SUMMARY
Parkinson’s disease (PD) is characterized by dopaminergic neurodegeneration in the nigrostriatal region of the brain; however, the neurodegeneration extends well beyond dopaminergic neurons. To gain a better understanding of the molecular changes relevant to PD, we applied two-dimensional LC-MS/MS to comparatively analyze the proteome changes in four brain regions (striatum, cerebellum, cortex, and the rest of brain) using a MPTP-induced PD mouse model with the objective to identify nigrostriatal-specific and other region-specific protein abundance changes. The combined analyses resulted in the identification of 4,895 non-redundant proteins with at least two unique peptides per protein. The relative abundance changes in each analyzed brain region were estimated based on the spectral count information. A total of 518 proteins were observed with significant MPTP-induced changes across different brain regions. 270 of these proteins were observed with specific changes occurring either only in the striatum and/or in the rest of the brain region that contains substantia nigra, suggesting that these proteins are associated with the underlying nigrostriatal pathways. Many of the proteins that exhibit significant abundance changes were associated with dopamine signaling, mitochondrial dysfunction, the ubiquitin system, calcium signaling, the oxidative stress response, and apoptosis. A set of proteins with either consistent change across all brain regions or with changes specific to the cortex and cerebellum regions were also detected. One of the interesting proteins is ubiquitin specific protease (USP9X), a deubiquination enzyme involved in the protection of proteins from degradation and promotion of the TGF-β pathway, which exhibited altered abundances in all brain regions. Western blot validation showed similar spatial changes, suggesting that USP9X is potentially associated with neurodegeneration. Together, this study for the first time presents an overall picture of proteome changes underlying both nigrostriatal pathways and other brain regions potentially involved in MPTP-induced neurodegeneration. The observed molecular changes provide a valuable reference resource for future hypothesis-driven functional studies of PD.
Keywords: mass spectrometry, mouse brain proteome, Parkinson’s disease, MPTP, LC-MS/MS, mouse model
INTRODUCTION
Parkinson’s disease (PD) represents a chronic, progressive neurodegenerative movement disorder characterized by the loss of dopaminergic neurons in the substantia nigra (SN) and accumulation of intraneuronal Lewy body inclusions (1-3). Since the first description of PD in 1817 (4), extensive studies have been made including the identification of 13 PD associated genetic loci (5, 6), which provide basic insights into the pathogenesis of the disease. Current hypotheses regarding mechanisms underlying the etiology and pathogenesis of PD, include abnormal protein aggregation, mitochondrial dysfunction, oxidative stress, and failure of the ubiquitin-proteasome system (4), and were mainly derived from studies using postmortem tissues or animal models; most notably, the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-models (7-10). As a selective neurotoxin, MPTP primarily targets the nigrostriatal dopaminergic system via the high affinity dopamine transporter (11, 12), where it induces the generation of excessive reactive oxygen species (ROS) and the selective destruction of dopaminergic neurons, thereby decreasing the dopaminergic projection into the striatum (7-10). The MPTP model has become the most commonly used PD model for elucidating molecular cascades of neurodegeneration (7); however, the exact etiology and mechanisms leading to the development of PD still remain unclear. Moreover, the neurodegeneration extends well beyond dopaminergic neurons (7, 13). Therefore, identification of MPTP-induced molecular changes within different brain regions is essential for defining molecular mechanisms underlying PD neurodegeneration.
Significant efforts have recently been focused on the application of proteomics technologies in order to profile protein abundance changes in brain tissues and biofluids in patients or animal models of neurodegenerative diseases (14, 15). Studies using Two-dimensional electrophoresis (2DE) coupled with mass spectrometry (MS) in PD patients, parkin-deficient mice, as well as MPTP-treated mice have consolidated the view that mitochondrial dysfunction and oxidative stress are involved in PD pathogenesis (16-21). We have also recently applied LC-MS and microarrays to profile protein and transcript abundance changes in the striatum of PD mouse models induced by MPTP and methamphetamine (METH) treatment for the purpose of identifying molecular changes associated with PD (22). However, since most of the studies were limited to only a single brain region, such as substantia nigra or striatum, without comparison with other brain regions, it is unclear whether the observed molecular changes are specific to nigrostriatal pathways. Moreover, the molecular changes in other brain regions may potentially be associated with neurodegeneration in PD have yet to be identified.
In this work we identify spatially resolved proteome changes induced by MPTP by comparatively analyzing four brain regions, i.e., striatum, cerebellum, cortex, and the rest of the brain tissue (ROB). Off-line strong cation exchange (SCX) LC fractionation was coupled with high resolution capillary reversed-phase LC-MS/MS to enhance the overall proteome coverage over the previous single dimensional LC-MS based study (22). The results reveal a total of 518 proteins with significant abundance changes induced by MPTP in the different brain regions; 270 of these proteins show abundance changes specific to the nigrostriatal region. Many of the observed abundance changes are associated with the dopamine signaling pathway, mitochondrial dysfunction, the ubiquitin system, calcium signaling, and apoptosis.
EXPERIMENTAL PROCEDURES
MPTP Mouse Model
Adult C57BL/6J male mice (8 weeks, 21–27 g), obtained from Jackson Laboratories (Bar Harbor, ME), were administrated four intraperitoneal (i.p.) injections of MPTP-HCl (Sigma-Aldrich, St. Louis, MO) in saline (15 mg/kg per injection) at 2 h intervals; the control mice were injected with the same volume of sterilized saline only. The cortex, cerebellum, striatum and rest of brain were dissected from 4 MPTP and 4 control mice 7 days after the injection, and snap-frozen immediately in liquid nitrogen until future processing. Animal procedures were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The loss of dopamine in striatum was assessed by using high performance liquid chromatography (HPLC) as previously described (22) and ~88% decrease in striatal dopamine level was observed following the MPTP treatment.
Protein Digestion
All tissues were homogenized in 50 mM NH4HCO3 (pH 7.8), and protein concentration was determined by a BCA assay (Pierce, Rockford, IL). Aliquots of homogenates from the same brain region from four mice were pooled for protein digestion. Approximately, 300 μg pooled sample was subsequently treated with 50% (v/v) trifluoroethanol (TFE) (Sigma-Aldrich) for 2 h at 60 °C, 5 mM tributylphosphine (TBP) (Sigma) for 0.5 h at 60 °C, 40 mM iodoacetamide (IAA) for 1 h at 37 °C, and then diluted five-fold with 50 mM NH4HCO3 prior to digestion with sequencing-grade trypsin (Promega, Madison, WI) for 3 h at 37 °C at a 1:50 trypsin to protein ratio (w:w) with 1 mM CaCl2 added during the digestion. The digested sample was then cleaned up with a SPE C-18 column (Supelco, Bellefonte, PA), then dried down using a speed-vac concentrator.
Strong Cation Exchange (SCX) Fractionation
The dried peptides were dissolved in 25 mM NH4HCO3, and subjected to SCX fractionation using a 200 × 2.1 mm (5 μm particles, 300 Å pore size) Polysulfoethyl A column (PolyLC, Columbia, MD) on an automated HPLC (Agilent, Palo Alto, CA) at a flow rate of 0.2 mL/min. Mobile phase solvents consisted of (A) 10 mM ammonium formate, 25% acetonitrile, pH 3.0 and (B) 500 mM ammonium formate, 25% acetonitrile, pH 6.8. Once the column was loaded, isocratic conditions at 100% A were maintained for 10 min. Peptides were separated applying a gradient from 0 to 50% B over 40 min, followed by a gradient of 50–100% B over 10 min. The gradient was then held at 100% solvent B for another 10 min. Twenty-five fractions were collected for each sample. Following lyophilization, the fractions were dissolved in 25 mM NH4HCO3 and stored at −80 °C.
Capillary LC-MS/MS Analysis
Each of the 25 SCX fractions was further analyzed using a fully automated custom-built capillary HPLC system coupled online with an LTQ ion trap mass spectrometer (ThermoFinnigan, San Jose, CA) using an in-house manufactured electrospray ionization interface. The reversed-phase capillary column was slurry packed using 3 μm Jupiter C18 particles (Phenomenex, Torrance, CA) in a 75 μm (inside diameter) × 65 cm fused silica capillary (Polymicro Technologies, Phoenix, AZ). The mobile phases consisted of A (0.2% acetic acid and 0.05% TFA in water) and B (0.1% TFA in 90% acetonitrile). An exponential gradient was employed during the separation, which started with 100% A that gradually increased to 60% B over the course of 100 min. The instrument was operated in data-dependent mode with an m/z range of 400–2000. The five most abundant ions from the MS analysis were selected for MS/MS analysis using a normalized collision energy setting of 35%. Dynamic exclusion was used to avoid repetitive analysis of the same abundant precursor ion.
MS/MS Data Analysis
The MS/MS data were searched against the mouse International Protein Index (IPI) database with a total of 51,252 total protein entries (Version 3.19, released on July 25, 2006, available on-line at http://www.ebi.ac.uk/IPI) with the decoy database searching option (for assessing false positive rate) using X!Tandem 2 software (23). The search parameters used were: 3 Da tolerance for precursor ion masses and 1 Da for fragment ion masses with no enzyme restraint and a maximum of three missed tryptic cleavages. Modifications for the static carboxamidomethylation of cysteine and dynamic oxidation of methionine were applied during the database search.
The following criteria were used for initial filtering of the raw X!Tandem results: Peptide_Expectation_Value_Log(e) < −2 with partial or full tryptic cleavage. Such criteria were established for mouse brain tissue using sequence-reversed database searching that provides 3.7% of false positive peptide identifications at the final identified unique peptide level (24). The reversed human protein database was created by reversing the order of the amino acid sequences for each protein, and the false positive rate (FPR) for peptide identifications was estimated by dividing the number of unique peptides identified from the reversed database search (NR) by the number of unique peptides identified from the normal database search (NN), i.e. FPR = NR/NN.
To remove redundant protein entries, ProteinProphet™ software (v1.7) was used as a clustering tool to group similar or related protein entries into protein groups (25). Peptides that passed the filtering criteria were assigned the identical probability score of 1 and entered into the software program (done exclusively for cluster analysis) to generate a final list of non-redundant proteins/protein groups. One protein IPI number was randomly selected to represent each corresponding protein group that consists of a number of database entries. Only those proteins or protein groups with two or more unique peptide identifications were considered to be confident protein identifications.
Abundance ratios of proteins were calculated based on the spectral count of proteins from each region in the format of fold-changes, i.e., if abundance ratio of [MPTP]/[Ctrl] ≥1, fold change equals [MPTP]/[Ctrl]-1, and if abundance ratio of [MPTP]/[Ctrl] <1, fold change equals 1-[Ctrl]/[MPTP]. To identify statistically significant abundance changes, proteins identified from the analyses for each brain region were divided into three groups based on their relative abundance levels: the high-abundance group (mean spectral count between two biological conditions greater than 10); the medium-abundance group (mean spectral count between 5 and 10); and the low-abundance group (mean spectral count less than 5), similar to the method previously described (26). Standard deviations (σ) of fold-changes between two replicate 2D-LC-MS/MS analyses were calculated for each protein abundance group based on the data from the previous study (26) . A fold change greater than 4σ was required for a protein to be considered to be significant for each protein group. The final cutoffs of fold-changes are 5, 1.75, and 0.6 for the low-, medium-, and high-abundance groups, respectively. Only those proteins with total spectral counts of peptide observation above 5 were retained for the analysis of abundance changes.
Western Blot Verifications of Selected Proteins
15 μg proteins were separated on a 4-12% NuPAGE Novex Bis-Tris gel (1 mm thick, Invitrogen, CA), and transferred to a 0.2 uL Nitrocellulose membrane (Invitrogen). The membranes were blocked in 10% nonfat milk in Tris-buffered saline (TBS: 50 mM Tris, 150 mM NaCl, pH 7.4) at room temperature for one hour, followed by different primary and secondary antibody incubations. Antibodies were used at the following dilutions: Beta tubulin (mouse, Abcam, MA), 1:20000; Usp9x (rabbit, provided by Dr. Stephen Wood (27)), 1:2000; Atp6v1f (Rabbit, Santa Cruz Biotechnology, CA), 1:200; Goat anti-rabbit IgG – HRP (Invitrogen), 1:5000; Rabbit anti-mouse HRP-IgG (Pierce), 1:20000. Immunoblotting bands were detected by Lumi-Imager F1 (Boehringer Mannheim Corporation, Indianapolis, IN).
RESULTS
LC-MS/MS Profiling of Four Brain Regions
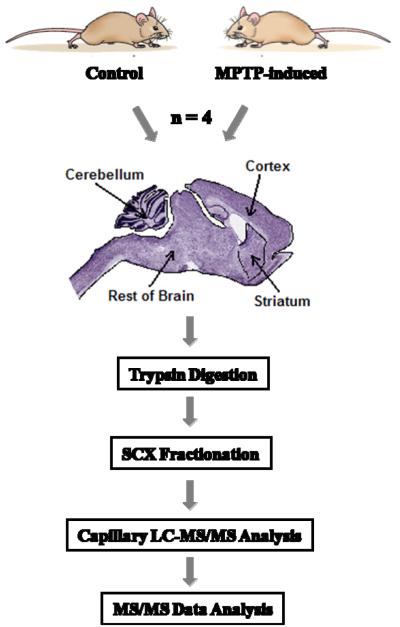
To achieve in-depth proteome coverage of the mouse brain regions, we applied offline SCX fractionation coupled with high resolution capillary reversed-phase LC-MS/MS for comparative analyses. As shown in the experimental workflow (Figure 1), samples from 4 mice in each experimental condition were collected. The whole brain was dissected into four regions, i.e., striatum, cerebellum, cortex, and “rest of the brain tissue” (ROB) that contains the substantia nigra (SN), with the purpose of identifying region-specific as well as multi-regional proteome changes. Samples from the four biological replicates per condition were pooled prior to trypsin digestion; the same amount of resulting peptides for all regions and conditions were fractionated into 25 fractions by SCX, and submitted for LC-MS/MS analysis. A total of 200 LC-MS/MS analyses were performed for all SCX fractions. After database searching against the mouse IPI protein database using the X!Tandem algorithm (23), 51,120 unique peptides were confidently identified with a false positive rate (FPR) of ~3.7% based on decoy database searching (24, 28), covering 6,980 non-redundant protein groups after removing possible redundant proteins by ProteinProphet (25). 4,895 (70%) proteins with two or more unique peptides were considered to be confidently identified proteins. The proteome coverage of each brain region is summarized in Table 1. 23,894 unique peptides and 3,928 proteins were identified from the striatum region; and the other three regions have the similar trends and numbers. In addition, there are 2,945 proteins in common across all four regions. All identified peptides and proteins are listed in Supplementary Tables 1 and 2.
Figure 1.
Flowchart showing the experimental workflow.
Table 1.
Number of peptides and proteins identified from the four brain regions.
| Regions | Unique peptides | Unique proteinsa | Significant proteinsb |
|---|---|---|---|
| Striatum (Str) | 23,894 | 3,928 | 192 |
| Rest of Brain (ROB) | 25,744 | 4,194 | 131 |
| Cerebellum (CB) | 24,084 | 4,171 | 133 |
| Cortex (Cor) | 22,940 | 3,515 | 124 |
Proteins identified with more than 2 unique peptides.
Proteins that show significant changes in abundance as induced by MPTP treatment.
Spatial Protein Abundance Distribution
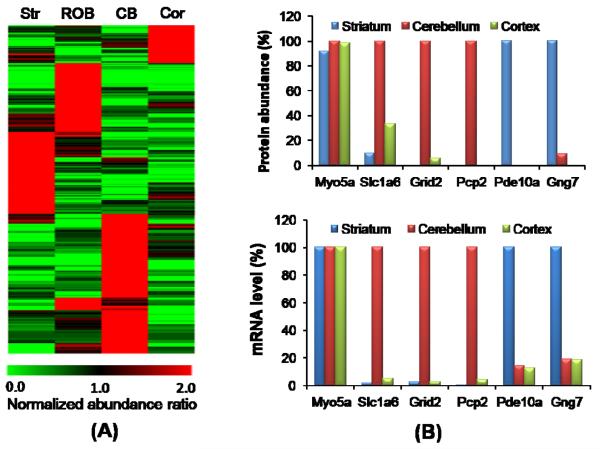
The spatial abundance distribution of each protein in the four analyzed brain regions was evaluated on the basis of spectral count. To illustrate the normal spatial abundance distribution, we only used data from control brain tissues for this analysis. Figure 2A shows the heatmap of ~500 proteins that are primarily localized in one or two regions among the four analyzed regions (Supplemental Table 3). Figure 2B illustrates the comparison of protein abundance patterns with mRNA abundance patterns for selected proteins. The spatial distribution patterns of protein abundances and mRNA abundances available from the Allen Brain Atlas (http://www.brain-map.org/) correlate well. Moreover, a number of these proteins are known as region-specific markers. For example, two highly-expressed proteins that were only identified in the cerebellum, cerebellin (CBLN1) and Purkinje cell protein-2 (PCP2), are associated with Purkinje cell synaptic structures (29, 30). Here we find these proteins are highly expressed in the cerebellum and absent in other brain regions (Figure 2B). CBLN1 is not shown in Figure 2B due to the lack of mRNA abundance information. Phosphodiesterase 10a (PDE10a) is highly expressed in the striatum, consistent with its known localization to striatal medium-sized spiny projection neurons and the response of these neurons to cortical stimulation (31, 32). Similarly, gamma 7 G-protein subunit (GNG7) is exclusively identified in the striatum where it is presumptively coupled to dopamine receptors (33).
Figure 2.
(A) Heatmap of ~500 region-specific proteins from control brain samples. Protein abundances were displayed with a normalized abundance ratio scale where the spectral count for each protein in a given region was divided by the average spectral count across the four regions. All proteins had at least five total spectral counts and the normalized abundance ratio was >3 in at least one region. (Str: striatum; ROB: rest of brain; CB: cerebellum; Cor: Cortex) (B) Selected localized proteins showing good agreement with mRNA abundance distributions available in the Allen Brain Atlas (http://www.brain-map.org/). The ROB is not included because the average data for the rest of brain region is less quantitative. The highest region abundance of each protein is set as 100%. Myosin-5A (MYO5A) is a general protein expressed the same across all brain regions.
MPTP-Induced Protein Abundance Changes
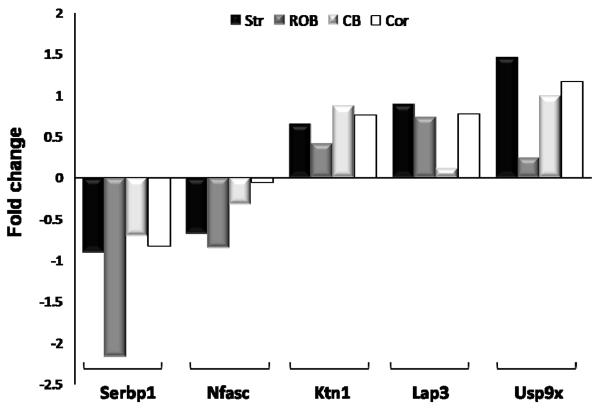
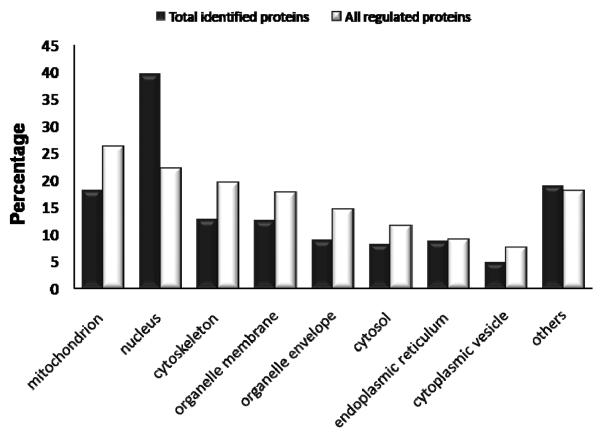
To identify region-specific protein abundance changes induced by MPTP treatment, spectral count data for each brain region were used as a semi-quantitative measurement for identifying relative abundance changes. After applying a stringent filtering criterion requiring fold-changes greater than four-fold of the standard deviation (4σ), 192, 131, 124 and 133 proteins were observed with significant abundance changes after MPTP treatment in the striatum, ROB, cortex and cerebellum, respectively, giving a total of 518 proteins with significant changes (Supplemental Table 4). 462 proteins change significantly in only one specific region after MPTP inducement. The striatum region appears to contain more proteins with significant changes than the other three regions, which is consistent with previous reports that MPTP mainly induces loss of neuron axon terminals in the striatum. Besides, region-specific protein abundance changes, we also observe a small set of proteins with consistent abundance changes across multiple brain regions with selected examples shown in Figure 3. Only one protein, plasminogen activator inhibitor 1 RNA-binding protein (Serbp1), has a consistent significant change across all four regions. A comparison of the cellular component distributions between total identified proteins and all the significantly changed proteins shows that a relatively large portion of proteins that respond to MPTP treatment are localized in mitochondria and cytoskeleton (Figure 4).
Figure 3.
MPTP induced protein abundance changes for selected proteins across the four regions. Abundance changes were displayed as fold changes where the fold change is [MPTP]/[Ctrl]-1 if abundance ratio of spectral count ≥1, or the fold change is 1-[Ctrl]/[MPTP] if abundance ratio <1. Region annotations: Str: striatum; ROB: rest of brain; CB: cerebellum; Cor: Cortex.
Figure 4.
Comparison of the cellular component distributions between the total identified proteins (black) and all regulated proteins (white) that show significant MPTP-induced abundance changes. The number of proteins in each category is expressed as a percentage of the total.
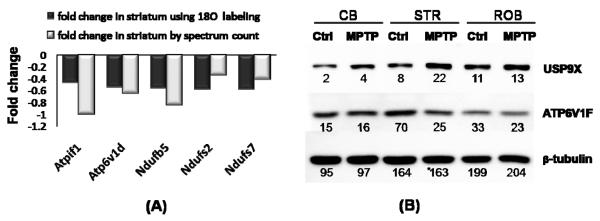
As a means to assess data quality, we were able to map our data to 41 proteins with previously reported changes in protein or gene expression associated with PD. As shown in Table 2, 37 proteins agree in the direction of the reported changes; and 12 of these proteins are known to be specifically changed in the nigrostriatal pathways due to MPTP treatment. For example, down-regulation of CSE1L, HNRPDL, COPE, ATP6V1E1 and BCAS1, and up-regulation of RAN, VIM, RPS4X, EIF4A2, UBB, THY1 and BCR were also observed in the mRNA analysis of SN from human PD subjects (34). Acetylcholinesterase precursor (ACHE) and breast carcinoma amplified sequence 1 (BCAS1), which were known to be inactivated by MPTP (35) and downregulated in SN in PD (34) were both decreased in striatum after MPTP treatment. Figure 5 further shows the fold changes of five proteins (ATPIF1, ATP6V1D, NDUFB5, NDUFS2, NDUFS7) associated with mitochondrial dysfunction that were observed with significant changes in both our previous LC-MS study in striatum using 18O stable isotope labeling (22) and the present study (Figure 5A). Western blot validations on two selected proteins, ubiquitin specific protease (USP9X) and vacuolar ATP synthase subunit F (ATP6V1F), were performed and the observed changes correlate with the proteomics spectral count data (Figure 5B).
Table 2.
Observed fold changes for selected proteins reported to be associated with PD.
| IPI reference | Gene symbol |
Protein name | Fold changea | References | |||
|---|---|---|---|---|---|---|---|
| Str | ROB | CB | Cor | ||||
| IPI00226140.4 | Maobb | amine oxidase b | 2.5 | −0.75 | 2 | 2 | (42) |
| IPI00129191.1 | Nos1 | nitric-oxide synthase | −0.25 | 0.8 | 8 | −0.11 | (77) |
| IPI00111013.1 | Ctsdb | cathepsin d | 1.5 | 0.25 | −0.083 | −0.25 | (22) |
| IPI00130992.1 | Capns1b | calpain small subunit 1 | 2 | −0.33 | 0.17 | 1.0 | (22) |
| IPI00129577.1 | Pdcd8b | programmed cell death protein 8 | 4.56 | 0 | 0.14 | −1.0 | (78) |
| IPI00331092.6 | Rps4x | 40s ribosomal protein s4 | 0 | 1.25 | −0.125 | 0.83 | (34) |
| IPI00400432.2 | Eif4a2 | eukaryotic initiation factor 4a-ii | 0.55 | 0.86 | −1.11 | −0.375 | (34) |
| IPI00227299.5 | Vimb | Vimentin | 1.0 | 0 | −0.05 | 0.17 | (34) |
| IPI00139518.3 | Ubb | ubiquitin b | −1.0 | 2.4 | 0.575 | −0.2 | (34) |
| IPI00109727.1 | Thy1 | thy-1 membrane glycoprotein | 0.63 | −0.46 | 0.33 | −0.31 | (34) |
| IPI00119024.3 | Arl6ip5 | pra1 family protein 3 | 0.43 | −0.4 | −5 | 1.0 | (34) |
| IPI00230124.4 | Fabp3b | fatty acid-binding protein | 1.0 | 2 | 4.56 | 1.0 | (79) |
| IPI00113517.1 | Ctsbb | cathepsin b | 1.1 | 0.05 | −0.8 | −0.04 | (48) |
| IPI00128567.1 | Acheb | acetylcholinesterase | −1.33 | 0 | 0 | 0 | (35) |
| IPI00626662.2 | Aldh1a1b | retinal dehydrogenase 1 | −1.22 | −0.45 | 0.29 | −1.0 | (80) (22) |
| IPI00118787.1 | Atp6v1d | vacuolar atp synthase subunit d | −0.57 | −0.5 | 0.17 | 0.55 | (22) |
| IPI00127598.1 | Atpif1 | atpase inhibitor | −1.0 | −0.17 | 0.3 | 0.25 | (22) |
| IPI00132531.1 | Ndufb5 | nadh dehydrogenase 1 beta | −0.8 | −0.67 | 0.4 | 1.33 | (22) |
| IPI00128023.3 | Ndufs2 | nadh-ubiquinone oxidoreductase 49 kda subunit |
−0.27 | 0.05 | −0.35 | 0.64 | (22) |
| IPI00120232.1 | Ndufs7 | nadh-ubiquinone oxidoreductase 20 kda subunit |
−0.33 | 0 | −2 | −0.33 | (22) |
| IPI00131814.1 | Ddcb | aromatic-l-amino-acid decarboxylase | −11.22 | 3.0 | 0 | 0 | (37) |
| IPI00138131.1 | Thb | tyrosine 3-monooxygenase | −4 | −3.0 | 0 | 0 | (38, 39) |
| IPI00131614.1 | Sncb | beta-synuclein | −0.6 | 0.32 | 0.12 | 0.71 | (81, 82) |
| IPI00131771.2 | Cox6cb | cytochrome c oxidase polypeptide VIc | −1.25 | 0.67 | −1.5 | −0.11 | (47) |
| IPI00119517.2 | Cask | peripheral plasma membrane protein cask | −1.0 | 0.1 | 0.44 | −1.375 | (83) |
| IPI00756676.1 | Bcas1 | breast carcinoma amplified sequence 1 | −0.67 | 0.09 | −0.44 | −0.4 | (34) |
| IPI00755892.1 | Hnrpdl | heterogeneous nuclear ribonucleoprotein D- like |
0.083 | 0 | −0.73 | 0.125 | (34) |
| IPI00130840.6 | Cope | coatomer subunit epsilon | 0 | −1.22 | 1.22 | −5.67 | (34) |
| IPI00480341.2 | Gps1 | cop9 signalosome | −0.67 | 0.5 | 1.0 | −2.67 | (34) |
| IPI00108143.1 | Hnrph2 | heterogeneous nuclear ribonucleoprotein h | −1.57 | 0.27 | −0.13 | −0.47 | (22) |
| IPI00134621.2 | Ran | gtp-binding nuclear protein ran | 0.5 | −0.14 | 0.25 | −0.14 | (34) |
| IPI00755653.1 | Bcr | breakpoint cluster region | 0.57 | −0.14 | −0.33 | 0 | (34) |
| IPI00112414.1 | Cse1l | exportin-2 | −0.5 | 0.22 | −0.4 | 0 | (34) |
| IPI00119115.1 | Atp6v1e1 | vacuolar atp synthase subunit e | −0.17 | −0.03 | 0.5 | −0.065 | (34) |
| IPI00308938.5 | Capn2 | calpain-2 catalytic subunit | 3.44 | 0.2 | 0.33 | 1 | (22) |
| IPI00331422.4 | Cars | cysteinyl-trna synthetase | 0.5 | 2 | 4.56 | −0.25 | (84) |
| IPI00135130.3 | Slc1a4 | neutral amino acid transporter a | −1 | 1.33 | 1 | 1.5 | (85) |
| IPI00136912.1 | Srmc | spermidine synthase | −4 | 0 | 0 | 0.2 | (22) |
| IPI00129685.1 | Tpt1c | translationally-controlled tumor protein | −1.5 | 0.29 | −0.4 | 0.18 | (34) |
| IPI00136967.2 | Gria2c | glutamate receptor 2 precursor | −1.67 | 0.17 | 1.0 | −0.86 | (86) |
| IPI00224045.1 | Chatc | choline o-acetyltransferase | 2.5 | 0.11 | 0 | 0 | (47) |
Significant fold changes were highlighted by gray color.
Proteins known to be specifically changed in the nigrostriatal pathways due to the MPTP-treatment.
Observed fold-changes of the protein disagree with previously reported changes.
Figure 5.
(A) Fold changes observed for selected mitochondrial proteins in the striatum. Fold-changes from 18O labeling is detailed in a previous paper[22]. (B) Western blot analysis of ubiquitin carboxyl-terminal hydrolase (USP9X) and vacuolar ATP synthase subunit F (ATP6v1f) in three different mouse brain regions before and after MPTP treatment. The spectral count for a protein in each brain region is labeled under each gel band. USP9X is observed to be up-regulated in all three regions. ATP6v1f is observed to be down-regulated in STR and ROB regions with MPTP treatment. The results agree with the proteome data. (STR: striatum; CB: cerebellum)
Nigrostriatal Specific Protein Abundance Changes
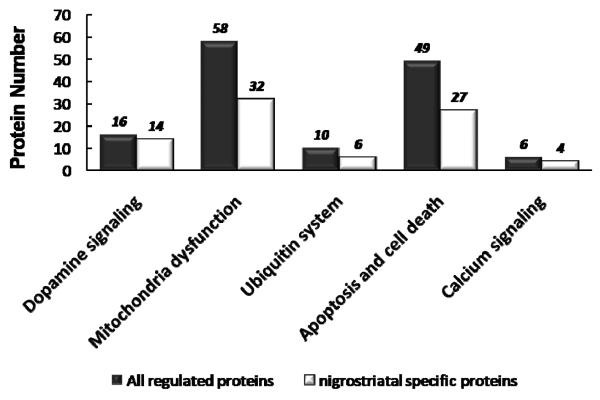
Proteins potentially associated with dopaminergic neurodegeneration underlying the nigrostriatal pathways can be identified by examining the patterns of protein abundance changes across the four analyzed regions. Potential nigrostriatal specific changes would occur in the striatum and/or the ROB region (containing SN) but without significant changes in the cerebellum and cortex regions. Following this data analysis, we observed a total of 270 proteins with significant nigrostriatal specific abundance changes among the 518 total regulated proteins (See Supplemental Table 5). Functional analysis of these proteins using the Ingenuity Pathways Analysis (IPA) tool (36) revealed that many of the proteins are associated with a number of functional categories, including mitochondrial dysfunction, dopamine synthesis and signaling, apoptosis, the ubiquitin system, and calcium signaling. Figure 6 shows the comparison of the number of nigrostriatal specific or total regulated proteins in each functional category. As shown, nigrostriatal specific proteins contributes to the main portion (>60%) of all functional categories, which is consistent with the knowledge that SN and striatum are the major sites of neurodegeneration in PD.
Figure 6.
Comparison of the number of nigrostriatal-specific and total significant proteins in each functional category. Nigrostriatal-specific proteins contribute to the main portion (>60%) of all functional categories.
Functional Implications for Regulated Proteins
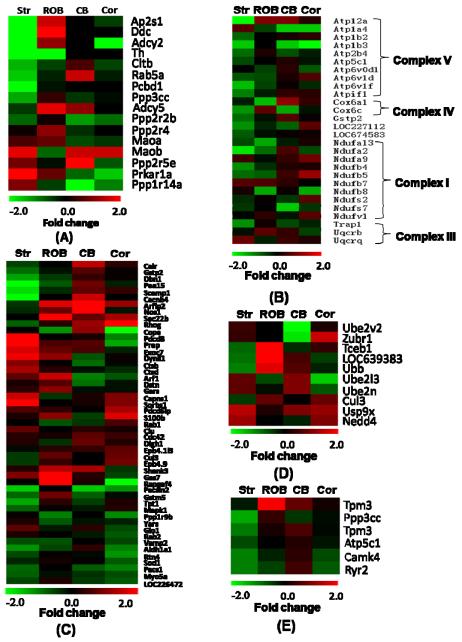
Table 3 lists the regulated proteins that are associated with different functional categories. The detailed protein abundance information is available in Supplementary Table 6. The abundance change patterns for each category are further displayed as heatmaps in Figure 7.
Table 3.
Significant proteins associated with different functional categories as shown in Figure 7.
| IPI reference | Gene symbol |
Protein name |
|---|---|---|
| Dopamine signaling | ||
| IPI00118022.1 | Ap2s1 | AP-2 complex subunit sigma-1 |
| IPI00131814.1 | Ddc | aromatic-l-amino-acid decarboxylase |
| IPI00377680.3 | Adcy2 | adenylate cyclase 2 |
| IPI00138131.1 | Th | tyrosine 3-monooxygenase |
| IPI00228978.2 | Cltb | clathrin light chain B |
| IPI00132410.1 | Rab5a | RAS-related protein |
| IPI00223800.4 | Pcbd1 | pterin-4-alpha-carbinolamine dehydratase |
| IPI00108318.1 | Ppp3cc | protein phosphatase 3 |
| IPI00343381.5 | Adcy5 | adenylate cyclase 5 |
| IPI00317759.4 | Ppp2r2b | protein phosphatase 2A |
| IPI00118723.3 | Ppp2r4 | serine/threonine-protein phosphatase |
| IPI00169711.1 | Maoa | amine oxidase A |
| IPI00226140.4 | Maob | amine oxidase B |
| IPI00224697.1 | Ppp2r5e | serine/threonine-protein phosphatase |
| IPI00119575.3 | Prkar1a | protein kinase |
| IPI00126719.1 | Ppp1r14a | protein phosphatase 1 |
| Mitochondria dysfunction | ||
| IPI00268145.4 | Atp12a | potassium-transporting atpase alpha chain |
| IPI00378485.2 | Atp1a4 | sodium/potassium-transporting atpase alpha-4 chain |
| IPI00123704.1 | Atp1b2 | sodium/potassium-transporting atpase subunit beta-2 |
| IPI00124221.1 | Atp1b3 | sodium/potassium-transporting atpase subunit beta-3 |
| IPI00378453.3 | Atp2b4 | ATP2B4 protein |
| IPI00313475.1 | Atp5c1 | ATP synthase gamma chain |
| IPI00313841.1 | Atp6v0d1 | vacuolar ATP synthase subunit D |
| IPI00118787.1 | Atp6v1d | vacuolar ATP synthase subunit D |
| IPI00315999.4 | Atp6v1f | vacuolar ATP synthase subunit F |
| IPI00121443.1 | Cox6a1 | cytochrome C oxidase, subunit VI A |
| IPI00131771.2 | Cox6c | cytochrome C oxidase polypeptide VIC |
| IPI00555023.1 | Gstp2 | glutathione S-transferase P 1 |
| IPI00672194.2 | LOC227112 | ATP synthase, H+ transporting, mitochondrial F0 complex |
| IPI00125460.1 | LOC674583 | ATP synthase coupling factor 6 |
| IPI00230715.4 | Ndufa13 | NADH dehydrogenase 1 alpha subunit 13 |
| IPI00315302.4 | Ndufa2 | NADH dehydrogenase 1 alphasubunit 2 |
| IPI00120212.1 | Ndufa9 | NADH dehydrogenase 1 alphasubunit 9 |
| IPI00132390.4 | Ndufb4 | NADH dehydrogenase 1 beta subunit 4 |
| IPI00132531.1 | Ndufb5 | NADH dehydrogenase 1 beta subunit 5 |
| IPI00133215.2 | Ndufb7 | NADH dehydrogenase 1 beta subunit 7 |
| IPI00387430.1 | Ndufb8 | NADH dehydrogenase 1 beta subunit 8 |
| IPI00128023.3 | Ndufs2 | NADH-ubiquinone oxidoreductase |
| IPI00120232.1 | Ndufs7 | NADH-ubiquinone oxidoreductase |
| IPI00130460.1 | Ndufv1 | NADH-ubiquinone oxidoreductase |
| IPI00132762.1 | Trap1 | heat shock protein 75 kDa |
| IPI00132347.1 | Uqcrb | ubiquinol-cytochrome C reductase binding protein |
| IPI00224210.4 | Uqcrq | ubiquinol-cytochrome C reductase |
| Apoptosis and cell death | ||
| IPI00123639.1 | Calr | calreticulin |
| IPI00555023.1 | Gstp2 | glutathione S-transferase P 1 |
| IPI00135475.2 | Dbn1 | drebrin |
| IPI00121013.1 | Pea15 | astrocytic phosphoprotein |
| IPI00278804.1 | Scamp1 | secretory carrier-associated membrane protein 1 |
| IPI00131720.3 | Cacnb4 | calcium channel subunit beta-4 |
| IPI00321942.4 | Arfip2 | arfaptin-2 |
| IPI00129191.1 | Nos1 | nitric-oxide synthase |
| IPI00114368.2 | Sec22b | vesicle-trafficking protein |
| IPI00116558.1 | Rhog | RHO-related GTP-binding protein |
| IPI00130840.6 | Cope | coatomer subunit epsilon |
| IPI00129577.1 | Pdcd8 | programmed cell death protein 8 |
| IPI00761930.1 | Prep | prolyl endopeptidase |
| IPI00227970.1 | Exoc7 | exocyst complex component 7 |
| IPI00121623.1 | Dynll1 | dynein light chain 1 |
| IPI00113517.1 | Ctsb | cathepsin B |
| IPI00111013.1 | Ctsd | cathepsin D |
| IPI00221613.4 | Arf1 | ADPribosylation factor 1 |
| IPI00127942.3 | Dstn | Destrin |
| IPI00112555.3 | Gars | glycyl-trna synthetase |
| IPI00130992.1 | Capns1 | calpain small subunit 1 |
| IPI00468140.2 | Sorbs1 | sorbin and sh3 domain-containing protein 1 |
| IPI00323483.3 | Pdcd6ip | programmed cell death 6-interacting protein |
| IPI00222557.4 | S100b | protein s100-b |
| IPI00466424.4 | Rab1 | member ras oncogene family |
| IPI00320420.3 | Clu | Clusterin |
| IPI00120193.1 | Cdc42 | cell division control protein 42 |
| IPI00125861.2 | Dlgh1 | disks large homolog 1 |
| IPI00125501.1 | Epb4.1l3 | band 4.1-like protein 3 |
| IPI00467383.2 | Cul3 | cullin-3 |
| IPI00125328.3 | Epb4.9 | Dematin |
| IPI00351827.5 | Shank3 | Shank3 |
| IPI00120345.1 | Gas7 | growth-arrest-specific protein 7 |
| IPI00270362.2 | Rapgef4 | RAP guanine nucleotide exchange factor 4 |
| IPI00125880.1 | Pacsin2 | protein kinase c and casein kinase substrate in neurons protein 2 |
| IPI00114380.1 | Gstm5 | glutathione S-transferase MU 5 |
| IPI00129685.1 | Tpt1 | translationally-controlled tumor protein |
| IPI00119663.2 | Mapk1 | mitogen-activated protein kinase 1 |
| IPI00420651.3 | Ppp1r9b | neurabin-2 |
| IPI00314153.4 | Yars | tyrosyl-trna synthetase |
| IPI00321734.6 | Glo1 | lactoylglutathione lyase |
| IPI00137227.1 | Rab2 | RAS-related protein |
| IPI00229703.5 | Vamp2 | vesicle-associated membrane protein |
| IPI00626662.2 | Aldh1a1 | retinal dehydrogenase 1 |
| IPI00469392.2 | Rtn4 | reticulon-4 |
| IPI00130589.7 | Sod1 | superoxide dismutase |
| IPI00321922.2 | Pacs1 | phosphofurin acidic cluster sorting protein 1 |
| IPI00750911.1 | Myo5a | Myo5a protein |
| IPI00130368.4 | LOC226472 | glutathione S-transferase, mu 4 |
| Calcium signaling | ||
| IPI00459570.1 | Tpm3 | tropomyosin 3, gamma |
| IPI00108318.1 | Ppp3cc | protein phosphatase 3 |
| IPI00278611.5 | Tpm3 | tropomyosin alpha-3 chain |
| IPI00313475.1 | Atp5c1 | ATP synthase gamma chain |
| IPI00132526.1 | Camk4 | calcium/calmodulin-dependent protein kinase type IV |
| IPI00338309.3 | Ryr2 | cardiac Ca2+ release channel |
| Ubiquitin system | ||
| IPI00402913.1 | Ube2v2 | ubiquitin-conjugating enzyme e2 variant 2 |
| IPI00378681.4 | Zubr1 | CG14472-PA |
| IPI00323130.3 | Tceb1 | transcription elongation factor B polypeptide 1 |
| IPI00108590.2 | LOC639383 | ubiquitin A-52 residue ribosomal protein |
| IPI00139518.3 | Ubb | ubiquitin B |
| IPI00128760.1 | Ube2l3 | ubiquitin-conjugating enzyme |
| IPI00165854.3 | Ube2n | ubiquitin-conjugating enzyme e2n |
| IPI00467383.2 | Cul3 | cullin-3 |
| IPI00108388.1 | Usp9x | ubiquitin carboxyl-terminal hydrolase FAF-x |
| IPI00462445.2 | Nedd4 | E3 ubiquitin-protein ligase |
Figure 7.
Heat maps of abundance changes for specific protein functional categories. (A) Dopamine signaling pathway; (B) Mitochondrial dysfunction; (C) Apoptosis and cell death; (D) the ubiquitin system; (E) Calcium signaling. Relative protein abundances are displayed as fold changes (MPTP/control).
Dopamine Signaling
Of the observed 19 genes involved in dopamine signaling, 16 of them were changed significantly in MPTP treated mouse brains as shown in Figure 7A. Furthermore, fourteen of those sixteen genes with significant abundance changes were found to be specific to the striatum and the ROB regions. Protein phosphatase 1 (PPP1R14A) is the only protein which decreases significantly in the cerebellum. Serine/threonine-protein phosphatase 2a (PPP2R5E) increases significantly in both striatum and cerebellum regions. Proteins known to be associated with PD, dopa decarboxylase (DDC), tyrosine hydroxylase (TH), and monoamine oxidase B (MAOB), were detected in both the striatum and the ROB regions, which were not detected in our previous study (22). DDC is an enzyme implicated in the dopamine synthesizing metabolic pathway (37). Following the hydroxylation of tyrosine to form L-dihydroxyphenylalanine (L-DOPA) that is catalyzed by TH (38, 39), DDC decarboxylates L-DOPA to form dopamine. Both DDC and TH were found downregulated in the striatum region. MAOB, an enzyme involved in the activation of MPTP and degradation of dopamine (40, 41), was upregulated in the striatum as previously reported (42). Moreover, four other proteins in the dopamine signaling pathway (ADCY2, CLTB, RAB5A, PPP3CC) and known to be associated with neurological diseases were also observed to be down-regulated in the striatum region (43-46).
Mitochondrial Dysfunction and Oxidative Phosphorylation
58 genes associated with mitochondrial dysfunction, including subunits of each mitochondrial complex (complex I, II, III, IV and V), were detected with significant abundance changes across all four regions after MPTP treatment, suggesting that a considerable degree of mitochondrial dysfunction in the brain is induced by MPTP. Among these, changes of 32 proteins were observed in striatum and ROB regions specifically. Figure 7B shows the abundance changes for 28 selected mitochondria proteins across each complex of mitochondria dysfunction. Some of these genes were also observed with similar down-regulation in our previous study (22) (see Figure 4). In addition, a number of these genes agreed with previous mRNA expression data. For example, cytochrome c oxidase subunit VIc (COX6C), which was previously reported to be down-regulated at the mRNA level after MPTP treatment (47), was also found to be down-regulated in protein abundance in the striatum region. Moreover, ten and eleven genes were observed with significant changes only in the cerebellum or cortex region, respectively. For example, protein abundances of aldehyde dehydrogenase family 3 (ALDH3A2), fumarylacetoacetate hydrolase domain containing 1 (FAHD1), propionyl-CoA carboxylase alpha chain (PCCA) decreased significantly only in the cerebellum; while sodium/potassium-transporting ATPase (ATP1A4, ATP1B3), vacuolar ATP synthase subunit D (ATP6v0D1), and ES1 protein homolog (D10JHU81E) were down-regulated in the cortex. Five proteins with considerable abundance changes, including dodecenoyl-coenzyme A delta isomerase (DCI), nucleoside diphosphate kinase B (NME2), LAP3 protein (LAP3), glutathione S-transferase P 1 (GSTP2) and ATP synthase, H+ transporting, mitochondrial F0 complex (LOC227112), were detected not only in the striatum, but also in the cerebellum and cortex regions. LOC227112 was observed with consistent down-regulation not only in striatum and ROB, but also in the cerebellum.
Apoptosis and Cell Death
A number of proteins functionally associated with apoptosis and cell death were detected with significant abundance changes. A total of 49 proteins in this family were measured with significant changes across four regions, with 27 of them specific to the striatum or ROB regions. Some of the genes were also previously reported with changes in mRNA abundances (see Supplementary Table 5). For example, the gene expression of cathepsin D precursor (CTSD) has been reported to increase Alzheimer’s disease (48), and similar up-regulation was observed in our previous proteomic study (22). A number of known pro-apoptotic proteins, including clusterin (CLU), cathepsins (CTSD, CTSB), calpain small subunit 1 (CAPNS1), prolyl endopeptidase (PREP), and programmed cell death protein 8 (PDCD8), were detected with significant up-regulation in the striatum or ROB region, which is consistent with the increased loss of dopamine neurons. Anti-apoptosis proteins, astrocytic phosphoprotein PEA-15 (PEA15) and translationally-controlled tumor protein (TPT1), were observed to have significant abundance decreases following MPTP treatment. Eighteen genes changed significantly in the non-nigrostriatal regions after MPTP treatment. For example, calreticulin (CALR), programmed cell death 6-interacting protein (PDCD6IP), and protein S100-B (S100B) were up-regulated in the cerebellum or cortex. Superoxide dismutase (SOD1) and glutathione S-transferase (LOC226472) were significantly down-regulated only in the cortex region. Several antioxidant proteins, including glutathione S-transferase (GSTM5, GSTP2, LOC226472) and superoxide dismutase (SOD1), were found to be down-regulated in different regions, suggesting a decrease in anti-oxidant capacity. Moreover, we observed a significant increase in the abundance of neuronal nitric oxide synthase (nNOS in cerebellum and ROB), which could contribute to an increase in reactive nitrogen species (RNS) in the cells.
The ubiquitin system
The ubiquitin system is one of the major pathways involved in the turnover and clearance of mis-folded cellular proteins. The dysfunction of this system, such as mutations in parkin (an E3 ligase) (49) and ubiquitin carboxy-terminal hydrolase L1 (UCHL1) (50), has been causally implicated in familiar PD (51, 52). A total of 149 proteins involved in the ubiquitin system were identified in this study using the KEGG PATHWAY database. Ten proteins were found with significant abundance changes in different brain regions (Table 3), and some of these changes have also been observed in other studies, for example, ubiquitin B (UBB) (34) and E3 ubiquitin-protein ligase (NEDD4) (53).
Calcium signaling
Several upregulated or downregulated proteins were observed in the calcium signaling pathway, including the downregulation of calcium/calmodulin-dependent protein kinase type IV (CAMK4), protein phosphatase 3 (PPP3CC), tropomyosin 3 (TPM3), ATP synthase (ATP5C1) in striatum. The results support the hypothesis that calcium signaling plays a role in PD.
DISCUSSION
Recent studies of PD have led to significant advances in our understanding of the pathogenesis of the disease (54-56). It has been generally recognized that the molecular mechanisms of neurodegeneration in PD involve complex interactions between environmental and genetic factors (57). Moreover, the neurodegeneration that occurs in PD extends well beyond dopaminergic neurons in the substantia nigra (SN) and striatum (7-10). In this work, we performed extensive region-specific protein abundance profiling by dividing the whole brain into four-different regions (i.e., striatum, cerebellum, cortex, and ROB), thus allowing both nigrostriatal and other region-specific proteome changes to be identified. In-depth proteome coverage was achieved by applying offline SCX fractionation coupled with capillary reversed-phase LC-MS/MS for comparative analysis of each of the brain regions from both control and MPTP treated mice.
Although the MPTP treatment leads to significant dopaminergic neuron or neuron terminal loss in SN and striatum, the effect of cell loss is corrected by using same amount of protein samples between both MPTP and control conditions for analysis. Our results revealed 518 proteins with significant abundance changes in different brain regions after MPTP treatment. The majority (462 out of 518) of these proteins were observed with significant abundance changes in a single brain region, which is not surprising because each of the brain regions have very different functions. We were able to identify 270 proteins that are potentially associated with nigrostriatal pathways by observing significant abundance changes in the striatum and/or ROB, but not in the cerebellum and cortex. As anticipated, since it is known that MPTP mainly induces a loss of neuron axon terminals in the striatum, the striatum region does show more proteins that have substantial changes than the other three regions. In addition to dopaminergic neuron death, proteins observed with significant changes only in the cerebellum and cortex are of potential interest to the study other types of neurodegeneration in these regions in addition to the dopaminergic neuron death.
Functional analysis of the regulated proteins revealed a number of interesting functional categories, including mitochondrial dysfunction, dopamine synthesis and signaling, apoptosis, the ubiquitin system, and calcium signaling. This observation is consistent with recent results from both animal models and genetic models, which suggest that many of these factors that contribute significantly to PD pathogenesis (54-56). Our results revealed region-specific protein abundance changes associated with these factors. It is not surprising to observe that the majority (>60%) of proteins associated with these functional categories show nigrostriatal-specific abundance changes.
The MPTP treatment leads to ~88% decrease in dopamine levels in the striatum as measured by HPLC; therefore, one would expect a significant disruption of the dopamine synthesis and signaling pathway. However, in our previous LC-MS study (22), no proteins associated with this pathway were detected due to the sensitivity limitation of one dimensional LC-MS. In this study, we observed 16 proteins involved in the dopamine signaling pathway that exhibited significant changes after MPTP treatment. The majority of these proteins displayed changes only in the striatum and ROB regions. For example, two proteins involved in dopamine synthesis, DDC and TH, had significant decreased abundances, which are consistent with the loss of dopamine neurons.
MPTP is known to cross the blood-brain barrier and being metabolized by monoamine oxidase B (MAOB) to MPP+ in glial cells. Once converted to MPP+, it is taken up by dopaminergic neurons via the dopamine re-uptake system (dopamine transporter, DAT). MPP+ is concentrated in the mitochondria where it inhibits the complex I of the respiratory chain, leading to ATP depletion and increased generation of reactive oxygen species (ROS) such as superoxide anion and hydroxyl radical, leading to the cell death of dopaminergic neurons (55, 58). Mitochondrial dysfunction indeed contributes to a wide range of human pathologies, including multiple neurodegenerative diseases, ischaemia-reperfusion injury in stroke and heart attack, diabetes and the cumulative degeneration associated with aging. It causes cell damage and death by compromising ATP production, disrupting calcium homeostasis and increasing oxidative stress. Furthermore, mitochondrial damage can lead to apoptotic cell death by causing the release of cytochrome c and other pro-apoptotic factors into the cytoplasm (55). Consistent with our previous observation of protein abundance changes associated with mitochondrial dysfunction (22), the current study considerably expanded the coverage of proteins associated with this function. In total we observed ~58 proteins with significant abundance changes that cover all five subunits of each complex (complexes I, II, III, IV and V). Our data further consolidates the pivotal function of mitochondria in PD neurodegeneration. Some proteins associated with oxidative stress response were also observed with significant abundance changes, which include glutathione S-transferase MU 5 (GSTM5), endoplasmic reticulum protein (ERP29), carbonyl reductase 1 (CBR1), cullin-3 (CUL3) and neuronal growth regulator 1 (NEGR1).
In addition to mitochondrial dysfunction and oxidative stress, we also identified numerous proteins functionally associated with apoptosis and cell death. Mitochondria are known to be pivotal in controlling apoptosis since they house many apoptogenic molecules that are released into the cytoplasm at the onset of apoptosis (55). These include cytochrome c, apoptosis-inducing factor and various caspases. In general, we observed up-regulation for pro-apoptotic proteins, and down-regulation for anti-apoptotic proteins, supportive of the increased cell death caused by MPTP. The increased apoptosis due to MPTP induction is also reflected by the decreased anti-oxidant capacity shown by the down-regulation of a number of known antioxidant proteins and the increased expression of neuronal nitric oxide synthase.
Our data also provide several lines of evidence that MPTP induced neurodegeneration may not be limited to the nigrostriatal regions. First, significant changes for a number of proteins associated with apoptosis and antioxidant functions were observed specifically in the cortex and cerebellum. For example, calreticulin (CALR), nitric-oxide synthase (NOS1) and programmed cell death 6-interacting protein (PDCD6IP), protein S100-B (S100B) were up-regulated in the cerebellum or cortex only. Similarly, a number of proteins associated with mitochondria dysfunction such as NADH dehydrogenase 1 (NDUFA9), NADH-ubiquinone oxidoreductase (NDUFS2, NDUFV1), sodium/potassium-transporting ATPase (ATP1a4, ATP1b3), beta-synuclein (SNCB) were also observed to have significant changes only in the cortex or cerebellum. As the cerebellum does not contain dopaminergic neurons, the observed effects of MPTP might derive from the glial cells or be related from other regions of the brain.
Several proteins from the ubiquitin system were observed with significant abundance changes in different brain regions. One interesting protein is ubiquitin-specific protease USP9X (2559 amino acid) that has a completely different sequence from a known Parkinson’s disease gene, UCHL1 (223 amino acid) (50). USP9X was significantly upregulated in the striatum, cerebellum and cortex with Western blot validations (Figure 6). USP9X, also known as FAM, is a mammalian homolog of Drosophila fat facets (59), which is an enzyme required for eye development and early embryogenesis (60, 61). USP9X deubiquitination prevents the proteasomal degradation of some substrates such as Af-6 (62) and beta-catenin (63), but also regulates the activity and intracellular localization of other substrates, such as epsin (64) and MARCH 7 (65). Therefore, a possible way for USP9X to be involved in PD is that during the development of PD, USP9X affects the degradation of alpha synuclein, leading to toxic aggregation. A recent study (66) reported that USP9X activates SMAD4 by deubiquitination at K519, resulting in the activation of SMAD2/3, and promotion of TGFβ pathway, which will induce cell apoptosis (67). The involvement of USP9X in apoptosis is confirmed by the data from the homolog fat facets, which is an enhancer of grim-reaper induced apoptosis in Drosophila (68). Considering that TGFb1 (69) and TGFb2 (70) were associated with neurodegenerative diseases, USP9X is possibly a novel protein involved in the development of PD by both promoting the TGFβ pathway and inducing the apoptosis of cells. In addition, USP9X is reported to interact with Huntingtin protein (71) raising the possibility of a general role for USP9X in neurodegeneration.
Calcium signaling is known to regulate multiple neuronal functions, including synaptic transmission, plasticity, and cell survival (72). High cytosolic Ca2+ can induce oxidative stress and excitotoxicity and has long been suspected to play a role in PD (73, 74). Recent evidence indicates that neuronal calcium signaling is abnormal in many neurodegenerative disorders, including AD and PD (75). Elevated intracellular Ca2+ participates in SN cell loss (76). Our observation of abundance changes in proteins associated with calcium signaling provides further evidence of its potential role in PD.
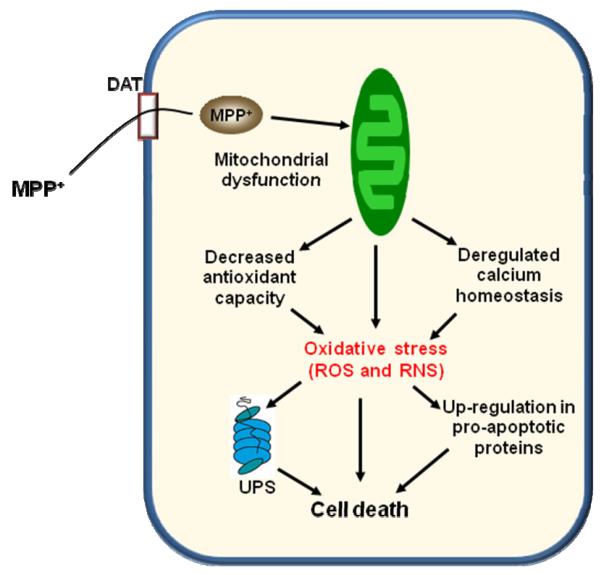
To summarize, our extensive proteome survey provides a comprehensive picture of protein abundance changes in different brain regions induced by MPTP treatment, the most commonly used mouse model for PD. The observed protein abundance changes are also supportive of the general mechanism of MPTP induced neurodegeneration as shown in Figure 8. Briefly, MPP+ induces mitochondria dysfunction, which leads to a decreased antioxidant capacity and the generation of excessive ROS and RNS. The oxidative stress condition leads to an up-regulation of pro-apoptotic proteins or factors, down-regulation of anti-apoptotic proteins, and disruption of the ubiquitin system, all of which eventually lead to cell death. Although our data supports that neurodegeneration mainly occurs in the nigrostriatal region, the study does provide additional novel information potentially associated with the neurodegeneration outside this region. It is not clear that all changes observed are associated with neurodegeneration. It may well be that some changes in protein expression represent the brain’s attempts to rectify trauma or detoxify the cells. We believe this dataset contains a rich source of novel proteins, e.g., USP9X, that may play an important role in neurological diseases and many of the proteins may represent interesting targets for further hypothesis-driven mechanistic studies.
Figure 8.
A general mechanism of MPTP induced neurodegeneration.
Supplementary Material
ACKNOWLEDGMENTS
Portions of this research were supported by the NIH Center of Proteomics Research Resource for Integrative Biology RR018522 (to R.D.S.), NIH grants R01 NS050148 (to D.J.S.) and the Pacific Northwest National Laboratory (PNNL) Directed Research Development program (to W.J.Q.). The experiment work was performed in the Environmental Molecular Sciences Laboratory, a U.S. Department of Energy (DOE) national scientific user facility on the PNNL campus. PNNL is multi-program national laboratory operated by Battelle for the DOE under Contract No. DE-AC05-76RLO 1830.
Abbreviations
- (PD)
Parkinson’s disease
- (SN)
Substantia nigra
- (MPTP)
1-Methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine
- (2DE)
Two-dimensional electrophoresis
- (IPI)
International Protein Index
- (SCX)
Strong cation exchange
- (SPE)
Solid phase extraction
- (TFE)
Trifluoroethanol
- (TBP)
Tributylphosphine
- (ROS)
Radical oxygen species
- (RNS)
Radical nitrogen species
- (MPP)
1-Methyl-4-phenylpyridinium
- (DAT)
Dopamine transporter
- (ATP)
Adenosine-5′-triphosphate
REFERENCES
- 1.Jenner P, Olanow CW. Understanding cell death in Parkinson’s disease. Ann Neurol. 1998;44:S72–84. doi: 10.1002/ana.410440712. [DOI] [PubMed] [Google Scholar]
- 2.Olanow CW, Tatton WG. Etiology and pathogenesis of Parkinson’s disease. Annu Rev Neurosci. 1999;22:123–144. doi: 10.1146/annurev.neuro.22.1.123. [DOI] [PubMed] [Google Scholar]
- 3.Forno LS, DeLanney LE, Irwin I, Langston JW. Electron microscopy of Lewy bodies in the amygdala-parahippocampal region. Comparison with inclusion bodies in the MPTP-treated squirrel monkey. Adv Neurol. 1996;69:217–228. [PubMed] [Google Scholar]
- 4.Parkinson J. an essay on the shaking palsy. J. Neuropsychiatry Clin. Neurosci. 1817:223–236. doi: 10.1176/jnp.14.2.223. [DOI] [PubMed] [Google Scholar]
- 5.Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson’s disease. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 6.Belin AC, Westerlund M. Parkinson’s disease: a genetic perspective. FEBS J. 2008;275:1377–1383. doi: 10.1111/j.1742-4658.2008.06301.x. [DOI] [PubMed] [Google Scholar]
- 7.Dauer W, Przedborski S. Parkinson’s disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 8.Przedborski S, Vila M. The 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine mouse model: a tool to explore the pathogenesis of Parkinson’s disease. Ann N Y Acad Sci. 2003;991:189–198. [PubMed] [Google Scholar]
- 9.Forno LS, DeLanney LE, Irwin I, Langston JW. Similarities and differences between MPTP-induced parkinsonsim and Parkinson’s disease. Neuropathologic considerations. Adv Neurol. 1993;60:600–608. [PubMed] [Google Scholar]
- 10.Heikkila RE, Hess A, Duvoisin RC. Dopaminergic neurotoxicity of 1-methyl-4-phenyl-1,2,5,6-tetrahydropyridine in mice. Science. 1984;224:1451–1453. doi: 10.1126/science.6610213. [DOI] [PubMed] [Google Scholar]
- 11.Javitch JA, D’Amato RJ, Strittmatter SM, Snyder SH. Parkinsonism-inducing neurotoxin, N-methyl-4-phenyl-1,2,3,6 -tetrahydropyridine: uptake of the metabolite N-methyl-4-phenylpyridine by dopamine neurons explains selective toxicity. Proc Natl Acad Sci U S A. 1985;82:2173–2177. doi: 10.1073/pnas.82.7.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burns RS, Chiueh CC, Markey SP, Ebert MH, Jacobowitz DM, Kopin IJ. A primate model of parkinsonism: selective destruction of dopaminergic neurons in the pars compacta of the substantia nigra by N-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Proc Natl Acad Sci USA. 1983;80:4546–4550. doi: 10.1073/pnas.80.14.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hornykiewicz O, Kish SJ. Biochemical pathophysiology of Parkinson’s disease. Adv Neurol. 1987;45:19–34. [PubMed] [Google Scholar]
- 14.Fountoulakis M, Kossida S. Proteomics-driven progress in neurodegeneration research. Electrophoresis. 2006;27:1556–1573. doi: 10.1002/elps.200500738. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Goodlett DR. Proteomic approach to studying Parkinson’s disease. Mol. Neurobiol. 2004;29:271–288. doi: 10.1385/MN:29:3:271. [DOI] [PubMed] [Google Scholar]
- 16.Periquet M, Corti O, Jacquier S, Brice A. Proteomic analysis of parkin knockout mice: alterations in energy metabolism, protein handling and synaptic function. J Neurochem. 2005;95:1259–1276. doi: 10.1111/j.1471-4159.2005.03442.x. [DOI] [PubMed] [Google Scholar]
- 17.Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, Wacker M, Klose J, Shen J. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 18.Basso M, Giraudo S, Corpillo D, Bergamasco B, Lopiano L, Fasano M. Proteome analysis of human substantia nigra in Parkinson’s disease. Proteomics. 2004;4:3943–3952. doi: 10.1002/pmic.200400848. [DOI] [PubMed] [Google Scholar]
- 19.Jin J, Hulette C, Wang Y, Zhang T, Pan C, Wadhwa R, Zhang J. Proteomic identification of a stress protein, mortalin/mthsp70/GRP75: relevance to Parkinson disease. Mol Cell Proteomics. 2006;5:1193–1204. doi: 10.1074/mcp.M500382-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Jin J, Meredith GE, Chen L, Zhou Y, Xu J, Shie FS, Lockhart P, Zhang J. Quantitative proteomic analysis of mitochondrial proteins: relevance to Lewy body formation and Parkinson’s disease. Brain Res Mol Brain Res. 2005;134:119–138. doi: 10.1016/j.molbrainres.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Diedrich M, Mao L, Bernreuther C, Zabel C, Nebrich G, Kleene R, Klose J. Proteome analysis of ventral midbrain in MPTP-treated normal and L1cam transgenic mice. Proteomics. 2008;8:1266–1275. doi: 10.1002/pmic.200700754. [DOI] [PubMed] [Google Scholar]
- 22.Chin MH, Qian WJ, Wang H, Petyuk VA, Bloom JS, Sforza DM, Lacan G, Liu D, Khan AH, Cantor RM, Bigelow DJ, Melega WP, Camp DG, 2nd, Smith RD, Smith DJ. Mitochondrial dysfunction, oxidative stress, and apoptosis revealed by proteomic and transcriptomic analyses of the striata in two mouse models of Parkinson’s disease. J Proteome Res. 2008;7:666–677. doi: 10.1021/pr070546l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenyo D, Beavis RC. A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Anal Chem. 2003;75:768–774. doi: 10.1021/ac0258709. [DOI] [PubMed] [Google Scholar]
- 24.Qian WJ, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Camp DG, 2nd, Smith RD. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 25.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 26.Qian WJ, Jacobs JM, Camp DG, 2nd, Monroe ME, Moore RJ, Gritsenko MA, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Smith RD. Comparative proteome analyses of human plasma following in vivo lipopolysaccharide administration using multidimensional separations coupled with tandem mass spectrometry. Proteomics. 2005;5:572–584. doi: 10.1002/pmic.200400942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanai-Azuma M, Mattick JS, Kaibuchi K, Wood SA. Co-localization of FAM and AF-6, the mammalian homologues of Drosophila faf and canoe, in mouse eye development. Mech Dev. 2000;91:383–386. doi: 10.1016/s0925-4773(99)00294-4. [DOI] [PubMed] [Google Scholar]
- 28.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 29.Urade Y, Oberdick J, Molinar-Rode R, Morgan JI. Precerebellin is a cerebellum-specific protein with similarity to the globular domain of complement C1q B chain. Proc Natl Acad Sci USA. 1991;88:1069–1073. doi: 10.1073/pnas.88.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan J, Luo Y, Denker BM. Purkinje cell protein-2 (Pcp2) stimulates differentiation in PC12 cells by Gbetagamma-mediated activation of Ras and p38 MAPK. Biochem J. 2005;392:389–397. doi: 10.1042/BJ20042102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujishige K, Kotera J, Omori K. Striatum- and testis-specific phosphodiesterase PDE10A isolation and characterization of a rat PDE10A. Eur J Biochem. 1999;266:1118–1127. doi: 10.1046/j.1432-1327.1999.00963.x. [DOI] [PubMed] [Google Scholar]
- 32.Threlfell S, Sammut S, Menniti FS, Schmidt CJ, West AR. Inhibition of Phosphodiesterase 10A Increases the Responsiveness of Striatal Projection Neurons to Cortical Stimulation. J Pharmacol Exp Ther. 2009;328:785–795. doi: 10.1124/jpet.108.146332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danielson PE, Watson JB, Gerendasy DD, Erlander MG, Lovenberg TW, de Lecea L, Sutcliffe JG, Frankel WN. Chromosomal mapping of mouse genes expressed selectively within the central nervous system. Genomics. 1994;19:454–461. doi: 10.1006/geno.1994.1094. [DOI] [PubMed] [Google Scholar]
- 34.Kim JM, Lee KH, Jeon YJ, Oh JH, Jeong SY, Song IS, Lee DS, Kim NS. Identification of genes related to Parkinson’s disease using expressed sequence tags. DNA Res. 2006;13:275–286. doi: 10.1093/dnares/dsl016. [DOI] [PubMed] [Google Scholar]
- 35.Zang LY, Misra HP. Inactivation of acetylcholinesterase by 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine hydrochloride. Mol Cell Biochem. 2003;254:131–136. doi: 10.1023/a:1027376303043. [DOI] [PubMed] [Google Scholar]
- 36.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 37.Burkhard P, Dominici P, Borri-Voltattorni C, Jansonius JN, Malashkevich VN. Structural insight into Parkinson’s disease treatment from drug-inhibited DOPA decarboxylase. Nat Struct Biol. 2001;8:963–967. doi: 10.1038/nsb1101-963. [DOI] [PubMed] [Google Scholar]
- 38.Jackson-Lewis V, Jakowec M, Burke RE, Przedborski S. Time course and morphology of dopaminergic neuronal death caused by the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Neurodegeneration. 1995;4:257–269. doi: 10.1016/1055-8330(95)90015-2. [DOI] [PubMed] [Google Scholar]
- 39.Tatton WG, Kwan MM, Verrier MC, Seniuk NA, Theriault E. MPTP produces reversible disappearance of tyrosine hydroxylase-containing retinal amacrine cells. Brain Res. 1990;527:21–31. doi: 10.1016/0006-8993(90)91056-m. [DOI] [PubMed] [Google Scholar]
- 40.Heikkila RE, Manzino L, Cabbat FS, Duvoisin RC. Protection against the dopaminergic neurotoxicity of 1-methyl-4-phenyl-1,2,5,6-tetrahydropyridine by monoamine oxidase inhibitors. Nature. 1984;311:467–469. doi: 10.1038/311467a0. [DOI] [PubMed] [Google Scholar]
- 41.Trevor AJ, Singer TP, Ramsay RR, Castagnoli N., Jr. Processing of MPTP by monoamine oxidases: implications for molecular toxicology. J Neural Transm Suppl. 1987;23:73–89. doi: 10.1007/978-3-7091-8901-6_5. [DOI] [PubMed] [Google Scholar]
- 42.Damier P, Kastner A, Agid Y, Hirsch EC. Does monoamine oxidase type B play a role in dopaminergic nerve cell death in Parkinson’s disease? Neurology. 1996;46:1262–1269. doi: 10.1212/wnl.46.5.1262. [DOI] [PubMed] [Google Scholar]
- 43.Laifenfeld D, Patzek LJ, McPhie DL, Chen Y, Levites Y, Cataldo AM, Neve RL. Rab5 mediates an amyloid precursor protein signaling pathway that leads to apoptosis. J Neurosci. 2007;27:7141–7153. doi: 10.1523/JNEUROSCI.4599-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, Hosford D, Barnes MR, Briley JD, Borrie M, Coletta N, Delisle R, Dhalla D, Ehm MG, Feldman HH, Fornazzari L, Gauthier S, Goodgame N, Guzman D, Hammond S, Hollingworth P, Hsiung GY, Johnson J, Kelly DD, Keren R, Kertesz A, King KS, Lovestone S, Loy-English I, Matthews PM, Owen MJ, Plumpton M, Pryse-Phillips W, Prinjha RK, Richardson JC, Saunders A, Slater AJ, St George-Hyslop PH, Stinnett SW, Swartz JE, Taylor RL, Wherrett J, Williams J, Yarnall DP, Gibson RA, Irizarry MC, Middleton LT, Roses AD. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65:45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 45.Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, Haroutunian V, Fienberg AA. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carter CJ. eIF2B and oligodendrocyte survival: where nature and nurture meet in bipolar disorder and schizophrenia? Schizophr Bull. 2007;33:1343–1353. doi: 10.1093/schbul/sbm007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gomez-Gallego M, Fernandez-Villalba E, Fernandez-Barreiro A, Herrero MT. Changes in the neuronal activity in the pedunculopontine nucleus in chronic MPTP-treated primates: an in situ hybridization study of cytochrome oxidase subunit I, choline acetyl transferase and substance P mRNA expression. J Neural Transm. 2007;114:319–326. doi: 10.1007/s00702-006-0547-x. [DOI] [PubMed] [Google Scholar]
- 48.Yan S, Sloane BF. Molecular regulation of human cathepsin B: implication in pathologies. Biol Chem. 2003;384:845–854. doi: 10.1515/BC.2003.095. [DOI] [PubMed] [Google Scholar]
- 49.Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K, Suzuki T. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 50.Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein MJ, Jonnalagada S, Chernova T, Dehejia A, Lavedan C, Gasser T, Steinbach PJ, Wilkinson KD, Polymeropoulos MH. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 51.Giasson BI, Lee VM. Are ubiquitination pathways central to Parkinson’s disease? Cell. 2003;114:1–8. doi: 10.1016/s0092-8674(03)00509-9. [DOI] [PubMed] [Google Scholar]
- 52.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 53.Dil Kuazi A, Kito K, Abe Y, Shin RW, Kamitani T, Ueda N. NEDD8 protein is involved in ubiquitinated inclusion bodies. J Pathol. 2003;199:259–266. doi: 10.1002/path.1283. [DOI] [PubMed] [Google Scholar]
- 54.Eriksen JL, Wszolek Z, Petrucelli L. Molecular pathogenesis of Parkinson disease. Arch Neurol. 2005;62:353–357. doi: 10.1001/archneur.62.3.353. [DOI] [PubMed] [Google Scholar]
- 55.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 56.Olanow CW. The pathogenesis of cell death in Parkinson’s disease - 2007. Mov Disord. 2007;22:S335–S342. doi: 10.1002/mds.21675. [DOI] [PubMed] [Google Scholar]
- 57.Maraganore DM, de Andrade M, Lesnick TG, Farrer MJ, Bower JH, Hardy JA, Rocca WA. Complex interactions in Parkinson’s disease: a two-phased approach. Mov Disord. 2003;18:631–636. doi: 10.1002/mds.10431. [DOI] [PubMed] [Google Scholar]
- 58.Hasegawa E, Takeshige K, Oishi T, Murai Y, Minakami S. 1-Methyl-4-phenylpyridinium (MPP+) induces NADH-dependent superoxide formation and enhances NADH-dependent lipid peroxidation in bovine heart submitochondrial particles. Biochem Biophys Res Commun. 1990;170:1049–1055. doi: 10.1016/0006-291x(90)90498-c. [DOI] [PubMed] [Google Scholar]
- 59.Wood SA, Pascoe WS, Ru K, Yamada T, Hirchenhain J, Kemler R, Mattick JS. Cloning and expression analysis of a novel mouse gene with sequence similarity to the Drosophila fat facets gene. Mech Dev. 1997;63:29–38. doi: 10.1016/s0925-4773(97)00672-2. [DOI] [PubMed] [Google Scholar]
- 60.Chen X, Overstreet E, Wood SA, Fischer JA. On the conservation of function of the Drosophila fat facets deubiquitinating enzyme and Fam, its mouse homolog. Dev Genes Evol. 2000;210:603–610. doi: 10.1007/s004270000109. [DOI] [PubMed] [Google Scholar]
- 61.Pantaleon M, Kanai-Azuma M, Mattick JS, Kaibuchi K, Kaye PL, Wood SA. FAM deubiquitylating enzyme is essential for preimplantation mouse embryo development. Mech Dev. 2001;109:151–160. doi: 10.1016/s0925-4773(01)00551-2. [DOI] [PubMed] [Google Scholar]
- 62.Taya S, Yamamoto T, Kano K, Kawano Y, Iwamatsu A, Tsuchiya T, Tanaka K, Kanai-Azuma M, Wood SA, Mattick JS, Kaibuchi K. The Ras target AF-6 is a substrate of the fam deubiquitinating enzyme. J Cell Biol. 1998;142:1053–1062. doi: 10.1083/jcb.142.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taya S, Yamamoto T, Kanai-Azuma M, Wood SA, Kaibuchi K. The deubiquitinating enzyme Fam interacts with and stabilizes beta-catenin. Genes Cells. 1999;4:757–767. doi: 10.1046/j.1365-2443.1999.00297.x. [DOI] [PubMed] [Google Scholar]
- 64.Chen H, Polo S, Di Fiore PP, De Camilli PV. Rapid Ca2+-dependent decrease of protein ubiquitination at synapses. Proc Natl Acad Sci USA. 2003;100:14908–14913. doi: 10.1073/pnas.2136625100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nathan JA, Sengupta S, Wood SA, Admon A, Markson G, Sanderson C, Lehner PJ. The ubiquitin E3 ligase MARCH7 is differentially regulated by the deubiquitylating enzymes USP7 and USP9X. Traffic. 2008;9:1130–1145. doi: 10.1111/j.1600-0854.2008.00747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, Inui M, Moro S, Modena N, Argenton F, Newfeld SJ, Piccolo S. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136:123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 67.Valderrama-Carvajal H, Cocolakis E, Lacerte A, Lee EH, Krystal G, Ali S, Lebrun JJ. Activin/TGF-beta induce apoptosis through Smad-dependent expression of the lipid phosphatase SHIP. Nat Cell Biol. 2002;4:963–969. doi: 10.1038/ncb885. [DOI] [PubMed] [Google Scholar]
- 68.Wing JP, Schreader BA, Yokokura T, Wang Y, Andrews PS, Huseinovic N, Dong CK, Ogdahl JL, Schwartz LM, White K, Nambu JR. Drosophila Morgue is an F box/ubiquitin conjugase domain protein important for grim-reaper mediated apoptosis. Nat Cell Biol. 2002;4:451–456. doi: 10.1038/ncb800. [DOI] [PubMed] [Google Scholar]
- 69.Rota E, Bellone G, Rocca P, Bergamasco B, Emanuelli G, Ferrero P. Increased intrathecal TGF-beta1, but not IL-12, IFN-gamma and IL-10 levels in Alzheimer’s disease patients. Neurol Sci. 2006;27:33–39. doi: 10.1007/s10072-006-0562-6. [DOI] [PubMed] [Google Scholar]
- 70.Goris A, Williams-Gray CH, Foltynie T, Brown J, Maranian M, Walton A, Compston DA, Barker RA, Sawcer SJ. Investigation of TGFB2 as a candidate gene in multiple sclerosis and Parkinson’s disease. J Neurol. 2007;254:846–848. doi: 10.1007/s00415-006-0414-6. [DOI] [PubMed] [Google Scholar]
- 71.Kaltenbach LS, Romero E, Becklin RR, Chettier R, Bell R, Phansalkar A, Strand A, Torcassi C, Savage J, Hurlburt A, Cha GH, Ukani L, Chepanoske CL, Zhen Y, Sahasrabudhe S, Olson J, Kurschner C, Ellerby LM, Peltier JM, Botas J, Hughes RE. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wojda U, Salinska E, Kuznicki J. Calcium ions in neuronal degeneration. IUBMB Life. 2008;60(9):575–90. doi: 10.1002/iub.91. [DOI] [PubMed] [Google Scholar]
- 73.Beal MF. Excitotoxicity and nitric oxide in Parkinson’s disease pathogenesis. Annals of neurology. 1998;44:S110–114. doi: 10.1002/ana.410440716. [DOI] [PubMed] [Google Scholar]
- 74.Chan CS, Guzman JN, Ilijic E, Mercer JN, Rick C, Tkatch T, Meredith GE, Surmeier DJ. ‘Rejuvenation’ protects neurons in mouse models of Parkinson’s disease. Nature. 2007;447:1081–1086. doi: 10.1038/nature05865. [DOI] [PubMed] [Google Scholar]
- 75.Bezprozvanny I. Calcium signaling and neurodegenerative diseases. Trends Mol Med. 2009;15:89–100. doi: 10.1016/j.molmed.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sulzer D, Schmitz Y. Parkinson’s disease: return of an old prime suspect. Neuron. 2007;55:8–10. doi: 10.1016/j.neuron.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Y, Dawson VL, Dawson TM. Oxidative stress and genetics in the pathogenesis of Parkinson’s disease. Neurobiology of disease. 2000;7:240–250. doi: 10.1006/nbdi.2000.0319. [DOI] [PubMed] [Google Scholar]
- 78.Gorman AM, Ceccatelli S, Orrenius S. Role of mitochondria in neuronal apoptosis. Dev Neurosci. 2000;22:348–358. doi: 10.1159/000017460. [DOI] [PubMed] [Google Scholar]
- 79.Mollenhauer B, Steinacker P, Bahn E, Bibl M, Brechlin P, Schlossmacher MG, Locascio JJ, Wiltfang J, Kretzschmar HA, Poser S, Trenkwalder C, Otto M. Serum heart-type fatty acid-binding protein and cerebrospinal fluid tau: marker candidates for dementia with Lewy bodies. Neurodegener Dis. 2007;4:366–375. doi: 10.1159/000105157. [DOI] [PubMed] [Google Scholar]
- 80.Galter D, Buervenich S, Carmine A, Anvret M, Olson L. ALDH1 mRNA: presence in human dopamine neurons and decreases in substantia nigra in Parkinson’s disease and in the ventral tegmental area in schizophrenia. Neurobiology of disease. 2003;14:637–647. doi: 10.1016/j.nbd.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 81.Culvenor JG, McLean CA, Cutt S, Campbell BC, Maher F, Jakala P, Hartmann T, Beyreuther K, Masters CL, Li QX. Non-Abeta component of Alzheimer’s disease amyloid (NAC) revisited. NAC and alpha-synuclein are not associated with Abeta amyloid. Am J Pathol. 1999;155:1173–1181. doi: 10.1016/s0002-9440(10)65220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fan Y, Limprasert P, Murray IV, Smith AC, Lee VM, Trojanowski JQ, Sopher BL, La Spada AR. Beta-synuclein modulates alpha-synuclein neurotoxicity by reducing alpha-synuclein protein expression. Human molecular genetics. 2006;15:3002–3011. doi: 10.1093/hmg/ddl242. [DOI] [PubMed] [Google Scholar]
- 83.Dev KK, van der Putten H, Sommer B, Rovelli G. Part I: parkin-associated proteins and Parkinson’s disease. Neuropharmacology. 2003;45:1–13. doi: 10.1016/s0028-3908(02)00337-4. [DOI] [PubMed] [Google Scholar]
- 84.Ko HS, Kim SW, Sriram SR, Dawson VL, Dawson TM. Identification of far upstream element-binding protein-1 as an authentic Parkin substrate. The Journal of biological chemistry. 2006;281:16193–16196. doi: 10.1074/jbc.C600041200. [DOI] [PubMed] [Google Scholar]
- 85.Chassain C, Bielicki G, Durand E, Lolignier S, Essafi F, Traore A, Durif F. Metabolic changes detected by proton magnetic resonance spectroscopy in vivo and in vitro in a murin model of Parkinson’s disease, the MPTP-intoxicated mouse. Journal of neurochemistry. 2008;105:874–882. doi: 10.1111/j.1471-4159.2007.05185.x. [DOI] [PubMed] [Google Scholar]
- 86.Buchwald P, Juhasz A, Bell C, Patfalusi M, Kovacs P, Hochhaus G, Howes J, Bodor N. Influence of the N-acetylation polymorphism on the metabolism of talampanel: an investigation in fasted and fed subjects genotyped for NAT2 variants. Pharmazie. 2006;61:125–134. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.