Summary
Through trans-splicing of a 39-nt Spliced Leader (SL) onto each protein-coding transcript, mature kinetoplastid mRNA acquire a hypermethylated 5′-cap structure, but its function has been unclear. Gene deletions for three Trypanosoma brucei cap 2′-O-ribose methyltransferases, TbMTr1, TbMTr2, and TbMTr3, reveal distinct roles for four 2′-O-methylated nucleotides. Elimination of individual gene pairs yields viable cells, however attempts at double knockouts resulted in the generation of a TbMTr2−/−/TbMTr3−/− cell line only. Absence of both kinetoplastid-specific enzymes in TbMTr2−/−/TbMTr3−/− lines yielded substrate SL RNA and mRNA with cap 1. TbMTr1−/− translation is comparable to wildtype, while cap 3 and cap 4 loss reduced translation rates, exacerbated by the additional loss of cap 2. TbMTr1−/− and TbMTr2−/−/TbMTr3−/− lines grow to lower densities under normal culture conditions relative to wildtype cells, with growth rate differences apparent under low serum conditions. Cell viability may not tolerate delays at both the nucleolar Sm-independent and nucleoplasmic Sm-dependent stages of SL RNA maturation combined with reduced rates of translation. A minimal level of mRNA cap ribose methylation is essential for trypanosome viability, providing the first functional role for the cap 4.
Keywords: gene knockout, methyltransferase, ribose 2′-O-methylation, SL RNA, spliced leader, trans-splicing
Introduction
The Spliced Leader (SL) RNA of kinetoplastids, including human pathogens Trypanosoma brucei, Trypanosoma cruzi and Leishmania major, plays a central role in gene expression by providing the 5′ cap to mature mRNAs. By trans-splicing of the SL exon, all coding regions within polycistronic pre-mRNAs acquire a unique 5′ cap 4 structure (Bangs et al., 1992; Freistadt et al., 1988; Perry et al., 1987). Higher eukaryotic mRNA caps (Banerjee, 1980) vary from those containing only a m7G (cap 0) attached to the first-transcribed nucleotide by a 5′-5′ triphosphate linkage, to those methylated at the 2′-O-ribose of the first (cap 1) or first and second nucleotides (cap 2). By contrast, the hypermethylated kinetoplastid cap 4 also has 2′-O-ribose methylation of the third and fourth nucleotides and base methylations at the first (m26A) and fourth (m3U) positions (Bangs et al., 1992). In a mixed SL population, mRNA with mutated SL and undermethylated cap do not associate with polysomes, implicating cap 4 and/or the primary sequence of the SL in translation (Zeiner et al., 2003). The role of each cap 4 ribose methylation in the wildtype SL sequence could be addressed by deleting three 2′-O-ribose methyltransferases (MTases).
In trypanosomes the triphosphatase, TbCet1, and a bifunctional enzyme with guanylyltransferase and MTase domains, TbCgm1, form cap 0 on the SL RNA (Ruan et al., 2007; Takagi et al., 2007). The cap 0 structure has been linked to several post-transcriptional processes, including mRNA stability, intracellular transport, and translation (Furuichi and Shatkin, 2000), however the role(s) of additional downstream cap methylations remains obscure. Cap 2′-O-ribose methylation on Xenopus maternal transcripts correlates with translation initiation and enhances translation efficiency (Kuge et al., 1998). The majority of mammalian mRNA are cap ribose methylated (Banerjee, 1980) and the enzymatic activity has only been purified partially from humans (Langberg and Moss, 1981). In addition to translational control, functions for mRNA 2′-O-ribose methylation include enhancement of processing and trafficking of transcripts (Kuge et al., 1998; Reddy et al., 1992). The conservation of cap 1 2′-O-ribose MTases in viral genomes suggests a critical function, with cap ribose methylation increasing ribosome association (Muthukrishnan et al., 1976) and translational control (Brownlee et al., 1995). Furthermore, the loss of cap 1 MTase activity results in reduced replication and virulence of the West Nile virus (Zhou et al., 2007).
Three 2′-O-ribose MTases have been identified in T. brucei cap 4 formation, and are named for the ribonucleotide positions modified. TbMTr1 is a cap 1 MTase that modifies the SL RNA and U1 snRNA (Zamudio et al., 2007). This enzyme defines a large class of uncharacterized eukaryotic MTases with homologs in humans. It shares biochemical properties with the partially purified cap 1 MTase activity from HeLa cell extract, but acts with specificity for the first nucleotides of the SL in T. brucei (Mittra et al., 2008). TbMTr1 resides in a specialized complex containing the SLA1 H/ACA snoRNP for pseudouridine formation at SL position 28 and a kinetoplastid-specific protein named MTase associated protein or TbMTAP (Zamudio et al., 2009). The TbMTr1 complex performs Sm-complex independent SL modifications, including cap 1 ribose methylation and formation of pseudouridine at position 28, while downstream cap methylation at positions 2, 3 and 4 requires Sm-complex formation (Zamudio et al., 2007; Zeiner et al., 2004a).
TbMTr2 and TbMTr3 are related to the vaccinia virus cap 1 MTase VP39 and have evolved unique specificities to modify the second and third nucleotides, respectively (Arhin et al., 2006a; Arhin et al., 2006b; Hall and Ho, 2006; Zamudio et al., 2006). This MTase family is restricted to poxvirus and kinetoplastid genomes (Zamudio et al., 2006). TbMTr2−/− cell lines are viable and the SL RNA lacks cap 2 modification (Arhin et al., 2006b). TbMTr3−/− cells are viable and lack 2′-O-ribose methylation on the third and fourth nucleotide (Arhin et al., 2006a); TbMTr3 may modify both positions (Arhin et al., 2006a; Zamudio et al., 2006). Simultaneous suppression of TbMTr2 and TbMTr3 by RNA interference (RNAi) does not affect cellular growth and yields a predominately cap 1 phenotype on SL RNA (Zamudio et al., 2006).
The SL RNA is transcribed as a 3′-extended form due to attenuated transcriptional termination (Sturm et al., 1999) and requires multiple nucleases to achieve the mature length of 142 nt (Hitchcock et al., 2004). In a final polishing step, the last uridine is removed by the small-RNA exonuclease TbSNIP in the nucleoplasm (Zeiner et al., 2004b). Using cap 4 formation and 3′-end maturation as markers in the biogenesis pathway, TbMTr1−/− cells experience a delay in processing, with accumulation of cap 0 and 3′-end extended substrate SL (Zamudio et al., 2007).
Effector proteins distinguish the fully methylated form of the cap 4 from incomplete cap structures. The T. brucei nuclear cap-binding complex protein CBP20 binds with higher affinity to the mature cap 4 as compared to cap 0 (Li and Tschudi, 2005). In Leishmania, homologs of the cap-dependent translation initiation factor eIF4E distinguish among different SL RNA cap structures (Dhalia et al., 2005; Yoffe et al., 2006).
We examined the role of cap 4 ribose methylation in translation through manipulation of the TbMTase genes. Attempts to derive cells with complete loss of mRNA cap ribose methylation were unsuccessful, indicating an essential role in kinetoplastid biology. TbMTr2−/−/TbMTr3−/− substrate SL RNA had no defect in 3′-end maturation; in contrast TbMTr1−/− cells have 3′-extended SL RNA indicating a delay in a distinct step of SL processing or trans-splicing. The absence of kinetoplastid-specific cap 3 and cap 4 ribose methylation reduced translation rates, providing the first functional role for the hypermethylated cap 4 structure. TbMTr1−/− and TbMTr2−/−/TbMTr3−/− cells have distinct growth patterns under serum starvation conditions as compared to wildtype cells, suggesting distinct functions that are beneficial for survival.
Results
Establishment of a TbMTr2−/−/TbMTr3−/− line
The kinetoplastid cap 4 structure, with ribose methylation of the first four nucleotides, has been implicated in trans-splicing and translation. In TbMTr1−/− cells, nucleotide modification at position one is detected on mRNA, suggesting base modifications are occurring in the absence of cap 1 ribose methylation (Zamudio et al., 2007). However, the detection of base modification at position 4 is reduced severely in TbMTr3−/− cells due to the absence of 2′-O-ribose methylation at nucleotides three and four (Arhin et al., 2006a). Since individual knockout cell lines of the three T. brucei cap ribose MTase are viable, we aimed to test for functional redundancy by removing the three enzymes responsible for cap ribose modifications with the goal of creating a phenotypically mRNA cap 0 cell line.
The pKO vectors were used to establish cap ribose MTase knockout cells with the available drug markers for the diploid T. brucei. First, single knockout lines for each MTase were generated. Previously we had generated a TbMTr1−/− (Phleo/Pur) line in a wildtype genetic background (Zamudio et al., 2007). Here we created a TbMTr2−/− (Hyg/Neo) line (Figure 1A) (Southern blot data not shown) and TbMTr3−/− (Phleo/Pur) line (Figure 1A), as verified by loss of TbMTr3 alleles in Southern blots (Figure 1B, Lane 1–3). The validation of the latter targeting vectors confirmed similar knockout results (Arhin et al., 2006a; Arhin et al., 2006b) and enabled us to continue toward isolation of double and triple knockout lines.
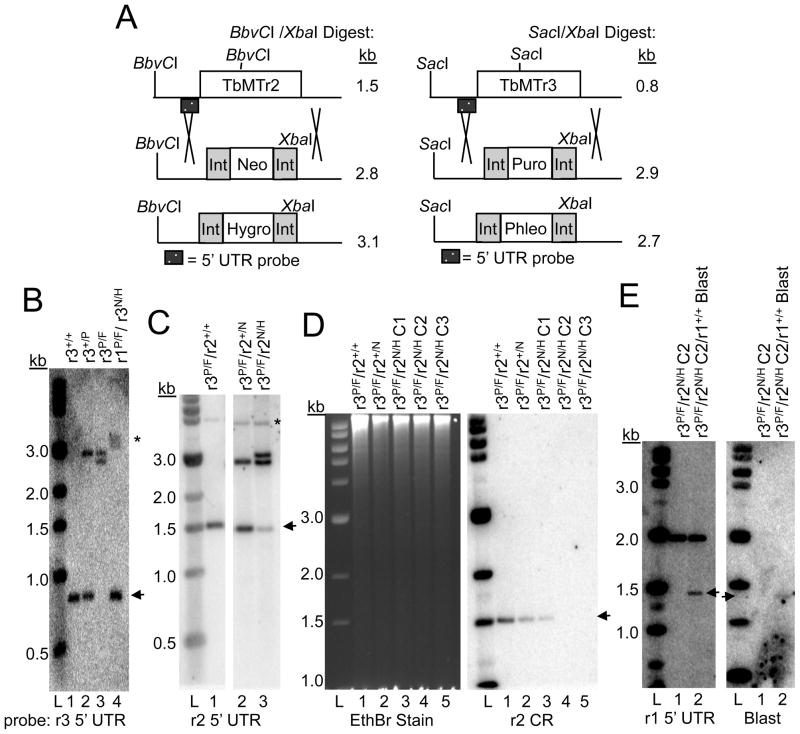
Fig. 1.
Establishment of cap ribose MTase double knockout cells. A. Schematic for Southern blot detection of gene disruption for TbMTr2 and TbMTr3. Restriction enzymes and probe location are noted for each. Int = processing sequences for drug markers. B. Southern blot for attempts to establish TbMTr3 knockout cells in wildtype (Lanes 1–3) and TbMTr1−/− genetic background (Lane 4). Resistance markers used: P = puromycin; F = phleomycin; N = neomycin; H = hygromycin. Blot of SacI/XbaI-digested genomic DNA probed for detection of the TbMTr3 5′ UTR. Arrowhead marks wildtype alleles in each blot. L = 1-kb molecular weight ladder (New England Biolabs). C. Detection of TbMTr2 knockouts in the TbMTr3−/− genetic background by probing with the TbMTr2 5′ UTR. Genomic DNA was digested with BbvCI and XbaI. * marks an extraneous, weakly cross-hybridizing band. D. Verification of loss of TbMTr2 in TbMTr2−/−/TbMTr3−/− clonal lines (C2, C3, Lanes 4–5) by probing for absence of TbMTr2 coding region (CR). Genomic DNA was digested with BbvCI and XbaI. Ethidium bromide staining of the gel served as a loading control. E. Southern blot of blasticidin resistant cell line established by a TbMTr1 blasticidin knockout vector in TbMTr2−/−/TbMTr3−/− double knockout cells. The blot was probed with 5′-UTR of TbMTr1 (left panel) and blasticidin (right panel).
Targeting of the TbMTr3 knockout vectors (Neo/Hyg cassettes) in TbMTr1−/− (Phleo/Pur) null-mutants was unsuccessful, with uncloned cell lines resistant to the four selection drugs still having intact TbMTr3 alleles (arrowhead; Figure 1B, Lane 4). In a portion of the population, larger bands containing the 5′ UTR are present, suggesting that aberrant recombination events have occurred. Clonal cell lines were obtained by limiting dilution, however all were positive for the TbMTr3 coding region, indicating that no TbMTr1−/−/TbMTr3−/− cells were present. Similarly, the targeting of TbMTr2 knockout vectors (Hyg/Neo) in the TbMTr1−/− (Phleo/Pur) genetic background resulted in the isolation of stable transfectants with intact TbMTr2 (data not shown).
Despite the inability to knockout a second cap MTase in the TbMTr1−/− lines, TbMTr2−/−/TbMTr3−/− cells were established by transfection first with a NEOR followed by HYGR TbMTr2 knockout vector in TbMTr3−/− (Phleo/Pur) cells (Figure 1C). In an uncloned population, the majority of cells had stoichiometric deletion of the TbMTr2 alleles, however a smaller fraction of the drug-resistant cells retained intact TbMTr2 (arrowhead; Figure 1C). Homogeneous populations of TbMTr2−/−/TbMTr3−/− double knockout cells were isolated by limiting dilution. The absence of the TbMTr2 coding region in clonal lines C2 and C3 was confirmed by Southern blot (Figure 1D, Lane 4–5).
The inability to establish TbMTr2 and TbMTr3 knockouts in the TbMTr1−/− genetic background suggests cap 2, cap 3, and possibly cap 4 Sm-dependent methylations affect viability in the absence of cap 1 Sm-independent ribose methylation. TbMTr1−/− cells have delays in SL RNA processing, with an accumulation of cap 0 and under-processed 3′-end SL RNA (Zamudio et al., 2007). As SL RNA is consumed at a high rate and required for maturation of every cellular mRNA, TbMTr1−/− cells may be vulnerable to additional stresses. The establishment of TbMTr2−/−/TbMTr3−/− cells provides a tool to determine the function of the kinetoplastid-specific cap 4 methylations and to test for functional redundancy. Possible roles for unique cap ribose methylation include enhancement of SL RNA biogenesis, trans-splicing or mRNA translation.
Establishment of TbMTr1/TbMTr2/TbMTr3 triple knockout line not successful
RNAi targeting of the three MTase resulted in knockdown of all three transcripts (data not shown), however the desired SL RNA cap 0 phenotype was not seen. Residual enzymatic activity present after RNAi induction may have prevented a growth or cap phenotype. To eliminate definitively the possibility of generating a cap 0 line, the generation of a triple MTase-knockout line in T. brucei cells was attemped.
A TbMTr1 knockout plasmid with a blasticidin resistance marker was transfected into the TbMTr2−/−/TbMTr3−/− cells. Five cell lines resistant to all five drugs were obtained. Southern blotting for the TbMTr1 allele and drug marker to confirm proper integration into the TbMTr1 genomic locus showed that the targeting construction generated a 1.4-kb band, not the expected 2.4-kb fragment (Figure 1E). To test if misintegration was dependent on the concentration of targeted DNA, titrations were performed using 1–10 μg of TbMTr1 knockout vector. Transfection with ≤2 μg DNA did not yield stable transfectants, while transfections with ≥5 μg DNA showed identical non-homologous integration events.
Because removal of even a single TbMTr1 allele in the absence of downstream cap ribose modifications was not possible, an important but unidentified function for these kinetoplastid-specific cap ribose methylations is indicated. The loss of multiple cap 4 ribose methylations may have an additive effect on cellular viability. In an effort to define a role for cap 2–4 ribose methylation, characterization of TbMTr2−/−/TbMTr3−/− cells was performed.
TbMTr2−/−/TbMTr3−/− cells display additive cap phenotypes
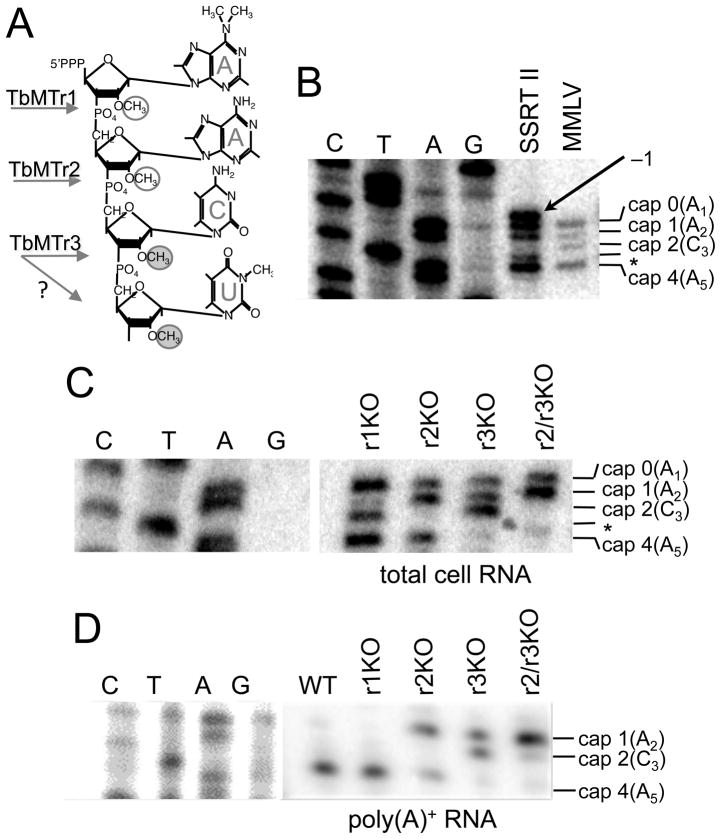
Cap 4 (Figure 2A) formation occurs sequentially from 5′ to 3′ (Mair et al., 2000). Cap 1 2′-O-ribose methylation by TbMTr1 is Sm protein-independent and occurs early in the SL RNA biogenesis pathway (Zamudio et al., 2009; Zeiner et al., 2004a). Subsequent cap 2 and cap 3 modifications are Sm-dependent and occur in the nucleoplasm where TbMTr2 and TbMTr3 are localized (Arhin et al., 2006a; Zamudio et al., 2006). Intermediates in cap 4 formation are detected by primer extension using a radiolabeled oligonucleotide complementary to the intron portion of substrate SL RNA. Since each of the cap methylations is sufficient to stop reverse transcriptase (RT) extension, the extension products can be mapped by comparison to the cognate sequencing ladder. The cap 3 termination product is not visualized by this assay, likely due to cap 3 and cap 4 being performed sequentially by TbMTr3 or by an unidentified TbMTr4 that is dependent on TbMTr3 activity. For this assay the RT choice is crucial, as false banding patterns are observed due to RT terminal transferase activity (Efimov et al., 2001; Schmidt and Mueller, 1999), making resolution with a parallel sequencing ladder essential (Figure 2B). The wildtype SL RNA cap profile in total RNA samples extended with Moloney murine leukemia virus (MMLV) RT showed four bands over a five nucleotide range corresponding to cap 0, cap 1, cap 2 and cap 4 intermediates. By contrast, Superscript (SS) RT II yielded five bands over a six-nucleotide range with the bands at −1 and +3 corresponding to a terminal transferase product. The strength of the shadowing effect at different positions, particularly at A1 and A5, indicates that sequence content influences terminal transferase efficiency.
Fig. 2.
TbMTr2−/−/TbMTr3−/− cells contain only cap 1 ribose methylated mRNA. A. Diagram of cap 4 structure with methylations circled and kinetoplasid-specific methylation noted with filled circles. The cap ribose methyltransferase for each nucleotide is indicated; ‘?’ Indicates the possibility of dual function for TbMTr3. B. Wildtype cap 4 profile on substrate SL RNA determined using radiolabeled TbSL40 oligonucleotides in total cell RNA samples with Superscript™ II (SSRT II) or Moloney murine leukemia virus (MMLV) RT. An asterisk (*) marks cap 3 intermediate not detected by primer extension; an arrow marks the spurious −1 extension produced by SS RT II. C. Cap 4 profile of SL RNA in cap ribose MTase knockout lines using TbSL40 on total cell RNA. r1KO = TbMTr−/−cells; r2KO = TbMTr2−/− cells; r3KO = TbMTr3−/−cells. D. Mature mRNA cap structures in cap ribose MTase knockout cells determined on poly(A)+ RNA using SL exon-specific primer. The contrast of the DNA sequencing ladder was adjusted in Photoshop.
To confirm the SL cap phenotype of TbMTr2−/−/TbMTr3−/− cells, primer extension analysis was performed with the MMLV reverse transcriptase. Each of the individual knockouts had loss of the corresponding cap modification (Figure 2C). TbMTr1−/− SL RNA showed caps 0, 2 and 4; a faint band corresponding to cap 1 may be due to the A1 base methylation that is detected in the mRNA by RNase T2 digestion (Zamudio et al., 2007). TbMTr2−/− SL RNA carried caps 0, 1 and 4, while TbMTr3−/− SL RNA possessed caps 0, 1 and 2. TbMTr2−/−/TbMTr3−/− SL RNA revealed predominantly cap 0 and cap 1 modifications with minor modification at nucleotide 4. This pattern is consistent with the combined individual phenotypes and characterized activities of these enzymes (Arhin et al., 2006a; Arhin et al., 2006b; Hall and Ho, 2006; Zamudio et al., 2006; Zamudio et al., 2007). The cap structure of mature mRNA was assayed on poly(A)+ RNA using primer extension with a radiolabeled primer complementary to the exon portion of the SL (Figure 2D). As observed previously TbMTr1−/− mRNA was modified at position 4 (Zamudio et al., 2007). For the single knockouts, TbMTr2−/− mRNA showed cap1 and cap 4, while TbMTr3−/− was cap 1 and cap 2. The TbMTr2−/−/TbMTr3−/− mRNA cap structure was predominantly cap 1. In all cap ribose MTase knockout cell lines, cap 0 SL was not detected on mRNA, implying that either downstream ribose methylation or base modifications occur or are required before trans-splicing, or function in mRNA stability.
The inability to establish full cap ribose-null cell lines suggests an essential role in kinetoplastid biology. As the loss of TbMTr1 cap 1 activity resulted in accumulation of substrate SL RNA in 3′-extended form (Zamudio et al., 2007), downstream methylations may also serve in processing through the biogenesis pathway or act during trans-splicing. TbMTr2 and TbMTr3 could be functionally redundant, thus additional SL RNA processing phenotypes were queried in TbMTr2−/−/TbMTr3−/− cells and compared to the individual knockout cell lines.
Complete 3′-end formation in TbMTr2−/−/TbMTr3−/− cells
Both the loss of cap 1 ribose methylation (Zamudio et al., 2007) and delay of cap 1 formation by disruption of TbMTAP (Zamudio et al., 2009), a component of the TbMTr1 SL RNA-processing complex, results in accumulation of cap 0 substrate SL RNA in a 3′-extended form.
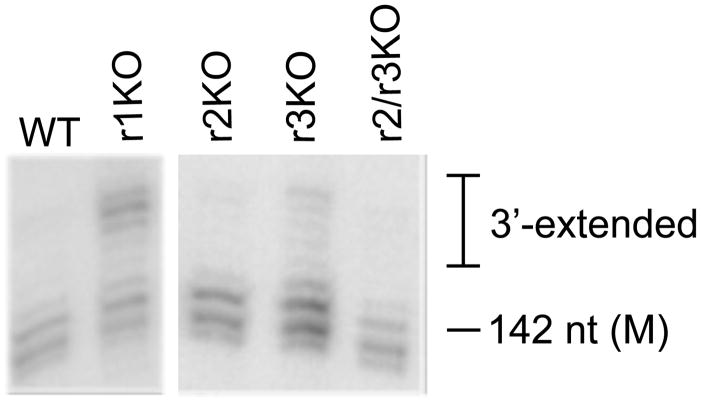
To determine if TbMTr2−/−/TbMTr3−/− substrate SL RNA is accumulating in underprocessed 3′-extended forms, high-resolution RNA blots were probed for substrate SL RNA in the cap MTase knockout lines (Figure 3). These blots show single-nucleotide resolution in which the primary SL transcripts display a range of sizes attributed to the attenuated termination of transcription by RNA polymerase II mediated by the downstream T tract (Sturm et al., 1999). Intermediates in 3′ processing (Zeiner et al., 2004b) and the mature 142-nt SL (Hitchcock et al., 2004) are visible as discrete species. Compared to the wildtype, TbMTr1−/−cells showed substantial accumulation of higher molecular weight bands, serving as a positive control for underprocessed substrate SL RNA. The SL RNA from individual TbMTr2−/− or TbMTr3−/− knockout lines and from TbMTr2−/−/TbMTr3−/− cells displayed 3′-processing comparable to wildtype, with no over-accumulation of 3′-extended forms. Normalization of total substrate SL levels from the individual and double knockout lines indicated that the TbMTr1−/−and TbMTAP−/− lines consistently showed overaccumulation of substrate relative to other small RNA markers (data not shown), but the amount of mature SL RNA did not differ significantly from wildtype levels.
Fig. 3.
TbMTr2−/−/TbMTr3−/− substrate SL RNA is not 3′-extended. High-resolution RNA blots of total cell RNA from cap MTase knockout cells were hybridized with TbSL40. See Experimental Procedures for details. M = mature length.
Normal 3′ processing of the SL RNA in TbMTr2−/−/TbMTr3−/− lines suggests that exonucleolytic trimming of the SL RNA occurs before cap 2 and cap 3 methylation, and is not dependent on these Sm-dependent modifications. To test for additional defects in TbMTr2−/−/TbMTr3−/− cells, the rate of translation was measured.
Decreased translation rates in under-methylated cell lines
Cap ribose methylation enhances translational efficiency (Kuge et al., 1998). To determine whether the kinetoplastid-specific cap 4 ribose methylations act in translation, in vivo translation rates were determined in the cap ribose MTase knockout cell lines.
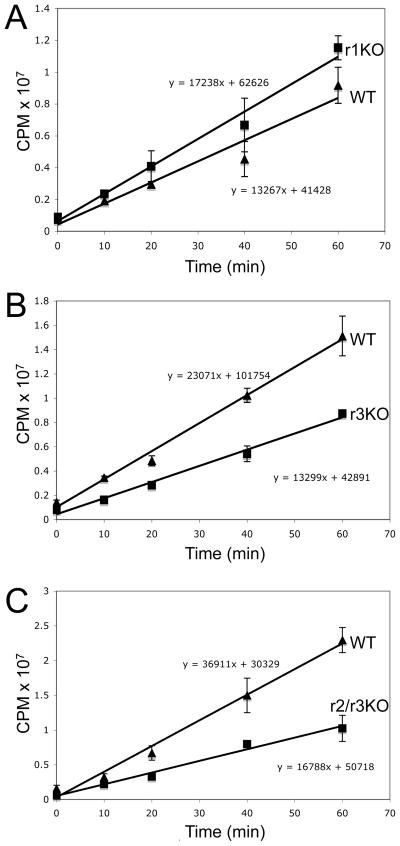
Kinetic analysis of [35S]-methionine incorporation into proteins over 60 min allowed the rate of translation to be determined in comparison to wildtype. Loss of cap 1 methylation in TbMTr1−/− cells did not produce a distinguishable difference in translation rate (Figure 4A). In contrast TbMTr3−/− cells showed a 57% rate of incorporation relative to wildtype cells (Figure 4B). Under similar conditions, TbMTr2−/−/TbMTr3−/− cells showed an incorporation rate 45% that of wildtype (Figure 4C).
Fig. 4.
Decreased translational efficiency due to loss of kinetoplastid-specific ribose cap methylation. A. Graph of [35S]-methionine incorporation into proteins in TbMTr1 knockout cells. B. Graph of [35S]-methionine incorporation in TbMTr3 knockout cells. C. Graph of [35S]-methionine incorporation in TbMTr2−/−/TbMTr3−/− double knockout cells. The bars represent the range of results from two independent experiments.
An alternative explanation for the reduced 35S-incorporation rate is decreased levels of mature mRNA in the knockout lines due to instability of mature mRNA. To test this possibility we quantified levels of the 40S ribosomal protein RPS23 and GAPDH mRNA from cellular equivalents of wildtype, TbMTr3−/− and TbMTr2−/−/TbMTr3−/− cells (Figure 5). Total cell RNA from 1.44 × 107 cells was resuspended in a volume of 60 μl each, yielding concentrations of: WT 0.395 μg/μl; TbMTr3KO 0.546 μg/μl; and TbMTr2/r3KO 0.646 μg/μl. The RPS23 mRNA levels were higher than wildtype in both knockout lines, on either a per-cell basis or relative to the absolute amount of RNA. The GAPDH mRNA levels were not significantly different in wildtype and TbMTr3−/− cells, however the increase was significant in TbMTr2−/−/TbMTr3−/− cells on the per-cell basis. The relative levels of the 7SL RNA were determined for the wildtype and TbMTr2−/−/TbMTr3−/− samples, and showed a nearly two-fold increase in TbMTr2−/−/TbMTr3−/−(data not shown). Using this alternative normalization standard, levels of RSP23 mRNA are approximately equal in these two samples, while the GAPDH levels are depressed to approximately 75% the level of wildtype in TbMTr2−/−/TbMTr3−/−. As the level of translation in the double knockout line was measured at 45% that of wildtype, the mRNA concentration in and of itself is not responsible for the reduction in translation.
Fig. 5.
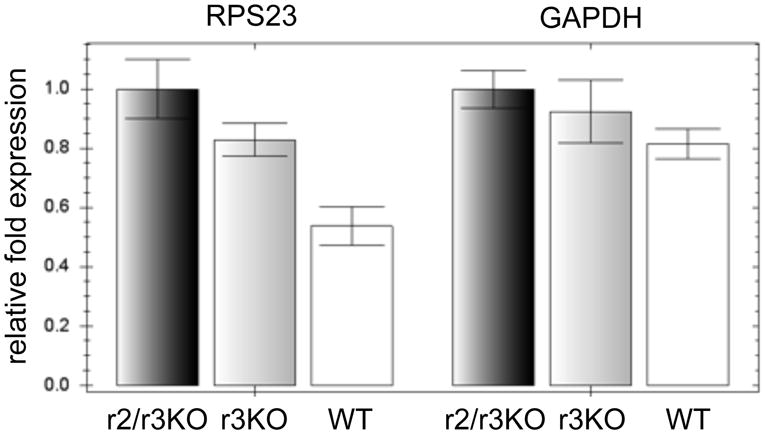
mRNA levels are not affected in MTase knockout lines on a per-cell basis. To compare endogenous mRNA levels, quantitative RT-PCR was used to measure RPS23 and GAPDH mRNA in wildtype (WT), TbMTr3−/− (r3KO) and TbMTr2−/−/TbMTr3−/− (r2/r3KO) cells. The bars represent the range of results from three independent reactions.
The representative mRNA levels indicate that reduced 35S-incorporation rates were not due to lower mRNA levels in the knockout lines. The decrease in 35S-incorporation rates in kinetoplastid-specific cap MTase knockout lines provides the first functional role for the cap 4 in kinetoplastid biology. Loss of TbMTr1 does not affect translation rate, indicating that the presence of kinetoplastid-specific methylations at positions 3 and 4 may compensate for the lack of cap 1. Alternatively, TbMTr1 cap 1 ribose methylation may facilitate efficient processing of SL RNA early in biogenesis, while the unique methylations function at later steps in mRNA maturation and translation.
Reduced growth of cap MTase knockout cell cultures
The consequence of delayed SL RNA processing or reduced translation rates is not evident in cell cultures maintained in mid-log growth under normal conditions. In search of growth differences under standard conditions, TbMTr1−/− and TbMTr2−/−/TbMTr3−/− cells were subjected to growth curve analysis without dilution.
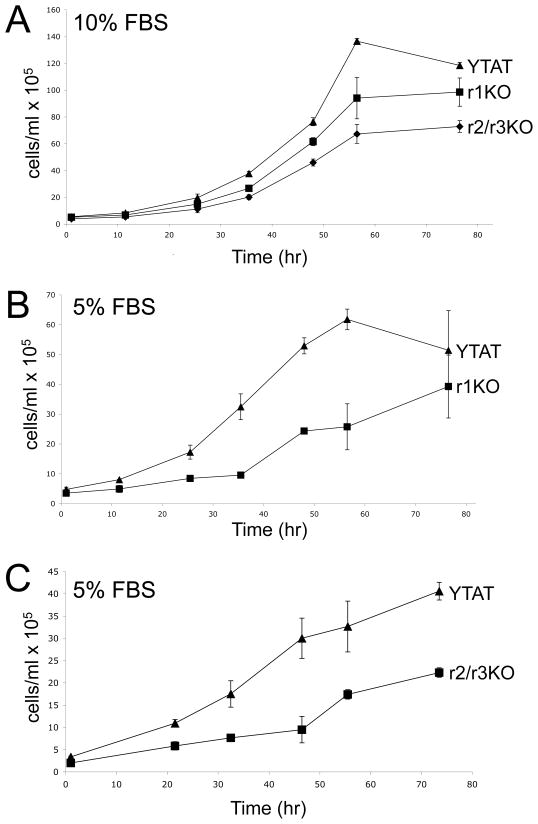
In comparison to wildtype cells, both TbMTr1−/− and TbMTr2−/−/TbMTr3−/− cells reached the stationary phase plateau of the growth curve at lower cellular concentrations, with TbMTr2−/−/TbMTr3−/− growing to the lowest final density (maximum density in cells/ml: WT=1.36 × 107, r1=9.85 × 106, r2/r3=7.28 × 106; Figure 6A). To determine differences in growth under stress conditions, growth curves were performed under low serum conditions. TbMTr1−/− (Figure 6B) and TbMTr2−/−/TbMTr3−/− (Figure 6C) cells were grown with 5% fetal bovine serum as compared to the normal 10%. In comparison to wildtype cells, TbMTr1−/− and TbMTr2−/−/TbMTr3−/− cells had prolonged lag phases and lower stationary phase concentrations with 5% FBS, again with TbMTr2−/−/TbMTr3−/− cells having the most pronounced differential.
Fig. 6.
Growth of TbMTr1−/− and TbMTr2−/−/TbMTr3−/− cells is compromised under serum starvation conditions. A. Wildtype (YTAT), TbMTr1−/−, and TbMTr2−/−/TbMTr3−/− growth curves in SM medium supplemented with 10% Fetal Bovine Serum (FBS). B. YTAT and TbMTr1−/−growth curves in SM medium supplemented with 5% FBS. C. YTAT and TbMTr2−/−/TbMTr3−/−growth curves in SM medium supplemented with 5% FBS.
Analysis of growth in normal and stress conditions suggests that the cap ribose MTase knockout cell lines are disadvantaged as compared to wildtype, a phenotype that may be attributed to defects in SL RNA processing and translation. Any cap MTase defective cells occurring spontaneously in the natural environment would be out-competed by the wildtype cap 4 population.
Discussion
We report the first functional role for unique cap 4 mRNA modifications in kinetoplastid protozoa. Failure to establish mutant cells lacking complete mRNA cap ribose methylation highlights their essential function. TbMTr3−/− and TbMTr2−/−/TbMTr3−/− cells have decreased translation rates. The additive effects of cap ribose MTase knockouts on substrate SL RNA consumption may contribute to the inability to establish cells lacking mRNA cap ribose methylation. The impact of the loss of these modifications is demonstrated by changes in stationary density and growth under serum starvation conditions.
Intracellular trafficking of RNA and translation initiation are facilitated by sets of proteins that bind to the 5′ cap. The recognition of m7G cap by cap-binding proteins occurs via a conserved mechanism of hydrophobic stacking interactions between two coplanar aromatic residues (Hodel et al., 1998). Caenorhabditis elegans has a family of cytosolic cap-binding proteins that distinguish between mRNAs with 5′ mono- or trimethylated caps (Jankowska-Anyszka et al., 1998). Similarly, the cap-binding proteins in trypanosomes have efficient binding to their unique cap 4 structure and its intermediates (Dhalia et al., 2005; Yoffe et al., 2006). In addition to mRNA processing and trafficking, the essential T. brucei 300-kDa nuclear cap-binding complex functions at early stages of trans-splicing (Li and Tschudi, 2005). The CBP20 cap-binding component of the complex binds the cap 4 structure preferentially, signifying recognition of the full cap 4 modifications as early as pre-assembly of the trans-spliceosome. In cap MTase knockouts, undermethylated substrate SL is incorporated into mRNA, but cap 0 structures are not detected. Whether cap 0 substrate SL can serve as a trans-splicing substrate or is stable on mature mRNA remains unclear.
By analysis of polysome association, mutagenesis studies in Leishmainia tarentolae implicate the full cap 4 or primary SL sequence in translation (Zeiner et al., 2003). Similar studies in Leptomonas collosoma show reduction in methylation and trans-splicing (Mandelboim et al., 2002). Here we have shown that in the absence of base changes in the SL the loss of cap 3 and/or cap 4 ribose methylations have a greater affect on translation rates than loss of cap 2. As TbMTr2−/−/TbMTr3−/− cells are viable, cap 1 methylated mRNA must associate with polysomes. Although not measured directly, no gross impairment of trans-splicing was evident in the T. brucei MTase knockout lines, as would be reflected in lower mRNA levels. Thus, the role the SL primary sequence in translation still requires investigation. The recognition of the full cap 4 structure in translation is supported by biochemical characterization of multiple eIF4E homologs in Leishmania major that distinguish between methylated cap structures (Dhalia et al., 2005; Yoffe et al., 2006). The decreased translation rates in TbMTr3−/− and TbMTr2−/−/TbMTr3−/− cells may reflect in vivo consequences of lower affinity cap binding by eIF4E protein during translation initiation.
As the 3′-most ribose and base methylations of cap 4 are unique to kinetoplastids, emphasis has been placed on determining their biological function for therapeutic targeting. Surprisingly, individual knockouts of cap 4 ribose MTases are viable; however redundancy or an ability of cap-binding proteins to function in the absence of single modifications may have masked the true function of full cap 4. The failure to establish T. brucei cells completely lacking cap ribose methylation argues for an essential function in mRNA maturation or translation. While a TbMTr2−/−/TbMTr3−/− growth phenotype was not observed in standard conditions, the abnormal growth under stress conditions presages that mutant cells may be compromised for survival in their natural environment. The extent of defects in the absence of cap ribose methylation requires further analysis in different lifestages, during differentiation, and in models of infection.
Full cap 4 may be preferred during mRNA processing and translation, but early cap modifications on SL processing intermediates could function specifically in SL RNA biogenesis. As the SL RNA accumulates in 3′-extended forms in TbMTr1−/− cells lacking only cap 1 ribose methylation and is distinct from TbMTr2−/− and TbMTr3−/− lines a separate function is indicated for cap 1 during SL RNA biogenesis. Co-transcriptional and cytoplasmic-trafficking-dependent SL RNA maturation may be dependent on cap 1. In either case the linkage of the Sm-protein-independent modification in the TbMTr1 complex and dependence of TbMTr2 and TbMTr3 activity on the Sm-protein complex binding, argue for two distinct stages in maturation (Zamudio et al., 2007; Zeiner et al., 2004a). The recognition of the SL cap 1 intermediate may facilitate proper transport of the SL RNA from a sub-nuclear processing center to other locales for subsequent maturation events, including 3′-end formation.
In Xenopus laevis, cap 1 ribose methylation acts to enhance translation (Kuge et al., 1998). The conservation of cap ribose methylation on eukaryotic mRNA supports the biological importance of these modifications. Differential mRNA cap ribose methylation states are observed in higher eukaryotic cells and likely reflect an additional level of post-transcriptional gene regulation. RNAi knockdown of a TbMTr1 family member in Caenorhabditis elegans caused maternal sterility (Rual et al., 2004), underscoring the importance of these modifications. The validation of cap ribose MTases in additional model systems will elucidate the extent of translation control by cap ribose methylation. Meanwhile, the extent of methylation in the kinetoplastid cap 4 provides a unique opportunity to unravel the subtleties of cap interaction with the translational machinery.
Experimental procedures
DNA Cloning
The pKO vectors (Lamb et al., 2001) for targeted gene disruption were used to establish MTase knockout cell lines. The drug cassette marker in the TbMTr1 pKO vector used to establish TbMTr1−/− cells (Zamudio et al., 2007) was replaced with blasticidin using ApaI and PacI sites. For TbMTr2 knockout vectors, the 5′UTR was amplified using MT417-5′-UTR-HindIII-F (AAA GCT TCC ATG CAG AAA TTG AG) and MT417-5′-UTR-EcoRI-R (TGA ATT CTT CAT TGA TGG TCT GTC G); the 3′UTR was amplified using MT417-3′-UTR-Xba-F (ATC TAG AAC CGT GAT TTG ATG) and MT417-3′-UTR-Not-R (TGC GGC CGC GAA TTA CAA CTT TC). For TbMTr3 knockout vectors, the 5′UTR was amplified using Fd-511-5′-UTR (GAT TTA AGC TTA GTG ACG CTC CAG) and Rv-511-5′-UTR (AAA TGG AAA CGC CTA CAC TCC C); the 3′UTR was amplified using Fd-511-3′-UTR (CTC TAG ATT TCC CCG CAG TTT ATT CCG) and Rv-511-3′-UTR (GGC GGC CGC ATA CCA CAG CAG GAA TC). The 5′ UTRs for each MTase were cloned in using HindIII and EcoRI. The 3′ UTRs were inserted with XbaI and NotI sites. Drug resistance marker for phleomycin, neomycin, hygromycin, puromycin, blasticidin and nourseothricin were cloned into the pKO vectors with ApaI and PacI sites as required.
DNA and RNA analyses
Total cell genomic DNA was isolated from stable cell lines obtained by clonal dilution using DNAzol (Invitrogen) as described by the manufacturer. For Southern blotting, 5 or 10 μg of genomic DNA was digested and resolved on 1% agarose gels, transferred and hybridized with [α-32P]-CTP incorporated random hexamer-primed probes (Amersham Biosciences) using the 5′ UTR for the probed MTase as the template.
Total cell RNA was isolated with TRIzol reagent (Invitrogen) as described previously (Zeiner et al., 2004b). High-resolution acrylamide RNA blotting, primer extension using Moloney murine leukemia virus or Superscript II RT, and DNA sequencing reactions were performed as described (Zeiner et al., 2004b) using the [γ-32P]-labeled oligonucleotides: exon specific, TbWTexon (CAA TAT AGT ACA GAA ACT G) or intron-specific SL RNA, TbSL40 (CTA CTG GGA GCT TCT CAT AC). For mRNA primer extensions to determine cap 4 structure, poly(A)+ RNA was isolated using the MicroPoly(A)Purist kit (Ambion). After transfer, the high-resolution blots of total cell RNA from 0.8 mm-thick 8 M Urea/8% polyacrylamide RNA gels were probed with [γ-32P]-labeled TbSL40 to detect substrate SL RNA or TbU3 (CCG GGC GGA GCC AGC AAC CTT C) for the U3 snoRNA. All blots were visualized on PhosphorImager screens (Amersham). Quantification of RNA blots was performed with ImageQuant software (Amersham).
For quantitative RT-PCR, cell concentration was determined using the Beckman-Coulter counter and 1.44 × 107 cells collect. Total RNA isolation and genomic DNA treatment was performed with the RNeasy Plus Mini Kit (Qiagen) and final RNA pellet resuspended in 60 μl. Using 1 μl of total RNA, cDNA was made using SS RT II and oligo dTs (Invitrogen) following the manufacturer’s protocol. To quantify the levels of specific mRNA transcripts, 1 μl of the cDNA reaction was amplified in SYBR green qPCR master mix following the manufacturer’s protocol (BioRad) using a C1000 thermocycler with the CXF96 real-time system (BioRad). TbGAPDH (Tb927.6.4280 and Tb927.6.4300) was amplified using primers GPRT5 (GGC TGA TGT CTC TGT GGT GGA) and GPRT6 (GGC TGT CGC TGA TGA AGT CG) and RSP23 (Tb10.70.7020 and Tb10.70.7030) using RPRT1 (AGA TTG GCG TTG GAG CGA AA) and RPRT2 (GAC CGA AAC CAG AGA CCA GCA).
T. brucei strains and cell culture
YTAT procyclic T. brucei was grown at 27 °C in SM medium (Cunningham, 1977) supplemented with 10% fetal bovine serum (FBS). For growth curve experiments under normal conditions, 1 × 105 cells/ml were seeded in 0.5 ml SM medium with 10% FBS in 24-well plates. Time points were taken and counted in triplicate with a Beckman Coulter counter. For monitoring serum starvation growth, 1 × 105 cells/ml were seeded in 5 ml SM medium supplemented with 5% FBS in 25 cm2 culture flasks. Transfections were performed as described previously (Wang et al., 2000). Selection was performed with hygromycin (20 μg/ml), puromycin (10 μg/ml), phleomycin (20 μg/ml), G418 (15 μg/ml), or blasticidin (15 μg/ml).
[35S]-methionine incorporation assay
The T. brucei cell culture was grown to mid-log phase (5 × 106 cells/ml) and counted with both a hemocytometer and Beckman-Coulter counter. 1 × 107 cells were collected by centrifugation at 2000 g and washed twice in SM medium lacking methionine (SM-m). The cell pellet was resuspended in 3 ml of SM-m and starved for methionine for 1 hr. Next, 200 μCi of [35S]-methionine (Specific Activity: >1000 Ci (37.0 Tbq)/mMole, 50 mM Tricine, 10 mM BME) was added and 0.5 ml samples collected in triplicate at 0, 10, 20, 40 and 60 min. Samples were centrifuged immediately at 13,000 g for 30 sec, the medium was removed, and proteins were TCA precipitated. The TCA pellet was transferred to 2.3 cm 3MM filter paper (Whatman) and washed with the following ice-cold solutions: SM-m, 10% TCA, 40 mM methionine, 95% ethanol and acetone using a vacuum trap. Filter papers were allowed to dry at room temperature, immersed in 10 ml of ScintiVerse (Fisher), and radioactive counts were ascertained in liquid scintillation counter (Beckman-Coulter).
Supplementary Material
Acknowledgments
We thank Jay Bangs for the pKO vectors; Sidney Harvey for access to the quantitative RT-PCR machine; Kent Hill and Katy Ralston for providing YTAT cells and primers for RT-PCR; Arnie Berk, Guillaume Chanfreau, Robert Hitchcock, and Isadora Ruvalcaba-Trejo for stimulating discussions. This work was supported by NIH grant AI056034. J.R.Z. was supported by USPHS National Research Service Award GM07104.
References
- Arhin GK, Li H, Ullu E, Tschudi C. A protein related to the vaccinia virus cap-specific methyltransferase VP39 is involved in cap 4 modification in Trypanosoma brucei. RNA. 2006a;12:53–62. doi: 10.1261/rna.2223406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhin GK, Ullu E, Tschudi C. 2′-O-Methylation of position 2 of the trypanosome spliced leader cap 4 is mediated by a 48 kDa protein related to vaccinia virus VP39. Mol Biochem Parasitol. 2006b;147:137–139. doi: 10.1016/j.molbiopara.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Banerjee AK. 5′-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol Rev. 1980;44:175–205. doi: 10.1128/mr.44.2.175-205.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs JD, Crain PF, Hashizume T, McCloskey JA, Boothroyd JC. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J Biol Chem. 1992;267:9805–9815. [PubMed] [Google Scholar]
- Brownlee GG, Fodor E, Pritlove DC, Gould KG, Dalluge JJ. Solid phase synthesis of 5′-diphosphorylated oligorobonucleotides and their conversion to capped m7Gppp-oligoribonucleotides for use as primers for influenza A virus RNA polymerase in vitro. Nucleic Acids Res. 1995;23:2641–2647. doi: 10.1093/nar/23.14.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham I. New culture medium for maintenance of tsetse tissues and growth of trypanosomatids. J Protozool. 1977;24:325–329. doi: 10.1111/j.1550-7408.1977.tb00987.x. [DOI] [PubMed] [Google Scholar]
- Dhalia R, Reis CR, Freire ER, Rocha PO, Katz R, Muniz JR, Standart N, de Melo Neto OP. Translation initiation in Leishmania major: characterisation of multiple eIF4F subunit homologues. Mol Biochem Parasitol. 2005;140:23–41. doi: 10.1016/j.molbiopara.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Efimov VA, Chakhmakhcheva OG, Archdeacon J, Fernandez JM, Fedorkin ON, Dorokhov YL, Atabekov JG. Detection of the 5′-cap structure of messenger RNAs with the use of the cap-jumping approach. Nucleic Acids Res. 2001;29:4751–4759. doi: 10.1093/nar/29.22.4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistadt MS, Cross GAM, Robertson HD. Discontinuously synthesized mRNA from Trypanosoma brucei contains the highly methylated 5′ cap structure, m7GpppA*A*C(2′-O)mU*A. J Biol Chem. 1988;263:15071–15075. [PubMed] [Google Scholar]
- Furuichi Y, Shatkin AJ. Viral and cellular mRNA capping: past and prospects. Adv Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MP, Ho CK. Functional characterization of a 48 kDa Trypanosoma brucei cap 2 RNA methyltransferase. Nucleic Acids Res. 2006;34:5594–5602. doi: 10.1093/nar/gkl573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock RA, Zeiner GM, Sturm NR, Campbell DA. The 3′-termini of small RNAs in Trypanosoma brucei. FEMS Microbiol Lett. 2004;236:73–78. doi: 10.1016/j.femsle.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Hodel AE, Gershon PD, Quiocho FA. Structural basis for sequence-nonspecific recognition of 5′-capped mRNA by a cap-modifying enzyme. Mol Cell. 1998;1:443–447. doi: 10.1016/s1097-2765(00)80044-1. [DOI] [PubMed] [Google Scholar]
- Jankowska-Anyszka M, Lamphear BJ, Aamodt EJ, Harrington T, Darzynkiewicz E, Stolarski R, Rhoads RE. Multiple isoforms of eukaryotic protein synthesis initiation factor 4E in Caenorhabditis elegans can distinguish between mono- and trimethylated cap structures. J Biol Chem. 1998;273:10538–10542. doi: 10.1074/jbc.273.17.10538. [DOI] [PubMed] [Google Scholar]
- Kuge H, Brownlee GG, Gershon PD, Richter JD. Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res. 1998;26:3208–3214. doi: 10.1093/nar/26.13.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JR, Fu V, Wirtz E, Bangs JD. Functional analysis of the trypanosomal AAA protein TbVCP with trans-dominant ATP hydrolysis mutants. J Biol Chem. 2001;276:21512–21520. doi: 10.1074/jbc.M100235200. [DOI] [PubMed] [Google Scholar]
- Langberg SR, Moss B. Post-transcriptional modifications of mRNA. Purification and characterization of cap I and cap II RNA (nucleoside-2′-)-methylatransferases from HeLa cells. J Biol Chem. 1981;256:10054–10060. [PubMed] [Google Scholar]
- Li H, Tschudi C. Novel and essential subunits in the 300-kilodalton nuclear cap binding complex of Trypanosoma brucei. Mol Cell Biol. 2005;25:2216–2226. doi: 10.1128/MCB.25.6.2216-2226.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair G, Ullu E, Tschudi C. Cotranscriptional cap 4 formation on the Trypanosoma brucei spliced leader RNA. J Biol Chem. 2000;275:28994–28999. doi: 10.1074/jbc.M004193200. [DOI] [PubMed] [Google Scholar]
- Mandelboim M, Estraño CL, Tschudi C, Ullu E, Michaeli S. On the role of exon and intron sequences in trans-splicing utilization and cap 4 modification of the trypanosomatid Leptomonas collosoma SL RNA. J Biol Chem. 2002;277:35210–35218. doi: 10.1074/jbc.M201910200. [DOI] [PubMed] [Google Scholar]
- Mittra B, Zamudio JR, Bujnicki JM, Stepinski J, Darzynkiewicz E, Campbell DA, Sturm NR. The TbMTr1 spliced leader RNA cap 1 2′-O-ribose methyltransferase from Trypanosoma brucei acts with substrate specificity. J Biol Chem. 2008;283:3161–3172. doi: 10.1074/jbc.M707367200. [DOI] [PubMed] [Google Scholar]
- Muthukrishnan S, Morgan M, Banerjee AK, Shatkin AJ. Influence of 5′-terminal m7G and 2′--O-methylated residues on messenger ribonucleic acid binding to ribosomes. Biochemistry. 1976;15:5761–5768. doi: 10.1021/bi00671a012. [DOI] [PubMed] [Google Scholar]
- Perry KL, Watkins KP, Agabian N. Trypanosome mRNAs have unusual “cap 4” structures acquired by addition of a spliced leader. Proc Natl Acad Sci USA. 1987;84:8190–8194. doi: 10.1073/pnas.84.23.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R, Singh R, Shimba S. Methylated cap structures in eukaryotic RNAs: structure, synthesis and functions. Pharmacol Ther. 1992;54:249–267. doi: 10.1016/0163-7258(92)90002-h. [DOI] [PubMed] [Google Scholar]
- Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, Vidal M. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J-p, Shen S, Ullu E, Tschudi C. Evidence for a capping enzyme with specificity for the trypanosome spliced leader RNA. Mol Biochem Parasitol. 2007;156:246–254. doi: 10.1016/j.molbiopara.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt WM, Mueller MW. CapSelect: a highly sensitive method for 5′ CAP-dependent enrichment of full-length cDNA in PCR-mediated analysis of mRNAs. Nucleic Acids Res. 1999;27:e31. doi: 10.1093/nar/27.21.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm NR, Yu MC, Campbell DA. Transcription termination and 3′-end processing of the spliced leader RNA in kinetoplastids. Mol Cell Biol. 1999;19:1595–1604. doi: 10.1128/mcb.19.2.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi Y, Sindkar S, Ekonomidis D, Hall MP, Ho CK. Trypanosoma brucei encodes a bifunctional capping enzyme essential for cap 4 formation on the spliced leader RNA. J Biol Chem. 2007;282:15995–16005. doi: 10.1074/jbc.M701569200. [DOI] [PubMed] [Google Scholar]
- Wang Z, Morris JC, Drew ME, Englund PT. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- Yoffe Y, Zuberek J, Lerer A, Lewdorowicz M, Stepinski J, Altmann M, Darzynkiewicz E, Shapira M. Binding specificities and potential roles of isoforms ofeukaryotic initiation factor eIF4E in Leishmania. Eukaryot Cell. 2006;5:1969–1979. doi: 10.1128/EC.00230-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamudio JR, Mittra B, Zeiner GM, Feder M, Bujnicki JM, Sturm NR, Campbell DA. Complete cap 4 formation is not required for viability in Trypanosoma brucei. Eukaryot Cell. 2006;5:905–915. doi: 10.1128/EC.00080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamudio JR, Mittra B, Foldynová-Trantírková S, Zeiner GM, Lukes J, Bujnicki JM, Sturm NR, Campbell DA. The 2′-O-ribose methyltransferase for cap 1 of spliced leader RNA and U1 small nuclear RNA in Trypanosoma brucei. Mol Cell Biol. 2007;27:6084–6092. doi: 10.1128/MCB.00647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamudio JR, Mittra B, Chattopadhyay A, Wohlschlegel JA, Sturm NR, Campbell DA. Trypanosoma brucei spliced leader RNA maturation by the cap 1 2′-O-ribose methyltransferase and SLA1 H/ACA snoRNA pseudouridine synthase complex. Mol Cell Biol. 2009;29:1202–1211. doi: 10.1128/MCB.01496-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiner GM, Sturm NR, Campbell DA. The Leishmania tarentolae spliced leader contains determinants for association with polysomes. J Biol Chem. 2003;278:38269–38275. doi: 10.1074/jbc.M304295200. [DOI] [PubMed] [Google Scholar]
- Zeiner GM, Foldynová S, Sturm NR, Luke_ J, Campbell DA. SmD1 is required for spliced leader RNA biogenesis. Eukaryot Cell. 2004a;3:241–244. doi: 10.1128/EC.3.1.241-244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiner GM, Hitchcock RA, Sturm NR, Campbell DA. 3′-end polishing of the kinetoplastid spliced leader RNA is performed by SNIP, a 3′->5′ exonuclease with a motley assortment of small RNA substrates. Mol Cell Biol. 2004b;24:10390–10396. doi: 10.1128/MCB.24.23.10390-10396.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007;81:3891–3902. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.