Abstract
Recent reports have demonstrated that Dicer, an RNase III endonuclease required for microRNA (miRNA) maturation, is aberrantly expressed in different types of cancer. Furthermore, Dicer has been reported to be regulated by the let-7 family of miRNA genes. We hypothesize that Dicer is aberrantly expressed in oral cancer cells due to altered expressions of let-7, and that Dicer contributes to the development and progression of the disease. Western blot examination of Dicer protein levels in four head and neck squamous cell carcinoma (HNSCC) cell lines, including two oral cancer cell lines, demonstrated that Dicer had between 4 to 24 fold higher expression levels when compared to normal human primary gingival epithelial cells. Furthermore, five of six oral cancer tissues analyzed by indirect immunofluorescence had increased Dicer protein expression, compared to normal gingival epithelial tissue. The Dicer mRNA levels were not found to correlate well with protein expression in the HNSCC cell lines, suggesting that Dicer protein expression was post-transcriptionally regulated. Analysis of let-7a and let-7b levels in HNSCC cell lines by real-time PCR demonstrated that let-7b, but not let-7a, was significantly reduced in the HNSCC cell lines compared to control cells. Lastly, transfection of oral cancer cells with chemically synthesized let-7b and small interfering RNAs targeting Dicer significantly inhibited cell proliferation up to 83% and >100%, respectively, as early as three days post-transfection. Together, these data demonstrate that elevated expression levels of Dicer in oral cancer cells correlate with down-regulation of let-7b and increased cell proliferation.
Introduction
Oral cancer ranks sixth worldwide in incidence and in the USA oral cancer represents 3% of the annually diagnosed malignancies in men (Jemal et al., 2009; Warnakulasuriya, 2009). Every year close to 36,000 Americans will be diagnosed with oral cancer and approximately 8,000 Americans will die (Jemal et al., 2009). Despite vast amounts of research and advances in the fields of oncology and surgery, mortality rates remain unchanged (Massano et al., 2006). Therefore, new therapeutic strategies are needed. In order to develop new therapies for treating oral cancer, new molecular insights into oral cancer biology are required.
RNAi is a post-transcriptional gene regulatory mechanism that can specifically silence gene expression by repressing translation and/or degrading mRNA by means of small non-coding double-stranded RNAs (dsRNAs) (Rana, 2007). Endogenous, small non-coding RNAs known as microRNAs (miRNAs) are a specific class of 19- to 25-nt non-coding evolutionary conserved RNAs that mediate gene expression at the post-transcriptional level by base pairing to partially complementary sites in the 3′-untranslated region (3′-UTR) of mRNAs (Rana, 2007). Human miRNAs regulate diverse cellular and molecular processes including cellular proliferation, differentiation, and apoptosis and are predicted to regulate >60% of all protein encoding genes within the human genome (Rana, 2007; Friedman et al., 2009). Based on the significant effects of miRNAs on gene expression, it is not surprising that miRNAs have also been implicated in the pathogenesis of cancer. Numerous studies have reported aberrant expression profiles of miRNAs in cancer, and miRNAs have been described as having oncogenic and tumor-suppressive properties (for review see (Calin and Croce, 2006; Dalmay, 2008)).
The biogenesis of miRNAs begins within the nucleus where miRNA genes are transcribed by RNA polymerase II into primary transcripts (pri-miRNAs) (Rana, 2007; Dalmay, 2008). The pri-miRNAs are then cleaved by the Drosha-DGCR8 complex into precursor miRNAs (pre-miRNAs) (Rana, 2007; Dalmay, 2008). Pre-miRNAs are 70-90-nt long molecules with a hairpin structure that are subsequently exported into the cytoplasm where they are further processed by Dicer (Rana, 2007; Dalmay, 2008). Dicer is a highly conserved RNase III type enzyme found in almost all eukaryotes that is essential for the RNAi and miRNA pathways (Rana, 2007). Dicer processes pre-miRNAs into mature ∼21 bp miRNA duplexes, which are subsequently incorporated into the RNA induced silencing complex (RISC) (Rana, 2007; Dalmay, 2008). There the passenger strand of the miRNA duplex is removed, allowing the guide strand to then target RISC to mRNAs containing partially complementary sequences in the 3′-UTR (Rana, 2007; Dalmay, 2008). Subsequently, the targeted mRNAs become either translationally repressed or degraded within cytoplasmic structures termed, GW/P-bodies (Jakymiw et al., 2007; Rana, 2007).
The discovery of RNAi has stimulated research on the role of this cellular process in the development and progression of cancer (Merritt et al., 2008). Although alterations in miRNA expression have been reported in cancer, the mechanisms of this dysregulation have not been fully elucidated (Calin and Croce, 2006). In some cases, genomic changes and changes in transcriptional regulation of miRNA expression have been found to correlate with changes in miRNA expression (Zhang et al., 2006; Blenkiron et al., 2007). Alternatively, global changes in miRNA expression in human cancers have also been linked to the dysregulation of genes required for miRNA biogenesis (Zhang et al., 2006; Blenkiron et al., 2007). Interestingly, during the last several years a number of reports have found Dicer to be aberrantly expressed in different types of cancer. More specifically, Dicer has been found to be overexpressed in prostate and precursor lesions of lung adenocarcinomas (Chiosea et al., 2006; Chiosea et al., 2007), or reduced in ovarian and lung cancer (Karube et al., 2005; Merritt et al., 2008). Furthermore, both low and high levels of Dicer have been correlated with poor prognosis in cancer patients (Karube et al., 2005; Chiosea et al., 2006; Chiosea et al., 2008; Merritt et al., 2008). The discrepancies in the dysregulation of Dicer expression among the various tumor types have been attributed to tissue specific differences and/or the degree of aggressiveness of the cancer (Grelier et al., 2009).
Head and neck squamous cell carcinomas (HNSCCs), including oral squamous cell carcinomas (OSCCs), also have aberrant miRNA expression levels (Tran et al., 2007; Chang et al., 2008; Wong et al., 2008; Childs et al., 2009; Henson et al., 2009; Li et al., 2009). Although alterations in miRNA expression levels in HNSCCs can be attributed in some cases to the loss or amplification of chromosomal material (Henson et al., 2009), there is also the possibility, as described above, that these alterations could be due to potential defects in the miRNA biogenesis machinery, such as abnormal Dicer expression levels. Despite the growing evidence that Dicer appears to be aberrantly expressed in cancer, the regulation of this gene remains unclear. Interestingly, several recent studies have demonstrated that Dicer expression can be regulated by let-7 miRNA (Forman et al., 2008; Selbach et al., 2008; Tokumaru et al., 2008). The let-7 family of miRNA genes are tumor-suppressor miRNAs that have been found to be down-regulated in lung, colorectal, and gastric cancers (Takamizawa et al., 2004; Akao et al., 2006; Yanaihara et al., 2006; Zhang et al., 2007). Therefore, the aim of this study was to examine whether Dicer is aberrantly expressed in OSCCs due to altered expressions of let-7, and whether Dicer plays an important role in oral cancer cell proliferation.
Materials and Methods
Cell Culture
Human tongue SCC cell lines CAL 27 and SCC-25 and human HNSCC cell lines FaDu and RPMI 2650 were purchased from American Type Culture Collection (ATCC, Manassas, VA). Each of the cell lines was cultured in ATCC specified complete growth media in a 37°C incubator with 5% CO2. Primary cultures of human gingival epithelial cells (pGECs) were obtained from gingival explants as described (Mao et al., 2007) and following Institutional Review Board (IRB) guidelines. The primary cells were cultured in keratinocyte growth medium (KGM; Cambrex, East Rutherford, NJ) at 37°C in 5% CO2. pGECS were used for experimentation between passages 4-6.
Western Blot
Cultured or transfected cells were washed with phosphate-buffered saline (PBS) and lysed using RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, and 1% Triton X-100) with Complete EDTA-free protease inhibitor (Roche, Palo Alto, CA, USA). The protein lysates were then resolved by SDS–PAGE on a 10% gel and transferred to nitrocellulose. The nitrocellulose membrane was cut just below the 100 kDa molecular weight protein marker into two pieces and blocked in 5% non-fat dried milk in PBS-Tween for 1 hour at room temperature. The top portion of the membrane containing the higher molecular weight proteins was incubated with mouse monoclonal anti-Dicer antibody (1:100; clone 13D6; Abcam, Cambridge, MA) and the bottom portion of the membrane containing the lower molecular weight proteins was incubated with mouse monoclonal anti-tubulin antibody (1:10,000; Sigma-Aldrich, St. Louis, MO) for 1 hour at room temperature. The membranes were then washed four times with PBS-Tween and incubated with horseradish-peroxidase-conjugated goat anti-mouse IgG (1:10,000; SouthernBiotech, Burmingham, AL) for 1 hour at room temperature. Immunoreactive bands were detected by the SuperSignal Chemiluminescent system (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions. To quantify Dicer protein expression levels in HNSCC cell lines and pGECs, ImageJ software (Abramoff et al., 2004) was used to measure the integrated density of the Dicer signal normalized to tubulin levels.
Indirect Immunofluorescence
After receiving IRB approval, six randomly selected formalin fixed paraffin-embedded oral carcinoma tissues and one normal gingival epithelial tissue specimen were obtained from the University of Florida Oral Pathology Biopsy Service archives and through Dr. Ikramuddin Aukhil in the Department of Periodontology, respectively. The staging and nodal status of the six patients with oral squamous cell carcinomas are listed in Table 1. 4-μm sections were cut and mounted on Superfrost Plus glass slides (Fisher Scientific, Pittsburgh, PA). The sectioned slides were deparaffinized using CitriSolv (Fisher Scientific, Pittsburgh, PA), hydrated by submersing in three separate concentrations of ethyl alcohol (100%, 95%, and 70%) and rinsed continuously in distilled water for 5 minutes. Afterwards, antigen retrieval was performed by incubating slides in a 1x Antigen Retrieval Citra Plus Solution (BioGenex, San Ramon, CA), according to the manufacturer's recommendation. Briefly, the slides were placed into the citrate-based antigen retrieval solution at 95°C for 20 minutes. The heated slides were cooled down to room temperature for another 20 minute interval, rinsed with distilled water for 5 minutes, and then blocked for 30 minutes with 1.5% Normal Horse Serum (Vector Laboratories, Burlingame, CA). Afterwards, the sections were incubated with rabbit polyclonal anti-Dicer (1:50; Sigma-Aldrich, St. Louis, MO) and mouse monoclonal anti-EGFR (1:50; Dako, Carpinteria, CA) antibodies for 1 hour at room temperature. After washing with PBS, the slides were incubated with the corresponding secondary fluorochrome-conjugated goat antibodies at room temperature for 1 h. Alexa Fluor 488 (1:400) and Alexa Fluor 568 (1:400) (Invitrogen, Carlsbad, CA) were the primary fluorochromes used. Finally, glass coverslips were mounted onto the slides using VECTASHIELD Mounting Medium with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA). Fluorescence images were acquired with a Zeiss (Thornwood, NY) Axiovert 200M microscope, using a 40× 0.75 NA objective and Zeiss AxioVision software. The images were analyzed using Adobe (San Jose, CA) Photoshop CS4 software. Both the cancerous and control regions were confirmed by a board certified Oral and Maxillofacial Pathologist.
Table 1.
Staging and Nodal Status of Patients with Oral Squamous Cell Carcinomasa
| OSCCb | Tumor Stage | Nodal status |
|---|---|---|
| 1 | III | Positive |
| 2 | I | Negative |
| 3 | II | Negative |
| 4 | I | Negative |
| 5 | I | Negative |
| 6 | III | Negative |
All biopsies were taken from primary tumors.
OSCC; oral squamous cell carcinoma
RNA Isolation and Real-time PCR
Total RNA including miRNAs were extracted and purified from cultured or transfected cells using the mirVana™ miRNA Isolation kit (Ambion/Applied Biosystems, Austin, TX), following the manufacturer's instructions. RNA was quantitated using a NanoDrop® ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE). For Dicer mRNA and 18S rRNA quantitation, total RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). For let7a, let-7b, and RNU6B quantitation, total RNA was reverse transcribed using TaqMan® specific RT primers and the TaqMan® microRNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). Afterwards, quantitative real-time PCR was performed in an Applied Biosystems (Foster City, CA) StepOne Real-Time PCR machine using pre-designed TaqMan® gene/miRNA specific assays for Dicer (Hs00229023_m1; assay location: nucleotide 2440/2496), 18S, let-7a, let-7b, and RNU6B (Applied Biosystems, Foster City, CA) combined with TaqMan® Fast Universal PCR Master Mix (Applied Biosystems, Foster City, CA), according to the manufacturer's instructions.
Transfections with miRNAs and siRNAs
The let-7b miRNA and small interfering RNA (siRNA) targeting Dicer (siDicer) were previously described (Hutvagner et al., 2001; Pauley et al., 2006) and were synthesized by Thermo Scientific Dharmacon Inc. (Lafayette, CO). The siRNA targeting GFP (siGFP; target sequence, 5′-GGC UAC GUC CAG GAG CGC ACC-3′) was commercially available and purchased from Thermo Scientific Dharmacon Inc. (Lafayette, CO). CAL 27 cells grown to 30-50% confluency were either mock transfected or transfected with 100 nM of siGFP, siDicer, or let-7b using Lipofectamine 2000™ (Invitrogen, Carlsbad, CA), according to the manufacturer's recommendation. Protein and mRNA extractions were performed 72 hours after transfection, after which the samples were further analyzed by Western blotting and real-time PCR, according to the protocols described above. For the cell proliferation assay the cells were trypsinized 24 hours post-transfection and re-seeded on a 96-well plate as described below.
Cell Proliferation Assay
Cell proliferation was quantitated using CellTiter 96® Aqueous One Solution Cell Proliferation Assay (Promega, Madison, WI) according to the manufacturer's protocol. Quantitation of cell number was achieved by re-seeding 5,000 cells on a 96-well plate 24 hours after transfection. At 3, 6, and 8 days post-transfection a Bio-Rad (Hercules, CA) Model 680 microplate reader was used to read the absorbance of each well at 490 nm. The absorbance values were then converted to number of cells based on a calculated standard curve.
Tumor Xenografts
After receiving IACUC approval, CAL 27 cells grown to 70% confluence were suspended in Dulbecco's Modified Eagle's Medium containing 10% fetal bovine serum / Matrigel (1:1; BD Biosciences, Bedford, MA) at 500,000 cells per 50 μl and injected submucosally in the floor of mouth of anesthetized eight-week-old NOD-SCID mice (NOD.CB17-Prkdcscid strain; Jackson Laboratory, Bar Harbor, ME). Oral tumors were grown for two weeks, after which the animals were sacrificed. The tumor tissue was then harvested and fixed in 10% formalin solution. Tissues were paraffin-embedded and 5-μm-thick sections were stained with hematoxylin and eosin (H & E). The sections were also processed for indirect immunofluorescence analysis as described above. H & E stained tissue sections were imaged with a Leica (Bannockburn, IL) DMLB2 microscope, using a 2.5× objective and Media Cybernetics (Bethesda, MD) QCapture Pro software.
Statistical Analysis
Comparisons between groups were performed using one-way ANOVA. A value of P < 0.05 was considered statistically significant.
Results
Dicer Overexpression in HNSCC Cell Lines
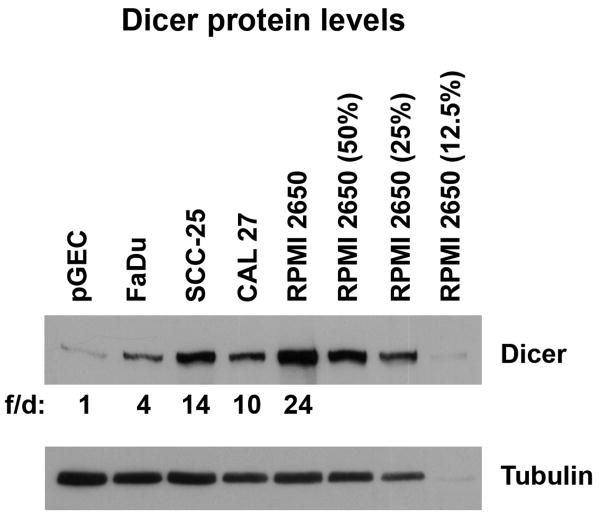
Dicer protein expression in several HNSCC cell lines was examined and compared to normal primary gingival epithelial cells (pGECs) by Western blot analysis (Fig. 1). Quantitation of Dicer expression levels using ImageJ software demonstrated that HNSCC cell lines (FaDu, SCC-25, CAL 27, and RPMI 2650) exhibited between 4 to 24 fold differences in Dicer protein levels compared to pGECs. More specifically, two OSCC-derived cell lines (SCC-25 and CAL 27), had between 10 to 14 times the level of Dicer compared to pGEC cells. Serial dilutions of RPMI 2650 cell lysates were also included to aid in visualizing the degree of Dicer overexpression.
Figure 1.
Overexpression of Dicer in head and neck squamous cell carcinoma cell lines. Western blot analysis of Dicer expression in several head and neck squamous cell carcinoma cell lines (FaDu, SCC-25, CAL 27, and RPMI 2650) compared to normal primary gingival epithelial cells (pGEC). Tubulin antibody was used to check for equal loading of samples. Serial dilutions of RPMI 2650 cell lysates (50%, 25%, and 12.5%) were included to aid in visualizing the degree of Dicer overexpression. The data is representative of two independent experiments. f/d; fold difference, quantitative measurement of the relative Dicer protein fold expression differences using ImageJ software.
Dicer Up-regulation in OSCCs
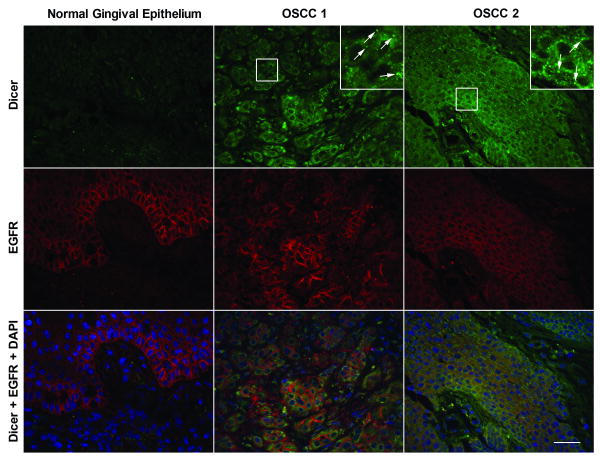
The aberrant expression of Dicer protein observed in HNSCC cell lines, including oral cancer cell lines, prompted us to examine the Dicer protein expression levels and cellular localization patterns on six randomly selected histological sections of OSCCs in comparison to normal gingival epithelial tissue. To help demarcate the basal epithelium from the underlying connective tissue within the normal tissue and to also help identify tumors of epithelial cell origin, the OSCCs were simultaneously stained for epidermal growth factor receptor (EGFR). EGFR is a known marker for epithelial-derived HNSCC cells, but it is also found to be expressed in normal proliferating epithelial cells and thus can be found in the growth zone (i.e. basal third region) of the normal oral epithelium (Hanahan and Weinberg, 2000; Herbst, 2004). Indirect immunofluorescence (IIF) analysis demonstrated cytoplasmic staining of Dicer and its up-regulation in OSCCs in comparison to that observed in normal epithelial tissue, which did not have any detectable levels of Dicer (Fig. 2). Interestingly, Dicer was also found to strongly localize to discrete cytoplasmic foci within the cancer cells (Fig. 2, inset panels, arrows). IIF analysis of the six different OSCCs demonstrated moderate to strong diffuse cytoplasmic and discrete cytoplasmic foci staining of Dicer in five of six samples examined in comparison to that observed in normal epithelial tissue. The two OSCCs that were most representative in terms of Dicer staining pattern of the six OSCCs examined and that had the strongest diffuse cytoplasmic and discrete foci expression of Dicer compared to normal tissue are shown in Figure 2.
Figure 2.
Dicer is up-regulated in oral squamous cell carcinomas. Indirect immunofluorescence analysis of Dicer (green) expression in formalin fixed paraffin-embedded normal human gingival epithelial and two oral squamous cell carcinoma (OSCC) tissues. EGFR (red) detection was used to help demarcate the normal basal epithelium from the underlying connective tissue and also help identify epithelial-derived cancerous regions in the OSCC tissues. Inset panels are higher (3×) magnification views of the boxed areas demonstrating the discrete cytoplasmic foci staining of Dicer (arrows). Nuclei (blue) were counterstained with DAPI. Scale bar: 25 μm.
The Cellular Localization of Dicer in a Mouse Model for Human Oral Cancer is Consistent with OSCCs
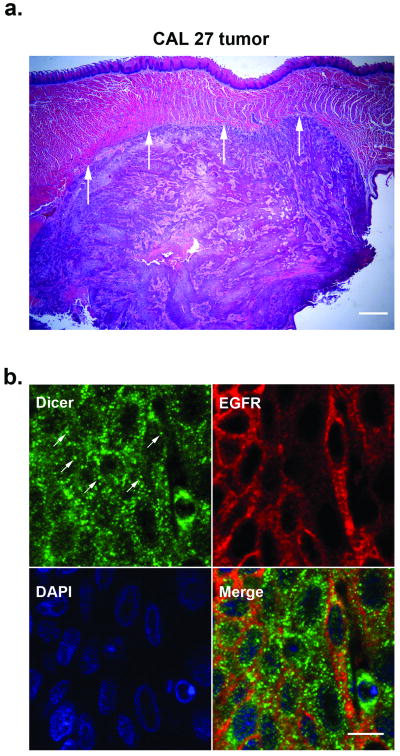
To confirm the Dicer staining pattern observed in OSCCs, we also analyzed Dicer localization in a mouse floor-of-mouth model for human oral cancer. The murine xenograft tumor was generated using the CAL 27 cell line. Hematoxylin and eosin-stained sections of the harvested tissue confirmed the presence of the murine CAL 27 xenograft tumor (Fig. 3a). Furthermore, IIF analysis of the CAL 27 xenograft tumor demonstrated Dicer localization within the cytoplasm and in discrete cytoplasmic foci consistent with staining observed in OSCCs (Fig. 3b). To help demarcate the tumor, the tissue was also costained for EGFR.
Figure 3.
The cellular localization of Dicer in a murine xenograft tumor is consistent with oral squamous cell carcinomas. (a) Hematoxylin and eosin-stained CAL 27 murine xenograft tumor. Arrows demarcate the boundary between the xenograft tumor and the overlying muscle tissue of the mouse tongue. Scale bar: 500 μm. (b) Indirect immunofluorescence analysis of Dicer (green) expression in formalin fixed paraffin-embedded xenograft tumor tissue. The discrete cytoplasmic foci staining of Dicer are indicted by arrows. EGFR (red) detection was used to help demarcate the xenograft tumor. Nuclei (blue) were counterstained with DAPI. Scale bar: 10 μm.
Of interest were the reproducible findings that Dicer strongly localized to discrete cytoplasmic foci in both human OSCCs and in the xenograft tumor. Because the Dicer foci strongly resembled GW/P-bodies, cytoplasmic structures that are linked to RNAi function (Eystathioy et al., 2002; Jakymiw et al., 2005; Jakymiw et al., 2007) and studied by our lab; we also costained Dicer with a GW/P-body marker in the xenograft tumor tissue. The staining, however, revealed that the cytoplasmic Dicer foci did not colocalize with GW/P-bodies (data not shown).
let-7b Levels are Down-regulated in HNSCC Cell Lines
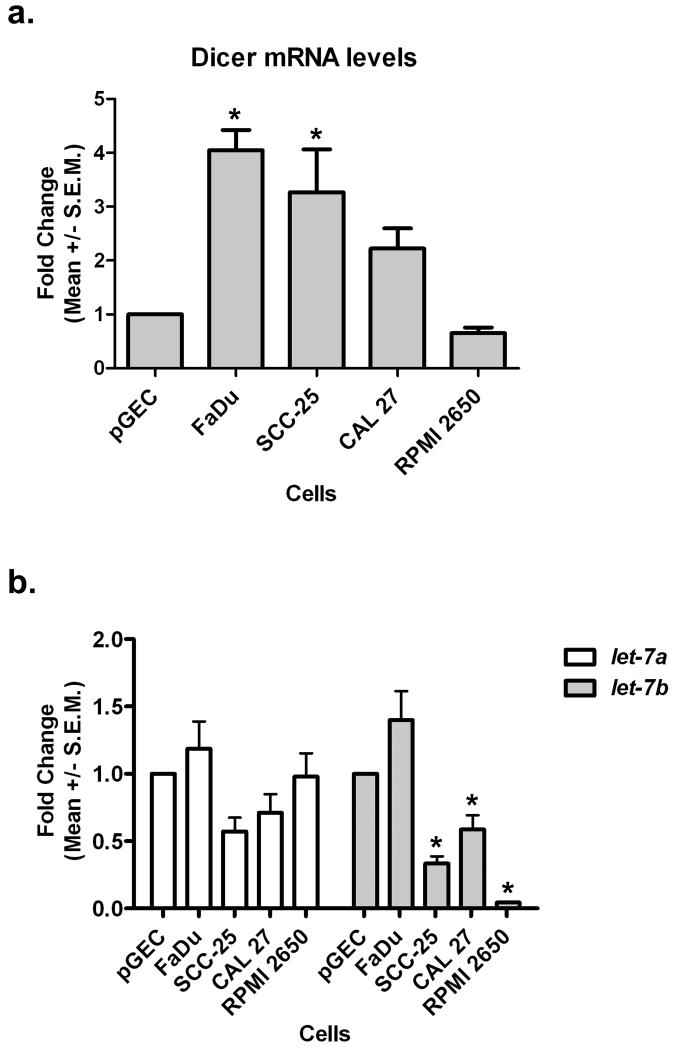
Having demonstrated that Dicer protein was overexpressed in HNSCC cell lines and in OSCCs; we next wanted to determine whether this up-regulation was potentially due to the aberrant expression of the Dicer transcript. Real-time PCR analysis of Dicer mRNA expression levels in the HNSCC cell lines, interestingly, demonstrated that the levels of Dicer mRNA were only significantly up-regulated in two (FaDu and SCC-25) out of the four HNSCC cell lines in comparison to pGECs (Fig. 4a). The fact that Dicer mRNA was not significantly overexpressed in the other two HNSCC cell lines (CAL 27 and RPMI 2650) even though the protein was (see Fig. 1), suggested that Dicer protein expression levels may be regulated at the post-transcriptional level. Because several recent reports have demonstrated that Dicer expression can be regulated by let-7a and let-7b (Forman et al., 2008; Selbach et al., 2008; Tokumaru et al., 2008), we performed real-time PCR to examine the expression levels of these two miRNAs in the HNSCC cell lines compared to pGECs (Fig. 4b). Let-7b, but not let-7a, was significantly found to be reduced in three (SCC-25, CAL 27, and RPMI 2650) out of the four HNSCC cell lines in comparison to pGECs. In particular, a ∼40-95% reduction in let-7b levels was observed in the HNSCC cell lines compared to control cells. Only FaDu cells exhibited no change in both let-7a and let-7b expression in comparison to pGECs.
Figure 4.
let-7b expression is reduced in head and neck squamous cell carcinoma cell lines. Real-time PCR analysis of (a) Dicer mRNA and (b) let-7 microRNA expression levels in several head and neck squamous cell carcinoma cell lines (FaDu, SCC-25, CAL 27, and RPMI 2650) compared to normal primary gingival epithelial cells (pGEC). Data shown were obtained from three biological replicates. S.E.M.; standard error measurement. *P < 0.05.
Dicer Depletion Inhibits Cell Proliferation of Oral Cancer Cells
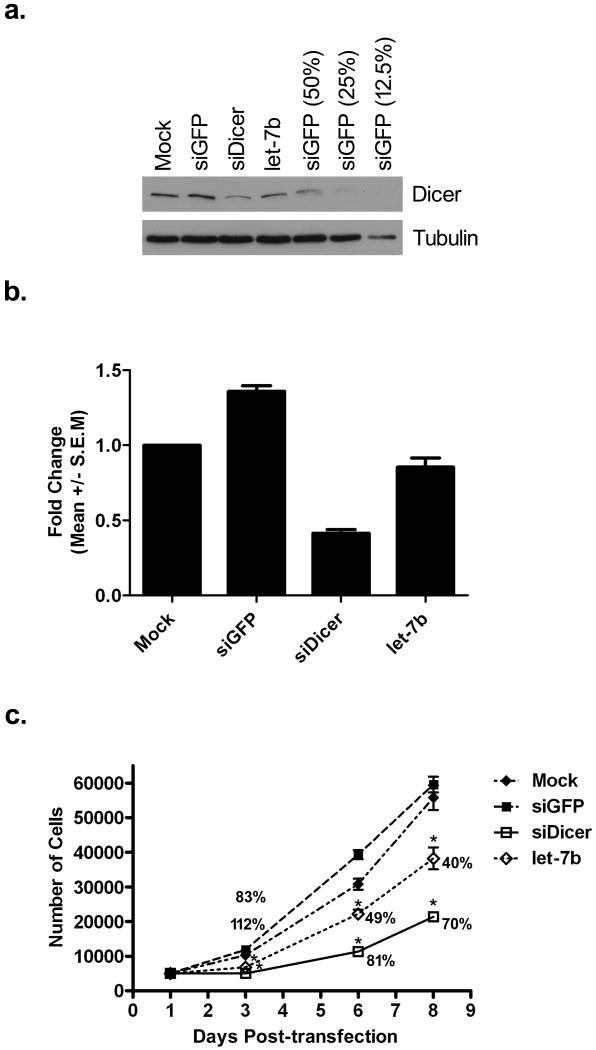
Due to the aberrant expression of let-7b in HNSCC cells, the apparent inverse correlation of let-7b levels with Dicer protein expression, and the fact that Dicer protein levels have been previously demonstrated to be specifically regulated by the direct targeting of let-7b (Forman et al., 2008; Selbach et al., 2008), we next wanted to confirm whether Dicer was similarly regulated by let-7b in oral cancer cells. Therefore, CAL 27 cells were either mock transfected or transfected with control non-targeting green fluorescent protein (GFP) siRNA (siGFP), an siRNA targeting Dicer (siDicer), or let-7b. The siDicer has been previously demonstrated to suppress Dicer levels (Hutvagner et al., 2001) and was therefore used as a positive control. Western blot analysis confirmed that siDicer and let-7b were capable of suppressing Dicer protein expression levels 72 hours post-transfection compared to siGFP and mock transfected cells (Fig. 5a). More specifically, based on serial dilutions of siGFP transfected cell lysates that were used to aid in quantitating the degree of Dicer overexpression, cells transfected with siDicer and let-7b exhibited ∼60% and ∼30% knockdown of Dicer protein relative to siGFP transfected cells, respectively. Furthermore, real-time PCR analysis of Dicer mRNA levels demonstrated that siDicer, but not let-7b, was capable of repressing Dicer mRNA levels (Fig. 5b). Having demonstrated that siDicer and let-7b were capable of suppressing Dicer protein levels, we next wanted to assess the effects of Dicer depletion on oral cancer cell proliferation. Therefore, a cell proliferation experiment was carried out where CAL 27 cells were either mock transfected or transfected with siGFP, siDicer, or let-7b, after which cell numbers were assayed 3, 6, and 8 days post-transfection (Fig. 5c). The growth curve showed that silencing of Dicer with either siDicer or let-7b significantly inhibited cell proliferation over a period of eight days compared to control siGFP or mock transfected cells. Furthermore, the growth inhibitory effect was greater for siDicer compared to let-7b. The largest average percent inhibition on cell proliferation was observed three days post-transfection with siDicer and let-7b transfected cells exhibiting a >100% and 83% inhibitory effect relative to control siGFP transfected cells, respectively. The inhibitory effect decreased over time, but was still effectual even up to eight days post-transfection with siDicer and let-7b transfected cells still maintaining a 70% and 40% inhibitory effect on oral cancer cell proliferation, respectively.
Figure 5.
Knockdown of Dicer inhibits cell proliferation of oral cancer cells. CAL 27 cells were either mock transfected or transfected with siGFP, siDicer, or let-7b. (a) Western blot analysis of Dicer protein levels 72 hours post-transfection. Serial dilutions of siGFP lysates (50%, 25%, and 12.5%) were used to help comparatively quantitate the degree of protein knockdown. Tubulin was used as a loading control. The experiment was repeated two additional times with similar results. (b) Real-time PCR analysis of Dicer mRNA levels 72 hours post-transfection. The data is representative of three independent experiments. (c) 5,000 cells were seeded on a 96-well plate 24 hours after either mock transfection or transfection with siGFP, siDicer, or let-7b. Cell numbers were assayed 3, 6, and 8 days post-transfection. The results represent the mean ± standard error measurement. Indicated at each time point are also the average percent inhibitory effects on cell proliferation for siDicer and let-7b relative to siGFP transfected cells. The data is representative of three independent experiments performed in triplicate. *P < 0.05 in comparison to control siGFP or mock transfected cells.
Discussion
Alterations of Dicer expression levels in various types of cancer have been observed (Karube et al., 2005; Chiosea et al., 2006; Chiosea et al., 2007; Chiosea et al., 2008; Merritt et al., 2008), but nothing has been reported about the status of Dicer in HNSCCs, in particular OSCCs until now. Furthermore, not much is known about the regulatory mechanisms that cause these alterations of Dicer expression levels in cancer. Therefore, this study explored the potential roles of Dicer and the regulatory mechanisms governing its expression levels in the pathogenesis of HNSCCs with a focus on oral cancer. Dicer protein was demonstrated to be overexpressed in all four HNSCC cell lines examined, including two tongue SCC-derived cell lines, relative to normal primary gingival epithelial cells. Furthermore, five of six OSCCs examined had increased Dicer protein expression, where Dicer was found to localize diffusely within the cytoplasm and in discrete cytoplasmic foci of the cancer cells, compared to cells within normal gingival epithelial tissue, which did not have any detectable levels of Dicer. The Dicer staining pattern observed in OSCCs was also confirmed in a mouse model of human oral cancer. Together, the above findings demonstrate that based on the samples analyzed, HNSCCs, including OSCCs, have increased Dicer protein levels and are consistent with several reports that have similarly found increased Dicer protein expression in other types of cancer, including prostate and ovarian carcinomas, and in SCCs of the lung (Chiosea et al., 2006; Chiosea et al., 2007; Flavin et al., 2008). It is important to note that a limitation of our study was the use of a small number of cell lines and tissues for Dicer expression analysis. Even though we found Dicer to be overexpressed in the HNSCC cell lines and tissues we examined, a more comprehensive study will be needed that will include testing of a greater number of cell lines and tissues to more conclusively ascertain the extent of high-level expression of Dicer in HNSCCs.
In the examination of Dicer mRNA in the HNSCC cell lines, only two of the cell lines (FaDu and SCC-25) had significant up-regulation of Dicer mRNA that could account for the increased protein expression. The other two cell lines (CAL 27 and RPMI 2650) did not have significant increases in Dicer mRNA, yet the Dicer protein levels were overexpressed. In fact, the RPMI 2650 cell line had reduced levels of Dicer mRNA compared to the control cells and yet had the highest level of Dicer protein expression of all the HNSCC cell lines analyzed. These findings suggest that Dicer expression is regulated at the post-transcriptional level. Interestingly, our data corroborate several studies that have similarly reported that Dicer mRNA does not correlate well with protein expression and that the regulation of Dicer expression seems to be largely at the post-transcriptional level (Grelier et al., 2009; Wiesen and Tomasi, 2009).
Recently, it was reported that Dicer expression was regulated by let-7 (Forman et al., 2008; Selbach et al., 2008; Tokumaru et al., 2008). To understand better the discrepancy in Dicer mRNA and protein expression levels observed in the HNSCC cell lines, the levels of let-7a and let-7b, two miRNAs previously found to regulate Dicer expression, were examined. Our analysis demonstrated that let-7b was significantly reduced in three out of the four HNSCC cell lines, including the two oral cancer cell lines, relative to control cells. This finding implied that the increased Dicer protein expression was due to the aberrant expression of let-7b in HNSCC cells. The fact that RPMI 2650 cells had extremely low levels of let-7b could explain why the protein was highly up-regulated even though the mRNA was reduced relative to normal cells. Furthermore, to validate let-7b regulation of Dicer expression levels, an exogenous mature let-7b was transfected into the CAL 27 oral cancer cell line which led to a reduction in Dicer protein levels, but not Dicer mRNA. These data suggested that let-7b-mediated reduction of Dicer protein was potentially due to translational repression and not mRNA degradation and confirmed a similar observation made by Selbach et al. who reported that Dicer was likely a direct translational target of let-7b (Selbach et al., 2008). Of note, although our study focused on the analysis let-7a and let-7b levels in HNSCC cell lines, there is the possibility that other let-7 family members may also contribute to the aberrant expression of Dicer in HNSCCs. Interestingly, let-7d has recently been reported to be reduced in primary human HNSCC tumors (Childs et al., 2009). Let-7d is a regulator of Dicer expression (Tokumaru et al., 2008) and could therefore also potentially contribute towards the overexpression of Dicer in HNSCCs.
Due to the fact that Dicer protein was increased in HNSCC cell lines and OSCCs, the biological consequence of Dicer in oral cancer cells was examined, in particular its role in cell proliferation. The addition of either an exogenous siRNA targeting Dicer or mature let-7b significantly inhibited the proliferation of oral cancer cells compared to control cells as early as three days post-transfection. The Dicer siRNA had a greater repressive effect on cell proliferation compared to let-7b. This was most likely due to the fact that the siRNA had a stronger knockdown effect on Dicer protein compared to let-7b. This is not unusual as miRNAs have been proposed to act as biological rheostats that make fine-scale adjustments to protein output (Baek et al., 2008). Regardless, our data demonstrate that Dicer appears to be a critical component for cell growth and supports earlier studies that have shown Dicer to be important in regulating cell cycle events and cell proliferation (Carmichael et al., 2004; Hatfield et al., 2005; Murchison et al., 2005). Moreover, let-7b appears to be an important regulator of Dicer levels and can affect the cell proliferation of oral cancer cells. Of note, let-7 has been reported to repress cell proliferation and is thought to be a master regulator of cell proliferation pathways (Johnson et al., 2007). Interestingly, Dicer was identified as one of several let-7 targets that can potentially influence cell division (Johnson et al., 2007).
Although our group and others have demonstrated that specific types of tumors have elevated levels of Dicer protein, there is evidence that also demonstrates that specific cancers can have decreased Dicer expression levels, such as in advanced lung adenocarcinomas (Karube et al., 2005). One simple explanation for these discrepancies could be due to tissue specific differences. Another possibility is that some of these studies focus primarily on the measurement of Dicer mRNA levels and not protein, and if one recalls, our data clearly demonstrated that Dicer mRNA does not correlate well with protein expression. Therefore, data obtained from PCR analyses need to be carefully interpreted because even though the level of Dicer mRNA may be found to be lower in comparison to normal cells, it does not necessarily mean the protein level will be similarly repressed. Take for example our findings for the RPMI 2650 cell line, where the Dicer mRNA was reduced compared to normal cells, yet the protein was still highly expressed. This example highlights the complexities of Dicer expression regulation and demonstrates that Dicer can be regulated by post-transcriptional regulatory mechanisms. This could also help explain some of the discrepancies observed in Dicer expression within the same tumor types. The fact that Dicer expression appears to be related to the aggressiveness and metastatic spread of cancer and that the Dicer gene is predicted to produce 14 mRNA variants (Grelier et al., 2009), further reaffirms the complexities associated with the regulation of the expression of this gene. Regardless, the fact that Dicer expression varies in different cancer types suggests that Dicer dysregulation appears to be important for cancer progression.
In conclusion, based on the samples analyzed, our present study demonstrated that Dicer protein was up-regulated in oral cancer cells and that its levels appeared to be regulated by let-7b. Moreover, Dicer knockdown inhibited cell proliferation of oral cancer cells. Together, this work implies that Dicer up-regulation in conjunction with let-7b down-regulation contributes to oral cancer progression. Silencing the expression of Dicer using RNAi strategies could be potentially used as an adjunct therapy to curb the proliferation of cancer cells.
Acknowledgments
Supported by: NIDCR, Grant number: K99DE018191; Bankhead-Coley Cancer Research Program Florida Department of Health, Grant number: 08BN-02; Andrew J. Semesco Foundation, Inc.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics Intern. 2004;11:36–42. [Google Scholar]
- Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, Tavare S, Caldas C, Miska EA. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Carmichael JB, Provost P, Ekwall K, Hobman TC. ago1 and dcr1, two core components of the RNA interference pathway, functionally diverge from rdp1 in regulating cell cycle events in Schizosaccharomyces pombe. Mol Biol Cell. 2004;15:1425–1435. doi: 10.1091/mbc.E03-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SS, Jiang WW, Smith I, Poeta LM, Begum S, Glazer C, Shan S, Westra W, Sidransky D, Califano JA. MicroRNA alterations in head and neck squamous cell carcinoma. Int J Cancer. 2008;123:2791–2797. doi: 10.1002/ijc.23831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs G, Fazzari M, Kung G, Kawachi N, Brandwein-Gensler M, McLemore M, Chen Q, Burk RD, Smith RV, Prystowsky MB, Belbin TJ, Schlecht NF. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol. 2009;174:736–745. doi: 10.2353/ajpath.2009.080731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, Dhir R. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiosea S, Jelezcova E, Chandran U, Luo J, Mantha G, Sobol RW, Dacic S. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–2350. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- Chiosea SI, Barnes EL, Lai SY, Egloff AM, Sargent RL, Hunt JL, Seethala RR. Mucoepidermoid carcinoma of upper aerodigestive tract: clinicopathologic study of 78 cases with immunohistochemical analysis of Dicer expression. Virchows Arch. 2008;452:629–635. doi: 10.1007/s00428-007-0574-5. [DOI] [PubMed] [Google Scholar]
- Dalmay T. MicroRNAs and cancer. J Intern Med. 2008;263:366–375. doi: 10.1111/j.1365-2796.2008.01926.x. [DOI] [PubMed] [Google Scholar]
- Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavin RJ, Smyth PC, Finn SP, Laios A, O'Toole SA, Barrett C, Ring M, Denning KM, Li J, Aherne ST, Aziz NA, Alhadi A, Sheppard BL, Loda M, Martin C, Sheils OM, O'Leary JJ. Altered eIF6 and Dicer expression is associated with clinicopathological features in ovarian serous carcinoma patients. Mod Pathol. 2008;21:676–684. doi: 10.1038/modpathol.2008.33. [DOI] [PubMed] [Google Scholar]
- Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelier G, Voirin N, Ay AS, Cox DG, Chabaud S, Treilleux I, Leon-Goddard S, Rimokh R, Mikaelian I, Venoux C, Puisieux A, Lasset C, Moyret-Lalle C. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J Cancer. 2009;101:673–683. doi: 10.1038/sj.bjc.6605193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- Henson BJ, Bhattacharjee S, O'Dee DM, Feingold E, Gollin SM. Decreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancy. Genes Chromosomes Cancer. 2009;48:569–582. doi: 10.1002/gcc.20666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 2004;59:21–26. doi: 10.1016/j.ijrobp.2003.11.041. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK. Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Jakymiw A, Pauley KM, Li S, Ikeda K, Lian S, Eystathioy T, Satoh M, Fritzler MJ, Chan EK. The role of GW/P-bodies in RNA processing and silencing. J Cell Sci. 2007;120:1317–1323. doi: 10.1242/jcs.03429. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S, Mitsudomi T, Takahashi T. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Huang H, Sun L, Yang M, Pan C, Chen W, Wu D, Lin Z, Zeng C, Yao Y, Zhang P, Song E. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res. 2009;15:3998–4008. doi: 10.1158/1078-0432.CCR-08-3053. [DOI] [PubMed] [Google Scholar]
- Mao S, Park Y, Hasegawa Y, Tribble GD, James CE, Handfield M, Stavropoulos MF, Yilmaz O, Lamont RJ. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell Microbiol. 2007;9:1997–2007. doi: 10.1111/j.1462-5822.2007.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massano J, Regateiro FS, Januario G, Ferreira A. Oral squamous cell carcinoma: review of prognostic and predictive factors. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006;102:67–76. doi: 10.1016/j.tripleo.2005.07.038. [DOI] [PubMed] [Google Scholar]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, Deavers MT, Mourad-Zeidan A, Wang H, Mueller P, Lenburg ME, Gray JW, Mok S, Birrer MJ, Lopez-Berestein G, Coleman RL, Bar-Eli M, Sood AK. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley KM, Eystathioy T, Jakymiw A, Hamel JC, Fritzler MJ, Chan EK. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 2006;7:904–910. doi: 10.1038/sj.embor.7400783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis. 2008;29:2073–2077. doi: 10.1093/carcin/bgn187. [DOI] [PubMed] [Google Scholar]
- Tran N, McLean T, Zhang X, Zhao CJ, Thomson JM, O'Brien C, Rose B. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun. 2007;358:12–17. doi: 10.1016/j.bbrc.2007.03.201. [DOI] [PubMed] [Google Scholar]
- Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45:309–316. doi: 10.1016/j.oraloncology.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Wiesen JL, Tomasi TB. Dicer is regulated by cellular stresses and interferons. Mol Immunol. 2009;46:1222–1228. doi: 10.1016/j.molimm.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Zhang HH, Wang XJ, Li GX, Yang E, Yang NM. Detection of let-7a microRNA by real-time PCR in gastric carcinoma. World J Gastroenterol. 2007;13:2883–2888. doi: 10.3748/wjg.v13.i20.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O'Brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]