiquitin moieties is extensively utilized to regulate the assembly of endosomal machinery as well as being a sorting signal for transmembrane proteins within the endosomal system (5–7). The crucial role for ubiquitination in the trafficking of endocytosed receptors has been documented for receptor-tyrosine kinases and a number of G protein-coupled receptors including the chemokine receptor CXCR4 (8–10). CXCR4 has been extensively studied as a co-receptor for the entry of T-trophic human immunodeficiency virus into CD4+ T-cells (11, 12). Along with its cognate ligand, Stromal cell-derived factor-1α (SDF-1/CXCL12), CXCR4 plays important roles in hematopoiesis, development, and organization of the immune system and stem cell homing (13–15). CXCR4 deregulation is associated with various pathologies, including human immunodeficiency virus infection, cardiovascular disease, and neurodegenerative diseases and is associated with metastatic disease in a number of cancers (16–20). Mutations that result in truncation of the C-terminal intracellular region of CXCR4 cause WHIM syndrome, a dominantly inherited immunodeficiency disease involving hypogammaglobulinemia and abnormal susceptibility to warts (19, 21). WHIM mutants of CXCR4 fail to undergo regulated internalization and are not effectively stabilized on the plasma membrane (22).
Upon stimulation with CXCL12, CXCR4 is rapidly internalized in a process requiring both ubiquitin and clathrin (23–26). CXCR4 has been reported to become monoubiquitinated by the HECT-family E3 ligase, atrophin-interacting protein 4 (AIP4/Itch),3 in a CXCL12-dependent manner, and is deubiquitinated by USP14 (27). These studies suggest that reversible ubiquitination is a key mechanism for regulating the trafficking and stimulation-induced degradation of CXCR4. CXCR4 is also known to be constitutively endocytosed in a ligand-independent manner (28, 29). Although CXCL12 mediated degradation of CXCR4 may be essential for signal attenuation, basal CXCR4 turnover was recently found to control steady-state abundance (28, 29).
Deubiquitinating enzymes mediate receptor-tyrosine kinase and G protein-coupled receptor homeostasis and degradation (30–36). The deubiquitinating enzyme, known as associated molecule with the SH3 domain of STAM (AMSH), has been implicated in the regulation of epidermal growth factor receptor, calcium-sensing receptor, and most recently, the δ-opioid receptor and the protease-activated receptor 2 (37–43). AMSH was first identified as an interacting partner of the SH3 domain of STAM (44) and later characterized as a Zn2+-dependent ubiquitin isopeptidase with a substrate preference for Lys-63-linked polyubiquitin chains (37, 45). AMSH is composed of multiple protein-protein interaction domains that have been shown to control its recruitment and localization (Fig. 3A). At the N terminus, AMSH contains a nuclear localization signal and a microtubule-interacting and transport (MIT) domain, shown to interact with several chromatin-modifying proteins (CHMPs) that are components of the endosomal sorting complex required for transport (ESCRT) III complex (38, 39, 44, 46–49). Mutations in AMSH that inhibit binding to CHMP3 result in the accumulation of ubiquitinated cargo on endosomes and inhibition of epidermal growth factor receptor degradation (38, 39). AMSH has a clathrin binding domain that interacts with the terminal domain of the clathrin heavy chain and is required for AMSH localization to endosomes (45, 50). AMSH contains a STAM-interacting motif (PPVVDRSLKPGA) that serves as a binding site for the SH3 domain of the STAM family of adaptor proteins (37, 44, 51, 52). STAM SH3 domain binding to this region has been shown to stimulate the deubiquitinating activity of AMSH in vitro (45, 52); however, a clear role for this interaction in a cellular context had not been previously demonstrated.
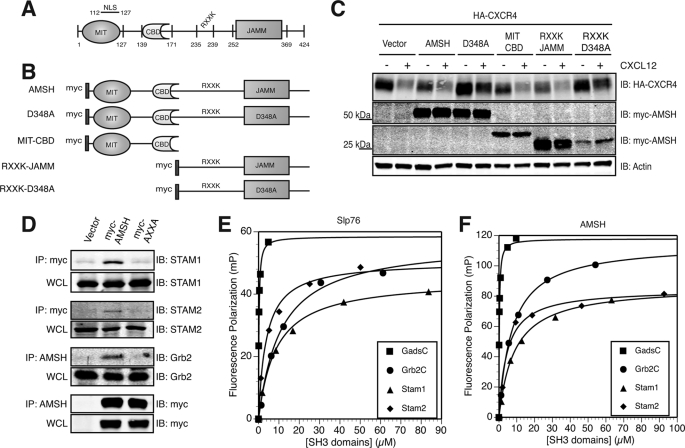
FIGURE 3.
The RXXK motif of AMSH interacts with a subset of SH3 domains and modulates CXCR4 stability. A, a schematic of domain organization of murine AMSH (GenBankTM accession no. BC025111). Amino acid boundaries of individual domains and motifs are as indicated; RXXK, conserved RXXK peptide sequence which interacts with SH3 domain. NLS, nuclear localization signal. B, shown are constructs of AMSH and mutants used in this study; D348A, catalytic domain mutant, MIT-CBD (amino acids 1- 187), RXXK-JAMM (amino acids 201–424), RXXK-D348A (amino acids 201–424). C, the RXXK motif of AMSH is required for AMSH-mediated regulation of CXCR4 stability. HEK293 cells co-transfected with HA-tagged CXCR4 and vector, wild type AMSH, D348A, MIT-CBD, RXXK-JAMM, or RXXK-D348A were treated with cycloheximide and vehicle (−) or CXCL12 (+) for 3 h. IB, immunoblot. D, the RXXK motif is essential for AMSH interaction with the adaptor proteins STAM1, STAM2, and Grb2. HeLa cells transiently expressing vector, wild type AMSH, or AXXA mutant were immunoprecipitated (IP) from Myc or AMSH (as indicated) to determine whether RXXK mutation to AXXA would alter the ability of AMSH to interact with endogenous binding partners. Corresponding input whole cell lysates (WCL) are shown. E and F, the RXXK motif of AMSH binds to Gads, Grb2, STAM1, and STAM2 SH3 domains with high affinity. Dissociation constants for the interactions between fluorescein-tagged peptides Slp76 (fluorescein-tagged APSIDRSTKPA) bound with affinities as shown to Gads-C (210 nm), STAM1 (8.6 μm), STAM2 (3.8 μm), Grb2-C (11 μm) (E), and AMSH (fluorescein-tagged PPVVDRSLKPG) bound with affinities as shown to Gads-C (240 nm), STAM1 (11 μm), STAM2 (4.8 μm), and Grb2-C (7 μm) (F). GST-SH3 domains were measured using fluorescence polarization (FP). Measurements (millipolarization units (mP)) were taken using Beacon 2000, and curve fitting was performed using DeltaGraph 5.7.5.
The substrate preferences of AMSH for Lys-63-linked polyubiquitin chains taken together with its known interaction partners implicate AMSH in the regulation of the ubiquitination status of proteins during endocytic trafficking. AMSH has been implicated in the endocytic trafficking of various receptors (37–42); however, a role for AMSH in the trafficking of CXCR4 has not been previously described. Endosomal sorting and degradation of CXCR4 are dependent on the adaptor protein hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs), arrestin-2, and on the AAA-type ATPase Vps4 (24, 25). Loss of both Hrs and arrestin-2 results in the accumulation of CXCR4 on enlarged early endosomes after activation with CXCL12. Vps4 has been implicated in the later-stage deubiquitination of CXCR4 and Hrs through what is assumed to be an indirect mechanism (24, 25).
Herein, we report a critical role for AMSH in the regulation of basal CXCR4 trafficking and steady-state homeostasis. We report that both AMSH depletion and overexpression of a catalytically inactive form of AMSH results in increased levels of CXCR4 protein. We identify conserved interactions between the RXXK motif in AMSH and the non-canonical SH3 domains of adaptor and scaffold proteins, including those of the STAM family with affinity approximating that of an optimal ligand. Recruitment of AMSH to the Hrs·STAM complex on sorting endosomes requires an intact RXXK, motif and our results suggest that AMSH functions at the sorting endosome to regulate the ubiquitination status of ESCRT-0 proteins. Because ubiquitination of endocytic machinery modulates their activity, we propose that AMSH acts to modulate ESCRT-0 function and subsequently affects the fate of endocytosed cargo as reflected in CXCR4 trafficking. Taken together, our studies on CXCR4 trafficking implicate AMSH as a key modulator of receptor fate through its action on the components of the endocytic machinery.
EXPERIMENTAL PROCEDURES
Cell Lines, cDNA Constructs, Antibodies, and RNAi
HEK293 (human embryonic kidney) and HeLa (human carcinoma) cells were grown in complete media consisting of Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum (SAFC Biosciences, Lenexa, KS), 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) in 5% CO2, 95% air at 37 °C. Stably transfected HeLa cells expressing HA-tagged CXCR4 were generated and selected by resistance for neomycin using G418 (Geneticin, Invitrogen) followed by fluorescence-activated cell sorting using a mouse anti-HA (F-7) antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The mammalian expression construct of murine AMSH was amplified by PCR and cloned into the pcDNA3.1(+) (Invitrogen) vector system at KpnI/ApaI. An N-terminal Myc epitope tag was inserted at the KpnI restriction site using annealed primer pairs. Point mutations at the catalytic aspartate residue and/or the RXXK recruitment motif were introduced by site-directed mutagenesis (QuikChange, Stratagene, La Jolla, CA). AMSH truncation mutants were cloned by PCR into the pcDNA3.1(+) vector system at BamHI/EcoRI. Glutathione S-transferase (GST) fusion constructs of SH3 domains from STAM1, STAM2, Grb2, and Gads were cloned into the pGEX-2TK vector at BamHI/EcoRI by Heather Schwartz (The University of Chicago, Chicago, IL). HA-tagged ubiquitin was provided by Suzanne D. Conzen (The University of Chicago, Chicago, IL). Cloning vectors for the bimolecular fluorescence complementation (BiFC) assay, pBiFC-VN173 and pBiFC-VC155 (53, 54), were provided by Chang-Deng Hu (Purdue University, West Lafayette, IN). AMSH and AXXA were cloned by PCR into pBiFC-VN173 at EcoRV/XbaI. STAM1 and Grb2 were cloned by PCR into pBiFC-VC155 at EcoRI/XhoI. The following primary antibodies were used for Western blotting, immunoprecipitation, and immunofluorescence microscopy: mouse anti-HA (Covance, Berkeley, CA), mouse anti-Myc 9E10 (Cell Signaling Technology, Inc., Danvers, MA), rabbit anti-STAM1 (Calbiochem (San Diego, CA) and Santa Cruz Biotechnology, Inc.), rabbit anti-STAM2 (Calbiochem), mouse anti-Grb2 (Cell Signaling Technology, Inc.), mouse anti-actin (Sigma), mouse anti-EEA1 (BD Biosciences), mouse anti-CD63 polyclonal (Developmental Studies Hybridoma Bank, Iowa City, IA), and rat anti-CXCR4 (BD Biosciences). Rabbit polyclonal Hrs anti-sera were generated against a GST fusion protein of murine Hrs (BC003239) amino acid residues 216–289. Rabbit polyclonal AMSH anti-sera were generated against the peptide MSDHGDVSLPPEDRV (amino acids 1–15). The following secondary antibodies were used: anti-mouse 800 (LI-COR, Lincoln, NE) anti-rabbit 680 (LI-COR), anti-mouse Alexa Fluor 488 (Invitrogen), anti-mouse Alexa Fluor 647 (Invitrogen), anti-rabbit Alexa Fluor 568 (Invitrogen), and anti-rat Alexa Fluor 488 (Invitrogen). The control siRNA oligo and the siRNA oligo targeting AMSH (UUACAAAUCUGCUGUCAUUUU) were purchased from Dharmacon Inc., Chicago, IL. The shRNA control in the pLKO.1 lentiviral vector was purchased from Sigma. To stably suppress AMSH, we used pLKO.1 shRNA constructs generated by The RNAi Consortium (TRC) (55). A pool of five different shRNA sequences (TRC0000073973–77) was purchased from Open Biosystems, Huntsville, AL. Lentiviral infections were performed as described (55). The shRNA, TRCN0000073975, provided the optimum knockdown of AMSH and was used for CXCR4 degradation experiments. CXCL12 was purchased from PeproTech (Rocky Hill, NJ).
Degradation Assay
HEK293 cells were seeded onto 100-mm tissue culture dishes and allowed to adhere overnight. Cells were co-transfected with HA-CXCR4 and Myc-AMSH or mutant AMSH using Lipofectamine 2000 (Invitrogen). 48 h post-transfection, cells were treated with vehicle (Dulbecco's modified Eagle's medium + 0.5% fetal bovine serum) or CXCL12 (25 nm) in the presence of 50 μg/ml cycloheximide (Sigma) for 3 h. Samples labeled 0′ represent cells not treated with cycloheximide or CXCL12. Cells were directly lysed in SDS sample buffer, and receptor levels were detected by immunoblot. For RNA interference experiments, HeLa cells were co-transfected with HA-CXCR4 and control siRNA or siRNA directed against AMSH at 50 nm concentration using Lipofectamine 2000 in the absence of serum. HeLa cells stably expressing HA-CXCR4 and shControl or shAMSH were lysed in SDS sample buffer, and receptor levels were detected by immunoblot.
Surface Biotinylation
For CXCR4 surface biotinylation experiments, HeLa cells were seeded onto 100-mm tissue culture dishes and allowed to adhere overnight. Cells were transiently co-transfected with HA-CXCR4 and control siRNA or siRNA-directed against AMSH using Lipofectamine 2000. 48 h post-transfection, cells were washed three times with ice-cold PBS. NHS-Biotin (Pierce) was dissolved in DMSO and added to HBSS on ice (Mediatech, Inc. Manassas, VA) for a final concentration of 2 mm NHS-Biotin. Cells were surface-biotinylated with the NHS-Biotin/HBSS mixture for 30 min at 4 °C and then washed 3 times in PBS containing 100 mm glycine to quench the reaction. Biotinylated cells were either lysed immediately or incubated at 37 °C for 3 h in the presence or absence of CXCL12. After treatment, cells were washed with ice-cold PBS and lysed on ice in 900 μl of surface biotinylation lysis buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 5% glycerol, 10 mm NaF, 5 mm EDTA, 1 mm CaCl2, 10 mm Na2VO4, protease inhibitors mixture (Roche Applied Science). Lysates were sonicated and clarified at 10,000 rpm for 15 min at 4 °C. Lysates were incubated with 50 μl of NeutrAvidin-agarose resin (Pierce) for 3 h at 4 °C to precipitate biotinylated material. Washed beads were eluted with SDS sample buffer, subjected to 10% SDS-PAGE, transferred onto nitrocellulose membrane, and immunoblotted with mouse anti-HA antibody to detect HA-CXCR4.
GST Pulldown Assays and Co-immunoprecipitation
Escherichia coli strain BL21 were transformed with GST fusion constructs and grown overnight at 37 °C in LB media. Cultures were diluted 1:50 and grown at 37 °C to an optical density (λ = 600 nm) of 0.6 and then induced with 1 mm isopropyl-1-thio-B-d-galactopyranoside (Denville Scientific, Inc., Metuchen, NJ.) for 3 h at 30 °C. The cells were harvested by centrifugation, resuspended in cell lysis buffer (50 mm Hepes, pH 7.5, 150 mm NaCl, 10% glycerol, 1% Triton X-100, 1.5 mm MgCl2, 1 mm EDTA, 100 mm NaF), and sonicated on ice. Lysates were clarified at 10,000 rpm for 15 min at 4 °C followed by incubation with GST fusion glutathione-Sepharose 4B beads (Thermo Fisher Scientific, Waltham, MA) for 4 h at 4 °C and washed with PBS. For AMSH binding experiments, 11 μg of GST fusion proteins were incubated with HeLa lysates transfected with AMSH or mutants. Reactions were incubated for 4 h at 4 °C. Washed beads were eluted with SDS sample buffer followed by Western blot with mouse anti-Myc antibody to detect AMSH binding. For co-immunoprecipitation of AMSH or AXXA with the endogenous adaptor proteins, STAM1, STAM2, Grb2, and Gads, HeLa cells were transfected with Myc-tagged AMSH constructs and incubated in complete media for 24 h. After incubation, cells were washed with ice-cold PBS and lysed in immunoprecipitation buffer (50 mm Tris-HCl, pH 7.5, 150 mm HCl, 0.5% Triton X-100, 10 mm NaF, 1 mm Na2VO4, 1 mm phenylmethylsulfonyl fluoride, protease inhibitors mixture). Cell lysates were clarified at 10,000 rpm for 15 min at 4 °C. Equal amounts of lysate were incubated with 3 μg of anti-Myc antibody overnight at 4 °C followed by incubation with protein G beads (Roche Applied Science) for 3 h at 4 °C. Washed beads were eluted with SDS sample buffer, separated by 10% SDS-PAGE, and visualized by immunoblotting with rabbit anti-STAM1 (Calbiochem), rabbit anti-STAM2, or rabbit anti-Grb2 antibodies to detect endogenous adaptor proteins.
Ubiquitination Assays
HeLa cells were seeded onto 100-mm tissue culture dishes and allowed to adhere overnight. Cells were co-transfected with HA-tagged ubiquitin and either vector, AMSH, or AMSH mutants using Lipofectamine 2000. 24 h post-transfection, cells were washed with ice-cold PBS and lysed on ice in 900 μl of immunoprecipitation buffer (as above). Equal amounts of lysate were incubated with 3 μg of rabbit anti-STAM1 (Santa Cruz) or rabbit anti-Hrs (see above) overnight at 4 °C. Protein G beads (Roche Applied Science) were added to lysates and further incubated for 3 h at 4 °C. Washed beads were eluted in SDS sample buffer, subjected to 10% SDS-PAGE, transferred onto nitrocellulose membranes, and immunoblotted with mouse anti-HA antibody (Covance) to detect ubiquitinated protein.
Fluorescence Polarization
The AMSH peptide, PPVVDRSLKPG (amino acids 230–240), was synthesized by Fmoc (N-(9-fluorenyl)methoxycarbonyl) solid-phase peptide synthesis and HPLC-purified. Fluorescein-labeled probes were prepared through the reaction of C-terminal peptides with 5-(and-6)-carboxyfluorescein succinimidyl ester (Molecular Probes, Invitrogen), purified by reverse-phase HPLC, and confirmed by mass spectrometry. The Slp-76 peptide, FL-APSIDRSTKPA (amino acids 232–242), was as previously described (56). Equilibrium dissociation constants (KD) were determined by fluorescence polarization using a Beacon 2000 Fluorescence Polarization System (Invitrogen). Binding studies were conducted at 22 °C using 5 nm fluorescein-labeled peptides dissolved in PBS containing 100 μg/ml bovine serum albumin and 1 mm dithiothreitol. Reaction mixtures were allowed to equilibrate for 10 min at room temperature before each measurement. Data fitting and dissociation constant determination was carried out as previously described (56).
Bimolecular Fluorescence Complementation and Immunofluorescence Microscopy
The bimolecular fluorescence complementation (BiFC) assay was used to study protein-protein interactions in living cells (53, 54). HeLa cells were seeded onto polylysine-coated glass coverslips (Corning) in 6-well plates and allowed to adhere overnight. Cells co-transfected with pBiFC-VN173-AMSH and pBiFC-VC155-STAM1 or pBiFC-VC155-Grb2 were incubated at 37 °C to allow adequate development of venus fluorescent protein (VFP) fluorescence (6–8 h) followed by fixation and permeabilization with methanol:acetone (1:1). Cells were immunostained with mouse anti-HA, rabbit anti-FLAG, rabbit anti-Hrs, mouse anti-EEA1 monoclonal, and mouse anti-CD63 monoclonal antibodies. Colocalization of CXCR4 with AMSH and endosomal markers was visualized using HeLa cells transiently co-transfected with HA-CXCR4 and AMSH or mutants. HeLa cells were seeded onto polylysine-coated glass coverslips in 6-well plates and allowed to adhere overnight. 24 h post-transfection with HA-CXCR4 and AMSH, cells were fixed and permeabilized with methanol:acetone (1:1). Cells were immunostained with rat anti-CXCR4, mouse anti-Myc, and rabbit anti-STAM1 (Santa Cruz) antibodies. Colocalization of AMSH or STAM1 with HA-ubiquitin was performed with transiently co-transfected cells expressing HA-ubiquitin and AMSH or mutants as before. In this experiment cells were fixed and permeabilized with methanol:acetone (1:1) followed by immunostaining with mouse anti-HA, rabbit anti-STAM1 (Santa Cruz), and rabbit anti-AMSH antibodies. Specimens were imaged by confocal fluorescence microscopy using a Leica TCS SP2 AOBS spectral microscope. The acquired images were analyzed using Image J software (National Institute of Health) (57, 58). The extent of colocalization in single cells was determined using the ImageJ-based JACoP (Just Another Colocalization Plugin) software (57, 58). Background was subtracted from each image before setting thresholds for each channel. We calculated Mander's coefficients for the overlap of green on red (tM1) and red on green (tM2). Only the tM1 is displayed in the figures. This coefficient ranges from 0 (indicating that no pixels within the threshold colocalize) to 1 (indicating that 100% of pixels colocalize). All quantitative image analysis was conducted in a blinded fashion. A minimum of 15 cells were analyzed for each experiment (biological replicate). In each experiment n represents the number of biological replicates. In total, 290 confocal paired (two-channel) images were collected and submitted electronically in a blinded fashion for analysis by the second author.
Statistical Analysis
Quantitative data were averaged across multiple independent experiments, with the number of experiments specified in the corresponding figure legend. The error bars represent the S.E. of the mean from multiple independent experiments. The statistical significance of the indicated differences was analyzed by Student's t test, with the relative significance specified by calculated p values listed in the figure legends.
RESULTS
AMSH Modulates CXCR4 Turnover
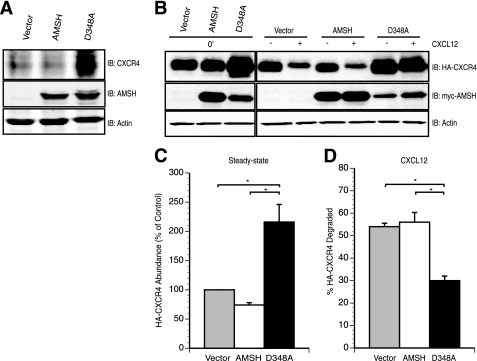
To ascertain whether AMSH plays a role in CXCR4 turnover and degradation, we examined the effects of a catalytically inactive mutant of AMSH on basal and agonist-promoted degradation of CXCR4. AMSH is a JAMM-domain isopeptidase, and aspartate 348 is part of the catalytic mechanism within the JAMM domain (37). Mutation of aspartate to alanine at residue 348 (D348A) results in a catalytically inactive protein (37). HeLa cells expressing high endogenous levels of CXCR4 were transiently transfected with vector control, wild type AMSH, or the D348A mutant. Overexpression of D348A resulted in increased basal levels of endogenous CXCR4 (Fig. 1A), suggesting a role for AMSH activity in the basal turnover of CXCR4. We next examined the role of AMSH in CXCR4 degradation using HEK293 or HeLa cells, established systems for the study of CXCR4 trafficking (23). Expression of D348A resulted in a 2-fold increase of CXCR4 in HEK293 cells compared with cells transfected with vector or wild type AMSH (Fig. 1, B and C). Furthermore, overexpression of D348A resulted in a significant reduction in agonist-induced degradation of HA-CXCR4 (Fig. 1, B and D). Although overexpression of D348A resulted in a decrease in agonist-induced degradation, CXCR4 degradation was still observed. These results suggest that AMSH catalytic activity plays a role in mediating CXCR4 steady-state levels.
FIGURE 1.
AMSH activity modulates CXCR4 turnover. A, overexpression of a catalytically inactive AMSH led to accumulation of endogenous CXCR4 under steady-state conditions. Whole cell lysates derived from HeLa cells transiently expressing pcDNA3.1 vector (vector), wild type AMSH, or D348A, a catalytic point mutant of AMSH, were immunoblotted (IB) against endogenous CXCR4, AMSH, and β-actin proteins as indicated. B–D, AMSH regulates both basal turnover and ligand-mediated degradation of CXCR4. B, HEK293 cells transiently co-transfected with HA-tagged CXCR4 (HA-CXCR4) and either vector, wild type AMSH, or D348A were treated with vehicle alone (0′) or in the presence of cycloheximide with (+) or without (−) CXCL12 for 3 h. C, shown is a graphical representation of HA-CXCR4 protein levels relative to the control cells. D, percent HA-CXCR4 degraded after a 3-h stimulation with CXCL12 relative to treatment with cycloheximide alone. Quantification of HA-CXCR4 levels by immunoblot was performed using LiCOR Odyssey infrared imager and software. Significance was assessed using Student's t test with the asterisks corresponding to p < 0.05, n = 3. All graphs were generated using Delta graph 5.7.5 software.
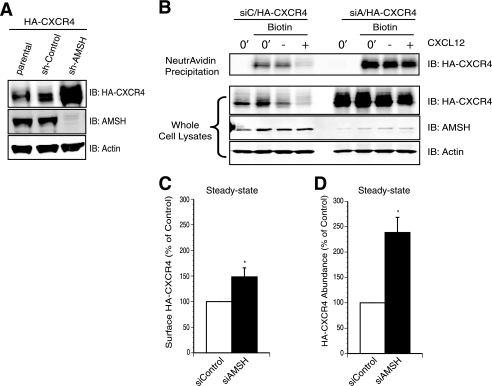
To confirm the role of AMSH in mediating CXCR4 stability, endogenous AMSH was depleted by RNA interference. HeLa cells stably expressing HA-CXCR4 were lentivirally transduced with shRNA targeting AMSH. Total HA-CXCR4 protein levels were greater in AMSH-depleted cells (Fig. 2A). To specifically follow the fate of the mature surface receptor, HeLa cells transiently co-transfected with HA-CXCR4 and siRNA targeting AMSH or non-targeting control were labeled by surface biotinylation, and degradation of cell-surface receptors was evaluated by NeutrAvidin affinity purification and Western blot. The protein levels of HA-CXCR4 were quantified under basal conditions and after stimulation. Cell surface biotinylation demonstrated a modest increase in CXCR4 protein at the cell surface compared with total protein levels in AMSH depleted cells (Fig. 2, B–D). These results suggest that the majority of the additional CXCR4 is intracellular. Furthermore, CXCR4 was still degraded in AMSH-depleted cells after stimulation with CXCL12, suggesting that the major role for AMSH is in the basal turnover of CXCR4.
FIGURE 2.
AMSH depletion results in increased abundance and intracellular accumulation of CXCR4. A, stable ablation of AMSH led to HA-CXCR4 accumulation under steady-state conditions. Whole cell lysates derived from HeLa cells stably expressing HA-CXCR4 and a lentiviral shRNA targeting either AMSH (sh-AMSH) or a non-targeting control shRNA (sh-Control) were immunoblotted (IB) against HA-CXCR4, AMSH, and β-actin proteins as indicated. B, AMSH regulated CXCR4 trafficking to the cell surface. Surface biotinylation of HeLa cells transiently express HA-CXCR4 and oligo-induced RNAi against AMSH (siAMSH/siA) or non-targeting control (siControl/siC). Cells were treated with vehicle alone (0′) or in the presence of cycloheximide with (+) or without (−) CXCL12 for 3 h. Biotinylated material was precipitated with NeutrAvidin-agarose resin, and HA-CXCR4 was followed by an immunoblot. Graphical representations of surface (C) and total cellular HA-CXCR4 abundance (D) are shown; asterisks correspond to p < 0.05, n = 3.
AMSH is composed of multiple domains that have been shown to play key roles in its recruitment and localization (Fig. 3A) (37–39, 44–48, 50, 51). To determine which domains or motifs within AMSH are essential for CXCR4 turnover, we tested AMSH mutants on CXCR4 stability. Fig. 3B shows the AMSH constructs used for overexpression experiments. Truncation mutants of AMSH were co-transfected along with HA-CXCR4 to determine which domains of AMSH are necessary for CXCR4 degradation. The truncation mutants, MIT-CBD and RXXK-JAMM, did not affect CXCR4 stability (Fig. 3C). However, RXXK-D348A showed an increased stability of CXCR4 under basal and stimulation conditions (Fig. 3C). These data implicate a key role for the RXXK motif in mediating CXCR4 down-regulation.
The RXXK Motif of AMSH Binds to a Subset of SH3 Domains with High Affinity
In a mammalian library screen of binding targets for non-canonical SH3 domains, we have previously identified a consensus PX(V/I)(D/N)RXXKP (RXXK) motif within AMSH (56). These SH3 domains have been identified as the C-terminal SH3 domains of the Grb2 family of adaptor proteins (denoted Grb2-C, Gads-C, and Grap-C) as well as those found in the endocytic adaptor proteins STAM1 and STAM2. AMSH has previously been shown to co-immunoprecipitate with overexpressed STAM1 and Grb2 (37, 44, 45, 51, 52), an interaction that depends on an intact STAM binding motif within AMSH (37, 44, 45, 51, 52). We have further characterized the SH3 domain binding site of AMSH by identifying the necessary residues for optimal binding and determining the binding affinities between AMSH and this subset of SH3 domains. To define the critical residues in the SH3 domain binding site of AMSH, peptide arrays were employed (59). The peptide corresponding to the SH3 domain binding site of AMSH was used as a template in which each residue in the sequence was substituted with each of the 20 natural amino acids. The resulting peptide libraries were probed with the GST-SH3 domains of STAM1 (Fig. supplemental S1A), Grb2-C, and Gads-C (data not shown). Although other studies have implicated proline 239 as important for binding (45), our results confirm an essential role for arginine 235 and lysine 238 while indicating a more minor role for prolines 231 and 239. Furthermore, the STAM1 SH3 domain did not accept prolines at positions −1 and +1 relative to the conserved arginine, indicating that embedding a conventional PXXP ligand within the context of the AMSH peptide is in fact highly disfavored for binding to the STAM1 SH3 domain. This is consistent with previous structural studies indicating that this region forms a 310 helix (60, 61).
The interactions between AMSH and the SH3 domains of the STAM and Grb2 families of adaptor proteins were confirmed using native proteins and revealed varying affinities and specificity for binding (Fig. 3, D–F, and supplemental Fig. S1B). To confirm the requirement of the arginine and lysine in the RXXK motif in the binding of AMSH to adaptor SH3 domains, co-precipitation experiments were performed using GST fusion proteins corresponding to the SH3 domains of STAM1, STAM2, Grb2-C, and Gads-C. Purified SH3 domains bound to glutathione-Sepharose beads were presented with cell lysates from HeLa cells transfected with vector, Myc-tagged AMSH, AXXK, KXXK, RXXA, or AXXA. Precipitation of AMSH and AMSH mutants was assayed by Western blot (supplemental Fig. S1B). The SH3 domains of STAM1 and STAM2 as well as the C-terminal SH3 domains of Grb2 and Gads were found to bind efficiently to wild type AMSH containing an intact RXXK motif. We also tested binding to the N-terminal SH3 domains of Gads and Grb2, which have been shown to bind conventional PXXP ligands. The N-terminal SH3 domain of Gads or Grb2 failed to bind to AMSH (data not shown). AMSH binding to the SH3 domains was not observed when arginine 235 was mutated to either alanine or lysine in most cases, although weak precipitation of the C-terminal SH3 domain of Gads was observed. Mutation of lysine 238 to alanine or mutation of arginine 235 and lysine 238 to alanines severely compromised binding to all of the SH3 domains tested (supplemental Fig. S1B). Results described above demonstrate the necessity of an intact RXXK motif of AMSH in supporting interactions with the non-canonical RXXK binding SH3 domains of the STAM and Grb2 families of proteins.
To detect interactions between full-length AMSH and endogenous adaptor proteins, Myc-tagged AMSH or the RXXK mutant, AXXA, was expressed in HeLa cells and immunoprecipitated, and co-immunoprecipitated endogenous STAM1, STAM2, and Grb2 were detected by Western blot. Each of the three adaptor proteins was successfully immunoprecipitated by AMSH. However, no interaction was observed when the RXXK motif was mutated to AXXA (Fig. 3D).
To determine binding affinities of the AMSH RXXK motif for the SH3 domains involved in this study, equilibrium dissociation constants were determined by fluorescence polarization for the interaction between a fluorescein-labeled synthetic peptide corresponding to the RXXK motif of AMSH (FL-PPVVDRSLKPG) and the SH3 domains of STAM1, STAM2, Grb2-C, and Gads-C (Fig. 3, E and F). The AMSH peptide was found to bind with high affinity to the Gads-C (Kd of 240 nm), STAM 1 (Kd of 11 μm), STAM2 (Kd of 4.8 μm), and Grb2-C (Kd of 7 μm). In our experiments, the Slp-76 peptide was found to bind with high affinity to the Gads-C (Kd of 210 nm), STAM1 (Kd of 8.6 μm), STAM2 (Kd of 3.8 μm), and Grb2-C (Kd of 11 μm). Incubation of a GST protein alone with the AMSH and Slp-76 peptides produced no discernible binding (data not shown). In both affinity and residue requirement, the motif in AMSH is almost identical to the Slp-76 RXXK motif that has been described as a near optimal binding motif for the SH3 domains under study (56). Our results confirm that AMSH, like Slp-76, preferentially binds a subset of SH3 domains that recognizes RXXK-containing ligands assuming a 310 helical conformation with similar affinities.
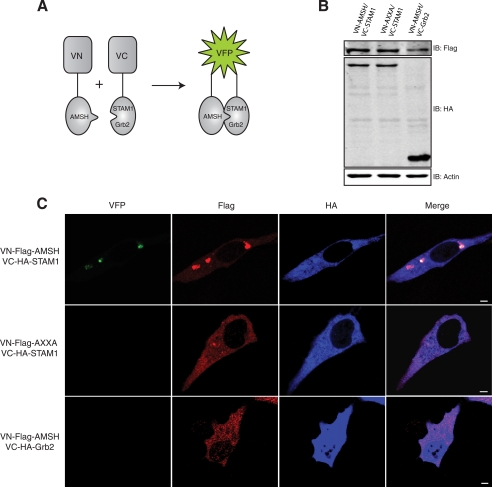
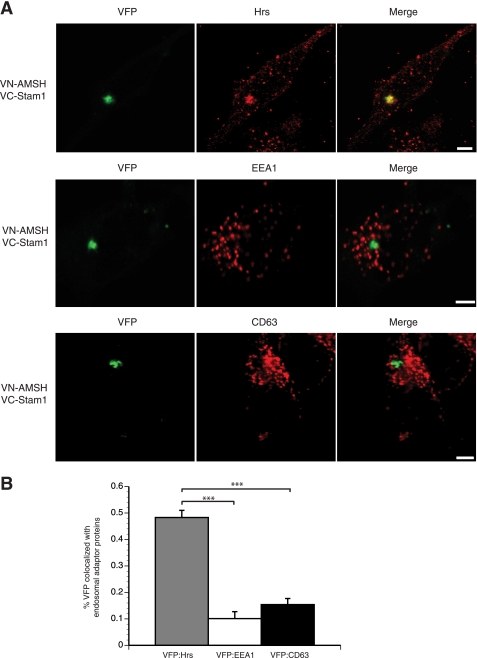
To determine whether AMSH forms a complex with our subset of SH3 domains on endocytic compartments, we employed the BiFC assay (53) to study the subcellular localization of the AMSH·STAM·Grb2 complex. In this assay, two fragments of the VFP are fused to putative interaction partners. The association of the two interaction partners allows formation of a BiFC complex, resulting in fluorescence. For BiFC analysis of the AMSH·STAM1 or AMSH·Grb2 complexes, AMSH was fused to the N-terminal fragment of VFP (VN-FLAG-AMSH), and STAM1 or Grb2 was fused to the C-terminal fragment of VFP (VC-HA-STAM1 or VC-HA-Grb2). Fig. 4A illustrates the BiFC technique and constructs used. HA and FLAG immunoblotting and immunofluorescent staining confirmed the expression of the VN and VC fusion proteins (Fig. 4, B and C). AMSH wild type, but not the AXXA mutant, forms a BiFC complex with STAM1 (Fig. 4C, top and middle panels). AMSH failed to form a BiFC complex with Grb2 (Fig. 4C, bottom panel), suggesting that this complex does not readily form in vivo. As the sorting endosome represents an intermediate between early and late endocytic compartments, the complex between AMSH and STAM was expected to reside between the early and late endosomes. Indeed, the AMSH·STAM1 complex colocalized with Hrs, a critical component of the sorting endosome (Fig. 5A, top panel). However, there was no significant overlap between AMSH·STAM1 BiFC and the early endosomal marker EEA1 or the multivesicular body marker CD63 (Fig. 5A, middle and bottom panels). Quantitative analysis revealed that colocalization of the AMSH·STAM1 BiFC complex with Hrs was significantly greater than that observed with both EEA1 and CD63 (Fig. 5B). Taken together, these results demonstrate that the RXXK motif is essential for formation of the AMSH·STAM complex on Hrs positive endosomes. Hrs and STAM form the ESCRT-0 complex on sorting endosomes. Thus, the AMSH·STAM complex appears to reside on sorting endosomes that represent an intermediate stage in the endocytic pathway.
FIGURE 4.
Subcellular localization of AMSH with STAM1 requires the RXXK/SH3 interaction. A, shown is a schematic representation of the BiFC assay. VN and VC represent the N-terminal and C-terminal fragments of the VFP, respectively, and are not singularly fluorescent. When the two fragments colocalize (green), fluorescence is observed. B and C, AMSH interacts directly with STAM1 via the RXXK motif but not with Grb2. B, HeLa cells transiently co-transfected with VN-AMSH/VC-STAM1 or VN-AXXA/VC-STAM1 or VN-AMSH/VC-Grb2 cells were lysed, and protein levels were detected by immunoblot (IB). C, cells were fixed and immunostained for FLAG (red) and HA (blue) to detect expression of VN-AMSH or VC-STAM1 and of VC-Grb2, respectively. Representative images are shown with scale bars corresponding to 5 μm.
FIGURE 5.
The AMSH·STAM1 interaction occurs on sorting endosomes. Direct interactions between AMSH and STAM1 occur at the Hrs-positive/sorting endosome and not at the early (EEA1-positive) or late (CD63-positive) endosome. A, HeLa cells co-transfected with VN-AMSH and VC-STAM1 were fixed and immunostained for Hrs, EEA1, or CD63 (red). Scale bars correspond to 5 μm. B, shown is a graphic representation of colocalization between VFP and endosomal proteins, where 0 indicates no overlap, and 1 indicates complete overlap; ***, p < 0.001 n = 3.
AMSH Mediates CXCR4 Trafficking and Degradation through Ubiquitination of ESCRT-0
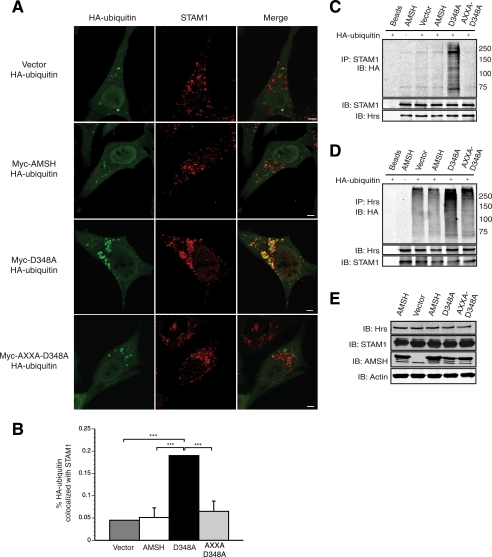
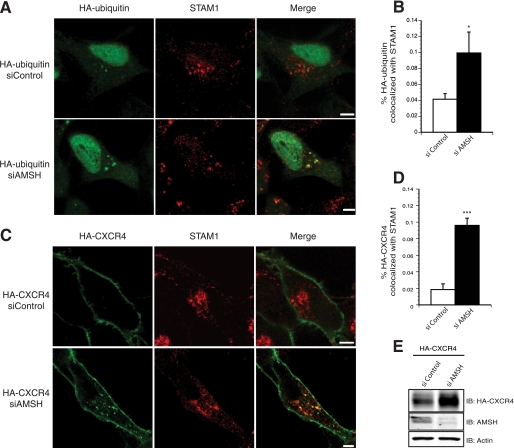
Our data suggest that AMSH mediates the basal turnover of CXCR4 that is dependent on its recruitment to early endosomes by the ESCRT-0 protein STAM. Hrs, the other component of ESCRT-0, has been shown to become ubiquitinated by the E3 ubiquitin ligase AIP4 in response to CXCR4 activation (24). The importance of ubiquitination in specifying function of endosomal proteins is well documented (35, 62, 63). Previous reports have indicated that loss of AMSH catalytic activity incurs hyperubiquitination of overexpressed STAM (45). To determine whether AMSH-mediated deubiquitination is involved in regulating ubiquitination of ESCRT-0, we tested the effects of AMSH and its mutants on steady-state ubiquitination and cellular abundance of Hrs and STAM1. Expression of D348A resulted in the accumulation of HA-ubiquitin on STAM1 positive endosomes (Fig. 6A, third panel). The accumulation of ubiquitin on enlarged endosomal structures was not observed in control or AMSH wild type-transfected cells (Fig. 6A, first and second panels). To assess the importance of AMSH recruitment by STAM in mediating ESCRT-0 ubiquitination, we mutated the arginine and lysine residues to alanine in the RXXK motif within the D348A mutant. This mutant failed to hyperubiquitinate STAM1-positive endosomes (Fig. 6A, fourth panel). Quantitative analysis revealed a significant increase of ubiquitin on STAM1 positive endosomes in D348A mutant cells (Fig. 6B). The ubiquitination status of STAM1 or Hrs was also assessed by immunoprecipitating endogenous STAM1 or Hrs from cell lysates co-expressing HA-ubiquitin and either vector, AMSH, D348A, or AXXA-D348A. Overexpression of wild type AMSH did not appear to significantly alter the prevalence of mono- or polyubiquitinated STAM1 or Hrs (Fig. 6, C and D). Conversely, overexpression of the D348A mutant resulted in hyperubiquitination of both STAM1 and Hrs (Fig. 6, C and D). Similar to the results in Fig. 6A, mutation of the RXXK motif of the D348A mutant reverses the hyperubiquitination signature of Hrs and STAM1 (Fig. 6, C and D). AMSH also binds to the SH3 domain of Grb2; therefore, we wanted to determine whether AMSH also mediates the ubiquitination status of Grb2. Although loss of AMSH catalytic activity results in the hyperubiquitination of ESCRT-0, it does not affect the ubiquitination status of Grb2 (supplemental Fig. S2) further implicating a specific role for the deubiquitinating activity of AMSH. Previous studies have shown a similar effect on overexpressed STAM ubiquitination (45, 49); however, our data show a different pattern of ubiquitination, which appears as a smear rather than a single band running at a higher molecular mass. This is an important distinction as the type of ubiquitination on ESCRT-0 adaptor proteins may play a role in protein-protein interactions and localization in the endocytic pathway. Specifically, STAM and Hrs can themselves become ubiquitinated, and this is important for their localization and functional activity (33, 64). An increase in HA-ubiquitin on STAM1-positive endosomes was also observed in cells depleted of AMSH (Fig. 7, A and B). AMSH knockdown results in a less drastic hyperubiquitination of ESCRT-0 than does expression of the D348A mutant (supplemental Fig. S3A). Neither D348A overexpression nor AMSH depletion resulted in a decrease in the cellular abundance of either Hrs or STAM1 (Fig. 6E and supplemental Fig. S3B). It has been shown previously that overexpression of D348A results in the redistribution of AMSH to endosomes (45) and accumulation of ubiquitinated cargo on both early and late endosomes (37, 38). Furthermore, redistribution to endosomes is dependent on an intact RXXK motif (45). To confirm the role of AMSH in mediating CXCR4 trafficking to sorting endosomes, we depleted endogenous AMSH using RNAi and measured CXCR4 colocalization on STAM1 positive endosomes. AMSH depletion leads to the accumulation of CXCR4 on STAM1-positive endosomes, suggesting that proper sorting of CXCR4 is dependent on AMSH (Fig. 7, C and D).
FIGURE 6.
AMSH regulates ESCRT-0 ubiquitination in a manner dependent upon the RXXK motif. The RXXK motif of AMSH is required in the spatial organization of AMSH to endosomes to deubiquitinate target protein cargo. A, HeLa cells co-transfected with HA-ubiquitin and vector, AMSH, D348A, or AXXA-D348A were fixed and immunostained with anti-STAM1 (red) and anti-HA (green). Representative images are shown with scale bars corresponding to 5 μm. B, quantification of colocalization between HA-ubiquitin and STAM1 from experiments in A; ***, p < 0.001, n = 2. C and D, the RXXK motif of AMSH is required for regulation of ubiquitin status of ESCRT-0 components. HeLa cells co-transfected with HA-ubiquitin and vector, AMSH, D348A, or AXXA-D348A were immunoprecipitated (IP) for endogenous proteins. STAM1 (C) and Hrs (D) and their ubiquitination status was assessed by immunoblot (IB). E, AMSH does not affect the stability of Hrs or STAM1. Corresponding whole cell lysates were analyzed by immunoblot against endogenous Hrs, STAM1, AMSH, and β-actin proteins.
FIGURE 7.
AMSH regulates ubiquitin dynamics and trafficking of CXCR4 on STAM-positive endosomes. AMSH ablation results in accumulation of ubiquitinated cargo and CXCR4 on endosomes marked by endogenous STAM1 protein. HeLa cells transfected with HA-ubiquitin (A) or HA-CXCR4 (C) in combination with either siControl or siAMSH were fixed and immunostained for HA (green) and STAM1 (red). Representative images are shown with scale bars corresponding to 5 μm. Graphic representations of colocalization between STAM1 and HA-ubiquitin (B) or HA-CXCR4 (D)are shown; *, p < 0.05, ***, p < 0.001, n = 2. E, whole cell lysates corresponding to C were immunoblotted (IB) against HA-CXCR4, AMSH, and β-actin as indicated.
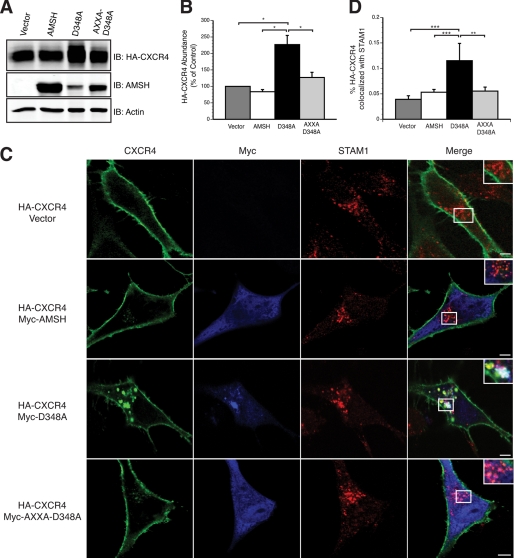
To assess the role of AMSH recruitment by STAM in the regulation of CXCR4 turnover and degradation, the effect of mutating this motif on D348A mutant overexpression was tested. Overexpression of wild type AMSH (Fig. 8A) or AMSH lacking an intact RXXK motif (data not shown) had no effect on CXCR4 levels. Conversely, overexpression of the D348A mutant resulted in a significant increase of basal CXCR4. Overexpression of the AXXA-D348A mutant failed to affect CXCR4 stability (Fig. 8, A and B). These results suggest that the RXXK motif is important for the recruitment of AMSH and subsequent catalytic activity. Previous studies have shown that CXCR4 is constitutively internalized and trafficked to early and recycling endosomes (28). Because an increase in CXCR4 abundance is seen in unstimulated cells where AMSH function has been perturbed, we examined the steady-state distribution of CXCR4 in cells overexpressing wild type or D348A using immunofluorescence microscopy. In the absence of stimulation, CXCR4 predominantly localizes to the plasma membrane in cells overexpressing vector alone or wild type AMSH. However, in D348A cells, CXCR4 colocalized with STAM1 on enlarged endosomes as well as to the plasma membrane (Fig. 8C). Upon stimulation, CXCR4 concentrates in enlarged endosomal structures in D348A-expressing cells (data not shown). The AXXA-D348A mutant lacking an intact RXXK motif failed to result in the accumulation of CXCR4 on enlarged endosomes (Fig. 8C). Quantitative analysis revealed a significant increase in CXCR4 colocalization with STAM1-positive endosomes in D348A cells (Fig. 8D). Our results suggest that AMSH recruitment via the RXXK motif is an important mediator of CXCR4 trafficking. Taken together, our results demonstrate that AMSH recruitment by STAM1 to early/sorting endosomes is an important mechanism in the basal turnover of CXCR4.
FIGURE 8.
AMSH recruitment via the RXXK motif mediates steady-state CXCR4 trafficking. AMSH mediated trafficking of CXCR4 through Stam1-positive endosomes requires the RXXK motif of AMSH and the direct interaction of that motif with STAM. A, HEK293 cells co-transfected with HA-CXCR4 and vector, AMSH, D348A, or AXXA-D348A and immunoblotted (IB) against HA-CXCR4, AMSH, and β-actin as indicated. B, shown is quantification of steady-state HA-CXCR4 protein abundance relative to the vector control for the corresponding immunoblot shown in panel A; *, p < 0.05, n = 3. C, HeLa cells transiently coexpressing HA-CXCR4 (green) and vector, AMSH, D348A, or AXXA-D348A (blue) were fixed and immunostained for endogenous STAM1 (red). Scale bars correspond to 5 μm. D, shown is a graphic representation of colocalization between CXCR4 and STAM1; ***, p < 0.001. **, p < 0.01, n = 2.
DISCUSSION
Reversible ubiquitination has been proposed as a key mechanism in regulating the trafficking of endocytic cargo (8, 32–35, 64), yet relatively little is known regarding the deubiquitinating enzymes involved or the mechanisms governing this process. The current study provides further evidence for a global regulatory role of AMSH in the endocytosis of surface receptors through modulation of endocytic adaptor proteins and describes a specific role for AMSH in the stability and trafficking of the chemokine receptor CXCR4.
We report that loss of AMSH catalytic activity by overexpression of a catalytic mutant results in a dramatic increase in CXCR4 basal levels and a decrease in CXCL12-mediated degradation (Fig. 1). AMSH depletion by RNAi-mediated knockdown also results in an increase in steady-state levels of CXCR4 (Fig. 2), further supporting a role for AMSH deubiquitinating activity in the regulation of receptor turnover. This is analogous to the general effect of AMSH on epidermal growth factor receptor, the δ-opioid receptor, and the protease-activated receptor 2 (37–39, 42). Although CXCR4 trafficking and degradation after CXCL12 activation has been the focus of numerous studies, less is known regarding the constitutive or basal endocytosis of CXCR4. CXCR4 is known to be constitutively endocytosed and recycled to the plasma membrane in a ligand-independent manner (28). This process is clathrin-dependent, with CXCR4 colocalizing with markers for early and recycling endosomes (28). Constitutive endocytosis and recycling of CXCR4 are important mechanisms for maintaining homeostatic control of cell surface levels to sense ligand. It is crucial for a cell to be able to rapidly up- and down-regulate cell surface levels of receptor in response to changing signals. Our results demonstrate that AMSH plays a role in modulating CXCR4 steady-state levels and that AMSH catalytic activity is critical for this regulatory mechanism.
A number of studies suggest that AMSH may be recruited to the endocytic pathway via multiple mechanisms. Specifically, AMSH can interact with clathrin through its clathrin binding domain (Fig. 3A), resulting in localization of AMSH to clathrin-coated pits and early endosomes (45, 50). Furthermore, AMSH binds to the SH3 domain of STAM1 through an integral STAM binding motif enabling the recruitment of AMSH to early/sorting endosomes (Fig. 5) (37, 44, 45, 51, 52). In addition to the recruitment of AMSH to early endocytic compartments, AMSH may be recruited to the multivesicular body via interactions with the ESCRT-III protein CHMP3 (38, 39, 44, 46–48). The integrity of these protein-protein interactions is presumably crucial for the spatial and temporal localization of AMSH to different functional compartments within the endocytic pathway. To determine which of these interactions influence the endocytosis and turnover of CXCR4, we examined several truncation mutants of AMSH. Mutational analysis revealed the importance of the STAM binding motif of AMSH for CXCR4 stability (Figs. 3C and 8). A truncation mutant of AMSH containing the STAM binding motif and a catalytically inactive JAMM domain (RXXK-D348A) results in a similar increase in CXCR4 levels observed with overexpression of the full-length catalytically inactive AMSH. This implies that the RXXK-D348A fragment alone is able to displace endogenous wild type AMSH by effectively competing for relevant protein-protein interactions. As the previously described function for this fragment is recruitment by STAM (37, 44, 45, 51, 52), our findings suggest that the interaction between AMSH and STAM may be critical for appropriate trafficking and degradation of CXCR4. Indeed, our results show that mutation of the STAM binding RXXK motif of AMSH within this mutant abrogates its ability to interfere with CXCR4 levels (Fig. 8). AMSH recruitment by STAM may be an important mechanism of CXCR4 endocytosis and stability and, therefore, important for a number of cellular processes involving functional CXCR4 including human immunodeficiency virus infection and stem cell homing.
Previous reports have established that the RXXK motif of AMSH binds a variant subfamily of SH3 domains found in proteins belonging to the Grb2 and STAM adaptor families (37, 44, 45, 51, 52, 56). Although the classical SH3 domains bind to proline-rich ligands that assume a polyproline type II helix (65), this subfamily of SH3 domains interacts with RXXK-containing ligands that assume a 310 helical conformation (60). To define the critical residues in the SH3 domain binding site of AMSH, we employed a number of biochemical techniques including co-immunoprecipitation and high density peptide array technology. The arginine and lysine residues in the RXXK motif are both critical for binding as loss of either residue is sufficient to disrupt interactions between AMSH and the non-canonical RXXK binding SH3 domains of the STAM and Grb2 protein families. Affinity of binding between AMSH and the above subset of SH3 domains were found to be in the 0.2–12 μm range. Affinities in this range suggest transient associations between these proteins in vivo, which is important for proteins involved in dynamic cellular process of endocytosis.
An interaction between AMSH and STAM reported here has also been previously described (37, 44, 45, 48, 52). Where this complex forms and whether the complex specifically localizes to endosomal compartments has never been demonstrated. To visualize the AMSH·STAM complex, we used bimolecular fluorescence complementation. Our results indicate that AMSH associates with STAM1 specifically on Hrs-positive endosomes, and this interaction is dependent on a functional RXXK motif in AMSH (Fig. 4–5). The AMSH·STAM1 BiFC complex does not appreciably colocalize with the early and late endosomal markers EEA1 and CD63, respectively (Fig. 5). This indicates that the AMSH·STAM1 interaction is an event specific to sorting endosomes decorated with the Hrs·STAM complex. As Hrs has been shown to be a central mediator of CXCR4 trafficking (24), the AMSH·STAM1 interaction provides a direct link between a deubiquitinating enzyme and the ESCRT-0 complex in the trafficking of CXCR4. The ESCRT-0 complex is thought to be involved in the initial selection of ubiquitinated cargo at the endosomal membrane (33). ESCRT-0 is composed of Hrs, STAM, and associated molecules. Both Hrs and STAM bind ubiquitin via ubiquitin-interacting motifs (66–68). STAM also contains a VHS domain reported to interact with ubiquitin (68). Ubiquitin binding by STAM and Hrs is thought to be essential for the efficient sorting of ubiquitinated membrane proteins (33, 64, 67, 69). In addition, STAM and Hrs can themselves become ubiquitinated, a modification shown to be important for their function at sorting endosomes (33, 64). Our results demonstrate that AMSH catalytic activity mediates the steady-state ubiquitination status of ESCRT-0 (Fig. 6). The ability of AMSH to deubiquitinate ESCRT-0 adaptors is dependent upon a functional STAM interacting motif in AMSH (Fig. 6). In agreement with published data implicating STAM as a substrate of AMSH (45, 48), our results also identify Hrs as a potential target of AMSH deubiquitinating activity. Neither STAM1 nor Hrs stability is measurably affected in D348A cells (Fig. 6E), indicating that the ubiquitin linkage cleaved by AMSH is not related to proteasomal degradation. Previous studies have shown that AMSH preferentially cleaves lysine-63 ubiquitin chains known to function in endocytosis (45), supporting our findings regarding stability of Hrs and STAM1.
CXCR4 trafficking and degradation is known to be regulated in part by Hrs (24). To determine whether the regulation of ESCRT-0, as mediated by AMSH, is an important mechanism for CXCR4 trafficking, we examined AMSH recruitment by STAM1 in the context of CXCR4 turnover. Overexpression of D348A, which cannot bind STAM1, failed to affect steady-state levels of CXCR4 (Fig. 8, A and B). Loss of AMSH catalytic activity or protein depletion by RNAi resulted in the accumulation of both CXCR4 and AMSH on enlarged STAM1-positive (Figs. 7 and 8) and Hrs-positive endosomes (data not shown). D348A accumulated on enlarged endosomes, as compared with wild type AMSH, which localizes diffusely in the nucleus and cytoplasm (Fig. 8C). This disparity in localization between AMSH and its catalytic mutant has been previously noted (45) and suggests that deubiquitination of endocytic cargo by AMSH is required for functional trafficking. One possible explanation for the relocalization of D348A to enlarged endosomes is that a catalytically inactive form of AMSH cannot dissociate from the endosomal membrane, resulting in not only its own accumulation but also that of bound STAM and ubiquitin chains, which can no longer be cleaved by AMSH. Failure of D348A to bind STAM1, however, did not lead to the accumulation of CXCR4 or AMSH on enlarged endosomes. This further supports a model where AMSH function at the sorting endosome is mediated by its interaction with STAM1 (Fig. 9), as we no longer see the accumulation of D348A or ubiquitin on the endosomal membrane.
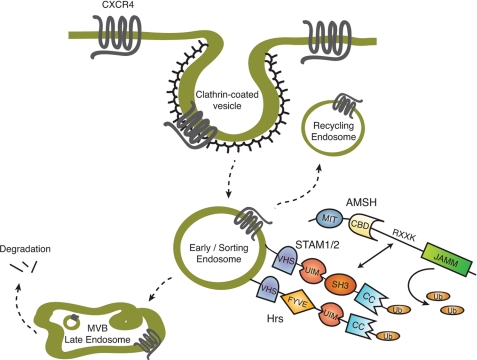
FIGURE 9.
Proposed model of AMSH function in the regulation of basal CXCR4 turnover. Constitutive endocytosis of CXCR4 via clathrin-coated pits results in trafficking of the receptor to the sorting endosome. At the sorting endosome, the ESCRT-0 complex, composed of Hrs and STAM, sort ubiquitinated (Ub) cargo toward either recycling or degradation. AMSH is recruited to the ESCRT-0 complex by direct interaction with STAM, mediated by the SH3 domain of STAM and the RXXK motif of AMSH. The RXXK/SH3 interaction is critical for AMSH-mediated deubiquitination of ESCRT-0 machinery. That ubiquitination of ESCRT machinery modulates the activity of individual components implicates AMSH as a regulator of endocytic adaptor protein function. As reflected in CXCR4 trafficking, this function of AMSH affects the fate of endocytosed receptors. UIM, ubiquitin-interacting motif; CC, coiled-coil; MVB, multivesicular body.
Collectively, our studies implicate AMSH in controlling endocytic trafficking through modulation of the ubiquitination status of ESCRT-0 proteins. This in turn affects the fate of endocytosed CXCR4 (Fig. 8). The recruitment of AMSH to ESCRT-0 on early/sorting endosomes requires an intact RXXK motif, which results in the deubiquitination of STAM1 and Hrs and potentially other targets on ESCRT-0-associated endosomes. This is required for proper sorting of receptors such as CXCR4, even in the absence of activating ligand. This demonstrates that AMSH recruitment by STAM1 is key for the regulation of constitutive or basal CXCR4 trafficking. Our findings not only demonstrate a role for AMSH in the trafficking of CXCR4, but they further build upon the model that AMSH is a global regulator of receptor endocytosis and degradation.
Supplementary Material
Acknowledgments
We thank Godfrey Getz, David Boone, and Raymond Hulse for helpful discussions and critical reading of the manuscript. We are grateful to Adriano Marchese (Loyola University of Chicago) for advice and assistance in setting up CXCR4 assays. Many thanks to Rebecca Dise for creation of the HA-CXCR4 stable HeLa cells and invaluable technical advice, Heather Schwartz for molecular cloning of GST-SH3 domain plasmids and general technical assistance, Ilana Berlin for invaluable technical and scientific discussion, Kate Higginbotham for general technical assistance, Christine Labno (The University of Chicago Comprehensive Cancer Center Microscopy Core) for training and technical assistance with fluorescence microscopy and assistance with quantitative analysis of images, and Dr. Richard Jones for synthesis and purification of the fluorescein-tagged PPVVDRSLKPG peptide.
This was work was supported by grants from the Leukemia Research Foundation, the Edward Mallinckrodt, Jr. Foundation, and a Concern Foundation CONquer canCER Now Award (to P. D. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental S1–S3.
- AIP4
- atrophin-interacting protein 4
- SH3
- Src homology 3
- AMSH
- associated molecule with the SH3 domain of STAM
- MIT
- microtubule-interacting and transport
- CHMP
- chromatin-modifying protein
- ESCRT
- endosomal sorting complex required for transport
- Hrs
- hepatocyte growth factor-regulated tyrosine kinase substrate
- HEK
- human embryonic kidney 293
- HeLa
- human carcinoma-derived Henrietta Lacks
- GST
- glutathione S-transferase
- BiFC
- bimolecular fluorescence complementation
- VFP
- venus fluorescent protein
- siRNA
- small interfering RNA
- shRNA
- short hairpin RNA
- HPLC
- high performance liquid chromatography
- CBD
- clathrin binding domain
- JAMM
- Jab1/MPN/Mov34 metalloenzyme catalytic domain
- HA
- hemagglutinin
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.von Zastrow M. (2003) Life Sci 74, 217–224 [DOI] [PubMed] [Google Scholar]
- 2.Sorkin A., Von Zastrow M. (2002) Nat. Rev. Mol. Cell Biol. 3, 600–614 [DOI] [PubMed] [Google Scholar]
- 3.Lefkowitz R. J. (1998) J. Biol. Chem. 273, 18677–18680 [DOI] [PubMed] [Google Scholar]
- 4.Tsao P., Cao T., von Zastrow M. (2001) Trends Pharmacol Sci 22, 91–96 [DOI] [PubMed] [Google Scholar]
- 5.Di Fiore P. P., Polo S., Hofmann K. (2003) Nat. Rev. Mol. Cell Biol. 4, 491–497 [DOI] [PubMed] [Google Scholar]
- 6.Haglund K., Di Fiore P. P., Dikic I. (2003) Trends Biochem. Sci 28, 598–603 [DOI] [PubMed] [Google Scholar]
- 7.Höller D., Dikic I. (2004) Biochem. Pharmacol 67, 1013–1017 [DOI] [PubMed] [Google Scholar]
- 8.Hanyaloglu A. C., von Zastrow M. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 537–568 [DOI] [PubMed] [Google Scholar]
- 9.Marchese A., Paing M. M., Temple B. R., Trejo J. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 601–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shenoy S. K. (2007) Circ. Res. 100, 1142–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bleul C. C., Farzan M., Choe H., Parolin C., Clark-Lewis I., Sodroski J., Springer T. A. (1996) Nature 382, 829–833 [DOI] [PubMed] [Google Scholar]
- 12.Oberlin E., Amara A., Bachelerie F., Bessia C., Virelizier J. L., Arenzana-Seisdedos F., Schwartz O., Heard J. M., Clark-Lewis I., Legler D. F., Loetscher M., Baggiolini M., Moser B. (1996) Nature 382, 833–835 [DOI] [PubMed] [Google Scholar]
- 13.Zou Y. R., Kottmann A. H., Kuroda M., Taniuchi I., Littman D. R. (1998) Nature 393, 595–599 [DOI] [PubMed] [Google Scholar]
- 14.Peled A., Grabovsky V., Habler L., Sandbank J., Arenzana-Seisdedos F., Petit I., Ben-Hur H., Lapidot T., Alon R. (1999) J. Clin. Invest. 104, 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tachibana K., Hirota S., Iizasa H., Yoshida H., Kawabata K., Kataoka Y., Kitamura Y., Matsushima K., Yoshida N., Nishikawa S., Kishimoto T., Nagasawa T. (1998) Nature 393, 591–594 [DOI] [PubMed] [Google Scholar]
- 16.Feng Y., Broder C. C., Kennedy P. E., Berger E. A. (1996) Science 272, 872–877 [DOI] [PubMed] [Google Scholar]
- 17.Walter D. H., Haendeler J., Reinhold J., Rochwalsky U., Seeger F., Honold J., Hoffmann J., Urbich C., Lehmann R., Arenzana-Seisdesdos F., Aicher A., Heeschen C., Fichtlscherer S., Zeiher A. M., Dimmeler S. (2005) Circ. Res. 97, 1142–1151 [DOI] [PubMed] [Google Scholar]
- 18.Balkwill F. (2004) Semin. Cancer Biol. 14, 171–179 [DOI] [PubMed] [Google Scholar]
- 19.Diaz G. A. (2005) Immunol. Rev. 203, 235–243 [DOI] [PubMed] [Google Scholar]
- 20.Diaz G. A., Gulino A. V. (2005) Curr. Allergy Asthma Rep. 5, 350–355 [DOI] [PubMed] [Google Scholar]
- 21.Gulino A. V. (2003) Curr. Opin. Allergy Clin. Immunol. 3, 443–450 [DOI] [PubMed] [Google Scholar]
- 22.Roland J., Murphy B. J., Ahr B., Robert-Hebmann V., Delauzun V., Nye K. E., Devaux C., Biard-Piechaczyk M. (2003) Blood 101, 399–406 [DOI] [PubMed] [Google Scholar]
- 23.Marchese A., Benovic J. L. (2001) J. Biol. Chem. 276, 45509–45512 [DOI] [PubMed] [Google Scholar]
- 24.Marchese A., Raiborg C., Santini F., Keen J. H., Stenmark H., Benovic J. L. (2003) Dev. Cell 5, 709–722 [DOI] [PubMed] [Google Scholar]
- 25.Bhandari D., Trejo J., Benovic J. L., Marchese A. (2007) J. Biol. Chem. 282, 36971–36979 [DOI] [PubMed] [Google Scholar]
- 26.Venkatesan S., Rose J. J., Lodge R., Murphy P. M., Foley J. F. (2003) Mol. Biol. Cell 14, 3305–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mines M. A., Goodwin J. S., Limbird L. E., Cui F. F., Fan G. H. (2009) J. Biol. Chem. 284, 5742–5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y., Foudi A., Geay J. F., Berthebaud M., Buet D., Jarrier P., Jalil A., Vainchenker W., Louache F. (2004) Stem Cells 22, 1015–1029 [DOI] [PubMed] [Google Scholar]
- 29.Futahashi Y., Komano J., Urano E., Aoki T., Hamatake M., Miyauchi K., Yoshida T., Koyanagi Y., Matsuda Z., Yamamoto N. (2007) Cancer Sci. 98, 373–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wojcik C. (2002) Trends Cell Biol. 12, 549. [DOI] [PubMed] [Google Scholar]
- 31.Urbé S., McCullough J., Row P., Prior I. A., Welchman R., Clague M. J. (2006) Biochem. Soc Trans 34, 754–756 [DOI] [PubMed] [Google Scholar]
- 32.Clague M. J., Urbé S. (2006) Trends Cell Biol. 16, 551–559 [DOI] [PubMed] [Google Scholar]
- 33.Williams R. L., Urbé S. (2007) Nat. Rev. Mol. Cell Biol. 8, 355–368 [DOI] [PubMed] [Google Scholar]
- 34.Komada M. (2008) Curr. Drug Discov. Technol. 5, 78–84 [DOI] [PubMed] [Google Scholar]
- 35.Mukhopadhyay D., Riezman H. (2007) Science 315, 201–205 [DOI] [PubMed] [Google Scholar]
- 36.Song L., Rape M. (2008) Curr. Opin. Cell Biol. 20, 156–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCullough J., Clague M. J., Urbé S. (2004) J. Cell Biol. 166, 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kyuuma M., Kikuchi K., Kojima K., Sugawara Y., Sato M., Mano N., Goto J., Takeshita T., Yamamoto A., Sugamura K., Tanaka N. (2007) Cell Struct. Funct. 31, 159–172 [DOI] [PubMed] [Google Scholar]
- 39.Ma Y. M., Boucrot E., Villén J., Affar el B., Gygi S. P., Göttlinger H. G., Kirchhausen T. (2007) J. Biol. Chem. 282, 9805–9812 [DOI] [PubMed] [Google Scholar]
- 40.Herrera-Vigenor F., Hernández-García R., Valadez-Sánchez M., Vázquez-Prado J., Reyes-Cruz G. (2006) Biochem. Biophys. Res. Commun. 347, 924–930 [DOI] [PubMed] [Google Scholar]
- 41.Reyes-Ibarra A. P., García-Regalado A., Ramírez-Rangel I., Esparza-Silva A. L., Valadez-Sánchez M., Vázquez-Prado J., Reyes-Cruz G. (2007) Mol. Endocrinol. 21, 1394–1407 [DOI] [PubMed] [Google Scholar]
- 42.Hislop J. N., Henry A. G., Marchese A., von Zastrow M. (2009) J. Biol. Chem. 284, 19361–19370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasdemir B., Murphy J. E., Cottrell G. S., Bunnett N. W. (2009) J. Biol. Chem. 284, 28453–28466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka N., Kaneko K., Asao H., Kasai H., Endo Y., Fujita T., Takeshita T., Sugamura K. (1999) J. Biol. Chem. 274, 19129–19135 [DOI] [PubMed] [Google Scholar]
- 45.McCullough J., Row P. E., Lorenzo O., Doherty M., Beynon R., Clague M. J., Urbé S. (2006) Curr. Biol. 16, 160–165 [DOI] [PubMed] [Google Scholar]
- 46.Zamborlini A., Usami Y., Radoshitzky S. R., Popova E., Palu G., Göttlinger H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19140–19145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsang H. T., Connell J. W., Brown S. E., Thompson A., Reid E., Sanderson C. M. (2006) Genomics 88, 333–346 [DOI] [PubMed] [Google Scholar]
- 48.Row P. E., Prior I. A., McCullough J., Clague M. J., Urbé S. (2006) J. Biol. Chem. 281, 12618–12624 [DOI] [PubMed] [Google Scholar]
- 49.Row P. E., Liu H., Hayes S., Welchman R., Charalabous P., Hofmann K., Clague M. J., Sanderson C. M., Urbé S. (2007) J. Biol. Chem. 282, 30929–30937 [DOI] [PubMed] [Google Scholar]
- 50.Nakamura M., Tanaka N., Kitamura N., Komada M. (2006) Genes Cells 11, 593–606 [DOI] [PubMed] [Google Scholar]
- 51.Kikuchi K., Ishii N., Asao H., Sugamura K. (2003) Biochem. Biophys. Res. Commun. 306, 637–643 [DOI] [PubMed] [Google Scholar]
- 52.Kim M. S., Kim J. A., Song H. K., Jeon H. (2006) Biochem. Biophys. Res. Commun. 351, 612–618 [DOI] [PubMed] [Google Scholar]
- 53.Hu C. D., Kerppola T. K. (2003) Nat. Biotechnol. 21, 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu C. D., Chinenov Y., Kerppola T. K. (2002) Mol. Cell 9, 789–798 [DOI] [PubMed] [Google Scholar]
- 55.Moffat J., Grueneberg D. A., Yang X., Kim S. Y., Kloepfer A. M., Hinkle G., Piqani B., Eisenhaure T. M., Luo B., Grenier J. K., Carpenter A. E., Foo S. Y., Stewart S. A., Stockwell B. R., Hacohen N., Hahn W. C., Lander E. S., Sabatini D. M., Root D. E. (2006) Cell 124, 1283–1298 [DOI] [PubMed] [Google Scholar]
- 56.Berry D. M., Nash P., Liu S. K., Pawson T., McGlade C. J. (2002) Curr. Biol. 12, 1336–1341 [DOI] [PubMed] [Google Scholar]
- 57.Rasband W. S. (1997–2000) ImageJ software, National Institutes of Health, Bethesda, MD [Google Scholar]
- 58.Abramoff M. D., Magelhaes P. J., Ram S. J. (2004) Biophotonics Int. 11, 36–42 [Google Scholar]
- 59.Frank R. (1992) Tetrahedron 48, 9217–9232 [Google Scholar]
- 60.Liu Q., Berry D., Nash P., Pawson T., McGlade C. J., Li S. S. (2003) Mol. Cell 11, 471–481 [DOI] [PubMed] [Google Scholar]
- 61.Harkiolaki M., Lewitzky M., Gilbert R. J., Jones E. Y., Bourette R. P., Mouchiroud G., Sondermann H., Moarefi I., Feller S. M. (2003) EMBO J. 22, 2571–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raiborg C., Stenmark H. (2009) Nature 458, 445–452 [DOI] [PubMed] [Google Scholar]
- 63.Acconcia F., Sigismund S., Polo S. (2009) Exp. Cell Res. 315, 1610–1618 [DOI] [PubMed] [Google Scholar]
- 64.Komada M., Kitamura N. (2005) J Biochem. 137, 1–8 [DOI] [PubMed] [Google Scholar]
- 65.Feng S., Chen J. K., Yu H., Simon J. A., Schreiber S. L. (1994) Science 266, 1241–1247 [DOI] [PubMed] [Google Scholar]
- 66.Raiborg C., Bache K. G., Gillooly D. J., Madshus I. H., Stang E., Stenmark H. (2002) Nat. Cell Biol. 4, 394–398 [DOI] [PubMed] [Google Scholar]
- 67.Urbé S., Sachse M., Row P. E., Preisinger C., Barr F. A., Strous G., Klumperman J., Clague M. J. (2003) J. Cell Sci. 116, 4169–4179 [DOI] [PubMed] [Google Scholar]
- 68.Mizuno E., Kawahata K., Kato M., Kitamura N., Komada M. (2003) Mol. Biol. Cell 14, 3675–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Raiborg C., Stenmark H. (2002) Cell Struct. Funct. 27, 403–408 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.