Abstract
Vascular system development involves a complex, three-dimensional branching process that is critical for normal embryogenesis. In the brain, the arterial systems appear to develop in a stereotyped fashion, but no detailed quantitative analyses of the mouse embryonic cerebral arteries have been described. In this study, a gadolinium-based contrast perfusion method was developed to selectively enhance the cerebral arteries in fixed mouse embryos. Three-dimensional magnetic resonance micro-imaging (micro-MRI) data were acquired simultaneously from multiple embryos staged between 10 and 17 days of gestation, and a variety of image analysis methods was used to extract and analyze the cerebral arterial patterns. The results show that the primary arterial branches in the mouse brain are very similar between individuals, with the patterns established early and growth occurring by extension of the segments, while maintaining the underlying vascular geometry. To investigate the utility of this method for mutant mouse phenotype analysis, contrast-enhanced micro-MRI data were acquired from Gli2-/- mutant embryos and their wild-type littermates, showing several previously unreported vascular phenotypes in Gli2-/- embryos, including the complete absence of the basilar artery. These results demonstrate that contrast-enhanced micro-MRI provides a powerful tool for analyzing vascular phenotypes in a variety of genetically engineered mice.
Keywords: angiogenesis, basilar artery, carotid artery, Gli2 mutant mice, vascular development
During mammalian embryogenesis, the vasculature forms as a highly complex three-dimensional (3D) system of interconnected blood vessels that provide the developing tissues with essential nutrients, growth factors, and oxygen. Studies using genetically engineered mice have provided critical data on the genetic and molecular factors controlling vascular development during embryogenesis and also during disease progression involving abnormal formation of blood vessels. In these studies, quantitative 3D analysis of vascular patterns is seldom performed since commonly used histologic methods are inherently two dimensional and prone to sectioning artifacts, making 3D reconstructions difficult. Volumetric imaging methods such as magnetic resonance micro-imaging (micro-MRI) could provide powerful new tools to study vascular patterning in both normal and mutant mouse embryos.
Vascular development during embryogenesis can be divided into two key processes: vasculogenesis and angiogenesis. The earliest step, vasculogenesis, involves the recruitment and assembly of endothelial-cell precursors, called angioblasts, into a primary vascular plexus (1). These primitive blood vessels are then extended and elaborated by the process of angiogenesis, which involves cell proliferation, differentiation of the arterial and venous components, and the establishment of the final 3D patterns of the vascular networks (2). While vasculogenesis is a process that occurs only during early embryonic development, angiogenesis is an ongoing process that can also occur after birth, depending on the needs of different tissues, which can be advantageous, as in angiogenesis after ischemia (3), or pathologic, as in tumor angiogenesis (4).
The development of the mammalian vasculature results in apparently stereotypical patterns in a number of vascular subsystems, including the cerebral arteries. However 3D vascular patterning during brain development has not been investigated in detail, in large part due to the lack of analytical tools. We are particularly interested in analyzing the 3D patterns of the cerebral arterial inputs and how these patterns change during normal development, and in mouse mutants after genetic alterations that disrupt vascular patterning. The primary vascular inputs into the developing brain are the basilar artery that perfuses the more posterior parts of the brain and the carotid arteries that supply blood to the more anterior brain regions. In this report, we demonstrate contrast-enhanced micro-MRI methods that enable 3D analysis of both cerebral arterial systems over a wide range of embryonic stages.
Materials and Methods
Animals
All procedures involving mice were approved by the Institutional Animal Care and Use Committee at New York University School of Medicine. Timed-pregnant ICR strain mice (Taconic Farms, Germantown, NY) were used for studies of normal embryonic development, where the stage was denoted as the embryonic day (E) following identification of a vaginal plug after overnight mating (E0.5 was defined as noon of the plug day). Embryos were analyzed at stages between E10.5 and E17.5. For mutant analysis, Gli2 heterozygous mutant mice were bred to generate Gli2-/- homozygous null mutant embryos, as well as Gli2+/- (heterozygous mutant) and Gli2+/+ (wild type) littermate controls. Polymerase chain reaction of embryo yolk sac DNA was used to identify genotypes, using primers for Neo and wild-type Gli2, as previously described (5).
Embryo Preparation for Micro-MRI
The procedure for vascular perfusion-fixation of mouse embryos was very similar to the protocol described previously by Smith et al. (6,7) and Smith (8). Pregnant mice were anesthetized using a ketamine (100 mg/mL):xylazine (20 mg/mL) mixture (0.2 mL/30g body weight, injected intraperitoneally). Individual embryos were surgically removed from the uterus after laparotomy, maintaining the vascular connections to the placenta and warmed in 37°C phosphate-buffered saline. Phosphate-buffered saline with heparin (5000 units/L) was perfused through the umbilical vein at a rate of 0.15 mL/min using an Ismatec Planetary Gear Driven Digital Pump (Cole-Parmer, Vernon Hills, IL), followed by perfusion of a fixative perfusion (2% [volume/volume] glutaraldehyde/1% formalin in phosphate-buffered saline at 300 mOsm/L), and finally the contrast agent, consisting of bovine serum albumin (BSA) conjugated to diethylenetriaminepentaacetic acid (DTPA) and chelated to gadolinium (BSA-DTPA-Gd; 1 mM), was dissolved in a 10% (wt/vol) gelatin solution (Sigma-Aldrich, St. Louis, MO) and perfused through the umbilical vein, allowing the contrast agent to completely fill the embryonic circulatory system. After perfusion, the umbilical vessels were securely tied with silk suture (Sofsilk 5-0; Fisher Scientific, Pittsburgh, PA), and the embryos were immersed in 4°C fixative to solidify the gelatin and to completely fix the tissues. Embryo samples were then maintained at 4°C until imaging with micro-MRI. Nontoxic food dyes were utilized to visualize the successive perfusions of phosphate-buffered saline, fixative, and contrast agent/gelatin under a dissection microscope (Nikon, Tokyo, Japan).
To selectively enhance the cerebral arteries, the BSA-DTPA-Gd contrast agent/gelatin solution was perfused through the umbilical artery rather than vein and stopped after 15 sec (E12.5), 25 sec (E15.5) or 45 sec (E17.5), when the embryo head first changed color, indicating that the contrast agent had reached the brain. The embryos were then immersed overnight in 4°C fixative and prepared for micro-MRI exactly as described above.
Micro-MRI of Mouse Embryos
MRI experiments were performed on a 7-T micro-MRI system, consisting of a Biospec Avance II console (Bruker Biospin MRI, Ettlingen, Germany) interfaced to a 200-mm horizontal bore, superconducting magnet (Magnex Scientific, Yarnton, UK) with an actively shielded gradient coil (Bruker BGA-9S; 90-mm inner diameter, 750-mT/m gradient strength, 100-μs rise time). A quadrature Litz coil (Doty Scientific, Columbia, SC) (inner diameter = 25 mm; length = 22 mm) was used to image the embryos mounted inside a 30-mL syringe (Fig. 1). To increase throughput, multiple embryos (Table 1) were glued into place (Krazy Glue; Elmer's Products Inc., Columbus, OH) in the 30-mL syringe, on each side of the plunger. The mouse embryo samples were then immersed in Fomblin (Solvay Solexis; Fisher Scientific), a liquid perfluoroether that matches magnetic susceptibility without producing background signal. Imaging was performed overnight with a 3D T1-weighted gradient echo sequence (echo time, TE = 5 ms; repetition time, TR = 50 ms; flip angle = 35°; field of view = 25.6-mm; matrix size = 512 (3); isotropic resolution = 50 μm; total imaging time = 14 h 35 min).
FIG. 1.
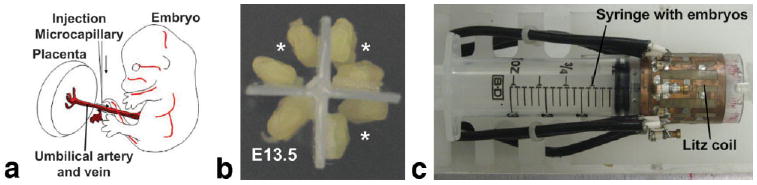
Contrast agent perfusion-fixation was used for vascular imaging. a: Embryos were carefully extracted from pregnant anesthetized female ICR mice and incisions were made in both the umbilical artery and vein, through which perfusion of fixative and contrast agent was performed with glass microcapillary needles. b: Multiple embryos were mounted onto the plunger of a 30-mL syringe and (c) imaged simultaneously in a quadrature Litz coil. In the example shown in (b), 6 E12.5 embryos (*) were mounted together with one E13.5 embryo.
Table 1.
Stage-Dependent Number of Embryos for Multiple Embryo Micro-MRI
| E10.5-12.5 | E13.5-15.5 | E16.5-18.5 | |
|---|---|---|---|
| Embryonic stage | |||
| Max number of embryos | 8 | 4 | 2 |
Image Analysis
Image analysis and segmentation were performed using the Analyze software package (V7.0; AnalyzeDirect, Overland Park, KS). First, 16-bit micro-MRI datasets were imported into Analyze to allow visualization and quantitative analysis. The following steps were then performed for each dataset: (i) 3D maximum intensity projections (MIPs) of the embryonic vasculature were computed using a user-defined threshold and automatically segmented using the “Tree Analysis” function, in which a 3D skeletal structure is automatically extracted from a volume of MR data. The tree can be displayed in conjunction with a volume-rendered image and edited, if necessary, to remove gaps due to insufficient vascular contrast in approximately 20% of the samples analyzed; (ii) segmentations were refined by adjusting the threshold for the computation of the MIPs; and (iii) quantitative data on the branching angles and segment lengths were obtained automatically using the “Stats” function, which allows the user to set parameters for the calculation of tree statistics, including calculations of branch lengths, angles, branching area, and brightness area product. For quantification, bilateral measurements of each parameter were averaged in each embryo before statistical analyses. Signal-to-noise ratio was measured as the mean signal within a region of interest in the brain, divided by the standard deviation of the signal in a region of interest in the background (Fomblin).
For coregistration analysis at E17.5, images were processed to form an average image representative, as described in previous reports (9-13). The average was formed through a series of registration steps, using software provided by the Mouse Imaging Centre (Toronto, Canada) and utilizing tools available from the Montreal Neurologic Institute (Montreal, Canada). Briefly, images were registered to a reference to remove extrinsic positional differences, and the image intensities were corrected for spatial nonuniformity and normalized (14). An unbiased average was created from an average of all the pairwise 12-parameter registrations. The average was subsequently refined through an iterative process of coregistering all images to the average at progressively finer scales, through six iterations at grid resolutions of 700, 600, 500, 240, 120 and 60 μm (11).
For quantitative analysis, the two-tailed Student's t test was used to test for statistical significance between measurements of embryos at different stages of development.
Histology
For confirmation of basilar artery phenotype, histologic analyses were performed. After micro-MRI, embryos were removed from Fomblin and washed for 10 min in 0.1% phosphate-buffered saline–Triton X-100 (Sigma-Aldrich). Following equilibration with 15% and 30% sucrose solution, embryos were embedded in optimal cutting temperature compound (Tissue-Tek; Sakura Finetechnical Co., Tokyo, Japan) and frozen for cryosectioning (16-μm sections). Immunohistochemistry for detection of CD31 used a standard protocol for staining of frozen sections using rat anti-CD31 antibody (BD Biosciences, San Jose, CA) at 1:100 dilution and biotinylated secondary antibody (biotinylated polyclonal goat anti-rat immunoglobulin; BD Biosciences). Tissues were then treated with streptavidin-horseradish peroxidase (BD Biosciences) followed by incubation in a 10 mg/mL 3,3′-diaminobenzidine tetrahydrochloride solution (Sigma-Aldrich). Images were acquired on a Leitz DM RXE microscope (Leica Microsystems, Wetzlar, Germany) with a charge-coupled device camera (Spot Insight; Diagnostic Instruments, Sterling Heights, MI).
Results
Contrast-Enhanced Micro-MRI Reveals Embryonic Vasculature
As previously reported (6,7,15), umbilical venous perfusion of the BSA-DTPA-Gd contrast agent/gelatin solution resulted in pronounced enhancement of the major embryonic vascular structures on 3D T1-weighted micro-MRI (Fig. 2). MIP processing was especially useful for visualizing the 3D vascular patterns (Fig. 2d), enabling analysis of the vasculature in fixed mouse embryos over a wide range of embryonic stages, starting at E10.5 (Fig. 3). Major blood vessels, including the aorta, cardinal vein/vena cava, and umbilical artery and vein, were readily identified from E10.5, while elaboration of the hepatic vasculature was apparent from E12.5, and the intersomitic vessels could be seen as early as E11.5. In the head, the major blood vessels were visualized starting at E11.5, especially the large veins in the facial region surrounding the brain.
FIG. 2.
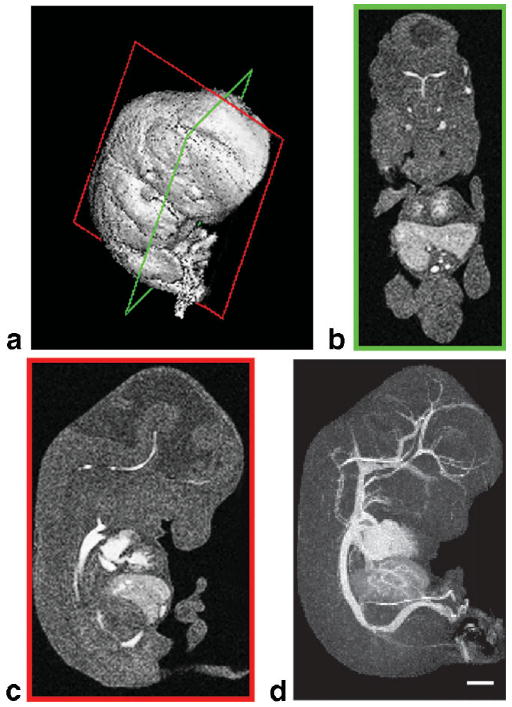
Image processing methods for visualizing the embryonic vasculature. a: After micro-MRI, surface rendering shows the full 3D dataset from an E12.5 mouse embryo. Coronal (green frame, b) and sagittal (red frame, c) sections demonstrate the contrast agent filling the embryonic vascular spaces. d: 3D MIP of the same data, viewed from the side, reveals the full extent of the contrast-enhanced vascular system. Scale bar = 1 mm.
FIG. 3.
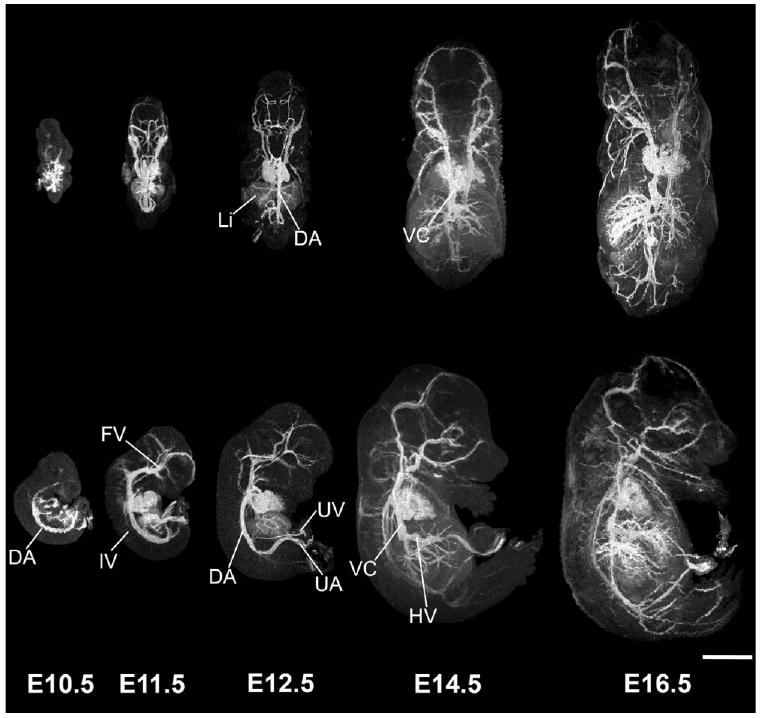
3D MIPs show the developing vasculature. Individual embryos, staged between E10.5 and E16.5 are viewed dorsally (top panels) and from the side (bottom panels), showing the increase in vascular pattern complexity at each developmental stage. Abbreviations: dorsal aorta, DA; vena cava, VC; intersomitic vessels, IV; facial vein, FV; umbilical vein, UV; umbilical artery, UA. Scale bar = 2.5 mm.
First-Pass Retrograde Perfusion to Enhance Cerebral Arteries
There are two main arterial input systems in the developing embryonic brain: the (medial) basilar artery and the (bilateral) carotid arteries. As expected from previous reports (6,7), anterograde (in the normal direction of blood flow) contrast perfusion through the umbilical vein resulted in enhancement of both arteries and veins (Figs. 2, 3). In the MIPs of the embryonic mouse brain, this tended to emphasize the larger draining veins surrounding the brain, making it difficult to identify and analyze the smaller cerebral arteries (Fig. 4a). To enable more effective visualization of the arterial systems, we developed a first-pass retrograde perfusion protocol to deliver the contrast agent more directly to the basilar and carotid arterial systems, stopping the perfusion before the contrast agent filled the larger venous vessels (Fig. 4b). This method was more effective at later embryonic stages (E17.5) compared to earlier stages (E12.5, E15.5; Fig. 5). This was likely due to the maturation of the cardiac valves at later embryonic stages, resulting in a more effective bypass of the heart and delivery of contrast agent to the brain during retrograde perfusion (Fig. 4). Despite partial venous labeling at the earlier stages, the first-pass retrograde perfusion method improved the visualization of the arterial anatomy in the brain from E12.5, enabling analysis of many arterial branches, including the basilar artery, anterior inferior cerebellar arteries, posterior cerebral arteries, middle cerebral arteries, posterior communicating arteries, internal carotid arteries, and anterior cerebral arteries (Fig. 5).
FIG. 4.
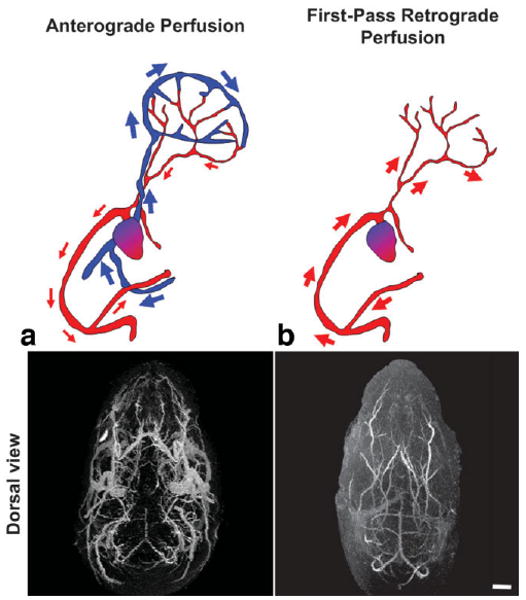
Modified perfusion protocol to enhance the cerebral arteries. a: Anterograde perfusion through the umbilical vein of an E17.5 embryo enhances both the venous and arterial blood vessels in the brain and head, as viewed from above (dorsal). b: First-pass retrograde perfusion through the umbilical artery of an E17.5 embryo selectively enhances the cerebral arteries. Scale bar = 1-mm.
FIG. 5.
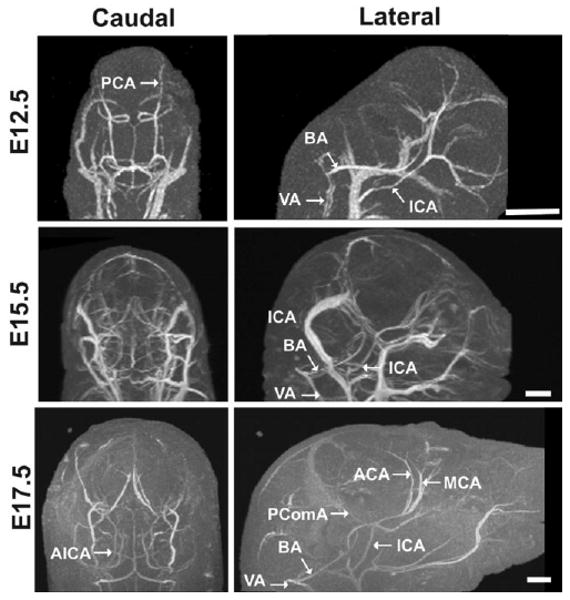
Micro-MRI demonstrated cerebral artery development from E12.5 to E17.5. Views from the back (caudal; left panels) and side (lateral; right panels) show the cerebral vasculature of E12.5 (top panels), E15.5 (middle), and E17.5 (bottom panels) embryos using the first-pass retrograde perfusion method. Venous structures were most effectively eliminated at E17.5, but the major cerebral arteries were easily visualized at all three stages. Abbreviations: basilar artery, BA; anterior inferior cerebellar artery, AICA; posterior cerebral artery, PCA posterior communicating artery, PComA; middle cerebral artery, MCA; anterior cerebral artery, ACA; internal carotid arteries, ICA; vertebral arteries, VA. Scale bars = 1 mm.
3D Analysis of Embryonic Cerebral Arteries
To examine the degree of geometric similarity between cerebral artery patterns in same stage mouse embryos, we performed 3D image coregistration at E17.5 (N = 6; Fig. 6). After image coregistration and averaging, the resulting brain images showed an increase in signal-to-noise ratio from 27 ± 10 (N = 6) in individual images compared to 46 in the averaged image. Since the volume chosen for registration was limited to the brain, the averaging was most effective on the brain and cerebral blood vessels and produced some blurring and distortion in regions of the head and face further removed from the brain. The resulting averaged MIPs emphasized the primary arterial vessels, including the carotid and basilar arteries and the vessels making up the circle of Willis. In contrast, many of the secondary arterial branches of both the basilar and carotid systems were not preserved in the averaged MIPs. Surprisingly, the larger venous vessels that were largely suppressed in individual MIPs after first-pass retrograde perfusion became visible in the average MIPs as a result of the increased signal-to-noise ratio after registration and averaging. This result suggests that, similar to the arteries, the primary venous patterns in the head and brain are also stereotyped.
FIG. 6.
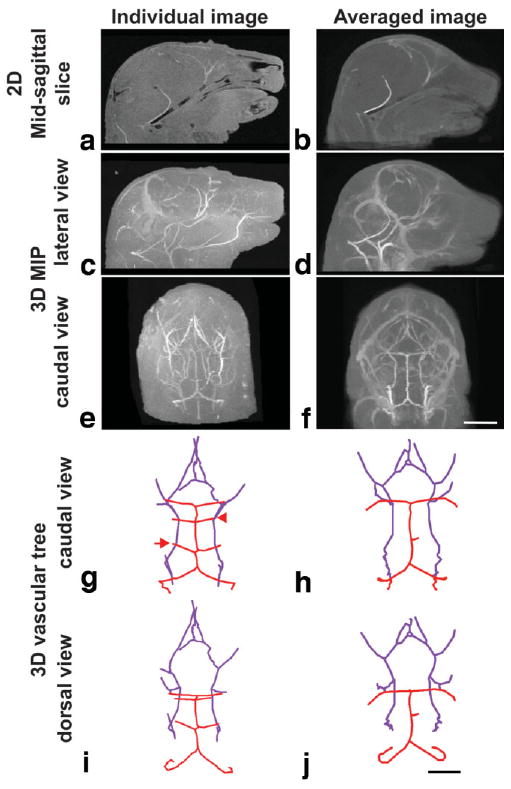
Image registration and averaging were performed to identify regions of similarity in the cerebral arteries. Differences are shown between an individual E17.5 embryo (left panels), and the registered/averaged image of six E17.5 embryos (right panels). Shown are 2D sagittal slices (a,b); lateral (c,d) and caudal (e,f) views of the MIPs and the extracted 3D vascular trees in caudal (g,h) and dorsal (i,j) views. In the vascular trees, the basilar artery system is shown in red, while the carotid artery system is in purple. Note the presence of the anterior inferior cerebellar artery (AICA, arrow) and the superior cerebellar artery (SCA, arrowhead) in the individual vascular trees not present in the registered/averaged trees. Scale bars = 250 μm. 1.75 mm (a–f); 1.5 mm (g–j).
To further assess the variability in the branching patterns between individual embryos, vascular “trees” were extracted automatically from the 3D MIPs of single embryos at each developmental stage (Fig. 7a). These 3D branching structures were then examined from different directions, and quantification of branch lengths and angles was performed. Analysis of six pairs of secondary branches revealed standard deviations of less than ± 20% for the lengths and angles at each embryonic stage (Fig. 7b,c). In general, cerebral arteries continuously grew in length between E12.5 and E17.5, with the largest changes occurring between E12.5 and E15.5 (33 ± 10% increase in length; Fig. 7d). Interestingly, the angles between branches were relatively constant throughout these stages of brain development, suggesting that the geometrical patterns are determined early and then grow by extending the segments between arterial branch points.
FIG. 7.
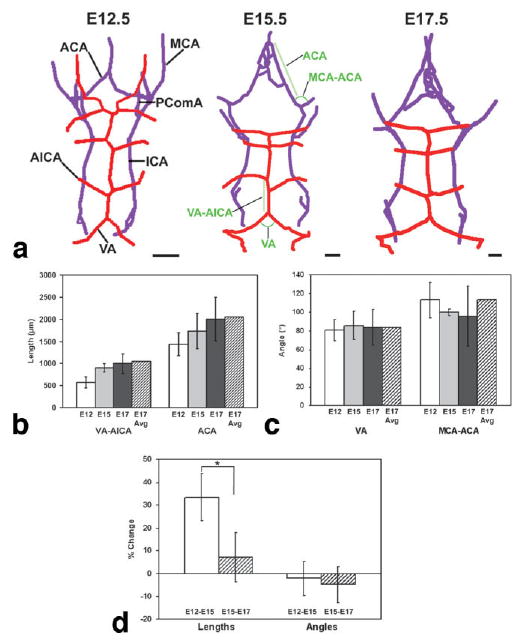
3D vascular trees were extracted and analyzed quantitatively. a: Vascular trees were extracted automatically, showing the basilar artery system (red) and the carotid artery system (purple) from a caudal (back) view at three stages: E12.5 (left); E15.5 (middle); and E17.5 (right). Abbreviations: anterior cerebral artery, ACA; anterior inferior cerebellar artery, AICA; internal carotid artery, ICA; middle cerebral artery, MCA; posterior communicating artery, PComA; vertebral artery, VA. Scale bars = 1 mm. Quantitative analysis of selected angles (b) and segment lengths (c) was performed at E12.5 (white bar), E15.5 (light gray bar) and E17.5 (dark gray bar), where the error bars indicate the standard deviations in each measurement. The lengths and angles shown in (b,c) are indicated in green on the middle panel of (a). The lengths and angles were also measured in the registered and averaged E17.5 images (hatched bars). c: Analysis of six lengths and six angles demonstrated a significant change in length between E12.5 and E15.5, compared to the same branches between E15.5 and E17.5 (*P < 0.05; N = 6), while there were no significant changes in the angles.
Contrast-Enhanced Micro-MRI Reveals Vascular Defects in Gli2-/- Mutants
To investigate the potential of these approaches for detecting and analyzing vascular phenotypes in mutant mouse embryos, we performed micro-MRI studies of E17.5 Gli2-/- homozygous mutants and their wild-type littermates (Fig. 8). 3D micro-MRI of Gli2-/- mutants clearly demonstrated the previously reported reduction of midbrain and cerebellum (data not shown) (16-18). After perfusion-fixation, micro-MRI also showed a leakage of the BSA-DTPA-Gd contrast agent into extra-vascular spaces in the Gli2-/- mutants, but not the wild-type mice, suggesting that the mutants have defects in vascular integrity, both in the brain and in tissues outside the brain (Fig. 8a,b). The extra-vascular leakage of contrast agent was also reflected in a decreased signal-to-noise ratio measurement in brain tissue (16 ± 5 [N = 4] in Gli2-/- mice, compared to 39 ± 10 [N = 4] in wild-type mice). This permeability phenotype reduced the contrast enhancement of blood vessels in Gli2-/- mice, especially inside the brain, but the carotid arteries were still visible as they entered the ventral side of the brain. Unexpectedly, micro-MRI revealed a complete absence of the vertebral and basilar arteries in Gli2-/- mutant embryos, close to the mid-hindbrain morphologic defects noted above (Fig. 8c,d). The absence of the basilar artery was confirmed by histologic analysis (Fig. 8e,f). In addition to the major basilar artery phenotype, we also observed differences in the patterning of the carotid arteries in Gli2-/- mutants (Fig. 8c,d). Specifically, the internal carotid arteries showed a significant constriction between the left and right arterial systems compared to wild-type littermates (569 ± 107 μm [N = 4] in Gli2-/- mice compared to 1513 ± 480 μm [N = 4] in wild-type mice), most likely a secondary effect resulting from the altered brain morphology.
FIG. 8.
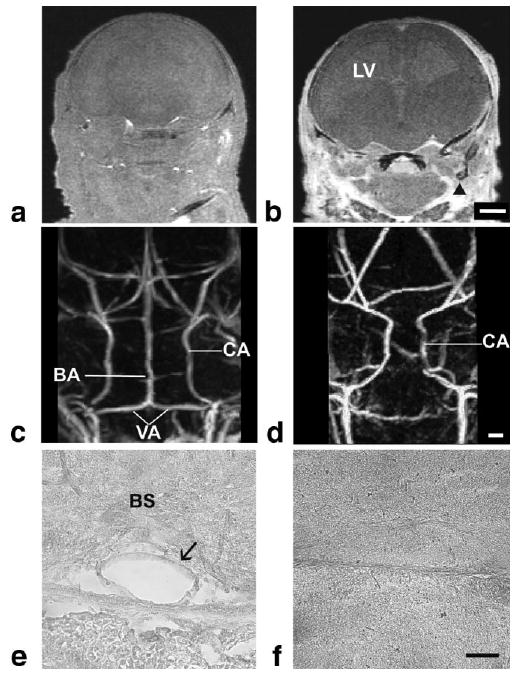
Gli2-/- mutant mice have severe vascular phenotypes. 3D coronal slices of E17.5 wild-type (a) and Gli2-/- (b) embryos show obvious leakage of contrast agent into extravascular spaces (arrow) in the mutants. Caudal views of the MIPs from wild-type (c) and Gli2-/- (d) mice, showing the carotid arteries (CA) in both, with a constriction in the carotid arteries as they enter the brain in mutant embryos and a complete absence of the basilar artery (BA). Histology was used to confirm the obvious presence of the BA in wild-type embryos (e, arrow) and its absence in Gli2-/- mutants (f). Abbreviations: basilar artery, BA; carotid artery, CA; vertebral arteries, VA; brain stem, BS; lateral ventricle, LV. Scale bar (a,b) = 1 mm; scale bar (c,d) = 275 μm; scale bar (e,f) = 50 μm.
Discussion
Previous reports have demonstrated the utility of micro-MRI as a tool for analyzing the embryonic mouse cardiovascular system, with the focus on the heart and adjacent blood vessels, using either (umbilical vein) contrast perfusion methods (6,7,15,19) or relying on endogenous contrast without contrast agent perfusion (20-23). Previous micro-MRI studies have used multiple-embryo imaging, acquiring data from up to 32 embryos simultaneously to dramatically increase the effective throughput for MRI-based phenotype analysis (15,20,21,23). In this study, multiple embryo micro-MRI was also employed to significantly improve the throughput of 3D data acquisition. In addition, the previously described contrast perfusion-fixation protocol was modified to selectively enhance the cerebral arteries, enabling the first detailed 3D analyses of the basilar and carotid arterial systems, the two major vascular inputs into the developing brain. Employing image processing and analysis tools, quantitative data were provided to show that there is a high degree of similarity in the 3D patterns of the carotid and basilar arteries and in the circle of Willis of late-stage mouse embryos. Quantitative analysis also demonstrated that vascular patterns are most likely determined at early stages of brain development and grow by extending branch segments, while preserving the underlying geometry of the branch points. Finally, contrast-enhanced micro-MRI was used to analyze previously unreported vascular phenotypes in Gli2-/- mutant mice.
Recently, optical projection tomography has been reported for the analysis of mouse vascular development (24,25). This method has the advantage of much higher spatial resolution compared to micro-MRI, as well as the ability to use fluorescently labeled antibodies to image molecular markers. However, optical projection tomography is limited to tissue penetration and sample size of approximately 1 cm3. This, together with the limited penetration of antibodies in whole embryo samples, makes optical projection tomography best suited to early embryonic stages (E8.5-10.5), providing complementary information to micro-MRI at later stages (≥E10.5). Another optical method that has been proposed for the analysis of mouse development is episcopic fluorescence image capture, in which a stack of images is acquired by photographing the face of the block containing an embryo after each section is cut (26). Episcopic fluorescence image capture is therefore a destructive method, but it allows for the acquisition of distortion-free, high-resolution 3D images for the analysis of gene expression patterns and embryonic anatomy the entire range of developmental stages. Micro-computed tomography has been used for 3D imaging of the brain vasculature in adult mice (27) and could likely be adapted for imaging the embryonic vasculature after umbilical perfusion with an X-ray opaque contrast agent. Contrast-enhanced micro-computed tomography offers high resolution (20 μm) for vascular imaging (28), similar to the highest resolutions reported for micro-MRI (29,30), but provides limited contrast between tissues and requires high radiation doses, precluding in vivo studies in developing embryos. Optical projection tomography and episcopic fluorescence image capture are both limited to ex vivo imaging, while micro-MRI has the potential to be applied in vivo in the future, albeit at lower resolution than the 50 μm used in these studies.
Unlike micro-computed tomography, optical projection tomography episcopic fluorescence image capture, or micro-MRI, ultrasound biomicroscopy is an established real-time, in vivo, micro-imaging approach that enables in utero analysis of mouse cardiovascular development and hemodynamic studies in embryonic blood vessels, including the cerebral vasculature (31). Ultrasound biomicroscopy currently offers a clear advantage over micro-MRI in image acquisition time, and ultrasound biomicroscopy contrast-enhancement is possible using intravascular injection of micro-bubble agents. At this point, robust methods for 3D acquisition and image segmentation are still required to enable effective volumetric analysis of the developing vasculature with ultrasound biomicroscopy.
The formation and development of the vascular system is critical for normal embryonic development, and analyses in genetically altered mice are needed to further understand the genetic factors underlying vascular patterning. Our studies demonstrate clearly the important roles that micro-MRI can play in these analyses. The Sonic Hedgehog (Shh) signaling pathway is critical for normal development of the nervous system, and recent studies have also suggested an important role during vascular development, in a number of vessels including the dorsal aorta, yolk sac, and coronary and lung vasculatures (36-40) (reviewed in Nagase et al. (37)). At this point, no detailed analyses have been reported on the roles during vascular development of the mouse Gli genes, the major effectors of Shh signaling. In the nervous system, Gli2 has been shown to be the main activator of Shh signaling (18,36-38), and Gli2-/- mice die at birth, having a reduced midbrain and cerebellum (5,16-18). Micro-MRI demonstrated the previously described brain defects, as well as obvious vascular permeability defects and a completely ablated basilar artery. The fact that this major vascular phenotype had been overlooked, despite numerous previous studies (16-18,36,37,41-44), highlights the fact that analysis of vascular defects is difficult with traditional histologic methods and that more effective 3D methods like micro-MRI are required.
One exciting possibility is the future extension of micro-MRI for in vivo vascular imaging in mouse embryos. Utilizing MRI acquisition methods gated to physiologic motion, we recently demonstrated 100-μm (isotropic) resolution micro-MRI for in utero imaging of the brain (45) and hearts (46) of live mouse embryos. In combination with contrast agents targeted to the surface of vascular endothelial cells, this might provide an effective approach for in vivo, contrast-enhanced micro-MRI of the vascular system after intravascular injection of contrast agents. This would allow unprecedented longitudinal imaging studies to further understand the complex and dynamic processes involved in establishing the final 3D patterns of the mammalian vascular system.
Acknowledgments
We thank Daniel Stephen for help and advice on breeding the Gli2 mutant mice and Dr. JoMichelle Corrales for sharing her observation that Gli2-/- mice might have cerebral vascular defects. We thank Drs. John Sled and Mark Henkelman at the Mouse Imaging Centre in Toronto, Canada, for providing analysis software used for image coregistration and averaging.
Grant sponsor: National Institutes of Health; Grant number: R01 HL078665.
References
- 1.Risau W, Flamme I. Vasculogenesis. Annu Rev Cell Dev Biol. 1995;11:73–91. doi: 10.1146/annurev.cb.11.110195.000445. [DOI] [PubMed] [Google Scholar]
- 2.Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438:932–936. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- 3.Silvestre JS, Mallat Z, Tedgui A, Levy BI. Post-ischaemic neovascularization and inflammation. Cardiovasc Res. 2008;78:242–249. doi: 10.1093/cvr/cvn027. [DOI] [PubMed] [Google Scholar]
- 4.Kerbel RS. Tumor angiogenesis. New Engl J Med. 2008;358:2039–2049. doi: 10.1056/NEJMra0706596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mo R, Freer AM, Zinyk DL, Crackower MA, Michaud J, Heng HH, Chik KW, Shi XM, Tsui LC, Cheng SH, Joyner AL, Hui C. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development. 1997;124:113–123. doi: 10.1242/dev.124.1.113. [DOI] [PubMed] [Google Scholar]
- 6.Smith BR, Johnson GA, Groman EV, Linney E. Magnetic resonance microscopy of mouse embryos. Proc Natl Acad Sci U S A. 1994;91:3530–3533. doi: 10.1073/pnas.91.9.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith BR, Linney E, Huff DS, Johnson GA. Magnetic resonance microscopy of embryos. Comput Med Imaging Graph. 1996;20:483–490. doi: 10.1016/s0895-6111(96)00046-8. [DOI] [PubMed] [Google Scholar]
- 8.Smith BR. Magnetic resonance imaging analysis of embryos. Methods Mol Biol. 2000;135:211–216. doi: 10.1385/1-59259-685-1:211. [DOI] [PubMed] [Google Scholar]
- 9.Kovacevic N, Henderson JT, Chan E, Lifshitz N, Bishop J, Evans AC, Henkelman RM, Chen XJ. A three-dimensional MRI atlas of the mouse brain with estimates of the average and variability. Cereb Cortex. 2005;15:639–645. doi: 10.1093/cercor/bhh165. [DOI] [PubMed] [Google Scholar]
- 10.Nieman BJ, Flenniken AM, Adamson SL, Henkelman RM, Sled JG. Anatomical phenotyping in the brain and skull of a mutant mouse by magnetic resonance imaging and computed tomography. Physiol Genomics. 2006;24:154–162. doi: 10.1152/physiolgenomics.00217.2005. [DOI] [PubMed] [Google Scholar]
- 11.Lerch JP, Carroll JB, Spring S, Bertram LN, Schwab C, Hayden MR, Henkelman RM. Automated deformation analysis in the YAC128 Huntington disease mouse model. Neuroimage. 2008;39:32–39. doi: 10.1016/j.neuroimage.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 12.Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr. 1994;18:192–205. [PubMed] [Google Scholar]
- 13.Collins DL, Holmes CJ, Peters TM, Evans AC. Automatic 3-D model-based neuroanatomical segmentation. Hum Brain Mapp. 1995;3:190–208. [Google Scholar]
- 14.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 15.Wadghiri YZ, Schneider AE, Gray EN, Aristizabal O, Berrios C, Turn-bull DH, Gutstein DE. Contrast-enhanced MRI of right ventricular abnormalities in Cx43 mutant mouse embryos. NMR Biomed. 2007;20:366–374. doi: 10.1002/nbm.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corrales JD, Rocco GL, Blaess S, Guo Q, Joyner AL. Spatial pattern of sonic hedgehog signaling through Gli genes during cerebellum development. Development. 2004;131:5581–5590. doi: 10.1242/dev.01438. [DOI] [PubMed] [Google Scholar]
- 17.Palma V, Ruiz i Altaba A. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development. 2004;131:337–345. doi: 10.1242/dev.00930. [DOI] [PubMed] [Google Scholar]
- 18.Blaess S, Corrales JD, Joyner AL. Sonic hedgehog regulates Gli activator and repressor functions with spatial and temporal precision in the mid/hindbrain region. Development. 2006;133:1799–1809. doi: 10.1242/dev.02339. [DOI] [PubMed] [Google Scholar]
- 19.Huang GY, Wessels A, Smith BR, Linask KK, Ewart JL, Lo CW. Alteration in connexin 43 gap junction gene dosage impairs conotruncal heart development. Dev Biol. 1998;198:32–44. doi: 10.1006/dbio.1998.8891. [DOI] [PubMed] [Google Scholar]
- 20.Schneider JE, Bamforth SD, Farthing CR, Clarke K, Neubauer S, Bhattacharya S. High-resolution imaging of normal anatomy, and neural and adrenal malformations in mouse embryos using magnetic resonance microscopy. J Anat. 2003;202:239–247. doi: 10.1046/j.1469-7580.2003.00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider JE, Bamforth SD, Farthing CR, Clarke K, Neubauer S, Bhattacharya S. Rapid identification and 3D reconstruction of complex cardiac malformations in transgenic mouse embryos using fast gradient echo sequence magnetic resonance imaging. J Mol Cell Cardiol. 2003;35:217–222. doi: 10.1016/s0022-2828(02)00291-2. [DOI] [PubMed] [Google Scholar]
- 22.Bamforth SD, Braganca J, Farthing CR, Schneider JE, Broadbent C, Michell AC, Clarke K, Neubauer S, Norris D, Brown NA, Anderson RH, Bhattacharya S. Cited2 controls left-right patterning and heart development through a Nodal-Pitx2c pathway. Nat Genet. 2004;36:1189–1196. doi: 10.1038/ng1446. [DOI] [PubMed] [Google Scholar]
- 23.Schneider JE, Bose J, Bamforth SD, Gruber AD, Broadbent C, Clarke K, Neubauer S, Lengeling A, Bhattacharya S. Identification of cardiac malformations in mice lacking Ptdsr using a novel high-throughput magnetic resonance imaging technique. BMC Dev Biol. 2004;4:16. doi: 10.1186/1471-213X-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharpe J, Ahlgren U, Perry P, Hill B, Ross A, Hecksher-Sorensen J, Baldock R, Davidson D. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- 25.Walls JR, Coultas L, Rossant J, Henkelman RM. Three-dimensional analysis of vascular development in the mouse embryo. PLoS ONE. 2008;3:e2853. doi: 10.1371/journal.pone.0002853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weninger WJ, Mohun T. Phenotyping transgenic embryos: a rapid 3-D screening method based on episcopic fluorescence image capturing. Nat Genet. 2002;30:59–65. doi: 10.1038/ng785. [DOI] [PubMed] [Google Scholar]
- 27.Dorr A, Sled JG, Kabani N. Three-dimensional cerebral vasculature of the CBA mouse brain: a magnetic resonance imaging and micro computed tomography study. Neuroimage. 2007;35:1409–1423. doi: 10.1016/j.neuroimage.2006.12.040. [DOI] [PubMed] [Google Scholar]
- 28.Marxen M, Thornton MM, Chiarot CB, Klement G, Koprivnikar J, Sled JG, Henkelman RM. MicroCT scanner performance and considerations for vascular specimen imaging. Med Phys. 2004;31:305–313. doi: 10.1118/1.1637971. [DOI] [PubMed] [Google Scholar]
- 29.Goddeeris MM, Rho S, Petiet A, Davenport CL, Johnson GA, Meyers EN, Klingensmith J. Intracardiac septation requires hedgehog-dependent cellular contributions from outside the heart. Development. 2008;135:1887–1895. doi: 10.1242/dev.016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petiet A, Kaufman MH, Goddeeris MM, Brandenburg J, Elmore SA, Johnson GA. High-resolution magnetic resonance histology of the embryonic and neonatal mouse: A 4D atlas and morphologic database. Proc Natl Acad Sci U S A. 2008;105:12331–12336. doi: 10.1073/pnas.0805747105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phoon CK, Turnbull DH. Ultrasound biomicroscopy-Doppler in mouse cardiovascular development. Physiol Genomics. 2003;14:3–15. doi: 10.1152/physiolgenomics.00008.2003. [DOI] [PubMed] [Google Scholar]
- 32.Byrd N, Becker S, Maye P, Narasimhaiah R, St-Jacques B, Zhang X, McMahon J, McMahon A, Grabel L. Hedgehog is required for murine yolk sac angiogenesis. Development. 2002;129:361–372. doi: 10.1242/dev.129.2.361. [DOI] [PubMed] [Google Scholar]
- 33.Lawson ND, Vogel AM, Weinstein BM. sonic hedgehog and vascular endothelial growth factor act upstream of the notch pathway during arterial endothelial differentiation. Dev Cell. 2002;3:127–136. doi: 10.1016/s1534-5807(02)00198-3. [DOI] [PubMed] [Google Scholar]
- 34.Vokes SA, Yatskievych TA, Heimark RL, McMahon J, McMahon AP, Antin PB, Krieg PA. Hedgehog signaling is essential for endothelial tube formation during vasculogenesis. Development. 2004;131:4371–4380. doi: 10.1242/dev.01304. [DOI] [PubMed] [Google Scholar]
- 35.Lavine KJ, White AC, Park C, Smith CS, Choi K, Long F, Hui CC, Ornitz DM. Fibroblast growth factor signals regulate a wave of hedgehog activation that is essential for coronary vascular development. Genes Dev. 2006;20:1651–1666. doi: 10.1101/gad.1411406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Tuyl M, Groenman F, Wang J, Kuliszewski M, Liu J, Tibboel D, Post M. Angiogenic factors stimulate tubular branching morphogenesis of sonic hedgehog-deficient lungs. Dev Biol. 2007;303:514–526. doi: 10.1016/j.ydbio.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 37.Nagase T, Nagase M, Machida M, Fujita T. Hedgehog signalling in vascular development. Angiogenesis. 2008;11:71–77. doi: 10.1007/s10456-008-9105-5. [DOI] [PubMed] [Google Scholar]
- 38.Ding Q, Motoyama J, Gasca S, Mo R, Sasaki H, Rossant J, Hui CC. Diminished sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development. 1998;125:2533–2543. doi: 10.1242/dev.125.14.2533. [DOI] [PubMed] [Google Scholar]
- 39.Matise MP, Epstein DJ, Park HL, Platt KA, Joyner AL. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development. 1998;125:2759–2770. doi: 10.1242/dev.125.15.2759. [DOI] [PubMed] [Google Scholar]
- 40.Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- 41.Lebel M, Mo R, Shimamura K, Hui CC. Gli2 and Gli3 play distinct roles in the dorsoventral patterning of the mouse hindbrain. Dev Biol. 2007;302:345–355. doi: 10.1016/j.ydbio.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 42.Corrales JD, Blaess S, Mahoney EM, Joyner AL. The level of sonic hedgehog signaling regulates the complexity of cerebellar foliation. Development. 2006;133:1811–1821. doi: 10.1242/dev.02351. [DOI] [PubMed] [Google Scholar]
- 43.Motoyama J, Milenkovic L, Iwama M, Shikata Y, Scott MP, Hui CC. Differential requirement for Gli2 and Gli3 in ventral neural cell fate specification. Dev Biol. 2003;259:150–161. doi: 10.1016/s0012-1606(03)00159-3. [DOI] [PubMed] [Google Scholar]
- 44.Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, Joyner AL. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development. 2000;127:1593–1605. doi: 10.1242/dev.127.8.1593. [DOI] [PubMed] [Google Scholar]
- 45.Deans AE, Wadghiri YZ, Berrios-Otero CA, Turnbull DH. Mn enhancement and respiratory gating for in utero MRI of the embryonic mouse central nervous system. Magn Reson Med. 2008;59:1320–1328. doi: 10.1002/mrm.21609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nieman BJ, Szulc KU, Turnbull DH. Three-dimensional, in vivo MRI with self-gating and image coregistration in the mouse. Magn Reson Med. 2009;61:1148–1157. doi: 10.1002/mrm.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]


