Abstract
High-throughput genetic and phenotypic analysis at the single cell level is critical to advance our understanding of the molecular mechanisms underlying cellular function and dysfunction. Here we describe a high-performance single cell genetic analysis (SCGA) technique that combines high-throughput microfluidic emulsion generation with single cell multiplex PCR. Microfabricated emulsion generator array (MEGA) devices containing 4, 32 and 96 channels are developed to confer a flexible capability of generating up to 3.4 × 106 nanoliter-volume droplets per hour. Hybrid glass-polydimethylsiloxane diaphragm micropumps integrated into the MEGA chips afford uniform droplet formation, controlled generation frequency, and effective transportation and encapsulation of primer functionalized microbeads and cells. A multiplex single cell PCR method is developed to detect and quantify both wild type and mutant/pathogenic cells. In this method, microbeads functionalized with multiple forward primers targeting specific genes from different cell types are used for solid-phase PCR in droplets. Following PCR, the droplets are lysed, the beads are pooled and rapidly analyzed by multi-color flow cytometry. Using E. coli bacterial cells as a model, we show that this technique enables digital detection of pathogenic E. coli O157 cells in a high background of normal K12 cells, with a detection limit on the order of 1:105. This result demonstrates that multiplex SCGA is a promising tool for high-throughput quantitative digital analysis of genetic variation in complex populations.
Traditional biological analyses probe large ensembles on the order of 103–106 cells, thereby revealing only the average genotypic and/or phenotypic characterization of the population. The advent of single cell analysis has revealed marked cellular heterogeneity in gene and protein expression,1–4 genetic/genomic alterations,5–7 and responsiveness to environmental and chemotherapeutic stimuli.8,9 A better understanding of cellular heterogeneity and the quantitative detection of rare mutants, such as circulating tumor cells present at an estimated frequency of 1 per 106–107 nucleated blood cells,10 demands high throughput single cell analysis techniques to both detect these very infrequent members of the population and to provide good statistical information regarding these stochastic events.
Microfluidics offers unprecedented capabilities for precisely manipulating small volumes and for enhancing reaction speed and efficiency by increasing relative analyte concentration, reducing diffusion time, and facilitating large-scale integration and automation. These capabilities suggest that microfluidic systems will be very valuable for ultrahigh throughput single cell analysis.11–14 Emulsion polymerase chain reaction (ePCR) provides another powerful tool for high-throughput genetic analysis because one can perform massively parallel single copy PCR reactions by partitioning statistically diluted targets (DNA or RNA) into small droplets dispersed in an oil phase.15,16 Conventional ePCR has found many applications, including next-generation sequencing,17 detection of rare mutations,18–20 and quantification of DNA methylation.21 However, ePCR also suffers from inherent limitations due to the use of mechanical agitation for emulsion generation, which produces high shear force and polydisperse droplet sizes. The transition to droplet microfluidics overcomes these limitations and has substantially improved the performance of emulsion-based bioassays.22–27
Microfluidic droplet generation technology enables the production of monodisperse droplets with precise control over the droplet size, while maintaining high-throughput. This capability not only enables uniform efficiency of enzymatic reactions or affinity binding,23 but also permits digital quantification of the absolute number of targets present in the initial sample.26,28 In addition, microfluidic encapsulation significantly reduces the damage to cells caused by vigorous mechanical agitation. Microfluidic operation is normally performed with a shear stress lower than 10 dynes/cm2, while vigorous mechanical agitation can potentially generate much higher shear stress that can disrupt cells.29 Our previous work indicated greater than 90% cell viability following microfluidic emulsification versus less than 80% for other encapsulation methods.25,30 Integrated microfluidic circuits also allow programmable in situ manipulation of droplets, such as droplet steering, trapping,31 and fusion,32 leading to a much broader spectrum of applications, including real time PCR,23 protein expression studies,27 and drug screening.33
Our previous work established a high-throughput single copy genetic amplification (SCGA) technique based on the use of a hybrid glass-PDMS-glass microdroplet generator (µDG) integrated with a three-valve diaphragm micropump for effective transport and encapsulation of large microbeads and cells into uniform nanolitre-volume droplets.25 To enable detection of extremely low frequency events in a vast population, herein we scale up the single-channel µDG to 4, 32, and 96-channel microfluidic emulsion generator array (MEGA) systems, further increasing the generation throughput up to 3.4 × 106 droplets per hour. In these MEGA systems, on-chip micropumps provide sufficient power to drive multiple droplet generators in parallel while the symmetrically designed microfluidic networks ensure even fluidic transport which is crucial for uniform droplet encapsulation. A new compact micropump composed of three coaxial ring-shaped valves is designed to enable the implementation of a 96-channel MEGA on a 4" wafer. This improvement simplifies device fabrication and operation and substantially reduces the dead volume.
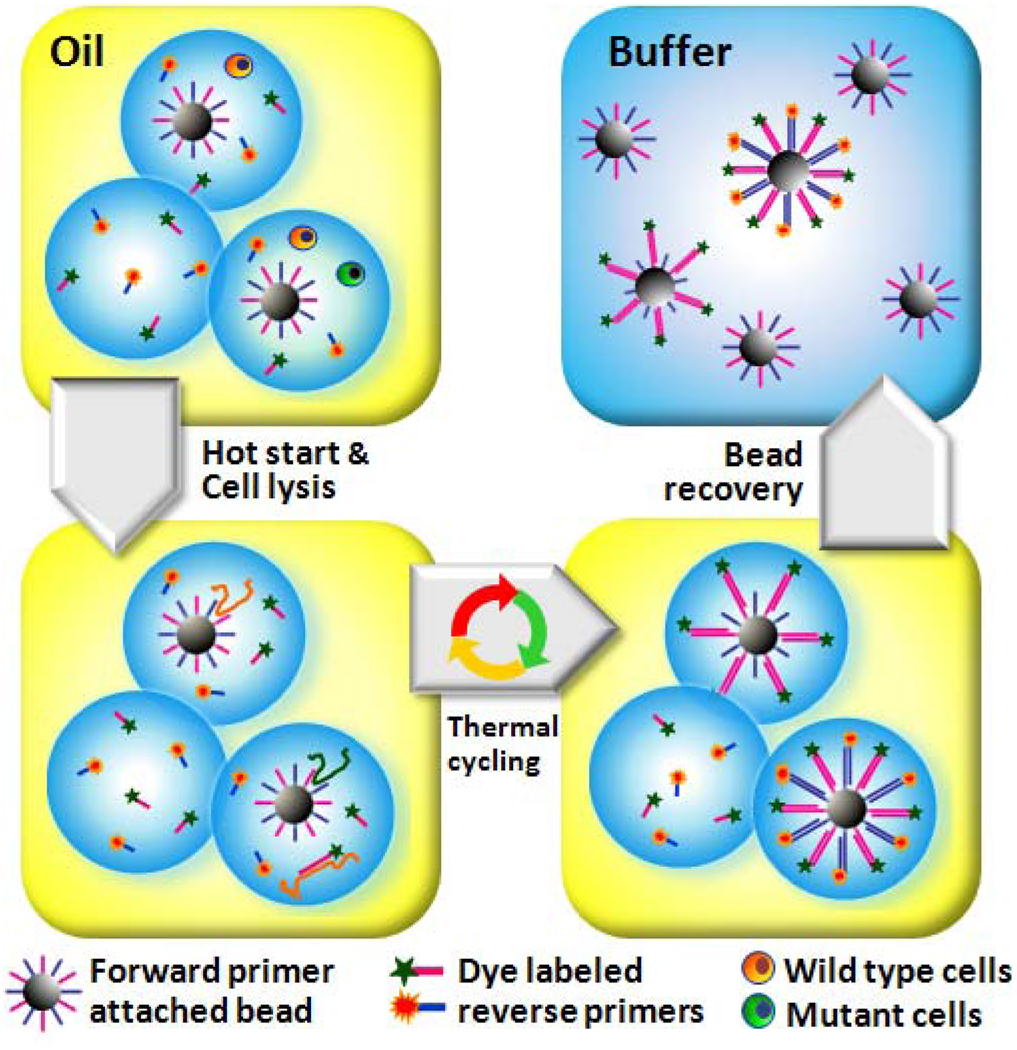
The integration of multiplex PCR with high-throughput emulsion generation is valuable for the analysis of genomic deletion, forensic genotyping, mutation and polymorphism analysis, and identification of pathogens.34 To detect and quantify both normal and mutant/pathogenic cells, herein we describe a multiplex single cell PCR approach, as illustrated in Figure 1, which allows efficient high-throughput PCR amplification of multiple target genes specific to different cell types. In this process, primer-linked beads and cells are diluted in the PCR mix such that isolated individual beads or cells are encapsulated into individual uniform reaction droplets dispersed in the carrier oil. Statistically, a small fraction of droplets will contain both one bead and one or more cells. Every bead is functionalized with forward primers for all targets, and the PCR mix contains reverse primers each labeled with a unique fluorescent dye. Thousands of such droplets, generated by MEGA chips within minutes, are collected in standard PCR tubes and thermally cycled in parallel. Each bead in a droplet containing only a single cell will carry one type of dye-labeled double-stranded amplicons after PCR, while a bead compartmentalized with different types of target cells will be labeled with multiple dyes. Post-PCR beads are recovered from the emulsion and rapidly analyzed by flow cytometry for multi-color fluorescent digital counting of each single cell detection event.
Figure 1.
Multiplex Single Cell/Copy Genetic Analysis (SCGA). Statistically dilute beads and templates are encapsulated into uniform nanoliter volume PCR-mix-in-oil droplets, which are then thermally cycled for PCR amplification. Each bead is functionalized with forward primers for all targets. PCR mix contains reverse primers each labeled with a unique fluorescent dye. Each bead in a droplet containing only a single target will carry one type of fluorescent amplicon after PCR, while a bead compartmentalized with two different templates or cells will be linked with multiple dye-labeled products. Following emulsion PCR, the droplets are broken and the beads are recovered and analyzed by flow cytometry for multi-color detection of the bound amplicons.
To demonstrate the utility of the multiplex SCGA technique for high-throughput single cell analysis of complex sample mixtures, we perform the detection and quantification of a major foodborne bacterial pathogen, E. coli O157:H7, which alone causes an estimated 73,000 infections and 61 deaths annually in the United States.35,36 High-throughput digital multiplex SCGA allows us to detect and quantify pathogenic E. coli O157:H7 cells in a background of normal E. coli K12 cells with a detection limit on the order of 1:105 within a 30 min microdroplet generation time. Such sensitivity is critical for many applications, such as food safety, where microbial pathogen detection needs to meet a zero tolerance policy for many foods.37 The results presented here also suggest that our technique has the potential for high-throughput single cell genotyping and quantitative detection of rare mutations in circulating fluids.
MATERIALS AND METHODS
MEGA Fabrication and Preparation
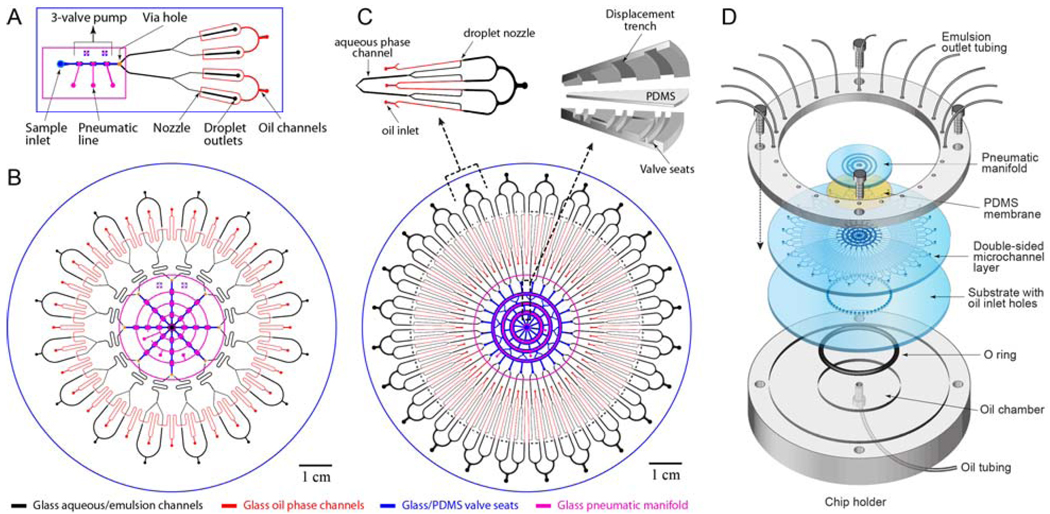
The four-layer MEGA chip shown schematically in Figure 2 is constructed from three 100-mm-diameter glass wafers and a thin PDMS membrane following a process similar to that described by Grover et al.38 The glass channels were coated with octadecyltrichlorosilane (OTS, Sigma-Aldrich) to render the surface hydrophobic. The devices were assembled with a custom plexiglass manifold (Figure 2D) which provides fluidic connections for oil infusion and emulsion collection.
Figure 2.
Microfluidic Emulsion Generator Array (MEGA) devices. (A) Layout of a glass/PDMS/glass hybrid 4-channel MEGA device with a pneumatically-controlled three-valve micropump integrated to drive four nozzles for droplet generation. (B) Design of a 32-channel MEGA device using an array of eight identical micropumps to operate 32 nozzles simultaneously. Two adjacent emulsion channels are combined to increase device density. (C) Layout of 96-channel MEGA on a 4" wafer composed of a single ring pump and 96 droplet generators. Inset: close-ups of a single repeating unit composed of four T-shaped nozzles (left) and the pump structure schematically showing three pairs of coaxial ring-shaped valves and displacement trenches (right). (D) Exploded view of the complete 4-layer 96-channel MEGA device and the plexiglass assembly module used to infuse oil and to collect the generated emulsion.
Cell Preparation
All cell culture and preparation were performed in a Class II biosafety cabinet (Labconco, Kansas City, MO) to avoid contamination. Two types of cells, E. coli K12 (ATCC #700926) and nontoxigenic E. coli O157 (ATCC 700728), were grown separately in Tryptic Soy Broth medium (TSB, Hardy Diagnostics K131, CA) overnight at 37 °C. E. coli K12 cells were transformed with a 3.9-kb pCR 2.1-TOPO vector (Invitrogen, Carlsbad, CA) to confer ampicillin resistance and grown with 1 mg/mL ampicillin added to the media. E. coli O157 was untreated and frequently tested for contamination by PCR. Cells were washed three times in 1× PBS and the final cell density was determined by using a hemacytometer.
Bead and PCR Preparation
All samples were handled in a UV-treated laminar flow hood (UVP, Upland, CA). Primers specific to the KI#128 island on the K12 genome and the OI#43 island on the O157 genome were designed to prevent cross-amplification between strains.39 Reverse primers were labeled with 6-FAM or Cy5 dye on the 5-prime end. 5′ amine modified forward primers were linked to agarose beads (34 µm mean diameter, Amersham Biosciences, NJ) via amine-NHS conjugation chemistry.25 The coupling reaction was performed at a ratio of ~1.5 µmol oligos per gram beads for pUC18 target. For E. coli cells, equimolar forward primers were used at a concentration of ~0.3 µmol oligo per gram beads. PCR mixes contain forward primer functionalized beads (40 beads/µL) and varied amounts of freshly prepared template DNA or cells.
Device Operation and PCR Quantification
A homemade pneumatic system controlled by Labview was used to operate the on-chip micropump. The microchip was prerun with a coating solution to minimize non-specific adsorption on glass, PDMS and tubing surfaces. PCR-mix-in-oil droplets were then generated at 5–8 Hz and collected into individual PCR tubes containing 40 µL microfine solution17 for subsequent thermal cycling. After emulsion PCR, beads were recovered from the droplets and analyzed using a multicolor flow cytometer (FC-500, Beckman Coulter). (See Supplemental Material for detailed methods)
RESULTS
MEGA Design
The 4-channel MEGA illustrated in Figure 2A is a multilayer device consisting of a bonded glass-glass microfluidic chip, a PDMS membrane, and a microfabricated manifold wafer. The glass-glass fluidic chip contains a microfabricated pattern of valve seats exposed on the top surface (blue), which is connected through a ‘via’ hole to an all-glass microfluidic network enclosed between two thermally bonded wafers. The assembly is then completed by contact bonding a manifold wafer (magenta) onto the fluidic chip with a PDMS membrane to form the micropump structure. The microfluidic network consists of the symmetrically bifurcated channels for the aqueous phase (black) and oil phase (red), which form four parallel crosses or droplet nozzles at the junctions. The 4-channel array defines a basic unit that is used to build up multiplexed MEGAs, such as the 32-channel MEGA with eight such arrays integrated onto a 4" wafer (Figure 2B). Further multiplexing of the MEGA on a 4" wafer is restricted by the number of individual pumps that can be symmetrically arranged in a circle. To achieve a higher density, we designed a new ring micropump composed of three pairs of coaxial ring-shaped valve seats connected by offset channels, as well as corresponding circular displacement trenches (Figure 2C, top right). This compact micropump, along with the T-shaped nozzle design (Figure 2C, top left) enables the implementation of a 96-channel MEGA on a 4" wafer (Figure 2C, bottom). In 32- and 96-channel MEGAs, oil channels are connected to the oil inlet holes drilled on the bottom substrate of the device.
A manifold module is employed to support MEGA devices, as sketched in Figure 2D, providing oil infusion and routing generated droplets into PCR tubes for thermal cycling. The bottom part of the manifold is designed to form a circular oil reservoir when sealed against the MEGA chip.
The oil pressure in the circular reservoir is uniformly distributed across the area on the bottom side of the chip, evenly infusing oil into channels via the symmetric inlet holes. This design minimizes the number of syringe pumps and tubing connections required for oil infusion.
Droplet PCR Performance of MEGAs
The symmetric design of the MEGA assures uniform fluidic transport across the array, leading to the generation of monodisperse droplets from all channels. A microphotograph in Figure 3A (left) demonstrates the generation of uniform 4 nL droplets by flow focusing at the cross-injectors of a 32-channel MEGA device. The droplets were collected from eight nozzles for the size measurement revealing a mean diameter of 198 µm with a size deviation of only 2.5% (Figure 3A, middle). Because of the pulsatile nature of the on-chip diaphragm pump, the droplet formation rate corresponds precisely with the pumping frequency of 5.6 Hz. At this speed, the 32-channel device offers a total throughput of 6.4 × 105 droplets per hour (dph).
Figure 3.
Characterization of droplet generation and PCR amplification using MEGAs. (A) 32-channel MEGA chip. Left: photo of the cross-shaped nozzles each generating ~4 nL droplets at 5.6 Hz (6.4 × 105 droplets per hour). Middle: size distribution of uniform droplets collected from eight nozzles in a device. RSD: relative standard deviation. Right: a representative flow cytometric analysis of 624 bp amplicons on beads amplified from pUC18 template (1 copy/droplet) in ~3 nL droplets generated from four nozzles in a device. (B) 96-channel MEGA. Left: image of droplet production at the T-shaped nozzles with a total throughput of 2.4 × 106 droplets per hour. Middle: size distribution of uniform ~2 nL droplets collected from sixteen nozzles in a device. Right: a representative flow cytometric histogram of beads carrying the FAM labeled PCR product from E. coli K12 at 0.2 cells per droplet (cpd) in 2.5 nL droplets. For all tests of droplet generation, the mock PCR mix containing ~100 beads per uL was used.
To characterize the performance of bead-based droplet PCR using the 32-channel MEGA, pUC18 DNA molecules were compartmentalized into ~3 nL droplets at 1 copy/droplet as the templates, from which a 624 bp product was amplified. The post-PCR beads recovered from each PCR tube were randomly mixed prior to flow cytometric analysis. The result obtained from 4 droplet generators is presented in Figure 3A (right), showing that 60.5% of the bead population (459 events) gains FAM signal due to the bead-bound amplicons, in line with the value expected from Poisson statistics (63.2%). The fluorescence intensity of the positive beads indicates an averaged PCR yield of ~150 attomoles of DNA product per bead, consistent with the yield obtained from our previous single-channel device.25 The results from other channels confirm that the 32-channel MEGA offers constant digital emulsion PCR performance as a result of the uniformity of droplet compartmentalization.
Incorporation of a compact ring-shaped micropump in the 96-channel MEGA maintains the generation performance while increasing throughput. Uniform droplets can be formed from a mock PCR mix containing agarose beads by flow shearing at the T-shaped injectors (Figure 3B, left, and Video S-1 in Supporting Information). The mean diameter of the droplets collected from 16 nozzles via 4 randomly chosen outlets was determined to be 162 µm (2.2 nL) with a deviation of only 3.8% (Figure 3B, middle). Furthermore, such size uniformity is preserved across a range of droplet volumes (1–5 nL; RSD < 5%). When the on-chip pump is operated at 7 Hz, the device produces up to 2.4 × 106 dph. To assess the encapsulation performance of the 96-channel MEGA, single cell emulsion PCR was performed targeting the KI#128 island on the K12 genome. Bacterial E. coli K12 cells and the forward primer linked beads were introduced at a statistical dilution of 0.2 cells and 0.1 beads per 2.5 nL droplet, respectively. After PCR and isolation, flow cytometric analysis (Figure 3B, right) shows that 15.1% of the total bead population (4423 beads) is strongly fluorescent, corresponding well to a theoretical value of 18.1% predicted by the Poisson distribution. This good agreement in yield indicates that successful single copy genetic amplification resulted from uniformly distributed cells and beads.
Pathogen Detection Using Multiplex SCGA
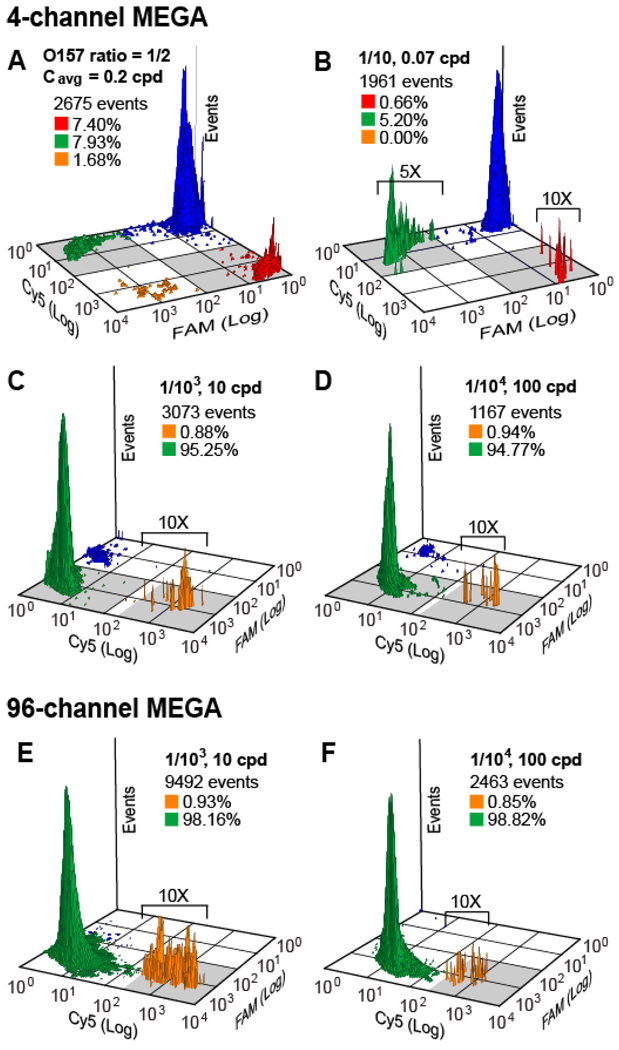
Multiplex single cell genetic analysis (SCGA) is demonstrated here by detecting pathogenic E. coli O157 cells in a background of normal E. coli K12 cells. Unique genes on the K12 genome (KI#128 island) and on the O157 genome (OI#43 island) are targeted by primers labeled with different fluorophores so that these two strains can be identified. For all pathogen detection, ~2.5 nL droplets were used and beads were introduced at ~0.1 beads per droplet (bpd). A 4-channel MEGA device was operated for ~18 min. to obtain ~3000 beads for flow cytometric analysis. A mixed E. coli bacterial sample containing 50% O157 cells was first analyzed at an average cell concentration (Cavg) of 0.2 cells per droplet (cpd) using a 4-channel MEGA device. As seen in Figure 4A, the flow cytometry profile shows four distinct bead populations: 212 FAM positive beads due to the amplicons from K12 (7.93%, green), 198 Cy5 positive beads for O157 (7.40%, red), 45 double positive beads due to coexistence of both cell types in a single droplet (1.68%, orange), and 2220 negative beads (82.99%, blue). The O157 cell ratio (O157 positive beads/total positive beads) is then determined to be 0.48, in good accord with the input O157 cell fraction of 0.5. Figure 4B presents the detection of a lower input O157 ratio of 1/10 at Cavg = 0.07 cpd: 5.20% of the beads are positive for K12, 0.66% are positive for O157, 0% are double positive beads. The experimental O157/K12 ratio of 1.1/10 is consistent with that expected.
Figure 4.
High-throughput digital multiplex detection of E. coli O157 in a background of E. coli K12 with various input ratios and average cell concentrations (Cavg) using MEGAs. (A, B) With Cavg≤ 0.2 cpd, flow cytometry profiles of beads show four distinct populations: negative (blue), FAM positive beads specific for K12 (green), Cy5 positive beads specific for O157 (red), and double positive beads for both cells (orange). Small populations are expanded along the event axis for better visualization, as indicated. The gray regions mark the population gating. The measured O157 ratios (O157 positive beads/total positive beads) are (A) 0.48 (expected: 0.5) and (B) 0.11 (expected: 0.1). (C, D) When Cavg≥ 10 cpd while keeping O157 cells at 0.01 cpd, double positive beads quantify O157 since a single O157 cell is co-compartmentalized into a droplet with multiple K12 cells. In this case the ratios of double positive beads divided by Cavg give the measured O157 ratios: (C) 0.92/103 (expected: 1/103) and (D) 0.98/104 (expected: 1/104). Up to 3000 beads can be analyzed using a 4-channel device for ~25 min. run time. Increasing Cavg to 100 cpd reduces the number of beads required, improving the detection sensitivity to 1/104 without excessively extending run time. (E, F) O157 detection using a 96-channel MEGA shows the measured O157 ratios consistent with the inputs: (E) 0.94/103 vs 1/103 and (F) 0.85/104 vs 1/104. Up to 104 events were processed within 5 min. run time. The capability offered by 96-channel MEGA (up to 3.4 × 106 droplets per hour) greatly increased analysis throughput and decreased processing time necessary for detecting a statistically significant population of a low-frequency sample. Bead concentration was 0.1 bpd for all cases.
Improving Throughput and Sensitivity
The multiplex SCGA process discussed above uses droplets/beads inefficiently because cells are highly dilute so that most droplets are empty. The detection of low frequency genetic variations requires high analysis throughput in order to obtain the statistically significant population for the target. One way to increase the process efficiency is to perform multiplex SCGA at elevated cell density, while still keeping the cells of interest statistically dilute. This is realistic because the target pathogen or mutant cells are typically present at very low relative concentrations compared to normal cells. Figure 4C demonstrates the use of a 4-channel MEGA device to detect E. coli O157 cells at a frequency of 1/103. By increasing Cavg from 0.2 cpd (Figure 4A) to 10 cpd, the effective density of O157 cells is raised from 0.0005 to 0.01 cpd, reducing the number of droplets/beads that must be processed by 50-fold. In this case, all beads should be FAM fluorescent due to co-encapsulated K12 cells and the presence of O157 cells in the droplets will modify only a fraction of beads with the Cy5 dye. As expected, a vast majority of beads (1846 out of 1961 events, 96.13%) are FAM positive and a small fraction of double positive beads (102 events, 0.88%) are observed. The measured O157 ratio is determined to be 0.92/103 (the ratio of double positive beads over all positive beads divided by Cavg), close to the input, 1/103. Multiplex SCGA preserves the quantitative performance for pathogen detection even when the average concentration is up to 100 E. coli cells per 2.5 nL droplet. As seen in Figure 4D, an experiment carried out at Cavg = 100 cpd records 11 O157 positive events in a total of 1167 events, giving an output of 0.98/104 in response to the input O157 ratio of 1/104.
When Cavg is higher than 100 cells per 2.5 nL droplet, non-specific amplification was found to be significant in the multiplex PCR assay, causing false positive scores for O157. To further improve the detection sensitivity, therefore, it is necessary to use highly multiplexed MEGA devices for high throughput droplet generation. A 96-channel MEGA device was evaluated for multiplex PCR detection of E. coli O157 cells, where the device was operated at ~7 Hz, allowing us to encapsulate up to 104 cells within a 5 min. microdroplet generation time. As shown in Figure 4E and F, the experiments at input ratios of 1/103 (10 cpd) and 1/104 (100 cpd) detect 88 and 21 O157 positive events, giving an output fraction of 0.94/103 and 0.85/104, respectively. A small number of negative events were observed in Figure 4C–F, which may be due to cell debris aggregates or unlysed cells. We have optimized the bead cleaning protocol for the 96-channel trials to remove cell debris and other interfering species. The high percentage of positive events (> 99%) in Figure 4E and F results from the combination of high efficiency droplet PCR and improved bead clean-up.
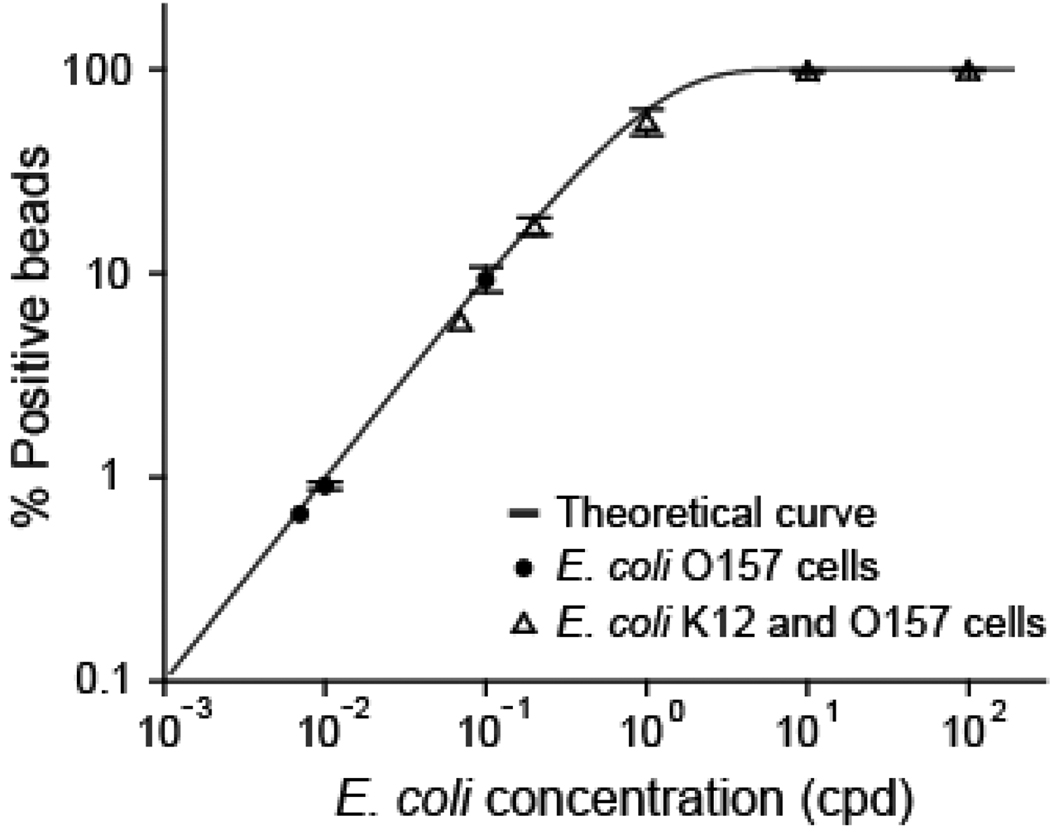
To verify that the observed performance is the result of digital quantification of each strain, we compare the percentage of positive beads obtained with various input ratios and average cell concentrations to that predicted by the Poisson distribution (Figure 5). The multiplex detection is seen to follow Poisson statistics even when individual O157 cells were detected within a high background of 100 cells per droplet. The good correspondence indicates successful single cell emulsion PCR, which allows digital quantification of the absolute cell concentration. For instance, with Cavg = 100 cpd and O157 cells diluted to 1/104 in a K12 background, the O157 cell density is 0.01 cpd and the detection resulted in 0.91% ± 0.04% double positive beads, consistent with the predicted ratio of 0.995%. Because of the presence of negative events, the average percentage of O157 positive beads is corrected to be 0.93% ± 0.05% (double positive beads divided by total positive beads), from which the O157 cell concentration is determined to be 3.7 ± 0.2 cells/µL (input 4 cells/µL).
Figure 5.
Comparison of the percentage of positive beads over total beads versus starting cell concentration. The red curve represents the value expected from Poisson statistics. The experimental data were obtained by detecting E. coli O157 cells in a background of E. coli K12 with overall cell concentrations of 0.07–100 cpd. By maintaining cells of interest (O157) at a statistically dilute concentration (0.007, 0.01 and 0.1 cpd), the multiplex detection follows Poisson statistics even in a background of high normal cell density, which enables performing single cell genetic analysis with highly improved throughput and efficiency. Error bars represent standard deviation (n≥ 3).
Limit of Detection and Dynamic Range
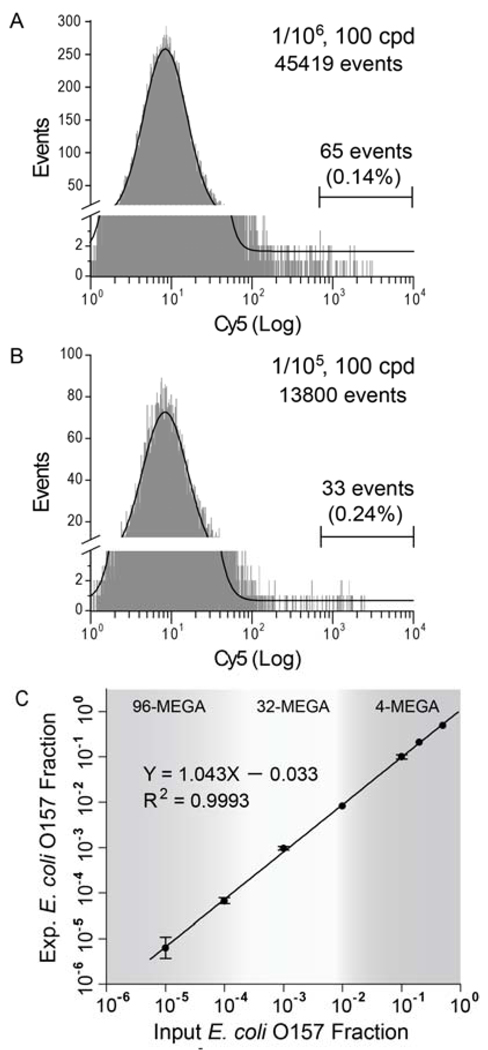
To determine the limit of detection (LOD) of multiplex SCGA for E. coli O157 detection, the assay was carried out at lower pathogenic ratios and Cavg = 100 cpd using a 96-channel MEGA. Figure 6A presents a representative analysis at an input ratio of 1/106 where the experimental output ratio (1.4/105) is one order of magnitude higher than that expected. In this case, to define a statistically significant population at the extremely low cell density (0.0001 cpd for O157 cells), a large volume of PCR mix (1500 µL) was used to produce ~8 × 105 droplets within 20 min., which leads to 45419 total events analyzed and 65 Cy5 positive events detected. The experimental readout at the input of 1/106 (1.8/105 ± 0.66/105, n = 4) is significantly above the expected value (p = 0.01), indicating that there is significant background signal. It is seen in Figure 6A that the peak for K12 cells (FAM positive only) tails off into the Cy5 positive region, suggesting that the background could be attributed to misamplification caused by the non-specific binding of O157 primers to the K12 templates. It is also possible that the false positive events stem from the buildup of trace environmental contamination in the lab during the analysis development process.
Figure 6.
Detection limit and dynamic range of multiplex SCGA for E. coli O157 detection at Cavg = 100 cpd. (A, B) Representative cytometric histograms (events vs. Cy 5 signal) obtained with input O157 ratios of 1/106 and 1/105, respectively, using a 96-channel MEGA. The measured O157 ratios (ratios of double positive beads divided by Cavg) are: (A) 2.4/105 (expected: 1/105) and (B) 1.4/105 (expected: 1/106), suggesting that the limit of detection of current assay is on the order of 1/105 due to non-specific amplification and/or environmental contamination. The solid lines represent the Gaussian fittings for the Cy5 negative populations. (C) Plot of experimental readout vs input O157 fraction showing a linear dynamic range for O157 fraction higher than 1/105. The color regions roughly mark the detection windows that meet the throughput offered by different MEGA devices within experimentally accessible run time. Error bars represent standard deviation (n≥ 3).
Figure 6B shows a representative result obtained with an input ratio of 1/105 which detects 33 Cy5 positive beads. At this pathogenic ratio, the experimental readouts (2.4/105 ± 0.91/105, n = 4) are in the same order as the expected value and significantly different from that obtained at 1/106 at p = 0.05 using the two-sample student’s t-test. By subtracting the background, the corrected value at the input ratio of 1/105 is determined to be 0.69/105 ± 0.34/105 (n = 4). Figure 6C summarizes the background corrected calibration of E. coli O157 detection as a function of the input O157 fraction, which shows a linear dynamic range for O157 fraction higher than 1/105 (y = 1.043× – 0.033). From this plot, the concentration detection limit is determined to be 3/105 with a 99% confidence.
DISCUSSION
High-Throughput MEGA Devices
Continuous-flow microfluidic droplet techniques rely on external mechanical agitation, such as magnetic stirring, to maintain the uniform dispersion of objects with a higher density than the carrier fluid (e.g. cells, beads, etc.) during droplet encapsulation.40 We found that the pulsatile nature of the integrated diaphragm pump effectively overcomes this problem of sedimentation. This observation was further confirmed by the reproducible results obtained for both single-copy DNA amplification and single-cell detection which agreed well with Poisson statistical analysis. However the generation rate from a single channel, determined by the frequency of pneumatically controlled pumping, is limited to ~ 10 Hz for nanoliter-volume droplets because of the response time of the PDMS membrane in the valve structure (tens of milliseconds).41
One of the advantages of the on-chip pump is that the microvalve structure can be precisely engineered to adjust pumping capacity, thereby conferring great flexibility in the multiplexing of droplet generators to increase throughput. Our results show that by increasing the valve dimensions for the single-channel device by 20%, the pump is able to drive four droplet generators in parallel without changing the pump actuation pressure/vacuum. This 4-channel design serves as a basic unit that is readily scalable to construct symmetric generator arrays for balanced fluidic transport, and thus uniform droplet formation. We demonstrate a 32-channel MEGA device containing an array of 8 such pumps. A maximum of 12 on-chip pumps and 48 droplet generators can be arranged on a 4" wafer to produce a device that generates up to 1.7 × 106 dph.
To further increase the throughput, we successfully implemented a new compact micropump composed of three coaxial ring-shaped valves, pushing the multiplexing limit on the 4" wafer to 96-channels. Utilizing the original pump array design would mandate a 6" device that would lead to increased dead volume and difficulty in fabrication and operation. Moreover, the operation of this integrated ring pump was found to be more robust than that of the array of individual pumps. In the pump array, bubbles trapped inside the valves of one or few pumps can cause unbalanced fluidic pressure applied to the individual generators, resulting in the production of polydisperse droplets. In contrast, the 96-channel MEGA system integrated with the ring pump shows greater tolerance to bubbles, because the larger volume of the ring-shaped valves reduces the impact of bubbles on pumping and the entire droplet generator array is driven by a single pump. The successful implementation of 96-channel MEGA offers a throughput of 3.4 × 106 dph, which greatly lowers the detection limit and decreased processing time necessary for detection of low-frequency events.
Multiplex SCGA
Multiplex single cell PCR is the cornerstone of the high-throughput SCGA technique, which maximizes the genetic information extracted from each single cell detection event. In this process, it is critical to compartmentalize single cells and beads into monodisperse nanoliter-volume droplets. Our results clearly demonstrate that the droplet uniformity enables strict Poisson statistical analysis to calibrate the performance of digital single cell PCR which is determined collectively by the statistical encapsulation of beads and cells, cell lysis, and PCR reaction. Such analysis permits digital quantification of the absolute number of targets in the initial sample. Other digital PCR techniques using agitation-based emulsification fundamentally lack this capability because of the extremely polydisperse droplets produced.18 In addition, the uniform droplets contain the same amount of reactants enabling a quantitative comparison of the PCR products. Thus one should be able to perform large-scale gene expression profiling at the single cell level, by transforming reverse transcription-PCR (RT-PCR) assays to the SCGA format.42
The multiplex SCGA displays great tolerance to PCR inhibition as the cell lysate and debris are significantly diluted in the large nanolitre droplets generated by MEGA and each reaction is independent. Efficient and specific multiplex PCR amplification can be achieved even when each droplet is loaded with up to 100 E. coli cells on average, which greatly increases the analysis throughput and hence the detection sensitivity, without excessively extending the droplet production time. This result indicates the feasibility of SCGA for the large-scale genetic analysis of larger and more complex mammalian cells. For instance, the analysis of cancer development and progression, circulating tumor cells, and stem cell differentiation, where single cell resolution may facilitate a deeper understanding of the biological mechanisms involved, can be envisioned.
Pathogen Detection
Most PCR-based microdevices reported the detection of only one bacterial strain by PCR, with detection limits ranging from a few to 104 bacterial cells.43–45 Recent work in our group showed that PCR reactions in a 250 nL microreactor can detect E. coli O157:H7 in a mixture with the K12 strain down to a ratio of 1:500,46 and that the detection limit of the microscale PCR can be as low as 1:1000 by using on-chip affinity cell preconcentration.39 Our quantitative digital format remarkably outperforms previous low-throughput microsystems with its capability to identify and quantify both E. coli O157 and K12 cells down to a pathogen-to-background ratio in the order of 1:105 (Figure 6). Such sensitivity makes the technique a promising candidate to achieve the level of detection and speed required for zero-pathogen-tolerance policy. Droplet generation using the 96-channel MEGA requires less than 30 min run time to achieve a limit of detection of 1:105. The entire procedure, including PCR thermal cycling, post-PCR cleanup, and flow cytometry takes approximately 4 hours and compares favorably to standard PCR-based detection assays while providing better sensitivity. In contrast, most commonly used methods today require at least two days to achieve this level of sensitivity since they rely on pathogen culturing.37 The SCGA technique can be further extended to detect multiple pathogenic microorganisms in one sample. Compared to the small beads used in the “BEAMing” technique, the large surface area of the microsphere used here allows multiple different primers to be conjugated with a density that supports efficient on-bead PCR reaction for each target. Thus our detection multiplicity is limited primarily by the number of fluorescent dyes that a flow cytometer can detect (commonly 5 colors) since that is a relatively standard degree of PCR multiplicity.
We believe that the detection limit of our multiplex SCGA can be further improved to beyond one in a million. Some nonspecific amplification was observed when the 2.5 nL droplets were loaded with 100 E. coli cells. Although the multiplex PCR conditions have been optimized to minimize non-specific amplification, it still can contribute to the false positive scores which limit the low-abundance detection to the level of 1:105 in this proof of concept work. In practical applications, we can perform droplet generation for longer periods of time at lower cell concentrations to minimize the effect of non-specific PCR amplification, and thus lower the detection limit. On the other hand, trace exogenous contamination from microorganisms and/or DNA present in air, reagents and instruments may cause considerable interference at these extremely low level detection limits. We did see false positive events occasionally in negative control experiments performed at a cell concentration lower than 1 cell/droplet. Sample preparation and experimental operation with more stringent environmental controls should prevent contaminations and lower the detection limit to 1:106 or lower.
CONCLUSION
The past few years have witnessed remarkable progress in the understanding and use of microemulsions for high throughput, sensitive biological analysis and next-generation sequencing. The results presented here show that the use of ultrahigh-throughput microfluidic platforms will enable large-scale quantitative genotypic studies of complex biological systems at the single molecule and single cell level. The multiplex SCGA technique enables the multi-parameter acquisition of digital genetic information to elucidate the importance of stochastic variations in biological populations and should lead to the detection and numeration of rare mutants for clinical diagnosis of diseases, such as chromosomal translocations in blood cells for the early detection of leukemia and lymphoma and the measurement of minimal residual disease and relapse.
Supplementary Material
ACKNOWLEDGMENT
We thank James Yang and Palani Kumaresan for their early contributions to this work. Microfabrication was carried out at the Microfabrication Laboratory at the University of California, Berkeley. This work was supported through the trans-NIH Genes, Environment and Health Initiative, grant U54ES016115. R. N. is supported by an NSF Graduate Research Fellowship, and J. S. is supported by the Canary Foundation and ACS Early Detection Postdoctoral Fellowship.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCE
- 1.Cai L, Friedman N, Xie XS. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 2.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 3.Toriello NM, Douglas ES, Thaitrong N, Hsiao SC, Francis MB, Bertozzi CR, Mathies RA. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20173–20178. doi: 10.1073/pnas.0806355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King KR, Wang SH, Irimia D, Jayaraman A, Toner M, Yarmush ML. Lab Chip. 2007;7:77–85. doi: 10.1039/b612516f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 6.Klein CA, Blankenstein TJF, Schmidt-Kittler O, Petronio M, Polzer B, Stoecklein NH, Riethmuller G. Lancet. 2002;360:683–689. doi: 10.1016/S0140-6736(02)09838-0. [DOI] [PubMed] [Google Scholar]
- 7.Fuhrmann C, Schmidt-Kittler O, Stoecklein NH, Petat-Dutter K, Vay C, Bockler K, Reinhardt R, Ragg T, Klein CA. Nucleic Acids Res. 2008;36:e39. doi: 10.1093/nar/gkn101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batchelor E, Loewer A, Lahav G. Nat. Rev. Cancer. 2009;9:371–377. doi: 10.1038/nrc2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li XJ, Ling V, Li PCH. Anal. Chem. 2008;80:4095–4102. doi: 10.1021/ac800231k. [DOI] [PubMed] [Google Scholar]
- 10.Pantel K, Brakenhoff RH, Brandt B. Nat. Rev. Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 11.Whitesides GM. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 12.Ottesen EA, Hong JW, Quake SR, Leadbetter JR. Science. 2006;314:1464–1467. doi: 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- 13.Sims CE, Allbritton NL. Lab Chip. 2007;7:423–440. doi: 10.1039/b615235j. [DOI] [PubMed] [Google Scholar]
- 14.Marcus JS, Anderson WF, Quake SR. Anal. Chem. 2006;78:3084–3089. doi: 10.1021/ac0519460. [DOI] [PubMed] [Google Scholar]
- 15.Nakano M, Komatsu J, Matsuura S, Takashima K, Katsura S, Mizuno AJ. Biotechnol. 2003;102:117–124. doi: 10.1016/s0168-1656(03)00023-3. [DOI] [PubMed] [Google Scholar]
- 16.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen ZT, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer MLI, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu PG, Begley RF, Rothberg JM. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diehl F, Li M, Dressman D, He YP, Shen D, Szabo S, Diaz LA, Goodman SN, David KA, Juhl H, Kinzler KW, Vogelstein B. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M, Diehl F, Dressman D, Vogelstein B, Kinzler KW. Nat. Methods. 2006;3:95–97. doi: 10.1038/nmeth850. [DOI] [PubMed] [Google Scholar]
- 20.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, Kinzler KW, Vogelstein B, Diaz LA. Nat. Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Chen WD, Papadopoulos N, Goodman SN, Bjerregaard NC, Laurberg S, Levin B, Juhl H, Arber N, Moinova H, Durkee K, Schmidt K, He YP, Diehl F, Velculescu VE, Zhou SB, Diaz LA, Kinzler KW, Markowitz SD, Vogelstein B. Nat. Biotechnol. 2009;27 doi: 10.1038/nbt.1559. 858-U118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teh SY, Lin R, Hung LH, Lee AP. Lab Chip. 2008;8:198–220. doi: 10.1039/b715524g. [DOI] [PubMed] [Google Scholar]
- 23.Beer NR, Hindson BJ, Wheeler EK, Hall SB, Rose KA, Kennedy IM, Colston BW. Anal. Chem. 2007;79:8471–8475. doi: 10.1021/ac701809w. [DOI] [PubMed] [Google Scholar]
- 24.Beer NR, Wheeler EK, Lee-Houghton L, Watkins N, Nasarabadi S, Hebert N, Leung P, Arnold DW, Bailey CG, Colston BW. Anal. Chem. 2008;80:1854–1858. doi: 10.1021/ac800048k. [DOI] [PubMed] [Google Scholar]
- 25.Kumaresan P, Yang CJ, Cronier SA, Blazei RG, Mathies RA. Anal. Chem. 2008;80:3522–3529. doi: 10.1021/ac800327d. [DOI] [PubMed] [Google Scholar]
- 26.Mazutis L, Araghi AF, Miller OJ, Baret JC, Frenz L, Janoshazi A, Taly V, Miller BJ, Hutchison JB, Link D, Griffiths AD, Ryckelynck M. Anal. Chem. 2009;81:4813–4821. doi: 10.1021/ac900403z. [DOI] [PubMed] [Google Scholar]
- 27.Huebner A, Srisa-Art M, Holt D, Abell C, Hollfelder F, Demello AJ, Edel JB. Chem. Commun. 2007:1218–1220. doi: 10.1039/b618570c. [DOI] [PubMed] [Google Scholar]
- 28.Kiss MM, Ortoleva-Donnelly L, Beer NR, Warner J, Bailey CG, Colston BW, Rothberg JM, Link DR, Leamon JH. Anal. Chem. 2008;80:8975–8981. doi: 10.1021/ac801276c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roubos JA, Krabben PM, Luiten RG, Verbruggen HB, Heijnen JJ. Biotechnol. Prog. 2008;17:336–347. doi: 10.1021/bp0001617. [DOI] [PubMed] [Google Scholar]
- 30.Yu S-H, Buchholz R, Kim S-K. Biotechnol. Tech. 1999;13:609–614. [Google Scholar]
- 31.Huebner A, Bratton D, Whyte G, Yang M, deMello AJ, Abell C, Hollfelder F. Lab Chip. 2009;9:692–698. doi: 10.1039/b813709a. [DOI] [PubMed] [Google Scholar]
- 32.Mazutis L, Baret JC, Griffiths AD. Lab Chip. 2009;9:2665–2672. doi: 10.1039/b903608c. [DOI] [PubMed] [Google Scholar]
- 33.Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML. Proc. Natl. Acad. Sci. U.S.A. 2009;106:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markoulatos P, Siafakas N, Moncany M. J. Clin. Lab. Anal. 2002;16:47–51. doi: 10.1002/jcla.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. Emerg. Infect. Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nugen SR, Baeumner AJ. Anal. Bioanaly. Chem. 2008;391:451–454. doi: 10.1007/s00216-008-1886-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batt CA. Science. 2007;316:1579–1580. doi: 10.1126/science.1140729. [DOI] [PubMed] [Google Scholar]
- 38.Grover WH, Skelley AM, Liu CN, Lagally ET, Mathies RA. Sensor Actuat. B-Chem. 2003;89:315–323. [Google Scholar]
- 39.Beyor N, Yi LN, Seo TS, Mathies RA. Anal. Chem. 2009;81:3523–3528. doi: 10.1021/ac900060r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koster S, Angile FE, Duan H, Agresti JJ, Wintner A, Schmitz C, Rowat AC, Merten CA, Pisignano D, Griffiths AD, Weitz DA. Lab Chip. 2008;8:1110–1115. doi: 10.1039/b802941e. [DOI] [PubMed] [Google Scholar]
- 41.Grover WH, Ivester RHC, Jensen EC, Mathies RA. Lab Chip. 2006;6:623–631. doi: 10.1039/b518362f. [DOI] [PubMed] [Google Scholar]
- 42.Zhong JF, Chen Y, Marcus JS, Scherer A, Quake SR, Taylor CR, Weiner LP. Lab Chip. 2008;8:68–74. doi: 10.1039/b712116d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lagally ET, Scherer JR, Blazej RG, Toriello NM, Diep BA, Ramchandani M, Sensabaugh GF, Riley LW, Mathies RA. Anal. Chem. 2004;76:3162–3170. doi: 10.1021/ac035310p. [DOI] [PubMed] [Google Scholar]
- 44.Liu RH, Yang JN, Lenigk R, Bonanno J, Grodzinski P. Anal. Chem. 2004;76:1824–1831. doi: 10.1021/ac0353029. [DOI] [PubMed] [Google Scholar]
- 45.Cady NC, Stelick S, Kunnavakkam MV, Batt CA. Sensor Actuat. B-Chem. 2005;107:332–341. [Google Scholar]
- 46.Thaitrong N, Toriello NM, Del Bueno N, Mathies RA. Anal. Chem. 2009;81:1371–1377. doi: 10.1021/ac802057f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.