Abstract
Quorum sensing has been implicated as an important global regulatory system controlling the expression of numerous virulence factors in bacterial pathogens. SmcR, a homologue of Vibrio harveyi LuxR, has been proposed as a quorum-sensing master regulator of Vibrio vulnificus, an opportunistic human pathogen. Previous studies demonstrated that SmcR is essential for the survival and pathogenesis of V. vulnificus, indicating that inhibiting SmcR is an attractive approach to combat infections by the bacteria. Here, we determined the crystal structure of SmcR at 2.1 Å resolution. The protein structure reveals a typical TetR superfamily fold consisting of an N-terminal DNA binding domain and a C-terminal dimerization domain. In vivo and in vitro functional analysis of the dimerization domain suggested that dimerization of SmcR is vital for its biological regulatory function. The N-terminal DNA recognition and binding residues were assigned based on the protein structure and the results of in vivo and in vitro mutagenesis experiments. Furthermore, protein-DNA interaction experiments suggested that SmcR may have a sophisticated mechanism that enables the protein to recognize each of its many target operators with different affinities.
Keywords: Bacterial Signal Transduction, Crystal Structure, Protein-DNA Interaction, Signal Transduction, Transcription Regulation, Quorum Sensing, Transcriptional Regulator SmcR, Vibrio vulnificus
Introduction
Many bacterial societies are based on chemical communication called quorum sensing. This process facilitates social networking in bacteria in response to environmental challenges that trigger alterations in gene expression or repression that are often critical for survival (1). Such “social behavior” results from the bacteria sensing and responding to fluctuations in the cell population, mediated by diffusible signaling molecules (often referred to as autoinducers, or AIs) that are synthesized within the bacterial cell.
The cell density-dependent regulation of bioluminescence in Vibrio harveyi is frequently used as a model for quorum sensing. The V. harveyi LuxR protein is the transcriptional activator of the luminescence operon, and its synthesis is controlled by the levels of three autoinducers, HAI-1, AI-2, and CAI-1, which are synthesized and detected by the LuxM/N, LuxS/PQ, and CqsA/S two-component signal transduction-resembling systems, respectively (2, 3). Homologues of LuxR, which controls expression of the genes involved in the quorum-sensing regulon, have been identified in various pathogenic Vibrio species (spp.).
The quorum-sensing system of the opportunistic human pathogen Vibrio vulnificus has been little studied (4). Recent evidence has indicated that the bacterium produces a signaling molecule, N-butanoyl-homoserine-lactone (C4-HSL), but the genes related to the signaling molecule seem to be different from those found in other Gram-negative bacteria (5). It was also predicted that V. vulnificus does not use the CqsA/S system. However, the bacterium has been known to express the LuxS/PQ, LuxU, LuxO, and key transcriptional regulator, LuxR, homologues, suggesting that one of the quorum-sensing systems in V. vulnificus is similar to the LuxS/PQ system in V. harveyi (4, 6–8). The SmcR protein, a LuxR homologue, has been proposed as a master quorum-sensing regulator in V. vulnificus. Many studies have demonstrated that SmcR regulates virulence genes and adaptive phenotypes (9–11).
In addition, a recent study based on a genome-wide search for DNA targets of SmcR suggested that at least 121 genes are under the control of this master regulator (12). Alignment of the DNA target sequences revealed a 22-bp consensus SmcR-binding sequence with a pseudo-2-fold symmetry pattern. The consensus sequence consists of an 8-bp inverted repeat, with a well conserved left half and a less well conserved right half. This result was comparable with the results of another genome-wide search, which showed that the V. harveyi LuxR recognizes a 21-bp consensus operator sequence with dyad symmetry (13).
The operator region (PvvpE) for transcription of the virulence factor gene, vvpE, has been extensively studied as a site regulated by SmcR in V. vulnificus (9, 12). The binding of SmcR to PvvpE induces the expression of vvpE, which encodes an elastolytic protease known to be associated with pathogenesis. The binding site was determined using a DNA footprinting technique, and the binding sequence showed a similar 22-bp consensus sequence pattern but with less well conserved sequences in the inverted repeat right half. Introducing the SmcR-recognition consensus sequence of the right half of the inverted repeat into the right half of PvvpE resulted in much higher binding affinity (12).
Such observations of the transcriptional regulation of SmcR have provided instructive information; however, the molecular details of the regulatory mechanisms of SmcR in V. vulnificus remain unclear. Recently, the structure of the Vibrio cholerae LuxR homologue protein, HapR, was determined, although the relationship between its structure and biological function remains to be fully investigated (14).
The goal of this study was to obtain molecular details of the regulatory mechanisms of SmcR in V. vulnificus based on its three-dimensional protein structure. Accordingly, the crystal structure of SmcR was determined at 2.1 Å resolution. On the basis of the crystal structure and supporting mutagenesis data in vivo and in vitro, we describe molecular insights into the transcriptional regulation mediated by SmcR.
EXPERIMENTAL PROCEDURES
General Genetic Materials and Bacterial Strains
The strains and plasmids used in this study are listed in supplemental Table S1. Escherichia coli strains were grown in Luria-Bertani (LB) medium or on LB containing 1.5% (w/v) agar at 37 °C. Unless mentioned otherwise, V. vulnificus strains were grown aerobically in LB medium supplemented with 2.0% (w/v) NaCl (LBS) at 30 °C. When required, antibiotics were added to the media as follows: 100 μg/ml ampicillin and 100 μg/ml kanamycin for both E. coli and V. vulnificus; 20 and 3 μg/ml chloramphenicol for E. coli and V. vulnificus, respectively; 10 and 3 μg/ml tetracycline for E. coli and V. vulnificus, respectively; and 100 μg/ml of streptomycin for E. coli. The oligonucleotides used are listed in supplemental Table S2. E. coli SM10λpir was used as a conjugal donor to transfer the broad host range vector, pJH0311 (15), containing wild-type or mutant smcR into V. vulnificus.
Preparation of the SmcR and Mutant Proteins
The smcR gene, encoding the transcriptional regulator SmcR protein, was amplified from the plasmid, pHS103 (9), by PCR using primers His-SmcR_F and His-SmcR_R, carrying the NcoI and XhoI restriction sites. The PCR product was then subcloned into the pHis-Parallel1 vector (16), a hexahistidine-tagged protein expression vector containing a recombinant tobacco etch virus protease cleavage site, to produce the plasmid pHis-smcR. E. coli BL21 (DE3) cells harboring the plasmid were grown in LB medium containing ampicillin at 37 °C until they reached an A600 of between 0.6 and 0.8. After being cooled to 25 °C, smcR expression was induced with 0.25 mm isopropyl-β-d-thiogalactopyranoside for 14 h. The expressed protein was then purified according to a previously described procedure (17). The purified protein was dialyzed against 20 mm Tris-HCl, pH 7.5, and 200 mm NaCl, concentrated to ∼15 mg/ml, and stored at −80 °C for use in the crystallization trials and biochemical experiments. The SeMet4-substituted SmcR protein was expressed in the methionine auxotroph E. coli B834 (DE3) strain (Novagen) in minimal medium supplemented with 50 mg/ml SeMet, under the same conditions as for the native protein expression. Purification of the SeMet substituted protein was identical to that for the native protein, except for the addition of 5 mm methionine to all buffers. Mutations were introduced into pHis-smcR using QuikChangeTM site-directed mutagenesis (Stratagene). The His6-tagged mutant proteins were overexpressed, purified by nickel-nitrilotriacetic acid-agarose chromatography, and used in electrophoretic mobility shift assays (EMSA).
Crystallization and Structure Determination
Crystallization trials were carried out using the sitting-drop technique at 21 °C. An initial SmcR crystal was produced under the following conditions: 0.2 m Li2SO4, 10% PEG 3000, and 0.1 m imidazole, pH 8.0. Subsequently, production of the crystal was optimized under the following conditions: 0.2 m Li2SO4, 7% PEG 3000, and 0.1 m imidazole (pH 7.2 to 8.0). Crystals appeared within a day and grew for a further 5 days. The SeMet-substituted SmcR crystal was produced under the same crystallization conditions as for the native protein. The SeMet crystals were transferred to a cryoprotectant solution containing 0.2 m Li2SO4, 30% PEG 3000, 0.1 m imidazole, pH 8.0, and 20% glycerol and then fished out and placed immediately in a −173 °C nitrogen gas stream. Single wavelength anomalous diffraction data were collected at 2.3 Å resolution on beamline 6C at the Pohang Accelerator Laboratory, Pohang, Korea. Because the SeMet crystals diffracted better than the native crystals, SeMet crystals were used to collect higher resolution data at 2.1 Å on the same beamline. All data were processed using the HKL2000 program suite (18).
The SeMet-substituted SmcR crystal belongs to the space group P212121. There were four molecules in the asymmetric unit with a packing density of ∼2.62 Å3/Da, corresponding to an estimated solvent content of ∼53.42%. The structure was determined by utilizing the anomalous signals from selenium atoms using the program SOLVE (19), which identified 18 sites. Density modification and subsequent automated model building were carried out using the program RESOLVE (20). The initial RESOLVE-built model was used as a guide to build the remainder of the protein manually into density-modified electron density maps with the program COOT (21). The refinement was performed with REFMAC5 (22). The SeMet-substituted crystal structure was solved at 2.1 Å resolution by molecular replacement with the program MOLREP (23) using the partially refined model of the SeMet crystal at 2.3 Å resolution. The refinement included the translation-liberation-screw procedure. The final refined model resulted in Rfree and Rcryst values of 0.251 and 0.198, respectively. No density was visible for the N-terminal five amino acid residues (Met-1 to Ile-4) of chain A, two residues (Met-1 and Asp-2) of chain C, and three residues (Met-1 to Ile-4) of chain D, nor for the three C-terminal residues (Glu-203 to Glu-205) of chain B, and two residues (His-204 and Glu-205) of chain D. These residues were not included in the model. The model contained 806 amino acid residues, including a cloning artifact (alanine) at the N terminus of the molecule B, 6 sulfate ions, and 453 water molecules, and satisfied the quality criteria limits of the program PROCHECK (24). The crystallographic data statistics are summarized in Table 1. The atomic coordinates and structure factor amplitudes of SmcR have been deposited in the Protein Data Bank (PDB) (25) under the accession code 3KZ9.
TABLE 1.
Crystallographic data collection and refinement statistics
The numbers in parentheses describe the relevant value for the highest resolution shell.
| Dataset | SeMet_SmcR_peak | SeMet_SmcR |
|---|---|---|
| Beamline (Pohang | 6C (MXII) | 6C (MXII) |
| Accelerator Laboratory) | ||
| Wavelength | 0.97944 | 1.23985 |
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| A | 78.56 Å | 78.50 Å |
| B | 99.10 Å | 99.03 Å |
| C | 129.18 Å | 129.06 Å |
| Resolution | 2.30 Å (2.38-2.30 Å) | 2.10 Å (2.18-2.10 Å) |
| No. of total reflections | 499,130 | 408,227 |
| No. of unique reflections | 45,577 | 59,111 |
| Redundancy | 11.0 (10.7) | 6.9 (6.2) |
| Completeness | 99.9% (99.2%) | 99.2% (95.6%) |
| Rsyma | 8.4% (38.8%) | 7.2% (47.5%) |
| I/σ(I) | 34.26 (5.56) | 26.97 (3.28) |
| Refinement | ||
| Resolution | 29.92-2.10 Å | |
| Reflections in work/test sets | 56,004/2,979 | |
| Rwork/Rfreeb,c | 19.8/25.1% | |
| Root mean square deviations | ||
| Bond lengths | 0.014 Å | |
| Bond angles | 1.329° | |
| Model composition | ||
| 806 residues | ||
| 453 waters | ||
| 6 SO4 | ||
| Geometry | ||
| Most favored regions | 95.5% | |
| Additional allowed regions | 4.5% | |
| Generously allowed regions | 0.0% | |
| PDB accession code | 3KZ9 | |
a Rsym = Σ|Ii − 〈I〉|/ΣI where Ii is the intensity of the ith observation and 〈I〉 is the mean intensity of the reflections.
b Rwork = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|, where Fcalc and Fobs are the calculated and observed structure factor amplitude, respectively.
c Rfree = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|, where all reflections belong to a test set of randomly selected data.
In Vivo Dimerization
The in vivo dimerization of SmcR was assessed using a bacterial two-hybrid system that allows easy in vivo detection of functional interactions between two proteins (26). Briefly, SmcR was genetically fused to two complementary fragments, T25 and T18, that constitute the catalytic domain of Bordetella pertussis adenylate cyclase as follows: the smcR gene was subcloned into the PstI and KpnI sites of pUT18C and pKT25, resulting in the hybrid plasmids, pHS450 and pHS451, respectively. For the subcloning, smcR was amplified from the template plasmid, pHS103, using the primers SMCRT18F and SMCRT25R, and SMCRT25F and SMCRT25R for pUT18C and pKT25, respectively. Association of the two-hybrid proteins, in other words the dimerization of SmcR, results in functional complementation between the T25 and T18 fragments and leads to cAMP synthesis in the adenylate cyclase-deficient E. coli strain, BTH101. The cAMP protein then triggers the transcriptional activation of lacZ.
To assess the phenotype, we performed a β-galactosidase assay (see under “In Vivo Activity of SmcR”). Critical residues involved in the dimerization of SmcR in the C-terminal region were mutated using gene splicing by overlap extension (27) and site-directed mutagenesis. Primers used to create the site-directed mutants of the C-terminal region of SmcR are listed in supplemental Table S2. These mutated DNAs were introduced into the bacterial two-hybrid system vector pUT18C. The eight site-directed mutant plasmids created in this manner were designated as pSM01 to pSM08 (supplemental Table S1). In addition, pYJ03 and pYJ08 were constructed to assess the binding effect of SmcR to the vvpE operator region PvvpE, using the primers DH0601F and DH0601R, based on pSM03 and pSM08 as templates. All constructions were confirmed by DNA sequencing.
Western Immunoblotting
The purified His-tagged SmcR protein was used to raise a primary antibody by immunizing Sprague-Dawley rats on three occasions at 3-week intervals with 200 μg of the protein for each immunization. Western immunoblotting was performed according to the procedure previously described by Jeong et al. (28).
EMSA
The binding of SmcR and its mutants to the labeled DNA and electrophoretic analysis of the DNA-protein complexes were performed as described in a previous study (9). In brief, the radioactively labeled DNA probe containing the operator region PvvpE, which is activated directly by SmcR, was generated by PCR using 32P-labeled VVPE021 and unlabeled VVPE022 as primers. This probe (20 ng) was incubated with increasing amounts of SmcR or its mutants in reaction buffer (0.1 m KCl, 0.02 m Tris-Cl, pH 8.0, 3 mm MgCl2, 0.1 mm EDTA, and 0.1 mm dithiothreitol) at 37 °C for 30 min and then separated by electrophoresis.
In Vivo Activity of SmcR
To determine the in vivo activity of SmcR, two different reporter systems were used. For the repression activity, wild-type and mutant smcR genes in the pHis series plasmids were subcloned into pBAD24BS (supplemental Table S1). The resulting plasmids were designated as the pBSS series, and these plasmids were co-transformed with the reporter plasmid, pKS0710, into E. coli DH5α cells. The reporter plasmid was constructed by fusing the promoter region of VV2_1398 to the plasmid pHK0011 containing a promoterless luxAB luciferase gene (29). As described in the previous study (12), the promoter region of VV2_1398 was one of the targets under direct control by SmcR. The binding of SmcR to the promoter region resulted in a total repression of its activity (data not shown). E. coli carrying one of the pBSS plasmids and the pKS0710 reporter plasmid was used for the in vivo assay. Overnight cultures of each BSS strain were diluted 100-fold into fresh LB containing 0.2% l(+)-arabinose and appropriate antibiotics, and then grown at 37 °C for 3.5 h. The A600 and cellular luminescence of 100 μl of each culture with 0.01% (v/v) decanal were detected by an InfiniteTM M200 microplate reader (Tecan, Männedorf, Switzerland) and expressed in arbitrary relative light units as described previously (9).
For the V. vulnificus system, a series of plasmids expressing wild-type SmcR, or its mutants, were constructed as follows: a gene fragment containing wild-type smcR and the rrnB terminator was amplified by PCR using pBSS-WT as a template, with the primers SMCR0805 and SMCR0806 carrying KpnI and XbaI sites, respectively. The resulting fragment was ligated into the same restriction enzyme sites of pJH0311 (15), resulting in the plasmid pBSJH-WT. Five other members of the pBSJH series were constructed by exchanging the wild-type smcR gene with the mutated smcRs in the pHis series of plasmids by cutting and ligating at the NcoI and XhoI sites. These pBSJH series of plasmids in E. coli SM10λpir were then transferred into the V. vulnificus reporter strain, DH0602, by conjugation. This reporter strain is the smcR and lacZ double-knock-out mutant containing the vvpE gene fused to a promoterless lacZ on its chromosome. The activity of the SmcR variants was determined by measuring β-galactosidase activity. The β-galactosidase activity was determined by the chloroform/SDS method described previously by Miller (30).
Differential Scanning Calorimetry (DSC)
DSC experiments were performed with a DSC calorimeter from MicroCal (Northampton, MA) at a scan rate of 1.0 °C/min. Measurements were carried out at a protein concentration of 10 mg/ml. The buffer containing 50 mm Tris-HCl, pH 7.5, 300 mm NaCl, and 5% glycerol from the last step of protein dialysis was used in the reference cell of the calorimeter. Data were analyzed with MicroCal DSC standard analysis software.
Isothermal Titration Calorimetry (ITC)
The PvvpE (5′-atcttattgataaatctgcgtaaaaaa-3′) and pseudo-palindromic PvvpE-mod (5′-ttcttattgataaatttgtgaataaaa-3′) duplex DNAs were chemically synthesized. The SmcR binding affinities for both PvvpE and PvvpE-mod were analyzed by an ITC experiment. SmcR and duplex DNA were dialyzed extensively against 50 mm Tris, pH 7.0, and 300 mm NaCl, respectively. Samples were then degassed by vacuum aspiration for 15 min prior to loading, and titration was carried out at 25 °C. The calorimetric assay was performed using a VP-ITC (MicroCal Inc., Northampton, MA). The stirring speed was 300 rpm, and the thermal power was recorded every 10 s. We titrated 0.3696 mm (calculated as a dimer) of SmcR in the syringe against 0.0272 mm PvvpE in the reaction cell (∼1.6 ml) and 0.1537 mm SmcR was titrated against 0.0076 mm of PvvpE-mod. Thermogram analysis for the titration was performed using the Origin package (version 7) supplied with the instrument.
RESULTS
Overall Structure of SmcR
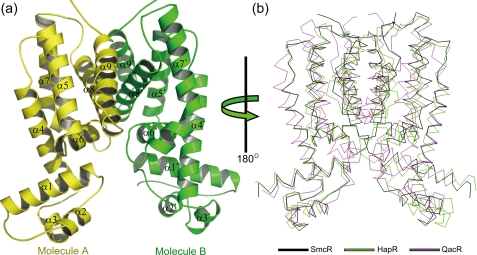
The crystal structure of SmcR was refined at 2.1 Å resolution. There are two homodimers in the asymmetric unit, arranged as molecules A and B in one homodimer and C and D in the other. The two molecules in each dimer are related by a 2-fold rotation axis (Fig. 1a). The refined model is entirely composed of α-helices. The structure of each SmcR molecule is essentially identical, with root mean square deviations of 0.95 Å (A and B), 0.43 Å (A and C), and 0.68 Å (A and D) of the Cα atoms. Superimposition of the Cα atoms of each dimer results in a root mean square deviations of 0.43 Å.
FIGURE 1.
Crystal structure of SmcR. a, ribbon representation of SmcR. b, structural comparison of TetR superfamily proteins. SmcR, the V. cholerae transcriptional regulator, HapR (PDB code 2PBX), and the S. aureus multidrug binding transcriptional repressor, QacR (PDB code 1JUP), are displayed in black, green, and magenta, respectively. The structures are rotated 180° about the y axis of a. Unless otherwise noted, figures were prepared using PyMOL.
The SmcR monomer is composed of nine α-helices: α1 (14–31), α2 (39–46), α3 (50–56), α4 (60–82), α5 (89–106), α6 (109–119), α7 (126–150), α8 (159–179), and α9 (183–196) and adopts a two-domain architecture, which includes the N-terminal DNA binding domain and the large C-terminal dimerization domain. The first three α-helices embrace a three-helix bundle that contains a canonical helix-turn-helix motif (α2 and α3). The dimerization domain of SmcR is formed by α-helices 4–9, with the α-helices 8 and 9 in each monomer forming an antiparallel four-helix dimerization motif. The overall fold of SmcR results in an Ω-shaped structure.
A search conducted using DALI (31) revealed that SmcR is structurally related to the TetR superfamily (Fig. 1b), which includes the V. cholerae transcriptional regulator, HapR (PDB ID code 2PBX; Z = 26.8) (14), and the Staphylococcus aureus multidrug binding transcriptional repressor, QacR (PDB ID code 1JUP; Z = 15.7) (32), as well as a number of other transcriptional regulators. The HapR protein is a homologue of LuxR, the quorum-sensing master transcriptional regulator of V. harveyi, and shows ∼70% amino acid identity with SmcR. QacR, however, is not related to LuxR or its homologues, and shows about 13% amino acid identity with SmcR.
Dimerization Domain and Its Biological Function
A recent study revealed that the dimerization of the quorum-sensing transcription factor, TraR (the LuxR homologue in the LuxI/LuxR-type quorum-sensing system of the plant pathogen Agrobacterium tumefaciens), enhances resistance to cellular proteases and maintains biological activity (33). The dimerization of quorum-sensing response regulators is known as a signaling molecule binding-dependent event (34, 35). In contrast, the dimerization of SmcR seems to occur in a signaling molecule-independent manner, although definitive evidence supporting this assumption has been lacking. In addition, the biological function of SmcR dimerization is uncertain.
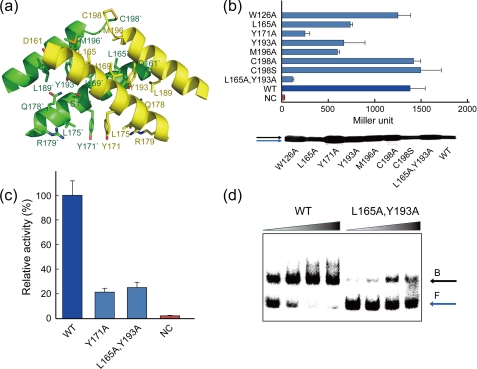
SmcR undergoes dimerization in the C-terminal domain. The C-terminal α-helices 8 and 9 in each monomer pack against one another to form an antiparallel, four-helix bundle at the subunit interface (Fig. 2a). Hydrophobic contacts by nonpolar residues are prominent at the dimerization interface and are involved in stabilizing the dimer structure. Residues involved in the interface are Asp-161, Leu-165, Gly-168, Ile-169, Tyr-171, Ser-172, Val-175, Gln-176, Arg-179, Leu-189, Tyr-193, Met-196, and Cys-198. In addition, Trp-126 on α-helix 7 stabilizes the dimerization by stacking against Arg-179 from the other molecule.
FIGURE 2.
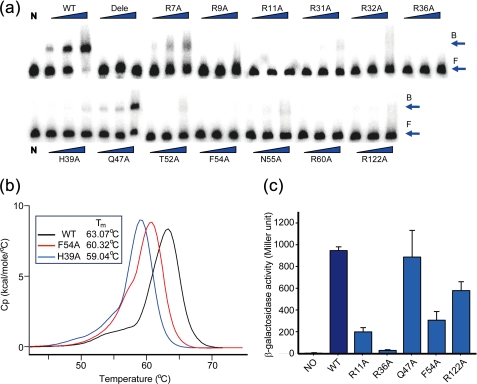
Dimerization of SmcR and its biological effects. a, C-terminal antiparallel helices of SmcR form a four-helix dimerization motif. The structure is rotated 90° about the y axis of Fig. 1a. Residues at the dimer interface are shown. b, site-directed mutagenesis analysis of the residues critical for dimerization using the bacterial two-hybrid system in E. coli BTH101. The phenotypes were assessed by their cellular β-galactosidase activities using cultures in the stationary growth phase (A600 = 2.0–3.0). Expression levels of all of the mutant proteins were compared by Western blot analysis. The black and blue arrows indicate the products of pHS451 and pHS450, respectively. NC indicates negative control as in supplemental Fig. S1 (also see text). WT, wild type. c, effects of the dimerization of SmcR on the transcriptional regulation of vvpE in V. vulnificus. Cellular β-galactosidase activities were measured using cultures in the stationary growth phase (A600 = 2.0–3.0). NC indicates negative control (DH0602 containing empty vector pJH0311). d, DNA binding effect of SmcR dimerization and its mutant to the vvpE regulatory region. A 200-bp DNA fragment from the upstream region of vvpE was radioactively labeled and then used as a DNA probe. The radiolabeled fragments (20 ng) were mixed with increasing amounts of wild-type or mutant SmcR (30, 60, 90, and 120 nm in 1st to 4th lanes, respectively), and then resolved on a 4% polyacrylamide gel. B, bound DNA; F, free DNA.
To improve our understanding of the dimerization of SmcR and its biological implications in vivo, we explored the ability of SmcR to dimerize using a bacterial two-hybrid system in E. coli. In this system, pHS450 was used as prey and pHS451 was used as bait (see in vivo dimerization under “Experimental Procedures”). The strain harboring the pUT18C and pHS451 plasmids was used as a negative control. We used pKT25-zip and pUT18C-zip encoding T25 and T18 fragments, respectively, fused in-frame with a DNA region encoding a 35-amino acid leucine zipper, as a positive control for complementation in this system. The positive control resulted in about 1900 units of β-galactosidase activity. This in vivo result confirmed the well known fact that leucine zipper proteins interact with each other (36). On the other hand, the negative control produced only about 30 units of activity. Finally, the SmcR-fused hybrid system showed strong lacZ transcriptional activity and resulted in about 1350 units of enzyme activity (supplemental Fig. S1).
Based on the SmcR structure, we performed site-directed mutagenesis of the residues at the dimerization interface. The ability of each mutant to dimerize was compared with wild-type SmcR using the bacterial two-hybrid system. Each mutated smcR, fused to pUT18C and pHS451, was co-transformed into BTH101. The results of the enzyme activity assay revealed that Leu-165, Tyr-171, Tyr-193, and Met-196 are critical for dimerization (Fig. 2b).
We confirmed the expression levels of all of the mutant proteins by Western blot analysis (Fig. 2b). The cellular levels of the wild-type and variants in BTH101 were fairly consistent, with only marginal differences.
To define the relationship between SmcR dimerization and transcriptional activity in V. vulnificus further, expression of the SmcR variants was induced in the strain DH0602. The vvpE operator region containing PvvpE was used for the system because vvpE has been well established as a gene that is under control of SmcR, which binds directly to PvvpE (9). The regulation activity was assessed by a β-galactosidase enzyme assay. The mutation on the dimer interface resulted in the loss of enzyme activity, indicating a lack of SmcR binding activity to PvvpE (Fig. 2c). This result further demonstrated the biological importance of SmcR dimerization.
In addition, we evaluated the effect of SmcR dimerization on binding to PvvpE. The double mutant SmcR (L165A/Y193A) was compared with wild type for DNA binding ability using EMSA. As seen in Fig. 2d, the mutant showed much weaker DNA binding ability than the wild-type SmcR. Therefore, this result demonstrated that the lack of transcriptional regulation activity of the mutants in V. vulnificus DH0602 could be ascribed to lower DNA binding affinity. Taken together, these results indicate that the dimerization of SmcR is vital for the exertion of its biological regulatory function.
DNA Binding Domain and Protein-DNA Interaction
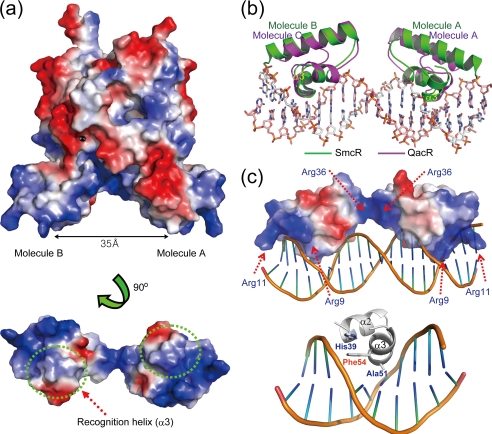
Although repeated experiments aimed at the preparation of the SmcR-PvvpE complex crystal using a number of DNA constructs, including artificially synthesized palindromic DNA, were not successful, we were able to speculate on the protein-DNA interaction based on the protein structure. The N-terminal domain of SmcR contains a cluster of three α-helices, with helices 2 and 3 forming the helix-turn-helix motif in each molecule. The second α-helices (recognition helix) in each of the helix-turn-helix motifs of the two molecules are separated by a distance of 35 Å. This distance almost exactly corresponds to one turn of a B-DNA double helix. We can speculate that if the recognition helix of one SmcR molecule binds in the major groove of DNA, the corresponding α-helix of the other molecule would also bind in the major groove one turn further along the DNA.
Furthermore, the surface electrostatic potential of the SmcR N-terminal DNA binding domain allows us to speculate on the mechanism of DNA recognition by SmcR (Fig. 3a). In general, each recognition helix in the dimer would sit in the major groove of the DNA helix through hydrophobic contact with DNA bases. The positively charged residues on the edge of both of the first α-helices in the dimer may be responsible for binding with the DNA phosphate backbone, whereas the region between the two subunits may be responsible for binding with the phosphate backbone in the minor groove of the DNA helix (Fig. 3c).
FIGURE 3.
DNA binding domain and DNA recognition of SmcR. a, electrostatic surface potential of the SmcR dimer. The DNA binding domain is rotated 90° about the x axis. The recognition helix of each subunit is indicated. b, superimposition of the DNA binding domains of SmcR and QacR complexed with DNA (PDB code 1JT0). SmcR (molecules A and B) and QacR (molecules A and C) are shown in green and magenta, respectively. c, putative DNA recognition residues. Basic residues predicted to bind with DNA sugar-phosphate backbones are displayed. DNA recognition residues on the helix-turn-helix motif are shown based on the QacR structure.
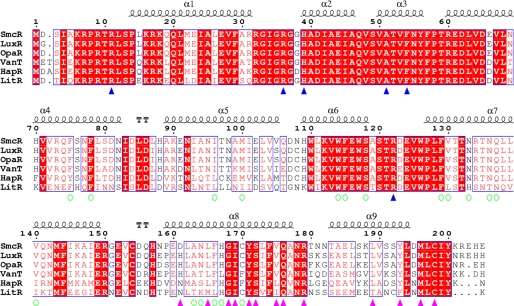
The DNA binding domain was superimposed with the Staphylococcus aureus multidrug binding transcriptional repressor QacR, which is complexed with its associated operator DNA (37). The DNA binding domain of molecule B of dimeric SmcR superimposed well with the corresponding domain of QacR, which sits in the major groove of DNA, whereas molecule A stays out of the major groove (Fig. 3b). This suggests that the SmcR-DNA binding event may undergo a conformational change. Nevertheless, based on the molecule B-DNA binding model, we could imagine a DNA-protein recognition event. In the QacR structure, Gly-37 and Tyr-40 on the recognition helix are critical for tight docking and contact with the major groove of DNA (37). In the superimposed model, Ala-51 and Phe-54 in SmcR correspond to the QacR residues (Fig. 3c). They may play a critical role in DNA recognition. In a report on HapR, a V. cholerae LuxR homologue (14), the corresponding residue (Phe-55), was shown to be critical for DNA binding in vitro and in vivo. On the other hand, the LuxR homologue proteins show strikingly conserved, positively charged residue distribution in the N-terminal region (Fig. 4). Residues such as Arg-9 and Arg-11 may contribute to interaction with the phosphate backbone in the minor groove of DNA as if both minor groove regions of DNA have been picked up with tongs.
FIGURE 4.
Structure-related functional sequence conservation between LuxR homologous proteins. Shown above the alignments are elements of the secondary structure of SmcR. The numbering shown is from SmcR. Blue triangles indicate the residues critical for binding with DNA. Open green circles represent residues involved in the formation of the putative ligand-binding pocket. Purple triangles represent residues located at the dimer interface. Biological sources and accession codes for the sequences are as follows: LuxR, V. harveyi (gi:107933356); OpaR, Vibrio parahemeolyticus (gi:28899290); VanT, Vibrio anguillarum (gi:18104604); HapR, V. cholerae (gi:87133250); and LitR, V. fischeri (gi:59712784). Sequence alignments were assembled using T-COFFEE software and visualized using ESPript software, both located on the ExPASy Proteomics Server.
The SmcR binding consensus sequence was identified as 5′-TTATTGATWWRWTWNTNAATAA-3′ (where W indicates A or T; R indicates G or A; and N indicates any nucleotide). Mutational analysis of the consensus sequence indicated that the 9th and 10th nucleotides from the center are the most important for SmcR binding (12). Interestingly, the positions of Arg-9 and Arg-11 are considered to be placed at or near the 9th or 10th nucleotides, which are predicted to be minor grooves of the DNA when superimposed with the DNA complexed with QacR protein.
A difference between the SmcR and QacR structures is the residue distribution between the subunits in the dimeric form. Positively charged residues are distributed in this region in SmcR (Fig. 3), whereas negatively charged residues are distributed in the equivalent region of QacR (data not shown). Those residues in SmcR are Arg-32, Arg-36, and Arg-60 in the DNA binding domain and Arg-122 in the dimerization domain. Such residues are thought to interact with the phosphate backbone in the minor groove of DNA. Interestingly, the SmcR model we built contains several sulfate molecules originating from the crystallization solution. They may represent the phosphate backbone in the DNA structure. Two sulfate molecules directly interact with Arg-9, Arg-32 and Arg-36, respectively (supplemental Fig. S2).
To evaluate the contribution of these residues to DNA binding, we first performed site-directed mutagenesis and analyzed the DNA binding affinity by EMSA using the PvvpE-containing operator region DNA. As shown in Fig. 5a, the wild-type SmcR bound to the DNA probe in a concentration-dependent manner, whereas most of the mutants, including the deletion mutant (Asp-2 to Arg-11), were unable to bind to the DNA probe. Arg-7 and Gln-47, which are indeed located far from the predicted DNA-binding site and were used as negative control residues, did not affect DNA binding. His-39 in the second helix is a residue involved in the hydrophobic packing of α-helices in the DNA binding domain (Fig. 3c). It is predicted that His-39 may either interact with the phosphate backbone of DNA or may be involved in maintaining the stabilization of Phe-54 by stacking its imidazole ring and the aromatic side chain of Phe-54. Mutations at the His-39 and Phe-54 positions resulted in the structural instability of SmcR as measured by DSC (Fig. 5b). Therefore, it is suggested that both His-39 and Phe-54 are critical for maintaining the structural integration of the DNA recognition motif.
FIGURE 5.
Mutational analysis of putative DNA-binding residues. a, electrophoretic gel mobility shift assay for binding of SmcR mutants to the vvpE regulatory region. A 200-bp DNA fragment from the upstream region of vvpE was radioactively labeled and then used as a DNA probe. The radiolabeled fragments (20 ng) were mixed with wild-type or mutant SmcRs (100, 150, and 200 nm in 1st to 3rd lanes, respectively) and were then resolved on a 4% polyacrylamide gel. N, no protein; WT, wild-type SmcR; Dele, deletion mutant (Asp-2 to Arg-11); B, bound DNA; F, free DNA. b, effect of putative DNA recognition residues on the stability of the helix-turn-helix motif. Tmvalue of each protein is indicated. c, effects of putative DNA-binding residues on the positive transcriptional activation of SmcR in the PvvpE::lacZ transcriptional fusion system in V. vulnificus DH0602. The phenotypes were assessed by cellular β-galactosidase activities. NO, DH0602 containing pJH0311.
We further evaluated the residues involved in DNA binding in vivo using the PvvpE region activated by the SmcR and lacZ transcriptional fusion assay system, and the resulting phenotypes can be quantified by β-galactosidase activity in V. vulnificus. Five mutants were selected for the experiment, and of these, the Q47A mutant was used as a positive mutant, which would have no effect on enzyme activity. As expected, SmcR had no effect on the regulation of β-galactosidase activity in the V. vulnificus reporter strain maintaining the Q47A mutant. A remarkable effect on enzyme activity was shown in the Arg-11, Arg-36, and Phe-54 mutants (Fig. 5c) that were completely defective for DNA binding in vitro (Fig. 5a). The Arg-122 mutant, which showed modest DNA binding ability in vitro, still possessed some remaining enzyme activity.
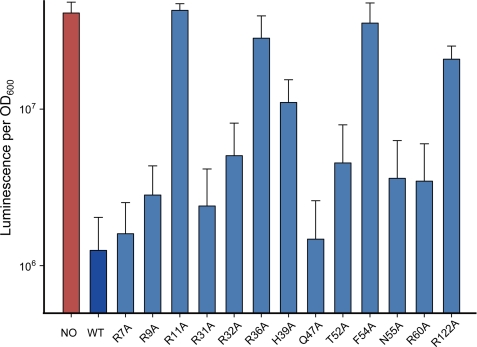
Interaction with a Negative Transcriptional Operator
We further performed interaction studies of negative regulation of VV2_1398 by SmcR. The operator region was used to assess the effect of the PvvpE-binding residues involved in the transcriptional activation of vvpE on the repression ability of SmcR using the luminescence assay system in E. coli. In this system, wild-type SmcR should not induce luminescence, due to its function as a repressor. As shown in Fig. 6, the strains carrying the R11A, R36A, and F54A mutations showed high luminescent activity. Other mutations such as H39A and R122A also affected the repression activity of SmcR. These observations are comparable with the EMSA results, and the β-galactosidase assay results in V. vulnificus, which revealed residues critical for positive transcriptional regulation by SmcR. Thus, the results suggest that SmcR may regulate its target genes through the same DNA binding mode for both positive and negative regulation.
FIGURE 6.
Effects of putative DNA-binding residues on the negative transcriptional repression of SmcR in the PVV2_1398::luxAB transcriptional fusion system in E. coli DH5α. Relative light units were calculated by dividing the luminescence by the A600 of each strain cultured as described in the text. NO, DH5α containing empty pBAD24BS vector and pKS0710; WT, wild type.
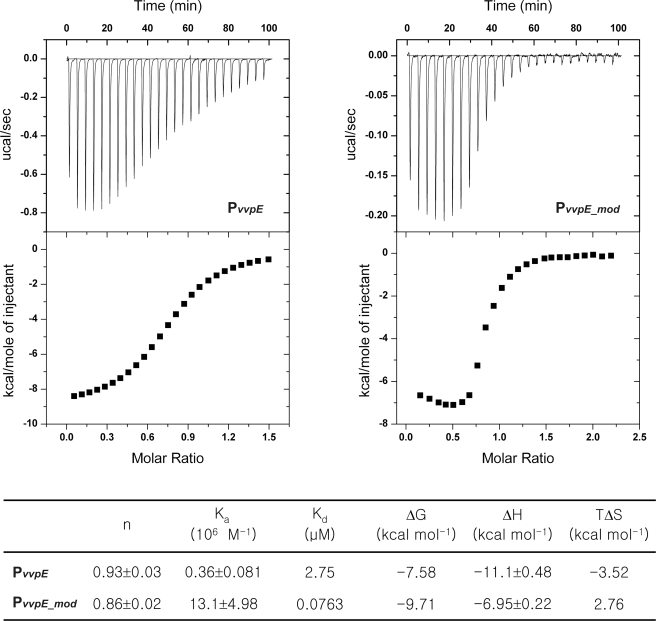
ITC Analysis of SmcR-DNA Interactions
ITC was used to accurately determine the binding parameters of SmcR with its target operator PvvpE, as well as the artificially synthesized pseudo-palindromic operator PvvpE-mod. Fig. 7 shows representative ITC results, including titration data and binding curves calculated using the best fit parameters. The ITC result revealed the binding stoichiometry of dimeric SmcR with duplex PvvpE, with a dissociation constant KD of 2.8 μm. The interaction of SmcR with pseudo-palindromic PvvpE-mod had a much higher affinity, with a KD of 76 nm (see “Discussion”).
FIGURE 7.
Isothermal titration calorimetry of SmcR and DNA interactions. Titrations of PvvpE and PvvpE-mod against SmcR were performed. Upper panel, raw data obtained from 25 automatic injections of 6-μl aliquots of SmcR against duplex DNA. Lower panel, integration plot of the data calculated from the raw data. Each set of duplex DNA is indicated in the panel. Thermodynamic values are shown in the table.
DISCUSSION
Biological Implications of SmcR Dimerization
The results of this study, obtained using the bacterial two-hybrid system in E. coli, verified that SmcR exists in a dimeric form in bacterial cells, a fact that could also be deduced during its purification and from its structure. From the results, it appears that the dimerization of SmcR is not mediated by a ligand, unlike the Vibrio fischeri LuxR homologues involved in the LuxI/LuxR type quorum-sensing systems, which form homodimers only in the presence of signaling molecules (34, 35). The in vitro and in vivo mutagenesis experiments for the residues at the dimer interface clearly demonstrated that the dimerization of SmcR is essential for its DNA binding and biological regulatory functions. Therefore, dimerization of the LuxR homologues from Vibrio spp. may also be important for their biological function.
On the other hand, although the ligand for SmcR has not yet been found, structural investigations revealed that the protein forms a putative ligand-binding site within the dimerization domain. The CASTp server (38) predicted a putative ligand-binding site in each SmcR monomer. The site forms an amphipathic pocket generated by Phe-75, Phe-78, Ile-96, Met-100, Trp-114, Phe-115, Ser-118, Phe-129, Val-130, Asn-133, Asn-136, Gln-137, Val-140, Ala-163, Asn-164, Phe-166, His-167, and Cys-170 (supplemental Fig. S3). The calculated volume of the pocket is 468.1 Å3. In our structure, an area of ambiguous electron density was observed at this site in each molecule. From a structural point of view, we do not rule out the involvement of a ligand that may control a biological function of SmcR in the quorum-sensing system of V. vulnificus.
DNA Recognition and Transcriptional Regulation by SmcR
In a previous study, it was suggested that SmcR is a master regulator of quorum sensing in V. vulnificus (12). Around 120 genes could be under the control of SmcR, which functions as both activator and repressor; however, the molecular mechanism governing the regulatory role of SmcR has not yet been studied in detail. Therefore, in this study, we tested the molecular recognition mode of SmcR for transcriptional activation using the PvvpE-fused lacZ transcriptional assay system (Fig. 5c), as well as for transcriptional repression using the negatively regulated VV2_1398 fused luminescence assay system (Fig. 6). The results showed the same DNA-binding mode of SmcR for both positive and negative regulation.
How then does SmcR recognize and regulate so many genes in a specific manner? In a previous study (12), the DNA recognition site of SmcR showed a 22-bp consensus sequence containing a pseudo-2-fold symmetry pattern with less sequence conservation in the right half of the repeat. The sequence conservation scored highest in the end sequences of the left half of the repeat. The low level of conservation in the right half of the repeat might be attributed to the different target protein binding affinities with SmcR. This may be an indication that SmcR can regulate a large variety of genes by controlling their binding affinities in a tightly regulated cell system. To demonstrate this unusual feature, we performed an ITC experiment and determined the binding affinity of SmcR with the chemically synthesized PvvpE and pseudo-palindromic PvvpE-mod (Fig. 7). The ITC result revealed an increased binding affinity of about 35-fold between PvvpE-mod and SmcR compared with that of PvvpE. Although PvvpE-mod showed much higher affinity with SmcR, our ongoing study concerning the regulation of SmcR in V. vulnificus showed that the activity of PvvpE is higher than that of PvvpE_mod in the in vivo PvvpE::lacZ transcriptional fusion reporter system.5 These results lend support to the hypothesis that SmcR may have different binding affinities for its target operators, which show sequence variation in the right repeat of the consensus sequence (12). Such variation in sequence and binding affinity may be the regulatory mechanism by which SmcR recognizes each of the ∼120 target genes with different affinities. The V. harveyi LuxR also appeared to have different binding affinities with its target-binding sites (13). To confirm this mode of regulation, an evaluation of the binding affinity with target operators under the control of SmcR should be performed.
In conclusion, we have made an effort to extend our knowledge of the transcriptional regulation mediated by SmcR, a homologue of the quorum-sensing master regulator, LuxR, in the pathogen V. vulnificus. Recent studies have demonstrated that LuxR homologues regulate multitranscriptional events in Vibrio spp. (3). Our study provides a platform for drug development against pathogenic Vibrios, as further investigations into the molecular mechanisms underlying transcriptional regulation by the quorum-sensing master regulator should lead to the development of novel strategies for new antibiotic therapies.
Supplementary Material
Acknowledgments
We thank Drs. Kyung-Jin Kim and Ghyung-Hwa Kim at the Pohang Accelerator Laboratory for help with data collection.
This work was supported by the 21st Century Frontier Microbial Genomics and Applications Center Program (to M. H. K.) and from the National Research Laboratory Program (R0A-2007-000-200390 to S. H. C.) funded by the Ministry of Education, Science, and Technology, Republic of Korea.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S3.
The atomic coordinates and structure factors (code 3KZ9) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
D. H. Lee and S. H. Choi, unpublished data.
- SeMet
- selenomethionine
- PDB
- Protein Data Bank
- DSC
- differential scanning calorimetry
- EMSA
- electrophoretic mobility shift assay
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1.Bassler B. L., Losick R. (2006) Cell 125, 237–246 [DOI] [PubMed] [Google Scholar]
- 2.Waters C. M., Bassler B. L. (2005) Annu. Rev. Cell Dev. Biol. 21, 319–346 [DOI] [PubMed] [Google Scholar]
- 3.Ng W. L., Bassler B. L. (2009) Annu. Rev. Genet. 43, 197–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milton D. L. (2006) Int. J. Med. Microbiol. 296, 61–71 [DOI] [PubMed] [Google Scholar]
- 5.Valiente E., Bruhn J. B., Nielsen K. F., Larsen J. L., Roig F. J., Gram L., Amaro C. (2009) FEMS Microbiol. Ecol. 69, 16–26 [DOI] [PubMed] [Google Scholar]
- 6.Kim S. Y., Lee S. E., Kim Y. R., Kim C. M., Ryu P. Y., Choy H. E., Chung S. S., Rhee J. H. (2003) Mol. Microbiol. 48, 1647–1664 [DOI] [PubMed] [Google Scholar]
- 7.Roh J. B., Lee M. A., Lee H. J., Kim S. M., Cho Y., Kim Y. J., Seok Y. J., Park S. J., Lee K. H. (2006) J. Biol. Chem. 281, 34775–34784 [DOI] [PubMed] [Google Scholar]
- 8.McDougald D., Rice S. A., Kjelleberg S. (2000) Gene 248, 213–221 [DOI] [PubMed] [Google Scholar]
- 9.Jeong H. S., Lee M. H., Lee K. H., Park S. J., Choi S. H. (2003) J. Biol. Chem. 278, 45072–45081 [DOI] [PubMed] [Google Scholar]
- 10.McDougald D., Rice S. A., Kjelleberg S. (2001) J. Bacteriol. 183, 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao C. P., Hor L. I. (2001) J. Bacteriol. 183, 1369–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee D. H., Jeong H. S., Jeong H. G., Kim K. M., Kim H., Choi S. H. (2008) J. Biol. Chem. 283, 23610–23618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pompeani A. J., Irgon J. J., Berger M. F., Bulyk M. L., Wingreen N. S., Bassler B. L. (2008) Mol. Microbiol. 70, 76–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Silva R. S., Kovacikova G., Lin W., Taylor R. K., Skorupski K., Kull F. J. (2007) J. Bacteriol. 189, 5683–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goo S. Y., Lee H. J., Kim W. H., Han K. L., Park D. K., Lee H. J., Kim S. M., Kim K. S., Lee K. H., Park S. J. (2006) Infect. Immun. 74, 5586–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheffield P., Garrard S., Derewenda Z. (1999) Protein Expr. Purif. 15, 34–39 [DOI] [PubMed] [Google Scholar]
- 17.Kim M. H., Kim Y., Park H. J., Lee J. S., Kwak S. N., Jung W. H., Lee S. G., Kim D., Lee Y. C., Oh T. K. (2008) J. Biol. Chem. 283, 31981–31990 [DOI] [PubMed] [Google Scholar]
- 18.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 19.Terwilliger T. C., Berendzen J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 571–579 [DOI] [PubMed] [Google Scholar]
- 20.Terwilliger T. C. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 22.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 23.Vagin A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 24.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 25.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karimova G., Pidoux J., Ullmann A., Ladant D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horton R. M., Ho S. N., Pullen J. K., Hunt H. D., Cai Z., Pease L. R. (1993) Methods Enzymol. 217, 270–279 [DOI] [PubMed] [Google Scholar]
- 28.Jeong K. C., Jeong H. S., Rhee J. H., Lee S. E., Chung S. S., Starks A. M., Escudero G. M., Gulig P. A., Choi S. H. (2000) Infect. Immun. 68, 5096–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeong H. S., Jeong K. C., Choi H. K., Park K. J., Lee K. H., Rhee J. H., Choi S. H. (2001) J. Biol. Chem. 276, 13875–13880 [DOI] [PubMed] [Google Scholar]
- 30.Miller J. H. (1976) Experiments in Molecular Genetics, pp. 352–355, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31.Holm L., Sander C. (1996) Science 273, 595–603 [DOI] [PubMed] [Google Scholar]
- 32.Schumacher M. A., Miller M. C., Grkovic S., Brown M. H., Skurray R. A., Brennan R. G. (2001) Science 294, 2158–2163 [DOI] [PubMed] [Google Scholar]
- 33.Pinto U. M., Winans S. C. (2009) Mol. Microbiol. 73, 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu J., Winans S. C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1507–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin Y., Luo Z. Q., Smyth A. J., Gao P., Beck von Bodman S., Farrand S. K. (2000) EMBO J. 19, 5212–5221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Shea E. K., Klemm J. D., Kim P. S., Alber T. (1991) Science 254, 539–544 [DOI] [PubMed] [Google Scholar]
- 37.Schumacher M. A., Miller M. C., Grkovic S., Brown M. H., Skurray R. A., Brennan R. G. (2002) EMBO J. 21, 1210–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J. (2006) Nucleic Acids Res. 34, W116–W118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.