Abstract
Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors, gefitinib and erlotinib, are effective therapies against mutant non-small cell lung cancers (NSCLCs). Treatment is limited by the development of resistance in part explained by the gain of a secondary EGFR mutation, T790M, at the gatekeeper residue. Irreversible EGFR inhibitors, including PF00299804, are effective in vitro and in vivo against EGFR mutant tumors that contain EGFR T790M and are currently under clinical development. In this study we generate models of resistance to PF00299804, using cell lines with EGFR T790M, and demonstrate that the PF00299804 resistant models develop focal amplification of EGFR that preferentially involves the T790M-containing allele. These PF00299804 resistant cell lines remain dependent on EGFR for growth as downregulation of EGFR by shRNA compromises their viability. We demonstrate that resistance to PF00299804 arises, at least in part, through selection of a pre-existing EGFR T790M amplified clone both in vitro and using a xenograft model in vivo. Our findings demonstrate that EGFR T790M is a common resistance mechanism to both reversible, and when amplified, the irreversible EGFR kinase inhibitors further emphasizing the need to develop more potent therapies against EGFR T790M. The findings can be used to guide studies of patient tumor specimens from ongoing clinical trials of irreversible EGFR kinase inhibitors.
Keywords: Epidermal growth factor receptor, drug resistance, EGFR T790M, amplification, tyrosine kinase inhibitor, non-small cell lung cancer
Introduction
Kinase inhibitors are effective clinical therapies against cancers where that target of the inhibitor is activated by an oncogenic mechanism(Demetri et al., 2006; Druker et al., 2001; Kantarjian et al., 2002). In lung cancer, EGFR kinase inhibitors, gefitinib and erlotinib, are effective clinical treatments for NSCLC patients whose tumors harbor activating mutations in EGFR(Inoue et al., 2006; Mok et al., 2008; Sequist et al., 2008). However, all patients will eventually develop resistance (herein referred to as acquired resistance) while being treated with gefitinib or erlotinib. EGFR T790M is the most common mechanism of acquired resistance to gefitinib and has been detected in 50% of NSCLC patients that acquire resistance and from cell line models that have been selected for gefitinib resistance(Balak et al., 2006; Engelman et al., 2007b; Kosaka et al., 2006). Although EGFR T790M is oncogenic by itself, when in cis with an EGFR activating mutation, the double mutant leads to significant enhancement of EGFR kinase activity and oncogenic transformation both in vitro and in vivo(Godin-Heymann et al., 2007; Vikis et al., 2007).
The mechanism by which EGFR T790M causes gefitinib resistance has also been elucidated. Unlike the analogous T315I mutation in ABL, which introduces a steric impediment for imatinib binding, EGFR T790M only modestly affects gefitinib binding but leads to a higher affinity for ATP similar to that of wild type EGFR(Yun et al., 2008). This observation also helps explain the observed pre-clinical efficacy of irreversible EGFR kinase inhibitors in cell line models harboring EGFR T790M(Engelman et al., 2007a; Kobayashi et al., 2005; Kwak et al., 2005). Irreversible EGFR kinase inhibitors are based on the same structural scaffold (4-anilinoquinazoline) as gefitinib and erlotinib but in addition contain an electrophilic motif that covalently binds Cys-797 of EGFR. The covalent binding allows irreversible EGFR inhibitors to achieve greater occupancy of the ATP-site relative to the reversible inhibitors providing the ability to inhibit EGFR T790M despite the increased ATP affinity conferred by the T790M mutation(Yun et al., 2008).
Several irreversible EGFR inhibitors, including HKI-272, BIBW2992 and PF00299804, have been evaluated in pre-clinical models and are currently under clinical evaluation(Engelman et al., 2007a; Kwak et al., 2005; Li et al., 2008). In a prior study, carcinogen exposed gefitinib sensitive PC9 cells were exposed to HKI-272 and EGFR T790M emerged as a resistance mechanism(Godin-Heymann et al., 2007). This parallels the clinical experience with HKI-272 where very little anti-tumor activity was observed both in gefitinib naïve and gefitinib resistant EGFR mutant NSCLC patients(Besse et al., 2008). In contrast, PF00299804 is effective in pre-clinical models harboring EGFR T790M and, unlike with HKI-272, tumor responses have already been observed in the phase I clinical trial in gefitinib/erlotinib resistant patients(Engelman et al., 2007a; Janne et al., 2008). However, very little is known about how lung cancers with established EGFR T790M develop resistance to irreversible EGFR inhibitors including PF00299804. In the current study we model resistance to PF00299804, using cell line models with EGFR T790M, and demonstrate that one mechanism of resistance to PF00299804 is by amplification of EGFR T790M.
Results
Gefitinib resistant PC9 cells contain EGFR T790M
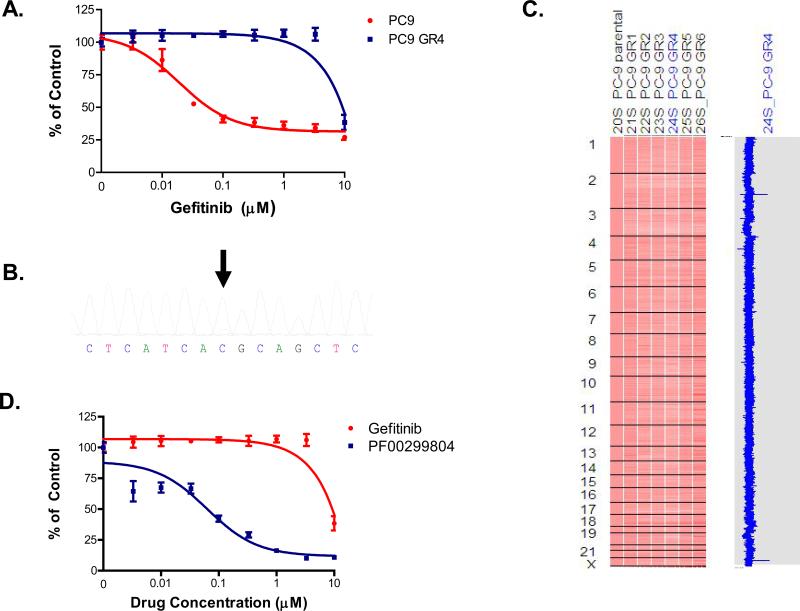
We first generated in vitro resistant clones of PC9 (EGFR delE746_A750) cells to gefitinib using previously described methods(Engelman et al., 2006; Engelman et al., 2007b). The PC9 cells were exposed to increasing concentrations of gefitinib starting at 10 nM, until they were able to freely proliferate in 1 μM gefitinib which occurred after 6 months of drug selection. This concentration was chosen because it is ~20 fold greater than the IC50 (50 nM) for growth inhibition and is the achievable plasma concentration in patients receiving gefitinib(Albanell et al., 2002). Six independent gefitinib resistant (GR) PC9 clones were isolated (Figure 1A).
Figure 1.
Gefitinib resistant PC9 cells contain EGFRT790M and are growth inhibited by PF00299804. A. Parental and resistant PC9GR4 cells were treated with gefitinib at the indicated concentrations, and viable cells were measured after 72 hours of treatment and plotted relative to untreated controls. B. Sequence tracing of EGFR exon 20 from PC9GR4 cells demonstrates a small peak (arrow) containing the mutant T allele (wild type ACG (T); mutant ATG (M)). C. The PC9GR clones do not contain other focal genomic changes. The PC9GR clones (right) were compared with the PC9 clones (1st column). The blue curve on the right indicates degree of amplification of each SNP from 0 (left) to 8 (right). D. PF00299804 inhibits the growth of PC9GR4 cells. The PC9GR4 cells were treated with increasing concentrations of gefitinib or PF00299804 and viable cells quantified using MTS assay.
To characterize the PC9GR clones we sequenced the EGFR kinase domain. All six clones contained the EGFR T790M (C to T) mutation as detected by direct sequencing (Figure 1B). We quantified the allele frequency of EGFR T790M using mass spectrometry and determined it to be 17-18% in the PC9GR clones (Figure S1). The irreversible EGFR inhibitor, PF00299804, which is effective in Ba/F3 cell line models harboring EGFR T790M(Engelman et al., 2007a), inhibited the growth of the PC9GR4 cells (Figure 1D). Similar results were obtained with the other PC9GR clones (data not shown). There were no significant genome-wide copy number changes in the PC9GR clones compared to the parental PC9 cells (Figure 1C)(Engelman et al., 2007b). Collectively our findings suggest that the resistant PC9GR cells develop EGFR T790M that accounts for resistance to gefitinib.
PF00299804 resistant PC9GR cells have enhanced baseline EGFR phosphorylation
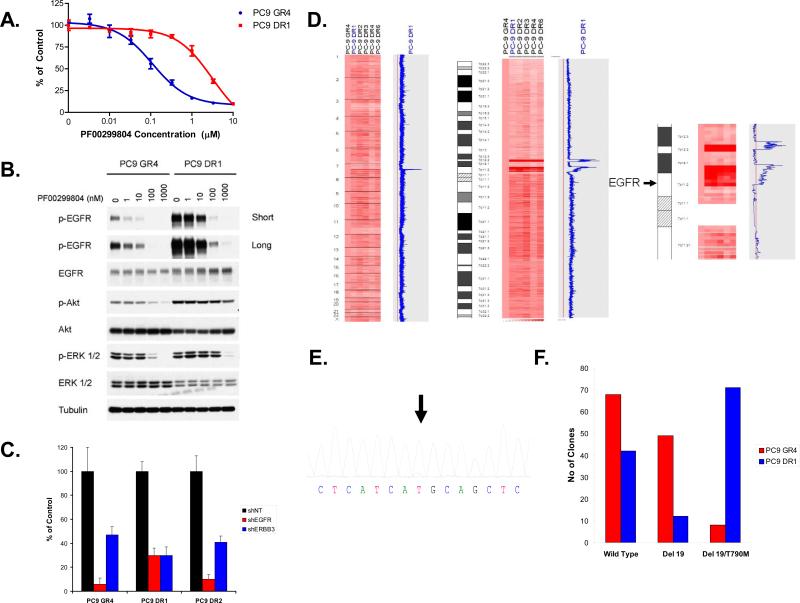
PF00299804 is currently undergoing clinical development in NSCLC patients that have developed acquired resistance to gefitinib or erlotinib. As cancers from many of these patients will contain EGFR T790M it will be important to understand how such cancers develop drug resistance to irreversible EGFR inhibitors including PF00299804. We thus modeled acquired resistance in vitro to PF00299804 using the gefitinib resistant EGFR T790M containing PC9GR4 (del E746_A750/T790M) cells. We exposed the PC9GR4 cells to increasing concentrations of PF00299804 until they were able to proliferate in 1 μM PF00299804 which occurred after only 1 month of drug selection. This concentration is ~ 10 fold greater than the IC50 (100 nM) for growth inhibition of PC9GR4 cells and 5 times greater than the mean steady state concentration of PF00299804 observed in NSCLC patients in the phase I clinical trial (Janne et al., 2008; Schellens et al., 2007). Five independent gefitinib/PF00299804 double resistant (DR; PC9GR4 (delE746_A750/T790M) cells made resistant to PF00299804) clones were isolated (Figure 2A).
Figure 2.
Generation and characterization of PC9DR cells. A. PC9GR4 and PC9DR cells were treated with PF00299804 at the indicated concentrations, and viable cells were measured after 72 hours of treatment. B. The PC9DR1 cells have higher baseline EGFR phosphorylation than the PC9GR4 cells and require greater concentrations of PF00299804 than PC9GR4 cells to inhibit EGFR, Akt and ERK1/2 phosphorylation. C. The PC9DR cells are EGFR dependent for growth. Control (NT; non-targeting), EGFR or ERBB3-specific shRNAs were introduced into parental or resistant cells and cell viability was measured using an MTS assay 6 days later. Viability is shown relative to cells expressing the control shRNA. Error bars indicate standard deviation. D. The PC9DR cells contain a focal amplification on chromosome 7 encompassing the EGFR locus. The PC9DR clones (right) were compared with the PC9GR4 cells (first column). The blue curve on the right indicates degree of amplification of each SNP from 0 (left) to 8 (right). Left, genome wide view; Center, chromosome 7 view; Right, detailed view of the pericentromeric region of chromosome 7. The genomic location of EGFR is indicated by the arrow. E. Sequence tracing of EGFR exon 20 from PC9DR1 cells demonstrates that the mutant allele (arrow) is the predominant allele. F. EGFR T790M is amplified in cis with delE746_A750 in PC9DR1 cells. RNA isolated from PC9GR4 or DR1 cells was subjected to RT-PCR, the resulting products cloned and the inserts sequenced.
We compared the effects of PF00299804 on phosphorylation of EGFR, Akt and ERK1/2 in the PC9GR4 to the PC9DR1 cells. Unlike the untreated PC9GR4 cells, the untreated PC9DR1 cells exhibited a much greater level of EGFR phosphorylation with only a very modest differences in the levels of total EGFR (Figure 2B). In the PC9GR4 cells EGFR phosphorylation was inhibited by lower (starting at 1 nM) concentrations of PF00299804 while in the PC9DR1 cells complete inhibition not observed until 1 μM of PF00299804 (Figure 2B). In both cell lines, inhibition of EGFR phosphorylation was accompanied by a concomitant inhibition of Akt and ERK1/2 phosphorylation. To determine whether the PC9DR cells were still dependent on EGFR signaling for their growth we downregulated EGFR and ERBB3 by short hairpin (sh)RNAs. We evaluated shRNAs against ERBB3 because EGFR mutant NSCLCs utilize ERBB3 to activate PI3K/Akt signaling(Engelman et al., 2005). The growth of all three cell lines (PC9GR4, PC9DR1 and PC9DR2 cells) was significantly inhibited by shRNAs targeting EGFR and ERBB3 but not by a control (non-targeting (NT)) shRNA (Figures 2C and S2). There was no effect of either shRNA against A549 cell line which is not EGFR dependent for its growth and viability (data not shown). Collectively, these findings suggest that the PC9DR cells remain EGFR dependent for their growth and survival.
PC9DR cells have a focal amplification in the EGFR T790M containing allele
In order to identify genomic basis for the differences in the PC9DR1 cells we compared genome wide copy number changes between the PC9DR and GR4 cells using SNP arrays(Engelman et al., 2007b). The PC9DR cells contained only 1 region of copy number gain, located on the short arm of chromosome 7 (7p11.2 to 7p12.2), encompassing EGFR, that was significantly different from the PC9GR4 cells (Figure 2D). We confirmed a 3-fold amplification in EGFR using quantitative PCR (QPCR) (Figure S3). However, a 3-fold increase in EGFR does not explain the more dramatic differences in EGFR phosphorylation in the PC9DR1 cells (Figure 2B). Prior studies have suggested that EGFR T790M, when in cis with an EGFR activating mutation, leads to an increase in EGFR kinase activity (Godin-Heymann et al., 2007; Vikis et al., 2007). We thus hypothesized that the amplification in the PC9DR cells preferentially affects the EGFR T790M containing allele. We sequenced EGFR in the PC9DR and noted a dramatic change in the T790M containing allele (Figure 2E) compared to the parental PC9GR4 cells (Figure 1B and S1). In addition, we used RT-PCR followed by cloning and evaluated 125 clones from PC9GR4 and PC9DR1 cells (Figure 2F). In the PC9DR1 cells, there was a 7 fold increase in the number of double-mutant clones (Figure 2F). All T790M containing clones were in cis with the exon 19 deletion mutation. Accordingly, we observed a 5 fold reduction in clones that contained the exon 19 deletion mutation alone compared to the PC9GR4 cells. Our findings suggest that the PC9DR cells contain an EGFR amplification which preferentially affects the EGFR T790M containing allele.
PC9DR cells contain amplified EGFR in both intra and extra-chromosomal regions
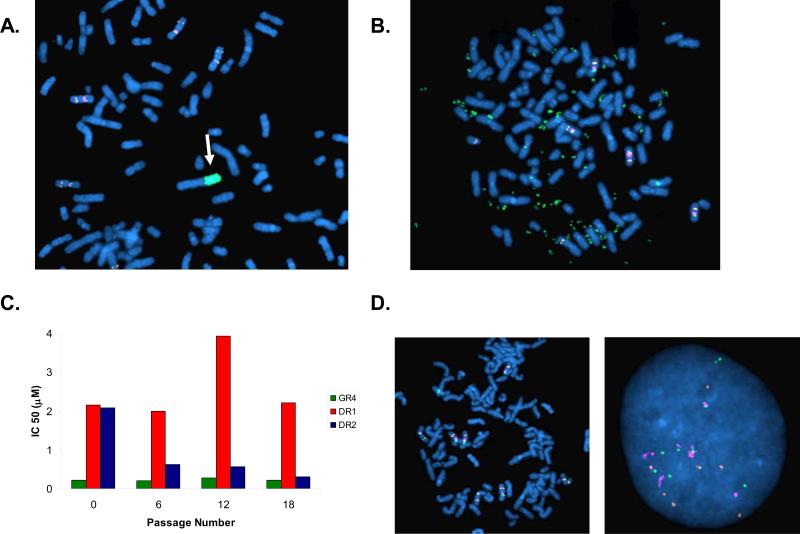
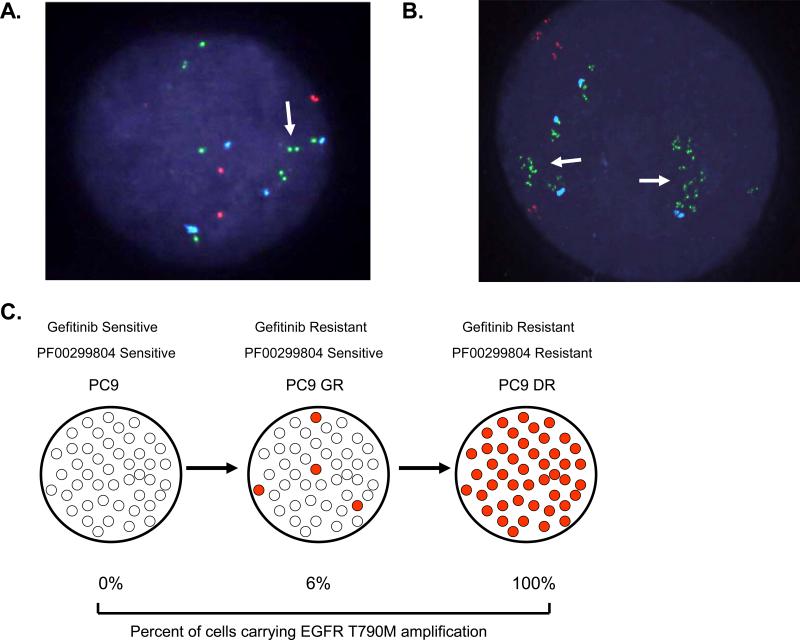
We used FISH to evaluate the EGFR locus in the PC9DR1 and DR2 cells. In the PC9DR1 cells, 13/74(18%) metaphases contained an intra-chromosomal amplification in EGFR (Figure 3A), while in the remaining 55/75(82%), EGFR was in extra chromosomal double-minutes (DMs) (Figure 3B). In contrast, in the PC9DR2 cells, all of the amplified EGFR (50/50 metaphases) were contained in DMs (Figure 3B), also evident using interphase FISH (Figures S4A and S4B). Amplification of the dihydrofolate reductase gene in DMs has previously been reported as a methotrexate resistance mechanism(Schimke et al., 1978). As DMs lack a centromere, they can be gradually lost from cells in the absence of a selection pressure(Kaufman & Schimke, 1981). We thus evaluated the change in IC50 to PF00299804 in the PC9DR1 and DR2 cells in the absence of PF00299804. While the IC50 in the PC9DR2 cells began to decline within 6 passages and was similar to the PC9GR4 cells by 18 passages, there was no significant change in the PC9DR1 cells (Figure 3C). FISH analysis of the PC9DR2 cells after 18 passages demonstrated a substantial decrease in the number of cells that contained DMs (9/50; 18%) including many cells without any evidence of EGFR amplification using FISH (Figure 3D). The PC9DR1 cells retained cells harboring an intrachromosomal amplification in EGFR (data not shown).
Figure 3.
EGFR amplification is detected in both intrachromosomal and extra-chromosomal locations in the PC9DR cells. A. Metaphase FISH analysis using PC9DR1 cells. A subset (18%) of cells contain an intrachromosomal focal amplification of EGFR (arrow). B. Metaphase FISH using PC9DR2 cells. The amplification of EGFR is exclusively located on double minutes. EGFR (CTD-2257H21; green); MET (RP-11-95I120; orange); CEP 7 (aqua). C. The PC9DR2 cells loose their resistance in the absence of PF00299804. PC9GR4, DR1 and DR2 cells were cultured in the absence of PF00299804 for 18 passages. The IC50 for PF00299804 was determined by MTS assay every 6 passages. D. Metaphase (left) and interphase (right) FISH analyses from PC9DR2 cells after 18 passages demonstrates loss of EGFR-containing double minutes.
PF00299804 resistant H3255 GR(L858R/T790M) cells develop an amplification of EGFR T790M
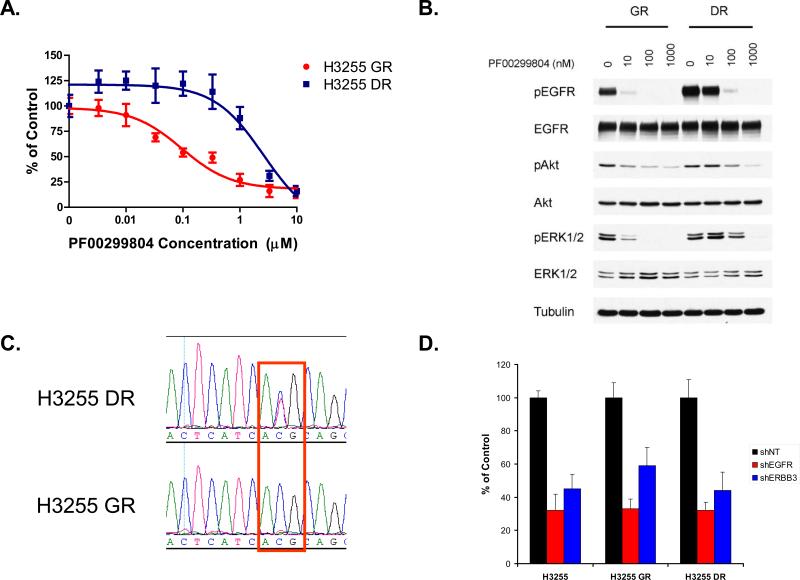
In order to determine whether EGFR T790M amplification was a unique feature to the PC9 cells we used another gefitinib resistant cell line, H3255GR(Engelman et al., 2006). The H3255GR cells contain T790M in cis with L858R but only in a minority (~3%) of the amplified alleles and thus difficult to detect by conventional Sanger sequencing methods(Engelman et al., 2006). We generated the H3255DR (H3255GR cells resistant to PF00299804) cells in an analogous manner to the PC9DR cells. The IC50 of PF00299804 in H3255DR cells was 3 μM (Figure 4A) which is greater than 10 times the mean plasma concentration (~ 200 nM) achieved in patients(Janne et al., 2008; Schellens et al., 2007). Analogous to the PC9DR cells, the untreated H3255DR cells exhibited increased EGFR phosphorylation compared to the untreated H3255GR cells (Figure 4B). PF00299804 was still able to inhibit phosphorylation of EGFR, Akt and ERK1/2 but only at higher concentrations similar to the PC9DR cells (Figure 4B). EGFR sequencing of H3255DR cells confirmed a now clearly visible EGFR T790M (Figure 4C) which is undetectable in the H3255GR cells. Quantitative mass spectrometry (Figure S1) demonstrated that EGFR T790M composed 45% (compared to 3% in H3255GR) of alleles. The H3255DR cells were still dependent on EGFR signaling for their growth as shRNA against EGFR and ERBB3 resulted in similar degree of growth inhibition as in the H3255 or H3255GR cells (Figures 4D and S5). FISH analyses of H3255DR cells demonstrated EGFR to be localized solely on chromosome 7 without evidence of double minutes (data not shown). Collectively, our findings suggest that two different gefitinib resistant cell line models (PC9GR and H3255GR) harboring EGFR T790M develop resistance to PF00299804 through amplification of EGFR T790M.
Figure 4.
The H3255DR cells contain an amplification in EGFR T790M. A. The H3255GR (L858R/T790M) cells were made resistant to PF00299804 by growing them in increasing concentrations of PF00299804. Parental and resistant H3255DR cells were treated with PF00299804 at the indicated concentrations, and viable cells were measured after 72 hours of treatment. B. The H3255DR cells have higher baseline EGFR phosphorylation than the H3255GR cells. C. Sequence tracing of EGFR exon 20 from H3255DR cells demonstrates that the mutant allele (T) is the predominant allele (wild type ACG (T); mutant ATG (M)). D. The H3255DR cells are EGFR dependent for growth. Control (NT; non-targeting), EGFR or ERBB3-specific shRNAs were introduced into H3255, H3255GR and H3255DR cells and cell viability was measured using an MTS assay 6 days later. Viability of cells is shown relative to cells expressing the control shRNA. Error bars indicate standard deviation.
Mechanism of EGFR T790M amplification
We next investigated the mechanism of EGFR T790M amplification. We noted that there was a substantial difference in the time required for development of gefitinib resistance in the PC9 cells (6 months) compared to subsequent PF000299804 resistance in the PC9GR cells (1 month). One possibility for these differences is that the PC9GR cells already contain a subpopulation of cells harboring an amplification of EGFR that is rapidly selected for by PF00298804. In order to determine this possibility, we used a high throughput FISH assay and scanned 2090 PC9GR4 cells for EGFR amplification. Although the vast majority (94%) contained no evidence of EGFR amplification (Figure 5A), we detected 125(6%) PC9GR4 cells harboring EGFR amplification (Figures 5B and 5C). On interphase FISH these cells are similar to the PC9DR2 cells which contain an EGFR amplification in 100% of the cells (Figure S4B). The findings of a large subpopulation of EGFR amplified cells, already present in the PC9GR4 cells, suggests that PF00299804 resistance develops as a result of selection of a pre-existing EGFR T790M amplified clone (Figure 5C).
Figure 5.
EGFR amplification pre-exists in a subset of PC9GR4 cells. High-throughput FISH analysis of PC9GR4 cells demonstrates that the majority of cells contain no EGFR amplification (A) while a subpopulation (6%) harbor clustered (arrow) EGFR amplification (B) similar in appearance to the PC9DR2 cells (see Figure S4B). EGFR (CTD-2257H21; green); MET (RP-11-95I120; orange); CEP 7 (aqua).
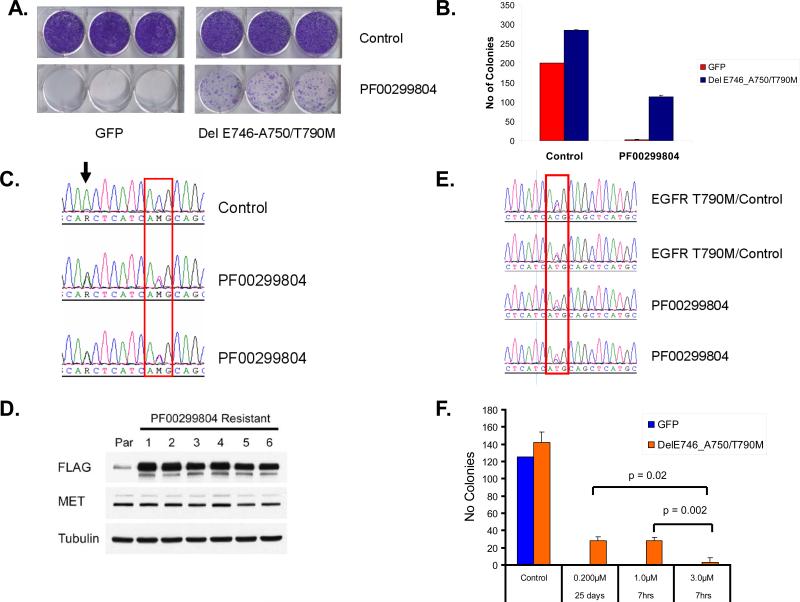
In order to further study the kinetics of drug resistance and to track the potential selection of EGFR T790M we introduced a C-terminal FLAG tagged EGFRdelE746_A750/T790M into the EGFR mutant NSCLC cell line HCC827. The HCC827delE746_A750/T790M cells are resistant to gefitinib but retain sensitivity to PF00299804 in a 3-day cell viability assay(Engelman et al., 2007a). We exposed HCC827GFP or HCC827delE746_A750/T790M cells to PF00299804 (100 nM) for 15 days and evaluated for resistant colonies (Figure 6A and 6B). There were no resistant colonies in the HCC827GFP cells while they were detected from HCC827delE746_A750/T790M cells (Figure 6A and 6B). We isolated 6 independent PF00299804 resistant colonies from HCC827delE746_A750/T790M cells followed by RT-PCR and EGFR sequencing (Figure 6C). In the resistant clones, the EGFR T790M allele is substantially increased compared to the untreated cells. Similarly, these clones express significantly greater amount of FLAG protein consistent with the increased expression of EGFR T790M (Figure 6D). Notably, these PF00299804 resistant cells do not express increased amount of MET protein (Figure 6D) which has previously been described as a resistance mechanism in the HCC827 cells but occurs only after several months of drug exposure(Engelman et al., 2007b).
Figure 6.
Selection for EGFR T790M expressing clones following exposure to PF00299804. A. HCC827 cells were transfected with either GFP or EGFR delE746_A750/T790M and the resulting cells were either untreated or exposed to 100 nM PF00299804. The cells were stained with crystal violet after 15 days of drug exposure. B. Quantification of colonies from A. Error bars indicate standard deviation. C. Sequence tracing of EGFR exon20 from untreated (control) or PF00299804 treated HCC827delE746_A750/T790M cells. The PF00299804 treated cells have a substantial increase in the mutant (T) allele. The delE746_A750/T790M transgene contains the G>A EGFR SNP (arrow) in cis and is concurrently amplified. D. The PF00299804 resistant HCC827delE746_A750/T790M cells express greater amount of EGFR T790M protein. The delE746_A750/T790M expression construct contains FLAG at the C-terminus. Cell extracts were immunoblotted to detect the indicated proteins. E. Sequence tracing of EGFR exon 20 from untreated (control) or PF00299804 treated xenografts. The PF00299804 treated tumors have an increase in the mutant (T) allele compared to the untreated tumors. F. Quantification of colonies using cells from A following continuous low dose (200 nM) or pulse (7hr) treatment with either 1 μM or 3 μM PF00299804. Error bars indicate standard deviation.
We next determined whether an increase in EGFR T790M could also emerge as a resistance mechanism to PF00299804 in vivo. We studied xenografts generated from HCC827delE746_A750/T790M that were either treated with vehicle (n=6) or PF00299804 (n=6) from two independent experiments (Figures S6A and S6B) including (n=4) from our prior study(Engelman et al., 2007a). We evaluated the tumors at the time of sacrifice from both groups, performed RT-PCR followed by EGFR sequencing. In 6/6 (100%) of the PF00299804 treated mice, the T allele (ATG; M), was more prevalent compared to the vehicle alone treated mice (Figures 6E and S6C). Collectively, these findings suggest that amplification of EGFR T790M as a mechanism of resistance to PF00299804 develops in cell line models that express EGFR T790M, either in vitro or in vivo, is a result of selection for either an EGFR T790M amplified or high expressing subclone.
Transient exposure to higher concentrations of PF00299804 is effective against EGFR T790M expressing cells
The current continuous dosing of PF00299804 leads to a plasma concentration of 200 nM in NSCLC patients which is insufficient to inhibit the growth of the PC9DR or HCC827DelE746_A750/T790M cells (Figures 2A and 6A) (Janne et al., 2008; Schellens et al., 2007). As these resistant cells are still EGFR dependent, we investigated whether transient exposure to higher doses of PF00299804 may be effective analogous to recent studies of dasatinib in chronic myeloid leukemia(Shah et al., 2008). Compared to continuous treatment with 200 nM, transient (7 hours followed by replacement with drug free media) treatment with 3 μM (the IC50 in PC9DR and H3255DR) resulted in significantly (p = 0.02; t-test) fewer resistant colonies (Figure 6F). This was not observed with 1 μM PF00299804 which is below the IC50 in PC9DR and H3255DR cells (Figures 2A, 4A and 6F).
Discussion
The development of acquired drug resistance to kinase inhibitors limits their effectiveness. Secondary mutations at the gatekeeper residue are among the most common mechanisms of acquired resistance to kinase inhibitors(Bradeen et al., 2006; Cools et al., 2003; Gorre et al., 2001; Shah et al., 2002; Tamborini et al., 2004). Several therapeutic strategies are currently being evaluated in pre-clinical models and in clinical trials to overcome this mechanism of drug resistance (Carter et al., 2005; Giles et al., 2007; Guo et al., 2007; Quintas-Cardama & Cortes, 2008). In lung cancer, EGFR T790M is the most common mechanism of acquired resistance to gefitinib and erlotinib and typically emerges within 9-13 months after initiating therapy(Inoue et al., 2006; Mok et al., 2008; Sequist et al., 2008). However, very little is known about how cancers that developed a gatekeeper mutation after treatment with gefitinib or erlotinib develop acquired resistance to second generation kinase inhibitors. In this study we used EGFR T790M lung cancer as a model to determine how resistance develops against agents aimed specifically at targeting the gatekeeper resistance mutation. Our findings suggest that although irreversible EGFR inhibitors may be transiently effective against cancers harboring EGFR T790M (Figure 1D), clones harboring amplified EGFR T790M will rapidly emerge in vitro and in vivo through selection of pre-existing EGFR T790M amplified or high expressing clones (Figures 5 and 6) leading to clinical drug resistance. These observations provide mechanistic insight into the origins drug resistance to EGFR targeted therapies. Amplification of the gatekeeper mutation containing allele is a unique mechanism of drug resistance and has not previously been described for other kinase inhibitors targeting drug resistant forms of mutant oncogenes. Furthermore, findings from this pre-clinical study can be used to guide future studies of patient tumor specimens, for evidence of EGFR T790M amplification, from ongoing clinical trials of irreversible EGFR kinase inhibitors.
Previous studies have demonstrated that EGFR T790M also emerges as a resistance mechanism to HKI-272 (used at clinically achievable concentrations; 200 nM) in carcinogen treated PC9 cells (Godin-Heymann et al., 2007). However, this concentration of HKI-272 do not effectively inhibit DelE746_A750/T790M (Yuza et al., 2007). We also find that HKI is not effective against the PC9GR4 cells (IC50 ~850 nM, data not shown). In contrast to the previous study, we modeled resistance to PF00299804 using the PC9GR4 cells that had already acquired a T790M in response to gefitinib. Unlike HK-272, PF00299804 effectively inhibits the growth of these cells (IC50 100 nM; Figure 1D). Similarly, there have been tumor responses in gefitinib/erlotinib resistant lung cancer patients in the phase I clinical trial of PF00299804 but no responses were observed in the corresponding trial with HKI-272(Janne et al., 2008; Wong et al., 2009). These differences between HKI-272 and PF00299804 are likely due to the lower potency of HKI-272 against the DelE746_A750/T790M mutant compared to PF00299804(Engelman et al., 2007a; Yuza et al., 2007). Importantly, we found that cancer cells became resistant to the more potent PF00299804 by selecting out a preexisting subset of cells with amplification of the T790M containing allele.
There are several potential strategies that emerge from these preclinical studies that could be further clinically tested. Although the clinical effects of irreversible EGFR TKIs may be transient, our study provides evidence that T790M amplification can revert (PC9DR2 cells; Figure 3C) in the absence of the selection pressure of the drug. Hence instead of continuous treatment, scheduled “drug holidays” or intermittent therapy may allow for prolonged cancer control by eliminating the continuous selection pressure and rapid emergence of drug resistant cancers. In addition, given the low achievable plasma concentrations using continuous dosing, intermittent administration at higher doses may transiently, given the irreversible nature of the agents, achieve plasma levels necessary to effectively inhibit amplified EGFR T790M. Even a transient but complete inhibition of EGFR phosphorylation may be sufficient to commit the cells to apoptosis and is supported by our pre-clinical studies (Figure 6F) and by recent studies using dasatnib in CML models(Shah et al., 2008). An alternative approach is to develop novel strategies, instead of irreversible EGFR inhibitors, to inhibit EGFR and/or EGFR signaling, including the use of inhibitors of HSP90 and/or EGFR downstream signaling (such as PI3K and MEK inhibitors) as these resistant cancers still remain dependent on EGFR signaling for their growth (Sawai et al., 2008; Shimamura et al., 2008). Finally, the prevention of the emergence of resistance may be a more effective clinical strategy than treating actual resistance. EGFR T790M can sometimes be detected in treatment naïve NSCLC tumors and its presence is associated with a shortened outcome following gefitinib treatment(Maheswaran et al., 2008). Thus treatment with an irreversible EGFR inhibitor instead of gefitinib or erlotinib may prevent the emergence of EGFR T790M and lead to a prolonged time to disease progression compared to currently clinically achievable (9-11 months) with gefitinib or erlotinib. In fact we have generated PF00299804 resistant PC9 cells and these resistant cells do not harbor EGFR T790M (data not shown) suggesting that this may also be a clinically effective strategy. Clinical studies of PF00299804 and BIBW2992 in gefitinib/erlotinib naïve EGFR mutant NSCLC patients are currently underway and it will be critical to determine whether EGFR T790M will also emerge as a mechanism of drug resistance in these patients.
Methods
Cell Culture and reagents
The EGFR mutant NSCLC cell lines PC9 (delE746_A750), H3255 (L858R) and H3255GR (L858R/T790M) have been previously characterized(Engelman et al., 2006; Ono et al., 2004; Paez et al., 2004).
PF00299804 was obtained from Pfizer. Gefitinib was obtained from commercial sources. Stock solutions of all drugs were prepared in DMSO and stored at -20°C.
Cell proliferation and growth assays
Growth and inhibition of growth was assessed by MTS assay according to previously established methods(Engelman et al., 2006; Mukohara et al., 2005; Paez et al., 2004). All experimental points were set up in six to twelve wells and all experiments were repeated at least three times. The data was graphically displayed using GraphPad Prism version 5.0 for Windows, (GraphPad Software; www.graphpad.com).
Antibodies and Western Blotting
Cells grown under the previously specified conditions were lysed in NP-40 buffer. Western blot analyses were conducted after separation by SDS/PAGE electrophoresis and transfer to nitrocellulose membranes. Immunoblotting was performed according to the antibody manufacturers’ recommendations. Anti-phospho-Akt (Ser-473), anti-total-Akt, and anti-EGFR antibodies were obtained from Cell Signaling Technology. The phosphor-EGFR(pY1068), total-ERK1/2, phospho-ERK1/2 (pT185/pY187) antibodies were purchased from Biosource International Inc. The total-Met (C-28) antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Generation of drug resistant cell lines
To generate drug resistant cell lines, NSCLC cells were exposed to increasing concentrations of either gefitinib or PF00299804 similar to previously described methods (Engelman et al., 2006; Engelman et al., 2007b). Individual clones from gefitinib resistant (GR) or gefitinib/PF00299804 double resistant (DR) cells were isolated and confirmed to be drug resistant. The H3255 gefitinib/PF00299804 double resistant (DR) cells were maintained as a pool as these cells did not grow as single colonies.
EGFR mutational analyses
Total RNA was isolated from cell lines or tumors using Trizol™ (Invitrogen, Carlsbad, CA) and purified using RNeasy™ minielute cleanup kit (Qiagen,Valencia, CA). cDNA was transcribed with Superscript II Reverse Transcriptase (Invitrogen Life technologies, Carlsbad, CA) and used as template for subsequent PCR based studies (Engelman et al., 2006; Engelman et al., 2007b). The PCR products were also cloned into a TOPO TA vector (Invitrogen, Carlsbad, CA), transformed into bacteria and the inserts from individual clones sequenced. The PCR primers and conditions are available upon request.
SNP analyses
SNP analyses were performed as previously described(Engelman et al., 2007b). Samples were processed for the Human Mapping 250K Sty single nucleotide polymorphism (SNP) array according to the manufacturer's instructions. Comparison of gene copy number differences was performed using the dChip software according to previously established methods(Engelman et al., 2007b; Zhao et al., 2005).
EGFR copy number analysis
The relative copy number for EGFR was determined using quantitative real time PCR as previously described (Engelman et al., 2007b). Quantification was based on standard curves from a serial dilution of normal human genomic DNA. All specimens were analyzed in triplicate. The PCR primers are available upon request.
FISH probes and hybridization
Bacterial artificial chromosome (BAC) clones CTD-2257H21 (EGFR (7p11.2 )) and RP11-95I20 (MET (7q31.2)) were purchased from Children's Hospital Oakland Research Institute (CHORI; Oakland, CA). DNA was extracted using a Qiagen kit (Qiagen Inc., Valencia CA) and labeled with Spectrum Green- or Spectrum Orange-conjugated dUTP by nick translation (Vysis/Abbott Molecular, Des Plaines, IL). The CEP7 probe (Vysis/Abbott Molecular, Des Plaines, Il) was used according to the manufacturer's instructions. Chromosomal mapping and hybridization efficiency for each probe set were verified in normal metaphase spreads (data not shown). Three color FISH assays were performed as previously described(Engelman et al., 2007b).
High throughput fluorescence in situ hybridization
A Bioview work station with Duet™ software (Bioview Ltd, Rehovot, Israel) was used to screen for evidence of EGFR amplification in the PC9 GR cells. Automatic scans were performed according to manufacturer's suggested guidelines after setting classification criteria for each FISH probe. Images were captured and classified in an automated fashion and manually reviewed to ensure accuracy. Cells that could not be scored were excluded from the analysis.
EGFR and ERBB3 shRNA constructs and lentiviral infection
EGFR and ERBB3 shRNA constructs cloned in pLKO.1 puro vector were described in (Engelman et al., 2005; Engelman et al., 2006). A vector containing a non-targeting (NT) shRNA was used as a control. Lentivirus production, titrations and infections were performed as in(Engelman et al., 2006; Rothenberg et al., 2008).
Mass spectrometric detection of EGFR T790M
PCR amplification, primer extension and mass spectrometry was carried out according to the manufacturer's recommended conditions (Sequenom, San Diego, CA). The PCR primers and conditions are available upon request. Allele frequencies of each sample in triplicate were generated using the Allelotyping method of Sequenom Spectro Aquire software. The data was curated to remove low intensity data, then the average allele frequency was calculated. Mutant allele frequencies were called as positive if the allele frequency was over two standard deviations of the background. The spectra of all positive mutant calls were reviewed to confirm the call.
Xenograft studies
The xenograft studies were performed using the HCC827DelE746_A750/T790M cells are previously described(Engelman et al., 2007a). PF00299804 was administered at 10 mg/kg/day by daily oral gavage. The experiment was terminated when the mean size of the control tumors reached 2000 mm3. The studies were performed in accordance with the standards of the Institutional Animal Care and Use Committee (IACUC) under a protocol approved by the Animal Care and Use Committee of the Beth Israel Deaconess Medical Center.
Supplementary Material
Acknowledgements
Supported by grants from the National Institutes of Health RO1CA114465-04 (P.A.J.), R01CA135257-01 (P.A.J., J.A.E., C.L.), R01CA137008-01 (J.A.E., P.A.J.), National Cancer Institute Lung SPORE P50CA090578 (P.A.J., J.A.E. and D.J.K.), American Cancer Society RSG0610201CCE (P.A.J., J.A.E.), and the Hazel and Samuel Bellin research fund (P.A.J.).
Footnotes
Conflict of Interest
Dr. Jänne receives royalties as a co-inventor on a patent awarded for the discovery of EGFR mutations, licensed to Genzyme Genetics, which was not involved in this study. Dr. Christensen is an employee of Pfizer.
References
- Albanell J, Rojo F, Averbuch S, Feyereislova A, Mascaro JM, Herbst R, et al. Pharmacodynamic studies of the epidermal growth factor receptor inhibitor ZD1839 in skin from cancer patients: histopathologic and molecular consequences of receptor inhibition. J Clin Oncol. 2002;20:110–24. doi: 10.1200/JCO.2002.20.1.110. [DOI] [PubMed] [Google Scholar]
- Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- Besse B, Eaton Kd, Soira JC, Lynch TJ, Miller V, Wong KK, et al. Neratinib (HKI-272), an irreversible pan-ErbB receptor tyrosine kinase inhibtor: preliminary results of a phase 2 tiral in patients with advanced non-small cell lung cancer. European Journal of Cancer. 2008;6:64. abstract 203. [Google Scholar]
- Bradeen HA, Eide CA, O'Hare T, Johnson KJ, Willis SG, Lee FY, et al. Comparison of imatinib mesylate, dasatinib (BMS-354825), and nilotinib (AMN107) in an N-ethyl-N-nitrosourea (ENU)-based mutagenesis screen: high efficacy of drug combinations. Blood. 2006;108:2332–8. doi: 10.1182/blood-2006-02-004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter TA, Wodicka LM, Shah NP, Velasco AM, Fabian MA, Treiber DK, et al. Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc Natl Acad Sci U S A. 2005 doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J, et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med. 2003;348:1201–14. doi: 10.1056/NEJMoa025217. [DOI] [PubMed] [Google Scholar]
- Demetri GD, van Oosterom AT, Garrett CR, Blackstein ME, Shah MH, Verweij J, et al. Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: a randomised controlled trial. Lancet. 2006;368:1329–38. doi: 10.1016/S0140-6736(06)69446-4. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Sawyers CL, Kantarjian H, Resta DJ, Reese SF, Ford JM, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344:1038–42. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Janne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, et al. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc Natl Acad Sci U S A. 2005;102:3788–93. doi: 10.1073/pnas.0409773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Mukohara T, Zejnullahu K, Lifshits E, Borras AM, Gale CM, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Gale CM, Lifshits E, Gonzales AJ, Shimamura T, et al. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res. 2007a;67:11924–32. doi: 10.1158/0008-5472.CAN-07-1885. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007b;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Giles FJ, Cortes J, Jones D, Bergstrom D, Kantarjian H, Freedman SJ. MK-0457, a novel kinase inhibitor, is active in patients with chronic myeloid leukemia or acute lymphocytic leukemia with the T315I BCR-ABL mutation. Blood. 2007;109:500–2. doi: 10.1182/blood-2006-05-025049. [DOI] [PubMed] [Google Scholar]
- Godin-Heymann N, Bryant I, Rivera MN, Ulkus L, Bell DW, Riese DJ, 2nd, et al. Oncogenic activity of epidermal growth factor receptor kinase mutant alleles is enhanced by the T790M drug resistance mutation. Cancer Res. 2007;67:7319–26. doi: 10.1158/0008-5472.CAN-06-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–80. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- Guo T, Agaram NP, Wong GC, Hom G, D'Adamo D, Maki RG, et al. Sorafenib inhibits the imatinib-resistant KITT670I gatekeeper mutation in gastrointestinal stromal tumor. Clin Cancer Res. 2007;13:4874–81. doi: 10.1158/1078-0432.CCR-07-0484. [DOI] [PubMed] [Google Scholar]
- Inoue A, Suzuki T, Fukuhara T, Maemondo M, Kimura Y, Morikawa N, et al. Prospective phase II study of gefitinib for chemotherapy-naive patients with advanced non-small-cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol. 2006;24:3340–6. doi: 10.1200/JCO.2005.05.4692. [DOI] [PubMed] [Google Scholar]
- Janne PA, Schellens JH, Engelman JA, Eckhardt SG, Milham R, Denis LJ, et al. Preliminary activity and safety results from a phase I clinical trial of PF-00299804, an irreversible pan-HER inhibitor, in patients (pts) with NSCLC. Journal of Clinical Oncology. 2008;26 Abstract 8027. [Google Scholar]
- Kantarjian H, Sawyers C, Hochhaus A, Guilhot F, Schiffer C, Gambacorti-Passerini C, et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med. 2002;346:645–52. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ, Schimke RT. Amplification and loss of dihydrofolate reductase genes in a Chinese hamster ovary cell line. Mol Cell Biol. 1981;1:1069–76. doi: 10.1128/mcb.1.12.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- Kosaka T, Yatabe Y, Endoh H, Yoshida K, Hida T, Tsuboi M, et al. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin Cancer Res. 2006;12:5764–9. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- Kwak EL, Sordella R, Bell DW, Godin-Heymann N, Okimoto RA, Brannigan BW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci U S A. 2005;102:7665–70. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Ambrogio L, Shimamura T, Kubo S, Takahashi M, Chirieac LR, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008 doi: 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–77. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok T, Wu Y-L, Thongprasert S, Yang C-H, Chu D, Saijo N, et al. Phase III, randomised, open label, first-line study of gefitinib vs. carboplatin/paclitaxel in clinically selected patients with advanced non-small cell lung cancer (IPASS) Annals of Oncology. 2008;19 [Google Scholar]
- Mukohara T, Engelman JA, Hanna NH, Yeap BY, Kobayashi S, Lindeman N, et al. Differential effects of gefitinib and cetuximab on non-small-cell lung cancers bearing epidermal growth factor receptor mutations. J Natl Cancer Inst. 2005;97:1185–94. doi: 10.1093/jnci/dji238. [DOI] [PubMed] [Google Scholar]
- Ono M, Hirata A, Kometani T, Miyagawa M, Ueda S, Kinoshita H, et al. Sensitivity to gefitinib (Iressa, ZD1839) in non-small cell lung cancer cell lines correlates with dependence on the epidermal growth factor (EGF) receptor/extracellular signal-regulated kinase 1/2 and EGF receptor/Akt pathway for proliferation. Mol Cancer Ther. 2004;3:465–72. [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Quintas-Cardama A, Cortes J. Therapeutic options against BCR-ABL1 T315I-positive chronic myelogenous leukemia. Clin Cancer Res. 2008;14:4392–9. doi: 10.1158/1078-0432.CCR-08-0117. [DOI] [PubMed] [Google Scholar]
- Rothenberg SM, Engelman JA, Le S, Riese DJ, 2nd, Haber DA, Settleman J. Modeling oncogene addiction using RNA interference. Proc Natl Acad Sci U S A. 2008;105:12480–4. doi: 10.1073/pnas.0803217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai A, Chandarlapaty S, Greulich H, Gonen M, Ye Q, Arteaga CL, et al. Inhibition of Hsp90 down-regulates mutant epidermal growth factor receptor (EGFR) expression and sensitizes EGFR mutant tumors to paclitaxel. Cancer Res. 2008;68:589–96. doi: 10.1158/0008-5472.CAN-07-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellens JH, Britten CD, camidge DR, Boss D, Wong S, Diab S, et al. First-inhuman study of the safety, tolerability, pharmacokinetics (PK), and pharmacodynamics (PD) of PF-00299804, a small molecule irreversible pan-HER inhibitor in patients with advanced cancer. Journal of Clinical Oncology, 2007 ASCO Annual Meeting Proceedings. 2007;19S Abstract 3599. [Google Scholar]
- Schimke RT, Kaufman RJ, Alt FW, Kellems RF. Gene amplification and drug resistance in cultured murine cells. Science. 1978;202:1051–5. doi: 10.1126/science.715457. [DOI] [PubMed] [Google Scholar]
- Sequist LV, Martins RG, Spigel D, Grunberg SM, Spira A, Janne PA, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–9. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- Shah NP, Kasap C, Weier C, Balbas M, Nicoll JM, Bleickardt E, et al. Transient potent BCR-ABL inhibition is sufficient to commit chronic myeloid leukemia cells irreversibly to apoptosis. Cancer Cell. 2008;14:485–93. doi: 10.1016/j.ccr.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–25. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- Shimamura T, Li D, Ji H, Haringsma HJ, Liniker E, Borgman CL, et al. Hsp90 inhibition suppresses mutant EGFR-T790M signaling and overcomes kinase inhibitor resistance. Cancer Res. 2008;68:5827–38. doi: 10.1158/0008-5472.CAN-07-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamborini E, Bonadiman L, Greco A, Albertini V, Negri T, Gronchi A, et al. A new mutation in the KIT ATP pocket causes acquired resistance to imatinib in a gastrointestinal stromal tumor patient. Gastroenterology. 2004;127:294–9. doi: 10.1053/j.gastro.2004.02.021. [DOI] [PubMed] [Google Scholar]
- Vikis H, Sato M, James M, Wang D, Wang Y, Wang M, et al. EGFR-T790M is a rare lung cancer susceptibility allele with enhanced kinase activity. Cancer Res. 2007;67:4665–70. doi: 10.1158/0008-5472.CAN-07-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, Fracasso PM, Bukowski RM, Lynch TJ, Munster PN, Shapiro GI, et al. A phase I study with neratinib (HKI-272), an irreversible pan ErbB receptor tyrosine kinase inhibitor, in patients with solid tumors. Clin Cancer Res. 2009;15:2552–8. doi: 10.1158/1078-0432.CCR-08-1978. [DOI] [PubMed] [Google Scholar]
- Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105:2070–5. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuza Y, Glatt KA, Jiang J, Greulich H, Minami Y, Woo MS, et al. Allele-Dependent Variation in the Relative Cellular Potency of Distinct EGFR Inhibitors. Cancer Biol Ther. 2007;6 doi: 10.4161/cbt.6.5.4003. [DOI] [PubMed] [Google Scholar]
- Zhao X, Weir BA, LaFramboise T, Lin M, Beroukhim R, Garraway L, et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 2005;65:5561–70. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.