Abstract
Heme degradation plays a pivotal role in the availability of the essential nutrient, iron, in pathogenic bacteria. A previously unannotated protein from Mycobacterium tuberculosis, Rv3592, which shares homology to heme-degrading enzymes, has been identified. Biochemical analyses confirm that Rv3592, which we have termed MhuD (mycobacterial heme utilization, degrader), is able to bind and degrade heme. Interestingly, contrary to previously reported stoichiometry for the Staphylococcus aureus heme degraders, IsdG and IsdI, MhuD has the ability to bind heme in a 1:2 protein to heme ratio, although the MhuD-diheme complex is inactive. Furthermore, the 1.75 Å crystal structure of the MhuD-diheme complex reveals two stacked hemes forming extensive contacts with residues in the active site. In particular, the solvent-exposed heme is axially liganded by His75 and is stacked planar upon the solvent-protected heme. The solvent-protected heme is coordinated by a chloride ion which is, in turn, stabilized by Asn7. Structural comparison between MhuD-diheme and inactive IsdG and IsdI bound to only one highly distorted metalloporphyrin ring reveal that several residues located in α-helix 2 and the subsequent loop appear to be responsible for heme stoichiometric differences and suggest open and closed conformations for substrate entry and product exit.
Keywords: Mycobacterium tuberculosis, heme binding protein, heme degradation, iron metabolism, X-ray crystallography
Introduction
Heme degradation is a metabolic role performed by diverse organisms, fulfilling various physiological functions. The identification of a mammalian microsomal heme oxygenase (HO) demonstrated the oxidative cleavage of heme to release biliverdin as its final product, along with free iron and carbon monoxide in equimolar amounts 1. In eukaryotes, this reaction is coupled with the conversion of biliverdin to bilirubin by biliverdin reductase 2, whereas prokaryotic HOs have been implicated in phycobilin and phytochrome biosyntheses 3. Additionally, increasing lines of evidence indicate that HOs from several bacterial pathogens play a major role in iron availability 3; host-heme degraded by HOs in Gram-positive and Gram-negative pathogens (i.e., HmuO from Corynebacterium diptheriae 4 and HemO from Neisseriae sp 5, respectively) provide an alternate source of iron, an essential element for growth, survival and pathogenicity.
Recently, a new family of heme degraders has been described in Staphylococcus aureus and Bacillus anthracis, as well as the non-pathogenic Bradyrhizobium japonicum 6; 7; 8. While these proteins do not share sequence or structural homology to canonical HOs, they are able to degrade heme. Furthermore, complementation studies show that S. aureus IsdI can restore growth in Corynebacterium ulcerans HO mutant (ΔHmuO) and demonstrate the ability of non-canonical heme degraders to function in vivo as heme oxygenases 7. The crystal structures of homodimeric IsdG and its homolog, IsdI, reveal overall topologies distinct from monomeric HOs albeit similar to monooxygenases involved in antibiotic synthesis in Streptomyces sp 9. Additionally, the heme-bound structure of a catalytically inactive IsdG mutant revealed that the α-meso edge of heme, which is buried in the active site of HOs 10; 11; 12; 13, is exposed to solvent 14. Together, these data imply that IsdG and IsdI undergo a different mechanism for heme degradation distinct from HOs.
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), is a bacterial pathogen responsible for approximately 8 million new infections and 2 million deaths per year worldwide 15. The ease of Mtb to enter latency and develop multi-drug resistance becomes a deadly combination for AIDS patients, and there is an urgent need to discover new anti-TB drugs. Similar to other pathogenic bacteria, the acquisition of iron in Mtb is required for infectivity and pathogenicity; as such, its iron acquisition pathways are well characterized 16; 17. Because heme biosynthesis and degradation are intricately linked with iron cycling 18 and Mtb possesses a biosynthetic pathway for heme 19, we hypothesized the presence of proteins that are able to catabolise heme. Therefore, proteins important for iron acquisition and/or heme degradation can potentially serve as new targets for anti-TB therapeutics. However, this effort has been hampered by the lack of identification and biochemical characterization of a heme degrading protein in Mtb.
In this study, we searched the Mtb genome 20 to identify proteins that may function as heme degraders. We have identified, for the first time, a putative Mtb heme degrader, Rv3592, which shares sequence homology with S. aureus IsdG and IsdI. Additionally, we show that Rv3592, which we term MhuD (mycobacterial heme utilization, degrader), is able to bind and degrade heme. Significantly, MhuD can bind two molecules of heme per monomer, which is different from the monoheme IsdG and IsdI although MhuD-diheme is inactive. Finally, we have also solved a 1.75 Å crystal structure of MhuD-diheme complex, which sheds light on the nature of heme binding with alternate conformations. The above results pave the foundation towards new TB therapeutics targeting heme degrading proteins.
Results
MhuD is homologous to bacterial heme degraders
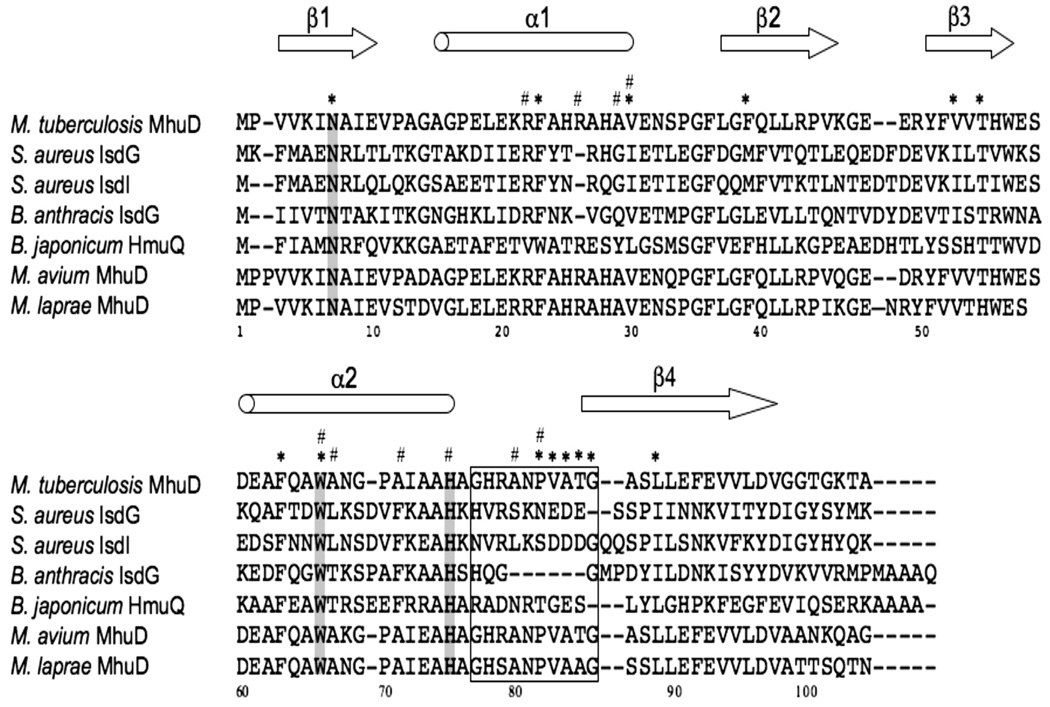
The existence and identity of a heme degrading protein in Mtb has not been established prior to this study. Concurrent presence of heme biosynthetic and degradation pathways in many organisms has been suggested to be important in maintaining cellular homeostasis by controlling the availability of heme and/or iron 18. Since Mtb possesses a biosynthetic pathway for heme 19, we hypothesized the presence of enzymes that are able to catabolise heme. To prove this hypothesis, we conducted extensive genome mining on heme degrading proteins in Mtb. A BLAST homology search of the entire Mtb genome 20 did not reveal the presence of a canonical HO gene. However, we identified a putative gene, Rv3592, which shares 46% and 43% sequence identity with S. aureus IsdG and IsdI, respectively, analogous heme degraders that are unrelated to HOs 7. Homologous proteins are found across other pathogenic and non-pathogenic bacteria, including B. anthracis IsdG 8 and B. japonicum HmuQ 6. Furthermore, Mtb MhuD is a conserved protein across all mycobacterial species, including Mycobacterium avium and Mycobacterium leprae. Multiple sequence alignments of these proteins show several key conserved residues that have been reported to be crucial for heme degrading activity in S. aureus IsdG (Asn7, Trp67 and His77, Fig. 1). In particular, mutational analyses of these residues have been shown to maintain heme binding albeit abolishing heme degradation 9. MhuD also contains these invariant residues corresponding to Asn7, Trp66 and His75, suggesting it could function as a heme degrading protein.
FIGURE 1.
Sequence alignment of Mtb MhuD with other heme degrading enzymes (i.e., S. aureus IsdG and IsdI, B. anthracis IsdG and B. japonicum HmuQ), as well as homologous mycobacterial proteins from M. avium and M. leprae. Cylinders and arrows define the regions of α-helices and β-strands, respectively. Residues that interact with the more solvent exposed or more solvent protected hemes are indicated by # or *, respectively. Conserved catalytic residues are shaded whereas loop residues that contribute to open and closed conformations are boxed.
MhuD binds and degrades heme
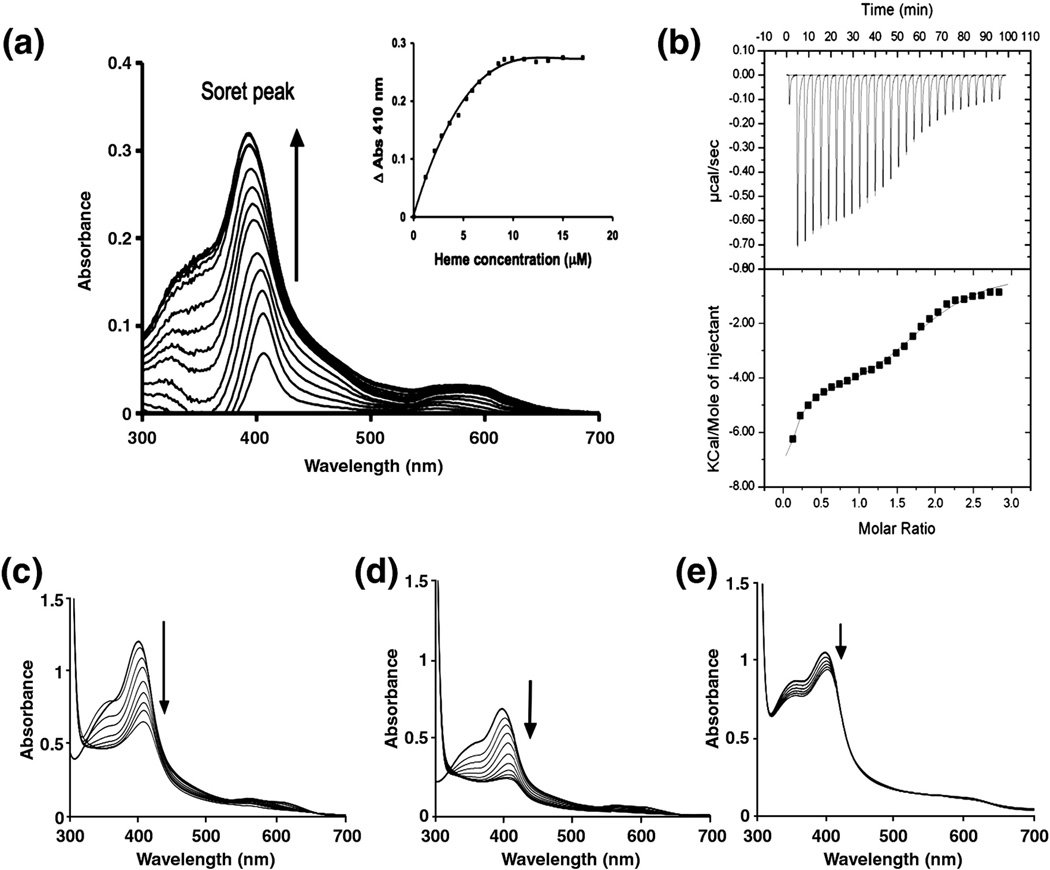
As a first report for mycobacteria, we found that MhuD both binds and degrades heme. To test our hypothesis that apo-MhuD is a novel protein involved in Mtb heme degradation, we investigated its ability to bind heme. Hemin was incrementally titrated into 5 µM purified apo-MhuD and the spectral range between 300 – 700 nm was measured using a dual-beam spectrophotometer. The resulting difference spectra are generated by subtracting the free heme spectra from the heme-titrated MhuD spectra. MhuD exhibits the hallmarks of a heme-binding protein with the appearance of a Soret peak at 410 nm, as well as a broad peak around 575 nm corresponding to the Q band region (Fig. 2A). Plotting the absorbance difference at 410 nm against heme concentrations reveals that heme binding is saturable (Fig. 2A inset).
FIGURE 2.
Mtb MhuD is a heme-degrading protein. (A) MhuD binds heme. Hemin (1 µM aliquots) was added to 5 µM apo-MhuD, resulting in the increase of the Soret peak at 410 nm (arrow). Inset: The change in absorbance at 410 nm versus heme concentration shows that heme binding is saturable. (B) Representative normalized heat signal (upper panel) and binding isotherm (lower panel) profiles of the binding of heme to MhuD, in which twenty-eight 10 µL injections of 400 µM hemin solution into 30 µM apo-MhuD were performed at 3.5 min intervals. The binding isotherm profile was fit to a sequential binding site model, whereby two heme binding sites per monomer were specified to be fitted in a sequential manner. (C–E) Heme degradation experiments, carried out in the presence of 0.5:1 molar ratio of catalase to heme-bound MhuD, were observed by the decrease of the Soret peak (arrow) by monitoring the spectral changes from 300 – 700 nm every 5 mins: (C) Cytochrome P450 reductase was added to 10 µM MhuD-heme at a 0.3:1 molar ratio and heme degradation is observed by the decrease in the Soret peak (arrow) with each addition of 10 µM NADPH. (D) 10 mM ascorbate was added to 5 µM MhuD-heme and heme degradation is observed. (E) 10 mM ascorbate was added to 5 µM MhuD-diheme and no heme degradation is observed. All experiments were performed in triplicate (A)–(E).
To gain further insight into MhuD heme binding, isothermal titration calorimetry (ITC) was used to investigate the heme:MhuD stoichiometry and to obtain binding constants. The ITC experiments in which apo-MhuD (in the cell) was titrated with hemin (in the syringe) generated a binding isotherm consistent with saturation of heme binding (Fig. 2B). Using the Origin software (MicroCal), we tried one (χ2 = 7.0 × 104) and two (χ2 = 3.35 × 104) binding site models and found that the best-fit parameters were obtained with the sequential binding (χ2 = 1.29 × 104) site model, whereby two heme binding sites per monomer were specified to be fit in a sequential manner. The calculated heme association constants (Ka) correspond to two heme sites of 1.2 ± 0.3 × 105 M−1 and 2.0 ± 0.3 × 105 M−1, respectively. Additional ITC experiments in which hemin (in the cell) was titrated with apo-MhuD (in the syringe) were performed. The data were fitted using a one-site binding model due to the initial high heme concentrations that obscured the observation of sequential binding (data not shown). When the number of binding sites was allowed to vary, the stoichiometry of heme to each MhuD monomer was 1.9 ± 0.2 heme molecules while the calculated Ka of 1.7 ± 0.3 × 105 M−1 is within the margins of error of the two affinities reported from the sequential binding mode. MhuD’s heme binding affinities are comparable to those of S. aureus IsdG and IsdI (i.e., 2 × 105 M−1 and 2.8 × 105 M−1, respectively) 4; 7, although the 2:1 (heme:protein) stoichiometry is a feature distinct from the 1:1 stoichiometry observed for all other heme degrading proteins reported to-date 4; 7; 8. Furthermore, MhuD’s ability to bind two molecules of heme per monomer (MhuD-diheme complex) is reinforced by the MhuD-diheme crystal structure as discussed below.
Because heme degradation has previously been shown to occur with monoheme-protein complexes 4; 7; 8, a 1:1 complex of heme to MhuD (MhuD-heme) was prepared to assess its ability to degrade heme. Briefly, a 1.5 molar excess of hemin was incubated with apo-MhuD and purified by size exclusion chromatography. The equimolar concentrations of protein and heme were verified by Bradford and pyridine hemochrome assays, respectively 21. Single turnover heme degradation reactions with MhuD-heme were monitored spectrophotometrically between 300 – 700 nm in the presence of suitable electron donors. While the cognate Mtb electron donor has not been identified, previous studies report that heme degrading activity may be observed with the addition of NADPH-cytochrome P450 reductase or ascorbate as electron donors 4; 7; 8. Incubation with either NADPH-cytochrome P450 reductase or ascorbate diminishes the Soret peak of MhuD-heme over time (Figs. 2C and 2D, respectively), indicative of heme degradation as observed for IsdG, IsdI and HmuQ 6; 8. The addition of catalase inhibits coupled oxidation of heme by exogenous hydrogen peroxide in myoglobin and cytochrome b5 22; 23, and is routinely used to study heme oxygenation by HOs, as well as heme-degrading proteins, IsdG and IsdI 7; 24. Therefore, to rule out non-enzymatic heme degradation, the above reactions were carried out in the presence of 5 µM catalase. These data also establish that, as with homologous proteins, monoheme-MhuD is able to degrade heme 6; 7; 8, and we are further investigating the nature of the degradation product. The above results strongly support that MhuD serves as a heme degrader in Mtb.
Heme stoichiometry-dependent MhuD activity
The heme degradation assays are based on well-established protocols, where the heme:MhuD ratio is 1:1. However, our ITC studies indicate that MhuD is capable of binding two hemes per monomer. To correlate MhuD’s heme degrading activity with heme binding ability, MhuD-diheme was prepared by incubating 4 molar excess of hemin with apo-MhuD, purified by size exclusion chromatography, and the 2:1 heme to protein stoichiometry was confirmed by Bradford and pyridine hemochrome assays 21. Contrary to the MhuD-heme study above, where incubation with ascorbate in the presence of catalase shows that the Soret peak of MhuD-heme decreases over time (Fig. 2D), the Soret peak of an equivalent protein concentration of MhuD-diheme with ascorbate and catalase diminishes very little in the same time period (Fig. 2E). Additionally, no heme degradation was observed when MhuD-diheme was incubated in the presence of NADPH-cytochrome P450 reductase. These results indicate that MhuD-diheme loses it ability to degrade heme, thus rendering it inactive. Herein, we encounter an interesting and important issue: a protein that has the capability of binding two heme molecules per monomer, yet only the monoheme-protein complex is active. The crystal structures described below help visualize such a difference caused by different heme stoichiometry.
Structural overview of MhuD-diheme
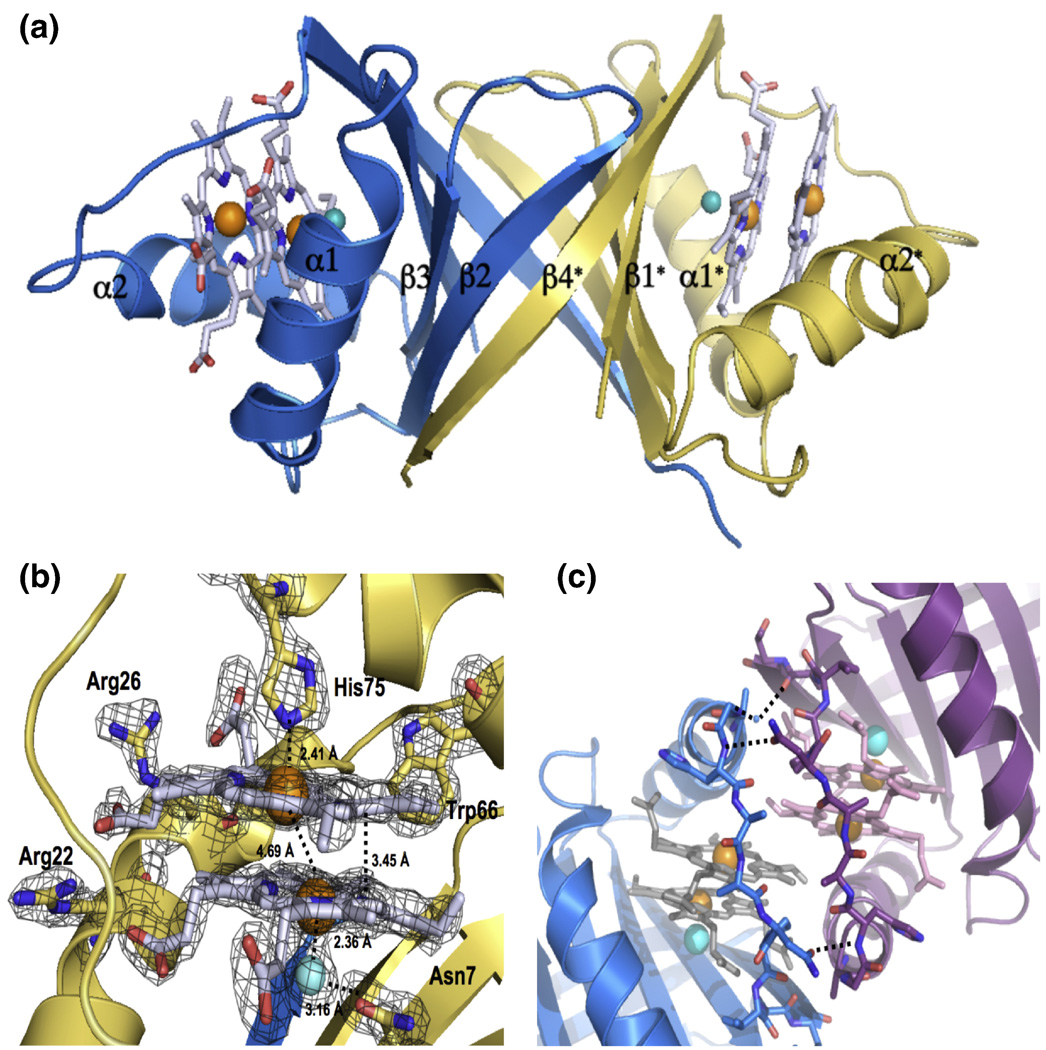
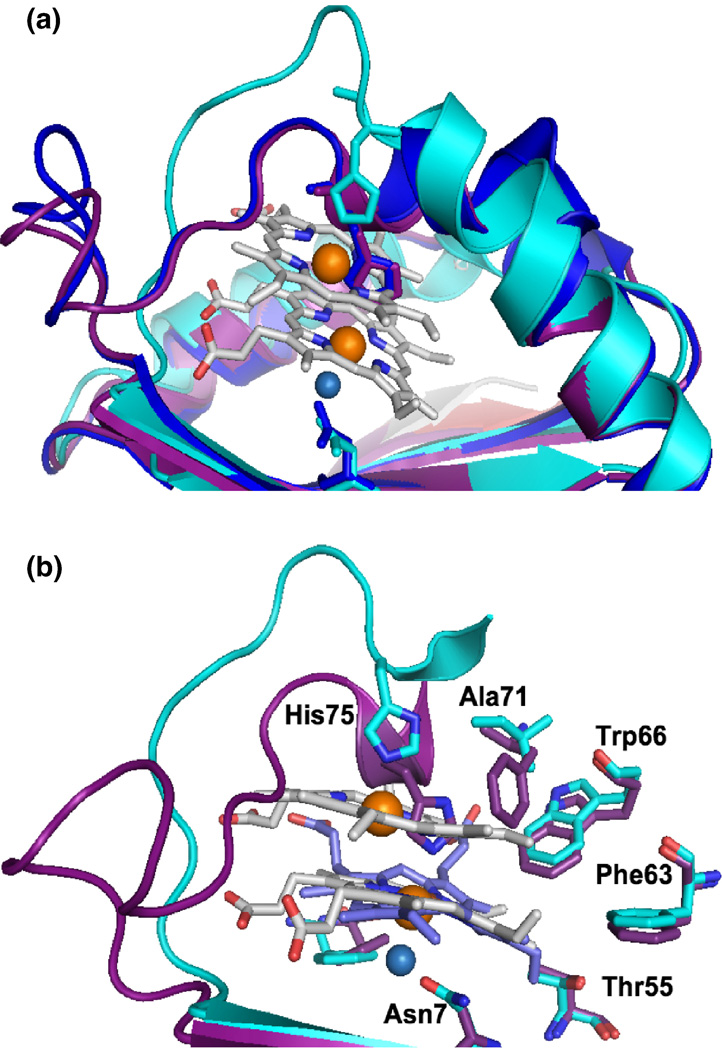
After screening over 1000 crystallization conditions, optimizing several of these and collecting > 100 synchrotron diffraction datasets, we solved the 3.5 Å crystal structure of apo-MhuD, in which there was no observable electron density for the loop region surrounding the proposed heme-binding region (residues 72–87 omitted) presumably due to its high flexibility to facilitate heme binding (data not shown). Despite thousands of crystallization trials, we could not crystallize MhuD-heme. As discussed below, the inability of crystallizing the monoheme complex may be an indication of protein flexibility necessary to conduct heme degradation. In contrast, we obtained crystals of MhuD-diheme complex which grew within 48 – 72 hours. The partial apo-MhuD structure was then used as a model for molecular replacement in solving the 1.75 Å MhuD-diheme crystal structure (Table 1). MhuD-diheme is a homodimer where each subunit is defined by a ferredoxin-like fold consisting of a βαββαβ secondary structure topology (Fig. 3A). The β2 and β4 strands from each monomer come together to form a central eight-stranded antiparallel β-barrel decorated with two α-helices from each monomer on opposite sides of the β-barrel. MhuD shares overall structural similarities with IsdG and IsdI 9; 14, as well as a monooxygenase, ActVA-Orf6, from Streptomyces coelicolor 25. Therefore, although MhuD has a similar protein fold as homologous monoheme structures, subtle conformational changes enable MhuD to bind two hemes.
Table 1.
X-ray diffraction data collection and atomic refinement statistics for apo- and hcmc-Rv3592 from M. tuberculosis.
| Apo-Rv3592 | Heme-Rv3592 | |
|---|---|---|
| Space Group | P21 | C2 |
| No of monomers per AS unit | 12 | 2 |
| Unit cell dimensions (Å) | 35.7 × 245.42 × 61.8 | 44.0 × 64.6 × 71.1, β = 90.1 |
| pH of crystallization condition | 6.5 | 7.0 |
| Data set | ||
| Wavelength (Å) | 0.97567 | 1.03317 |
| Resolution range (Å) | 50–3.1 | 50-1.75 |
| Unique reflections (total) | 19,537(58,736) | 19,968(147,615) |
| Completeness (%)¶ | 96.5(81.3) | 98.9 (98.6) |
| Rmerge¶,a | 14.8(40.4) | 6.6(16.5) |
| I/σ¶ | 7.8(1.4) | 28.1 (10.7) |
| Model refinement | ||
| Residues missing | 72–87 | 0 |
| Resolution range (Å) | 50-3.1 | 20-1.75 |
| No. of reflections (working/free) | 18746/1002 | 18936/1018 |
| No. of protein atoms | 7566 | 1596 |
| No. of water molecules | 0 | 236 |
| No of heme/dimmer | 0 | 4 |
| No. of chloride/dimer | 0 | 2 |
| Rwork/Rfreeb,% | 33.1/35.4 | 17.8/23.3 |
| Average B-factor all atoms (Å2) | 28.4 | 23.7 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.013 | 0.011 |
| Bond angles (degrees) | 1.737 | 1.616 |
| Ramachandran Plot | ||
| Most favorable region (%) | 90.2 | 93.7 |
| Additional allowed region (%) | 4.8 | 6.3 |
Statistics for the highest resolution shell are given in (brackets) unless otherwise stated.
Rmerge =Σ|I-<I>|/ΣI
Rwork=Σ|Fobs-Fcalc|/ΣFobsRfree was computed identically except where all reflections belong to a test set of 5% randomly selected data.
FIGURE 3.
Structure of MhuD-diheme. (A) Ribbon representation of dimeric MhuD-diheme complex (Chain A:blue, Chain B: gold). The hemes, two per monomer, are represented as stick models with carbons (white), nitrogen (blue) and oxygen (red) with Fe (orange spheres), and the Cl− ion (cyan spheres) coordinates the more solvent-protected heme. (B) Heme binding pocket with Fo-Fc omit map contoured at 3.0 σ (black mesh). Fe and Cl− spheres are colored orange and cyan, respectively. (C) Ribbon diagram of the two subunits at the crystallographic interface shown in blue and magenta, respectively. Stick representations for heme and residues in the interacting loop regions, in light grey for one subunit and pink in the other. Fe and Cl− spheres are colored orange and cyan, respectively. The water molecule is represented by a small blue sphere and hydrogen-bonding between residues in the loop regions by black broken lines.
Heme binding pocket
Each MhuD monomer binds two heme molecules within the active site (Figs. 3A and 3B). The positions of the four heme iron atoms in the MhuD dimer were verified from the anomalous signal by SHELX C/D/E 26. The porphyrin rings are stacked 3.45 Å apart with an inter-iron distance of 4.69 Å and oriented such that the propionate groups from one heme are rotated approximately 80° relative to those from the second heme. Besides observing good correlation of the modeled hemes within the experimental electron density map (Fig. 3B), we further validated their propionate positions by refining the structure with hemes placed in several alternate orientations which resulted in high negative densities in 2Fo–Fc maps.
The heme-binding pocket is lined with mainly hydrophobic residues; in all, twenty-one residues interact with the diheme molecules, including Asn7, Trp66 and His75 which have been observed to be critical catalytic residues for IsdG (Figs. 1 and 3B) 9. The iron atom in the solvent-exposed heme is pentacoordinated by His75 (2.41 Å) and further stabilized by an additional nine residues. The side chain of Arg26 hydrogen bonds with both propionate groups, as well as forms a salt bridge between propionate-7 and Nε Arg26 (2.62 Å). Additionally, propionate-7 is stabilized by the NH1 atom of Arg22 (3.13 Å). The solvent-protected heme interacts with fourteen residues in the active site including three residues, Val30, Trp66 and Pro82, which also interact with the solvent-exposed heme (Fig. 1). Due to crystallization conditions and consistency of bond lengths between chloride and iron ions from previous reports 14; 27; 28, the atom liganded to the solvent-protected heme iron is modeled as a Cl− ion (2.36 Å), which is in turn bonded to Asn7 (3.16 Å) (Fig. 3B). As with the solvent-exposed heme, propionate-7 in the solvent-protected heme is stabilized by the NH1 of Arg22 (2.98 Å) as well as backbone N atoms from Val83 (2.71 Å) and Ala84 (2.71 Å). Propionate-6 interacts extensively with the backbone N atom of Gly86 (2.92 Å).
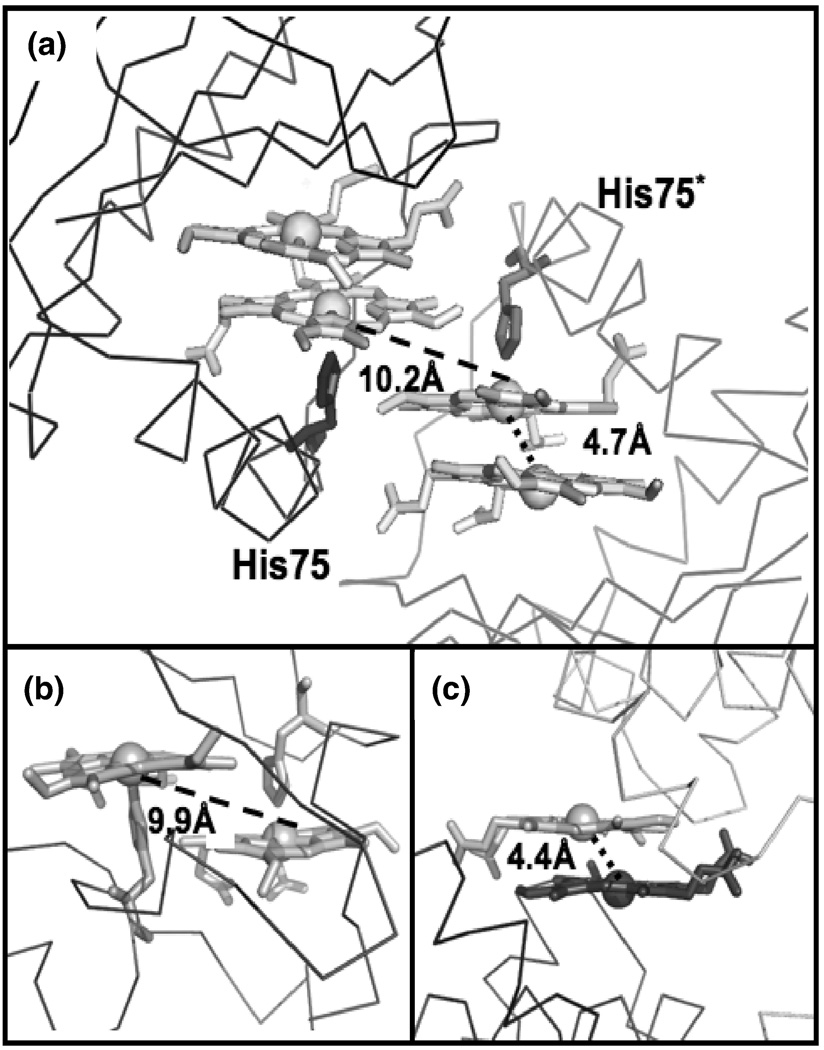
The diheme active site is located near a two-fold crystallographic symmetry axis that forms a protein-protein interface between two dimeric MhuD biological subunits (Fig. 3C). This interface is facilitated by Van der Waals contacts between residues 76–83 from both subunits, which are located in the loop regions following α-helix 2. The interaction within this region is further stabilized by a weak hydrogen bond between the carbonyl of His87 and the NH2 moiety of Asn80 in the adjacent loops, as well as a water-mediated hydrogen bond between a carbonyl to backbone N atom between the two subunits. Furthermore, the solvent-exposed hemes from each subunit stack nearly parallel upon each other (4.0 Å apart) with an inter-iron distance of 10.2 Å (Fig. 4A). Thus, at the crystallographic protein-protein interface between two biological subunits, there are four heme molecules planar stacked upon each other.
FIGURE 4.
Structural comparisons of multi-heme binding sites. (A) Line representation of the two subunits of MhuD-diheme at the crystallographic interface shown in light grey and dark grey, respectively. Hemes are in stick representation. (B) Active site of NapB (PDB code: 1JNI), polypeptide backbone is shown as a line representation whereas the diheme and coordinating histidines are shown as stick representations. (C) Line representation of the two subunit of ChaN (PDB code: 2G5G) at the crystallographic interface (shown in light grey and dark grey, respectively). Hemes are in stick representation.
Discussion
In contrast to previous studies of homologous bacterial heme-degrading enzymes 6; 7; 8, mycobacterial MhuD has the ability to bind two molecules of heme per monomer as indicated by the ITC data. The elucidation of the 1.75 Å X-ray crystal structure of MhuD-diheme revealed the presence of two heme molecules in the active site, contrary to the equimolar ratio of metalloporphyrin to protein observed in the structures of inactive heme-IsdG-N9A and non-iron CoPPIX-IsdI complexes 14. Four datasets collected from different crystals under a total of two conditions were analyzed and, in every instance, two heme molecules per monomer were determined, confirming that the structural stoichiometry is not the consequence of a single crystal. Moreover, MhuD-diheme is unlikely to be an artifact of crystallization since it was purified by size exclusion chromatography and no additional heme was added during crystallization.
Comparison with other diheme structures
There is only one other protein structure in the PDB with a similar diheme planar stacked conformation in its active site, NapB, a cytochrome subunit of nitrate reductase involved in electron transfer 29. As with the MhuD-diheme structure, NapB-diheme has nearly parallel heme planes stacked at Van der Waals distances although the inter-iron distance is twice as long (9.9 Å) compared to that of MhuD-diheme (4.7 Å), Figs. 4A and 4B, respectively. NapB’s longer inter-iron distance is due to the steric clash between one heme and the histidine coordinating the adjoining heme (Fig 4B); similar diheme-dihistidine interactions (10.2 Å) are also observed at the crystallographic interface between MhuD’s solvent-exposed hemes (Fig. 4A). Additionally, there is a striking similarity between the MhuD active-site diheme conformation and the heme-heme stacking interactions at the crystallographic dimer interface of ChaN, an iron-regulated lipoprotein that has been implicated in heme acquisition in the Gram-negative pathogen Campylobacter jejuni 30. In ChaN, the parallel stacked diheme conformation occurs between two heme molecules each associated with a monomer, where the heme planes are 3.5 Å apart and the inter-iron distance is 4.4 Å, although the propionates of each heme are rotated 180° whereas in the MhuD active-site an ~ 80° rotation is observed (Figs. 4C and 4A, respectively).
Heme-heme stacking at crystallographic interfaces, as observed in the structures of MhuD and ChaN 30, is not unprecedented. Similar stacking interactions and orientations are observed in the crystal structure of Gram-positive Streptococcus pyrogenes Shp, a cell surface protein involved in heme uptake 31. Additionally, a NEAT domain of IsdH, an S. aureus extracellular heme receptor involved in heme acquisition, have two crystallographically related hemes which appear to interact in a similar manner to that of MhuD 32. Furthermore, it was proposed that this diheme stacking between IsdH-NEAT domain monomers forms a multimeric complex that allows additional heme association, thus facilitating IsdH to transport more than one heme per monomer at a time. In sum, the heme-heme stacking in the MhuD-diheme structure reported herein is not only the first among its structural homologs, but also adds one more example to the novel heme-stacking geometry reported only once before.
Differences between the heme-binding pockets of MhuD-diheme compared to IsdG- and IsdI-metalloporphyrin structures
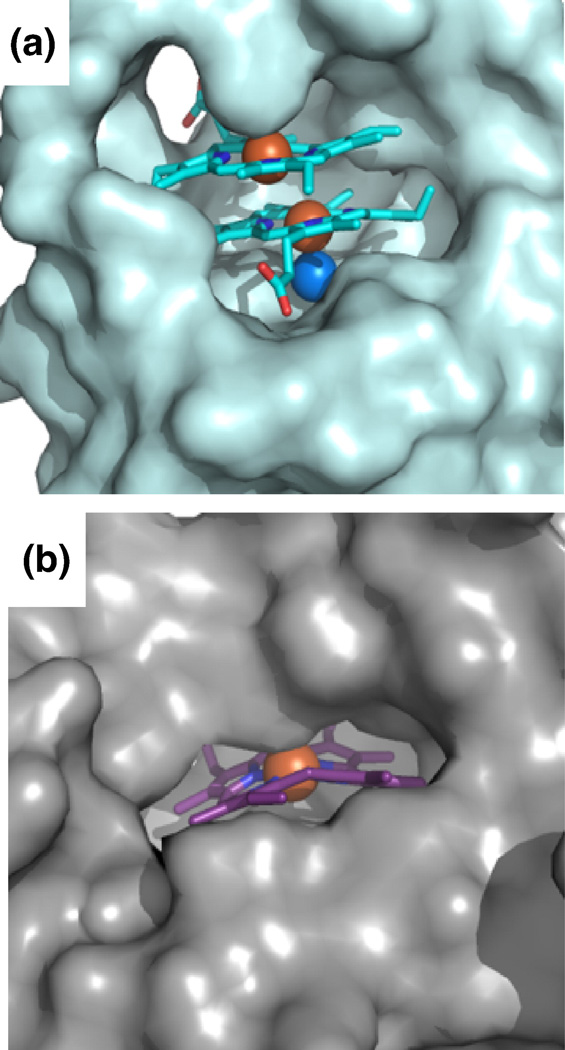
There are several distinct structural differences between MhuD-diheme and the structures of catalytically inactive heme-IsdG (heme-IsdG-N9A) and IsdI bound to a non-degradable substrate cobalt protoporphyrin IX (CoPPIX-IsdI) 14. Superimpositions of MhuD-diheme with heme-IsdG-N9A (rmsd. 2.2 Å) and CoPPIX-IsdI (rmsd 2.1 Å) show overall structural similarity (Fig. 5A). However, notably both IsdG and IsdI have only one metalloporphyrin per monomer, where their porphyrin rings occupy a similar location as MhuD’s solvent-protected heme although their propionate groups are rotated approximately 90° about the axis normal to the tetrapyrrole ring with respect to the solvent-protected heme in MhuD. In both IsdG and IsdI metalloporphyrin structures, the metal is axially coordinated by histidine (His77 and His76, respectively) as seen for the solvent-exposed heme of MhuD-diheme, while CoPPIX-IsdI has a Cl− ion as a distal axial ligand and Asn7 is coordinated to the Cl− ion, as observed for the solvent-protected heme of MhuD-diheme. Interestingly, the porphyrin rings in IsdG and IsdI are more distorted than those of MhuD. The distortions of the dihemes in MhuD (both 0.7 Å), analyzed with normal-coordinate structural decomposition 33, are not as severe as those in the porphyrin rings in IsdG (1.9 Å) and IsdI (2.3 Å), which are the largest observed to-date 14. These discrepancies are due to subtle structural differences: in MhuD, α-helix 2 is extended whereas the corresponding residues in the S. aureus proteins consist of two α-helices kinked at a 45° angle (Fig. 5A). A closer inspection of the heme binding pockets by both amino acid sequence and structural alignments indicate that the difference can be attributed to the absence of a conserved phenylalanine in MhuD (Phe73, Phe72 and Ala71 in IsdG, IsdI and MhuD, respectively). In IsdG and IsdI, this phenylalanine is involved in an edge-to-face aromatic interaction with the conserved catalytic tryptophan (Trp67 and Trp66 in IsdG and IsdI, respectively) and π-π stacking interactions with the imidazole ring of the conserved catalytic histidine, thus restricting the motion of this catalytic residue, as well as the subsequent loop (Fig. 5B). Consequently, the loop region in both IsdG and IsdI precludes the existence of a second heme molecule, although similar conserved residues in the three proteins remain involved in substrate binding despite the presence of an additional heme in MhuD (Fig. 5B). Another distinct difference within the loop region is at its C-terminal leading up to β-strand 4: in MhuD, the hydrophobic residues make extensive contacts predominantly to the more solvent-exposed heme, whereas the corresponding residues (i.e., aspartates or glutamates) in the Isd heme-degraders are flipped out and exposed to solvent. Moreover, the solvent accessible surfaces of Isd hemes compared to that of MhuD dihemes are quite distinct. For MhuD, the solvent-exposed heme has ~220 Å2 (as calculated by AreaIMol 34) surface accessibility to solvent, whereas the solvent-protected heme has ~54 Å2 solvent accessibility (Fig. 6A). In the IsdG-heme structure, ~73 Å2 of the porphyrin surface is solvent exposed to the surface (Fig. 6B). These subtle structural differences in MhuD may be attributed to the heme binding residues in α-helix 2 and the subsequent loop region. Interestingly, these residues are invariant or conservatively substituted in all mycobacterial MhuD homologs to-date, thus hinting at a novel and evolutionarily conserved diheme binding feature in mycobacteria.
FIGURE 5.
Comparison of the heme binding sites between MhuD and Isd proteins. (A) Superposition of heme-degrading enzyme structures show that the α-helix 2 in MhuD (cyan) is extended whereas they are kinked by 45° in IsdG (magenta) and IsdI (blue), thus precluding the existence of a second heme. Furthermore, the loop region in MhuD that makes extensive contacts with the two hemes is flipped out in IsdG and IsdI. Porphyrin ring is represented as a stick model with carbons (white), oxygen (red) and nitrogen (blue). Fe and Cl− spheres are colored orange and marine, respectively. (B) Most of the residues within the active site of MhuD and IsdG are conserved except for Phe73 and Ala71 of IsdG and MhuD, respectively. Residue side-chains are represented by stick models with carbon (magenta and cyan for IsdG and MhuD, respectively), oxygen (red) and nitrogen (blue). Additionally, the porphyrin ring of heme (stick representation with carbon in light blue for IsdG) in IsdG is kinked and is located in the same position as the solvent-protected MhuD heme (carbon in white).
FIGURE 6.
Surface representation of hemes in MhuD and IsdG. (A) Surface representation of one subunit of MhuD (light cyan) with the dihemes in stick representation with carbon (cyan), oxygen (red) and nitrogen (blue). Fe and Cl− spheres are colored orange and marine, respectively. (B) Surface representation of one subunit of IsdG (light grey) with heme in stick representation (carbon in magenta).
Functional implications of the MhuD-diheme structure
Comparisons of the heme bound heme-degrading protein structures suggest potential open inactive (MhuD-diheme) and closed active (IsdG- and IsdI-metalloporphyrin) conformations. The apo structures of MhuD, IsdG and IsdI 9 are disordered near the heme-binding site, in the loop region following α-helix 2, suggesting a dynamic active-site pocket. Additionally, attempts to fit the apo version of the MhuD-diheme structure into the apo-MhuD dataset resulted in clashes between subunits arising from residues 80–90, yet again implying that this loop region is highly flexible. The extended α-helix 2 in MhuD-diheme and the subsequent loop regions for all three proteins (Figs. 5A and 5B) allow heme entry and product exit, which is not possible in the closed active conformation as observed in both IsdG and IsdI-metalloporphyrin structures, Fig. 6B 35. Flexibility of the heme-binding pocket has also been reported in HO-1, where the distal and proximal helices are further apart in the apo structure, leading to an increase in the size of the pocket 36. Additionally, the distal helix, which contains a conserved Gly-rich motif, has been shown to contribute to differences in the open and closed states of holo HO-1.
Protein-induced non-planar porphyrin rings have been reported to influence biological activity of proteins, including enzymatic reactivity 35. In contrast to HO-1-associated heme, the porphyrin rings in IsdG and IsdI are severely distorted, suggesting that distortion may be necessary for catalysis in the closed conformation of non-HO heme degraders 14. Incidentally, porphyrin ring distortion has also been proposed for a newly identified class of proteins, YfeX and EfeB, whereby iron extraction occurs without the oxidative cleavage of the porphyrin ring 37. Therefore, comparison of the MhuD structure with its homologs, as well as extensive biochemical analyses discussed above, imply the importance of protein flexibility in all families of heme degraders.
Biological Relevance
Similar to other pathogenic bacteria, Mtb requires the essential micronutrient iron for growth and hence, for its infectivity and pathogenicity. Because the process of iron acquisition is a good target for chemotherapeutic drug development, mycobacterial siderophore (iron-chelating molecules) biosynthetic pathways have been well characterized 16; 17. During macrophage-associated infection, the siderophore genes are transcriptionally up-regulated in response to limited iron availability by the activating protein, IdeR 38, and are eventually secreted to sequester iron from iron-containing host proteins (i.e., transferrin, lactoferrin and ferritin). Additionally, the transcriptional factor, sigma factor E, has been shown to control gene expression in the presence of macrophages; interestingly, MhuD is up-regulated 2.5 fold during macrophage infection 39, suggesting its potential role in Mtb virulence. Thus, MhuD may be crucial to produce available iron from heme during iron-deplete conditions. Alternatively, when iron supply is not limited, MhuD’s ability to bind two hemes per monomer could provide a venue for heme storage, a regulatory mechanism which may be analogous to mycobacterial ferritin proteins, BfrA and BfrB, for iron storage 17. This novel feature of Mtb may be an evolutionarily more economical strategy for mycobacteria to utilize the same protein scaffold to acquire double the amount of iron at the cost of being inactivated.
Several heme-uptake systems encoded within operons have been described for pathogenic bacteria, such as S. aureus (i.e., isd operon) and S. marcescans (i.e., has operon), where heme is extracted from hemoglobin through secreted or cell wall-anchored proteins 40; 41. MhuD is conserved across mycobacterial species and is predicted to be within its own operon or in an operon with its adjacent gene, Rv3593, encoding a probable lipoprotein 42; 43. However, none of the surrounding genomically-conserved encoded proteins share homology to other known proteins involved in heme uptake. Additionally, a genomic-wide search failed to identify a putative mycobacterial heme-uptake pathway 44. Nonetheless, this does not negate the possibility of exogenous heme acquisition. Since host (human) iron is most abundant as heme and a major role of bacterial heme oxygenases is their involvement in heme-iron acquisition 3, it is therefore conceivable that Mtb may possess a yet undiscovered mechanism to shuttle extracellular heme into its cytoplasm as yet another alternate source of iron.
Methods and Materials
Construction of the expression vectors for MhuD
Regions of the coding DNA spanning M. tuberculosis protein MhuD (Rv3592) were PCR amplified from M. tuberculosis H37Rv genomic DNA using KOD HotStart Polymerase Kit (Novagen) with 5’ and 3’ primers containing NdeI and HindIII restriction enzyme sites respectively. The PCR product was gel purified (Qiagen) and ligated into a linearized blunt vector, pCR-BluntII-TOPO (Invitrogen) and transformed into OneShot TOP10 E. coli cells (Invitrogen). Double-digestion with NdeI and HindIII were performed on Rv3592 from the blunt vector as well as the plasmid pET-22b (Novagen). Excised Rv3592 and pET-22b were ligated and transformed into E. coli BL21-GOLD (DE3) cells (Novagen).
Expression and Purification of apo MhuD
E. coli BL21-GOLD (DE3) cells harboring pET-22b-Rv3592 were grown aerobically at 37°C in LB medium containing 100 µg/mL ampicillin. At OD600nm of ~0.8, protein expression was induced by 1 mM isopropyl-β-D-thiogalactoside and cells were harvested after 4 hr by centrifugation at 5,000 rpm for 30 min and then washed with resuspension buffer (20 mM Hepes pH 7.8, 350 mM NaCl). Cells were lysed by sonication on ice in resuspension buffer containing phenylmethylsulfonyl fluoride (PMSF) and hen egg lysozyme and the suspension was then centrifuged at 12,000 rpm for 40 min before filtration (1 µm) to remove cell debris. The cell lysate was then loaded onto a Ni2+-charged HiTrap chelating column (5 mL) and washed with 50 mM Tris pH 7.4, 350 mM NaCl and 10 mM imidazole. Fractions of the eluted protein (between 50 and 100 mM imidazole) were collected and concentrated (Amicon, 5kDa MWCO) to 1 mL. The protein was further purified on an S75 gel filtration column equilibrated with 20 mM Tris pH 8 and 10 mM NaCl. The eluted protein was finally loaded onto an ion exchange column (HiTrap Q HP, 5 mL), yielding pure apo-MhuD, which eluted at 150 mM NaCl.
Reconstitution of apo-MhuD with heme
A 0.5 mM hemin solution was prepared by initially dissolving 3.3 mg hemin chloride in 500 µL 0.1M NaOH, to which 500 µL 1M Tris pH 8.0 was added and finally diluted with 50mM Tris pH 7.4, 150 mM NaCl to 10 mL 45. To reconstitute MhuD-heme and MhuD-diheme complexes, hemin was added to 0.1 mM apo-MhuD up to 1.5:1 and 4:1 molar ratios, respectively, at 4°C and excess hemin was removed with an S75 gel filtration column. Protein concentrations were determined by a modified Bradford assay using bovine serum albumin as a standard, and heme concentrations were determined by the pyridine hemochrome method 21.
Heme binding experiments
Aliquots of hemin (1 to 10 µM) were added into 5 µM apo-MhuD, 50 mM Tris pH 8.0 and 150 mM NaCl at 25 °C and heme binding was monitored by difference absorption spectroscopy (Varian Cary 3E) between 300 – 700 nm corresponding to the Soret and Q band regions 5 minutes after each titration. All experiments were performed in triplicate.
Isothermal Titration Calorimetry (ITC)
The association constant and stoichiometry of heme binding was assessed using VP-ITC (MicroCal). Briefly, 28 × 10 µL injections of 400 µM hemin solution were titrated into 1.5 mL 30 µM MhuD at 3.5 min intervals, with constant stirring at 307 rpm. The experiments, including the reference experiment in which hemin was titrated into buffer, were performed in triplicate in 50 mM Tris pH 7.4, 150 mM NaCl at 30 °C. All data were analyzed using the Origin software from MicroCal. After baseline correction with the blank experiment, a non-linear least square method was used to minimize χ2 values and obtain the best fit parameters for the binding affinities and stoichiometry. The best fit parameters were obtained from the sequential binding site model in which two binding sites were specified to be fitted in a sequential manner. The reverse reaction, where 400 µM MhuD was titrated into 30 µM hemin, was also performed under identical conditions and the best fit parameters were fitted using a one-site binding model.
Single Turnover Heme Degradation Experiments
The single turnover degradation reaction of MhuD-heme was investigated in two separate electron-donor reactions provided by either NADPH-cytochrome P450 reductase or ascorbate. Degradation of heme was spectrophotometrically (DU800, Beckman Coulter) monitored by 100 µL reactions carried out at 25°C 4; 5. These reactions were also carried out in the presence of catalase (0.5:1 catalase:MhuD-heme) from Aspergillus niger (Sigma-Aldrich) to rule out non-enzymatic degradation of heme. All reactions were performed in triplicate. (i) NADPH-cytochrome P450 Reductase assay: Human cytochrome P450 reductase (Sigma-Aldrich) was added to 10 µM MhuD-heme, 50 mM Tris pH 7.4 and 150 mM NaCl at a 0.3:1 molar ratio. Degradation was initiated by the addition of 10 µM increments of NADPH, up to a final concentration of 100 µM and spectral changes were monitored upon each addition. (ii) Ascorbate assay: 10 mM sodium ascorbate was added to 5 µM MhuD-heme or MhuD-diheme, 50 mM Tris pH 7.4 and 150 mM NaCl. Spectral changes were monitored every 5 minutes.
Crystallization, Data Collection, Structure Determination and Refinement
Purified apo-MhuD and MhuD-diheme were concentrated to 10 mg/mL and 15 mg/mL, respectively, in 50 mM Tris pH 7.4 and 150 mM NaCl for crystallization trials. Apo-MhuD crystallized in 0.1 M Bis-Tris pH 6.5, 25% PEG-3350 and MhuD-heme crystallized in 0.1 M Bis-Tris pH 5.0, 0.2 M NaCl, 20% PEG-3350, 10 mM triethylamine HCl. Crystals were swiped through 1:1 crystallization conditions and 40% glycerol, and diffraction data collected at 70K. Complete data sets were collected from single crystals. The apo-MhuD crystal diffracted to 3.5 Å with unit cell dimensions of 35.7 × 245.4 × 61.6 Å with 12 subunits per asymmetric units in space group P21, while the MhuD-diheme crystal diffracted to 1.75 Å with unit cell dimensions of 43.9 × 64.6 × 71.1 Å with one dimer per asymmetric unit in space group C2. Images were indexed, integrated and reduced using HKL2000 46. Despite the high resolution for MhuD-diheme, data was collected with a poor positioning of the detector, leading to a higher than expected signal to noise ratio (I/σ = 0.7). The initial phases for the apo-MhuD structure were determined by Phaser 47 using a structural model of TT1380, a conserved hypothetical protein from Thermus thermophilus (PDB code: 1IUJ) as the search model, whereas apo-MhuD was used as the search model for MhuD-diheme. Four strong heme iron sites per asymmetric unit were located from the anomalous using SHELXC/D/E 26. The additional phase information from the SAD calculation was combined with the molecular replacement model phases through SIGMAA and DM in CCP4 program suite 48, which improved the resulting electron density map. Model building was performed with a combination of Arp/Warp 49 and Coot 50 and refinement was carried out by RefMac5 51. The omit map was calculated by removing the heme coordinates from the model followed by refinement using CNS 52. Data collection and refinement statistics are presented in Table 1. The stereochemistry and geometry of each MhuD monomer was validated with PROCHECK 53 and ERRAT 54, and was found to be acceptable. Since both monomers were not identical, it was possible to model in residues 2–102 for chain A and only residues 2–99 for chain B. Furthermore, the sidechains of Arg79 (for chains A & B) and Asn81 (chain A) were modeled as alanines as there was no observable electron density. All molecular graphics were prepared with PyMol 55.
Acknowledgements
This work has been supported by a grant from the National and Californian American Lung Association (RG-78755-N to C.W.G) and the National Institutes of Health (AI081161 to C.W.G). The authors wish to thank Dr. John T. Belisle, Colorado State University, NIH, NIAID Contract NO1 AI-75320 for the generous supply of Mtb H37Rv genomic DNA. We thank Drs. Duilio Cascio and Michael Sawaya for invaluable support with data collection of apo-MhuD at the Advanced Light Source at Berkeley National Laboratories, and general crystallography. We would also like to thank all of the staff at Stanford Synchrotron Radiation Lightsource (SSRL) for their invaluable help in data collection. Portions of this research were carried out at the national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. We grateful to all the staff at SSRL for their invaluable help in data collection. Finally, we would like to thank Drs. Tom Poulos, Huiying Li, Sheryl Tsai, Carla Theimer and Morgan Beeby for invaluable discussions, and Marinor Concepcion and Christopher Chun for their assistance in the project.
Abbreviations used
- Mtb
Mycobacterium tuberculosis
- Rv number
Sanger center notation for each gene in Mtb
- TB
tuberculosis
- HO
heme oxygenase
- Isd
iron-regulated surface determinant
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Protein Data Bank accession number
The atomic coordinates and structure factors for the crystal structure of Mtb MhuD-diheme have been deposited with the Protein Data Bank (RCSB, http://www.rcsb.org/pdb) as entry 3HX9.
References
- 1.Tenhunen R, Marver HS, Schmid R. Microsomal heme oxygenase. Characterization of the enzyme. J Biol Chem. 1969;244:6388–6394. [PubMed] [Google Scholar]
- 2.Drummond GS, Kappas A. Prevention of neonatal hyperbilirubinemia by tin protoporphyrin IX, a potent competitive inhibitor of heme oxidation. Proc Natl Acad Sci U S A. 1981;78:6466–6470. doi: 10.1073/pnas.78.10.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frankenberg-Dinkel N. Bacterial heme oxygenases. Antioxid Redox Signal. 2004;6:825–834. doi: 10.1089/ars.2004.6.825. [DOI] [PubMed] [Google Scholar]
- 4.Wilks A, Schmitt MP. Expression and characterization of a heme oxygenase (Hmu O) from Corynebacterium diphtheriae. Iron acquisition requires oxidative cleavage of the heme macrocycle. J Biol Chem. 1998;273:837–841. doi: 10.1074/jbc.273.2.837. [DOI] [PubMed] [Google Scholar]
- 5.Zhu W, Wilks A, Stojiljkovic I. Degradation of heme in gram-negative bacteria: the product of the hemO gene of Neisseriae is a heme oxygenase. J Bacteriol. 2000;182:6783–6790. doi: 10.1128/jb.182.23.6783-6790.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puri S, O'Brian MR. The hmuQ and hmuD genes from Bradyrhizobium japonicum encode heme-degrading enzymes. J Bacteriol. 2006;188:6476–6482. doi: 10.1128/JB.00737-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skaar EP, Gaspar AH, Schneewind O. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J Biol Chem. 2004;279:436–443. doi: 10.1074/jbc.M307952200. [DOI] [PubMed] [Google Scholar]
- 8.Skaar EP, Gaspar AH, Schneewind O. Bacillus anthracis IsdG, a heme-degrading monooxygenase. J Bacteriol. 2006;188:1071–1080. doi: 10.1128/JB.188.3.1071-1080.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu R, Skaar EP, Zhang R, Joachimiak G, Gornicki P, Schneewind O, Joachimiak A. Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J Biol Chem. 2005;280:2840–2846. doi: 10.1074/jbc.M409526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirotsu S, Chu GC, Unno M, Lee DS, Yoshida T, Park SY, Shiro Y, Ikeda-Saito M. The crystal structures of the ferric and ferrous forms of the heme complex of HmuO, a heme oxygenase of Corynebacterium diphtheriae. J Biol Chem. 2004;279:11937–11947. doi: 10.1074/jbc.M311631200. [DOI] [PubMed] [Google Scholar]
- 11.Schuller DJ, Wilks A, Ortiz de Montellano PR, Poulos TL. Crystal structure of human heme oxygenase-1. Nat Struct Biol. 1999;6:860–867. doi: 10.1038/12319. [DOI] [PubMed] [Google Scholar]
- 12.Schuller DJ, Zhu W, Stojiljkovic I, Wilks A, Poulos TL. Crystal structure of heme oxygenase from the gram-negative pathogen Neisseria meningitidis and a comparison with mammalian heme oxygenase-1. Biochemistry. 2001;40:11552–11558. doi: 10.1021/bi0110239. [DOI] [PubMed] [Google Scholar]
- 13.Sugishima M, Omata Y, Kakuta Y, Sakamoto H, Noguchi M, Fukuyama K. Crystal structure of rat heme oxygenase-1 in complex with heme. FEBS Lett. 2000;471:61–66. doi: 10.1016/s0014-5793(00)01353-3. [DOI] [PubMed] [Google Scholar]
- 14.Lee WC, Reniere ML, Skaar EP, Murphy ME. Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. J Biol Chem. 2008;283:30957–30963. doi: 10.1074/jbc.M709486200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raviglione MC, Luelmo F. Update on the global epidemiology of tuberculosis. Curr Issues Public Health. 1996;2:192–197. [PubMed] [Google Scholar]
- 16.De Voss JJ, Rutter K, Schroeder BG, Barry CE., 3rd Iron acquisition and metabolism by mycobacteria. J Bacteriol. 1999;181:4443–4451. doi: 10.1128/jb.181.15.4443-4451.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ratledge C. Iron, mycobacteria and tuberculosis. Tuberculosis (Edinb) 2004;84:110–130. doi: 10.1016/j.tube.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Ryter SW, Tyrrell RM. The heme synthesis and degradation pathways: role in oxidant sensitivity. Heme oxygenase has both pro- and antioxidant properties. Free Radic Biol Med. 2000;28:289–309. doi: 10.1016/s0891-5849(99)00223-3. [DOI] [PubMed] [Google Scholar]
- 19.Parish T, Schaeffer M, Roberts G, Duncan K. HemZ is essential for heme biosynthesis in Mycobacterium tuberculosis. Tuberculosis (Edinb) 2005;85:197–204. doi: 10.1016/j.tube.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 21.Falk JE. Chemistry and Biochemistry of Porphyrins and Metalloporphyrins. In: Florkin M, Stotz EH, editors. Comprehensive Biochemistry. Vol. 9. Amsterdam: Elsevier Publishing Company; 1963. [Google Scholar]
- 22.Avila L, Huang HW, Damaso CO, Lu S, Moenne-Loccoz P, Rivera M. Coupled oxidation vs heme oxygenation: insights from axial ligand mutants of mitochondrial cytochrome b5. J Am Chem Soc. 2003;125:4103–4110. doi: 10.1021/ja029311v. [DOI] [PubMed] [Google Scholar]
- 23.Sigman JA, Wang X, Lu Y. Coupled oxidation of heme by myoglobin is mediated by exogenous peroxide. J Am Chem Soc. 2001;123:6945–6946. doi: 10.1021/ja015776u. [DOI] [PubMed] [Google Scholar]
- 24.Bhakta MN, Olabisi A, Wimalasena K, Wilks A. Catalytic turnover dependent modification of the Pseudomonas aeruginosa heme oxygenase (pa-HO) by 5,6-O-isopropyledine-2-O-allyl-ascorbic acid. J Inorg Biochem. 2008;102:251–259. doi: 10.1016/j.jinorgbio.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Sciara G, Kendrew SG, Miele AE, Marsh NG, Federici L, Malatesta F, Schimperna G, Savino C, Vallone B. The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis. Embo J. 2003;22:205–215. doi: 10.1093/emboj/cdg031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 27.Jamaat PR, Safari N, Ghiasi M, Naghavi SS, Zahedi M. Noninnocent effect of axial ligand on the heme degradation process: a theoretical approach to hydrolysis pathway of verdoheme to biliverdin. J Biol Inorg Chem. 2008;13:121–132. doi: 10.1007/s00775-007-0308-9. [DOI] [PubMed] [Google Scholar]
- 28.Kwak B, Rhee H, Park S, Lah MS. Synthesis and Characterization of [Mn(III)(6)(N-formylsalicylhydrazidate)(6)(MeOH)(6)]: A New Type of Macrocyclic Hexanuclear Manganese Cluster. Inorg Chem. 1998;37:3599–3602. doi: 10.1021/ic971538h. [DOI] [PubMed] [Google Scholar]
- 29.Brige A, Leys D, Meyer TE, Cusanovich MA, Van Beeumen JJ. The 1.25 A resolution structure of the diheme NapB subunit of soluble nitrate reductase reveals a novel cytochrome c fold with a stacked heme arrangement. Biochemistry. 2002;41:4827–4836. doi: 10.1021/bi012144b. [DOI] [PubMed] [Google Scholar]
- 30.Chan AC, Lelj-Garolla B, F IR, Pedersen KA, Mauk AG, Murphy ME. Cofacial heme binding is linked to dimerization by a bacterial heme transport protein. J Mol Biol. 2006;362:1108–1119. doi: 10.1016/j.jmb.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Aranda Rt, Worley CE, Liu M, Bitto E, Cates MS, Olson JS, Lei B, Phillips GN., Jr Bis-methionyl coordination in the crystal structure of the heme-binding domain of the streptococcal cell surface protein Shp. J Mol Biol. 2007;374:374–383. doi: 10.1016/j.jmb.2007.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe M, Tanaka Y, Suenaga A, Kuroda M, Yao M, Watanabe N, Arisaka F, Ohta T, Tanaka I, Tsumoto K. Structural basis for multimeric heme complexation through a specific protein-heme interaction: the case of the third neat domain of IsdH from Staphylococcus aureus. J Biol Chem. 2008;283:28649–28659. doi: 10.1074/jbc.M803383200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jentzen W, Song XZ, Shelnutt JA. Structural Characterization of Synthetic and Protein-Bound Porphyrins in terms of the Lowest-Frequency Normal Coordinates of the Macrocycle. J. Phys. Chem. B. 1997;101:1684–1699. [Google Scholar]
- 34.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 35.Shelnutt JA, Song XZ, Ma JG, Jia SL, Jentzen W, Medforth CJ. Nonplanar porphyrins and their significance in proteins. Chemical Society Reviews. 1998;27:31–41. [Google Scholar]
- 36.Lad L, Schuller DJ, Shimizu H, Friedman J, Li H, Ortiz de, Montellano PR, Poulos TL. Comparison of the heme-free and -bound crystal structures of human heme oxygenase-1. J Biol Chem. 2003;278:7834–7843. doi: 10.1074/jbc.M211450200. [DOI] [PubMed] [Google Scholar]
- 37.Letoffe S, Heuck G, Delepelaire P, Lange N, Wandersman C. Bacteria capture iron from heme by keeping tetrapyrrol skeleton intact. Proc Natl Acad Sci U S A. 2009;106:11719–11724. doi: 10.1073/pnas.0903842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gold B, Rodriguez GM, Marras SA, Pentecost M, Smith I. The Mycobacterium tuberculosis IdeR is a dual functional regulator that controls transcription of genes involved in iron acquisition, iron storage and survival in macrophages. Mol Microbiol. 2001;42:851–865. doi: 10.1046/j.1365-2958.2001.02684.x. [DOI] [PubMed] [Google Scholar]
- 39.Fontan PA, Smith I. Mycobacterium tuberculosis sigma factor E protects against environmental stress, immune responses. Microbe. 2009;4:119–123. [Google Scholar]
- 40.Cescau S, Cwerman H, Letoffe S, Delepelaire P, Wandersman C, Biville F. Heme acquisition by hemophores. Biometals. 2007;20:603–613. doi: 10.1007/s10534-006-9050-y. [DOI] [PubMed] [Google Scholar]
- 41.Reniere ML, Torres VJ, Skaar EP. Intracellular metalloporphyrin metabolism in Staphylococcus aureus. Biometals. 2007;20:333–345. doi: 10.1007/s10534-006-9032-0. [DOI] [PubMed] [Google Scholar]
- 42.Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacker M, Latendresse M, Paley S, Rhee SY, Shearer AG, Tissier C, Walk TC, Zhang P, Karp PD. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2008;36:D623–D631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roback P, Beard J, Baumann D, Gille C, Henry K, Krohn S, Wiste H, Voskuil MI, Rainville C, Rutherford R. A predicted operon map for Mycobacterium tuberculosis. Nucleic Acids Res. 2007;35:5085–5095. doi: 10.1093/nar/gkm518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cavallaro G, Decaria L, Rosato A. Genome-based analysis of heme biosynthesis and uptake in prokaryotic systems. J Proteome Res. 2008;7:4946–4954. doi: 10.1021/pr8004309. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida T, Kikuchi G. Purification and properties of heme oxygenase from pig spleen microsomes. J Biol Chem. 1978;253:4224–4229. [PubMed] [Google Scholar]
- 46.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 47.McCoy AJ, Grosse-Kuntsleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 49.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 52.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 53.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 54.Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeLano WL. The PyMOL Molecular Graphics System on World Wide Web. 2002 http://www.pymol.org.