Abstract
Many proteins contain conformationally flexible segments that undergo significant changes in the backbone conformation or completely lack a welldefined conformation. Previously, we have developed the generalized local propensity (GLP), a quantitative sequence-based measure of the protein backbone flexibility. In this paper, we present the CFP (Conformational Flexibility Profile) webserver that constructs the GLP flexibility profile for a usersubmitted sequence and uses this profile to identify segments with high backbone flexibility. The statistical significance of a flexible sequence segment is assessed using the discrete scan statistics based on the density of flexible residues observed in this segment.
Availability
CFP is publicly available at http://cfp.rit.albany.edu
Keywords: conformational variability, protein backbone, flexibility, local propensity, sequence
Background
Many proteins contain conformationally flexible segments. These segments undergo significant changes in backbone conformation, or are completely disordered (lack a well-defined structure) [1–3]. A quantitative representation of the conformational flexibility of the protein backbone is important for many applications. Previously, we developed generalized local propensity (GLP), a quantitative sequence-based measure of backbone flexibility [4]. The GLP can be used to construct sequence-based protein flexibility profiles, and provides an objective numeric threshold for defining conformationally flexible segments [5]. For a given sequence position k, the GLP measures the width of the context-dependent distribution of backbone conformations accessible to this position, glp(k) (see references [4–5] for details). If glp(k) ≥ 1, it indicates that sequence position k is conformationally flexible.
Here, we present the CFP (Conformational Flexibility Profile) web server that constructs the GLP flexibility profile for a user-submitted sequence and uses this profile to identify segments with high conformational flexibility. Below is a brief outline of the steps implemented in CFP: The GLP flexibility profile is constructed for the query sequence and then smoothed using a sliding window of size W1. Consecutive positions which have GLP above a threshold T1 are merged into seed flexible segments. Each seed flexible segment is extended by adding extension windows of size W2 until its average GLP drops below an extension threshold T2. An extension window is added only if its average GLP is above a certain threshold T3. This extension procedure is similar to that used in the SEG program [6].
The extended flexible segments are reported in the final table. If the number of flexible residues observed in a given final flexible segment is unusually high (p-value ≪ 0.05), then this segment is marked as statically significant. The significance of the number of flexible residues is estimated using the discrete scan statistic. This statistical procedure is the same as the one we previously implemented in the BIAS software to identify statistically significant clusters of userspecified amino acid types [7–8]. The web-server is publicly available at http://cfp.rit.albany.edu.
Methodology
Input
The only mandatory input is the query protein sequence. All other input fields have default values that can be modified by advanced users, if desired. These input fields are described below. Instructions for each field and general information about the methodology and the output format can be found by clicking a corresponding help hyperlink on the input page.
Smoothing window size
The size of the sliding window (W1) used to smooth the raw profile. High values of W1 tend to reveal long flexible segments and mask the short ones. Lower values tend to reveal short segments.
GLP threshold for seed segments
The threshold T1 used to identify seed flexible segments. Contiguous sequence positions that have values of the smoothed GLP profile above this threshold are merged into a seed flexible segment.
Extension threshold
Each seed segment with high flexibility is extended on both sides until its average GLP drops below this threshold (T2)
Extension window threshold
The ends of a seed flexible segment are extended if the extension window has the average GLP above this threshold (T3).
Extension window size
The size of the extension window (W2).
Hatshaped local smoother
Positions in the center of the smoothing window contribute more to the smoothed GLP score than positions at the ends of the window.
Equal weights smoother
The smoothed GLP score is the unweighted average computed over all positions in the window.
Minimum seed segment
Seed segments with length smaller than this threshold are not extended.
Maximum separation between merged segments
Flexible segments separated by this or smaller number of positions are merged into one.
Flexible residues
A set of flexible residues used in the scan statistics to estimate the statistical significance of flexible segments (G, H, D, N by default).
SWIS-PROT or PDB background frequencies
The amino acid frequencies of the SwissProt or Protein Databank are used to estimate the statistical significance.
X axis size, Y axis size
The size of X and Y axis of the plot in pixels.
Create a plot
Display the smoothed GLP profile in webbrowser.
Create a text file
Save the raw and smoothed GLP profiles in a text file.
Output
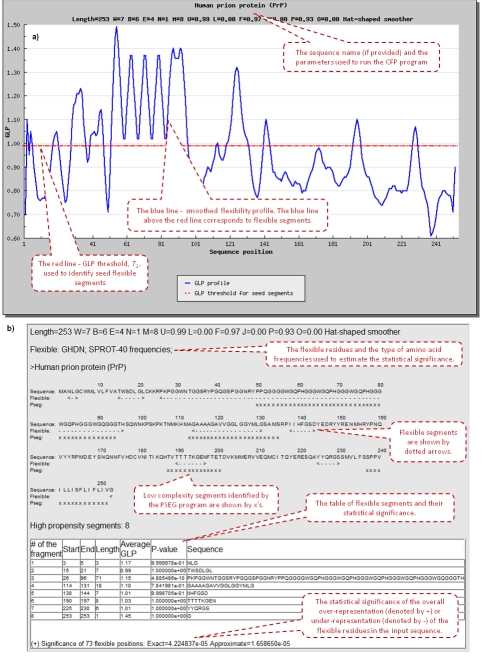
The CFP output consists of two parts. The first part shows the smoothed GLP plot of the input sequence (Figure 1A). The second part shows the detailed information for every flexible segment found in the input sequence and the pvalues that provide the estimates of the statistical significance (Figure 1B). If the pvalue for a given segment is less than 0.05, this segment has an unusually high density of residues with high degree of backbone flexibility.
Figure 1.
a) First part of the output the GLP flexibility plot; b) Second part of the output the summary of all flexible segments.
Acknowledgments
This work was supported by grant number R03LM009034 from the National Library of Medicine/NIH, and by grant number LM06789 from the National Library of Medicine/NIH.
Footnotes
Citation:Kuznetsov & Rackovsky, Bioinformation 4 (5): 176-178 (2009)
References
- 1.Alexandrov V, et al. Protein Sci. 2005;14:633. doi: 10.1110/ps.04882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kosloff M, Kolodny R. Proteins. 2008;7:891. doi: 10.1002/prot.21770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunker AK, et al. BMC Genomics. 2009;9:S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuznetsov IB, Rackovsky S. Protein Sci. 2003;12:2420. doi: 10.1110/ps.03209703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuznetsov IB, Rackovsky S. Protein Sci. 2004;13:3230. doi: 10.1110/ps.04833404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wootton JC, Federhen S. Methods Enzymol. 1996;266:554. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 7.Kuznetsov IB, Hwang S. Bioinformatics. 2006;22:1055. doi: 10.1093/bioinformatics/btl049. [DOI] [PubMed] [Google Scholar]
- 8.Kuznetsov IB. Bioinformatics. 2008;24:1534. doi: 10.1093/bioinformatics/btn233. [DOI] [PubMed] [Google Scholar]