Abstract
Regulatory T cells (Treg) are critical to the maintenance of immunological self-tolerance and immune homeostasis by suppressing aberrant or excessive immune responses. Treg specifically express the transcription factor Foxp3, which mediates the coordinate activation of genes such as CTLA-4 and GITR along with repression of T effector cytokines such as interleukin-2 and interferon-γ. Despite progress in understanding mechanisms of Foxp3-dependent gene activation, the molecular mechanism of Foxp3-dependent gene repression remains largely unknown. Herein we report the identification of Eos, a zinc-finger transcription factor of the Ikaros family, as a critical mediator of Foxp3-dependent gene silencing in Treg. Eos interacts directly with Foxp3 and is necessary for gene silencing without affecting expression of Foxp3 activated genes. We further demonstrate that Eos and its corepressor C-terminal binding protein 1 (CtBP1) are necessary for histone modifications and ultimately promoter methylation involved in selective gene silencing in Treg. Knockdown of Eos in Treg abrogates their ability to suppress immune responses in vitro and in vivo and endows them with partial effector function. This transcriptional control of Treg function through association between Foxp3 and Eos/co-repressor can potentially be exploited for immune-based therapies.
Naturally occurring CD4+CD25+ regulatory T cells (Treg), which specifically express the transcription factor Foxp3, are essential for maintaining immunologic self-tolerance and immune homeostasis(1–3). The suppressive activity of Treg involve the coordinate activation of many genes, including CTLA-4 and GITR by Foxp3 along with repression of genes encoding T effector cytokines such as interleukin-2 (IL-2) and interferon-gamma (IFN-γ)(4–7). Although it has been shown that Foxp3 interacts with the transcription factors NFAT (nuclear factor of activated T cells), AML1(acute myeloid leukemia-1)/Runx1(runt-related transcription factor 1), the histone acetyl transferase (HAT)/histone deacetyl transferase (HDAC) complex, and NF-κB(4, 8–10), the molecular mechanism by which Foxp3 mediates selective gene silencing in Treg is still largely unknown. Because Foxp3, unlike the other members (Foxp1, Foxp2) of the Foxp family, does not contain a canonical gene repression domain, we hypothesized that Treg must express a partner for Foxp3 responsible for its capacity to silence critical genes such as those encoding T effector cytokines among many others.
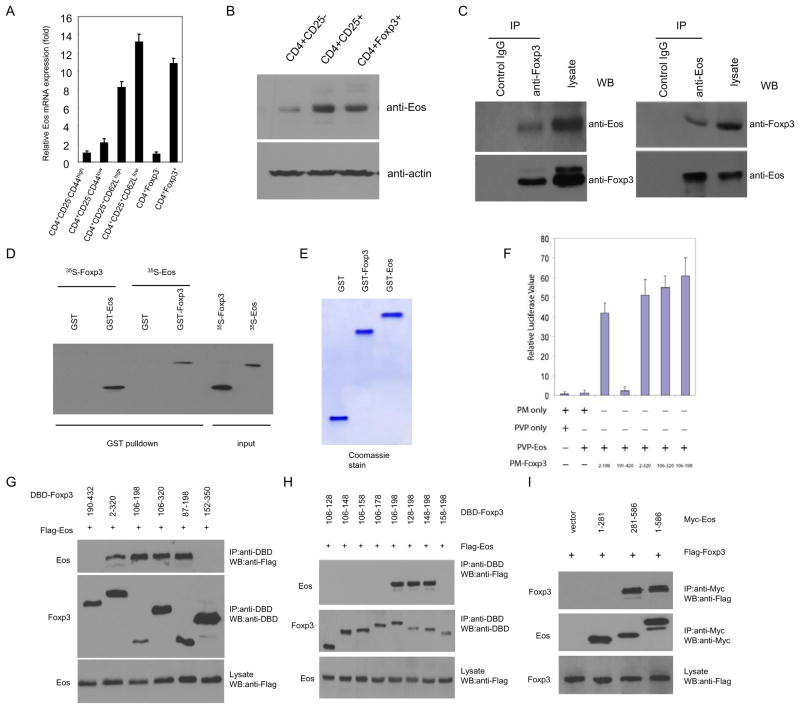
To gain insight into the molecular mechanisms of Treg development and function, we used an Affymetrix gene array to identify and characterize genes that are selectively expressed in anergic/ Treg cells but not in effector/memory T cells of the same origin and epitope specificity. To this end, gene chip comparison analysis was carried out on antigen specific CD4+ T cells under tolerizing vs. activating conditions both in vitro and in vivo(11). One of the genes that exhibited significantly higher expression in anergic/Treg conditions relative to activating conditions was Eos, a zinc finger transcription factor that belongs to the Ikarosfamily. We subsequently confirmed the gene chip result with real-time quantitative RT-PCR (qRT-PCR) and Western Blot. As shown in the Fig. 1A,B, Eos is highly expressed in the CD4+CD25+ and CD4+Foxp3+ Treg population. We further separated natural CD4+CD25+ Treg cells into two subpopulations: the CD4+CD25+CD62Lhi and CD4+CD25+CD62Llo. The latter subpopulation has been proposed to represent activated Treg (12). As shown in Fig 1A, while both CD4+CD25+CD62Lhi and CD62Llo cells express Eos, the CD4+CD25+CD62Llo cells express even higher levels of Eos than the CD4+CD25+CD62Lhi subset, suggesting that Eos expression is closely associated with activated Treg.
Fig. 1. Eos is highly expressed in subpopulation of CD4+ T cells, and it physically interacts with Foxp3 in Treg cells.
CD4+ T cells pooled from spleens and LN of C57BL/6 or GFP-Foxp3 mice were sorted into indicated subsets, and the expression of Eos was assessed by (A) real-time quantitative RT-PCR analysis using primers and internal fluorescent probes specific for Eos and 18s rRNA, and (B) Western blot analysis, respectively. (C) Endogenous interaction between Eos and Foxp3 in Treg cells. Treg cells were lysed, subjected to immunoprecipitation (IP) with anti-Foxp3 (left), anti-Eos (right) antibodies or control IgG. The immunoprecipitates were resolved on SDS-PAGE followed by Western blot analysis (WB) with anti-Eos and anti-Foxp3 antibodies as indicated, respectively. (D) In vitro interaction between Eos and Foxp3 was analyzed by GST pull-down assay. In vitro transcribed/translated [35S]-methionine-labelled Eos or Foxp3 was incubated with glutathione beads bound with bacterially expressed GST-Foxp3 or GST-Eos, respectively. GST alone was included as a negative control. After 30 mins incubation, the beads were washed 3 times with PBS and eluted with 5mM reduced glutamine. The eluted product was subjected to SDS-PAGE and autoradiograph. Input consisted of 10% of the [35S]-methionine-labelled proteins. (E) Coomassie blue staining. GST fusion protein purified from bacterially expressed GST-Foxp3 or GST-Eos were subjected to SDS-PAGE and stained with Coomassie blue. (F) Mapping of the Eos-interacting domain in Foxp3 with a mammalian two-hybrid system. The cDNA of Eos was fused to the VP16 AD in the pVP16 vector (Clontech), the cDNAs of different fragments of Foxp3 were fused to GAL4 DB in the pM vector. 293T cells were transfected with the indicated vectors together with pnull-RL (Renilla Luciferase, Promega) and pG5Luc, a luciferase reporter plasmid containing GAL4 promoter. After 24h, cells were harvested and luciferase activity was measured and normalized to Renilla luciferase activity. (G) Mapping of the Eos-interacting Domain in Foxp3 by coimmunoprecipitation. 293T cells were transfected with the indicated plasmids. After 24hr, cell extracts were immunoprecipitated with anti-DBD antibodies (Santa Cruz), the immune complexes were subjected to SDS-PAGE, and blotted with anti-Flag (top), anti-DBD (middle). The whole cell lysates were blotted with anti-Flag antibody (bottom). (H) The co-IP assay was applied to map the Eos-interaction domain in Foxp3. Numbers at top of each lane represent the peptide fragments of Foxp3 fused to the DBD epitope. (I) The co-IP assay was applied to map the Foxp3-interaction domain in Eos. Numbers at top of each lane represent the peptide fragments of Eos fused to the Myc epitope. The results are representative of one of three independent experiments.
Given that both Eos and Foxp3 are highly expressed in Treg cells and that Eos is known to have transcriptional repression activity (13), we speculated that Eos might interact with Foxp3 and mediate its gene silencing activity. To test this possibility, we carried out coimmunoprecipitation between HA-tagged Eos and non-tagged Foxp3 ectopically expressed in Jurkat T cells (a transformed human T cell line), which lack endogenous Foxp3. HA-Eos was found to coimmunoprecipitate with Foxp3 (fig. S1A). Similar experiments revealed that two other members (Ikaros and Helios) of the Ikaros family did not interact with Foxp3 (fig. S1B, C). Furthermore, we did not obseve the endogenous association between Foxp3 and Ikaros in purified natural Treg (Fig. S2A). While Ikaros was bound to the IL-2 promoter in Treg, binding diminished significantly upon TCR stimulation (Fig. S2B). These data argue against a role for Ikaros in Foxp3-dependent gene regulation in Treg cells, particularly when activated. In contrast, endogenous Foxp3 and Eos coimmunoprecipitate in lysates prepared from natural Treg cells (Fig. 1C). To verify that the interaction between Eos and Foxp3 is direct, we employed affinity purified recombinant GST-Eos fusion protein (Fig. 1E) to pull down [35S]-labeled Foxp3 that was produced by in vitro transcription and translation. GST-Eos alone, rather than GST, was bound to [35S]-Foxp3, suggesting a direct interaction between the two proteins in vitro (Fig. 1D). We also used affinity purified GST-Foxp3 fusion protein (Fig. 1E) to pull down [35S]-Eos in a reciprocal approach, which gave a similar result (Fig. 1D). Next we determined the Eos-interacting domain in Foxp3 by using co-immunoprecipitation. Human embryonic kidney 293T cells were co-transfected with constructs encoding tagged Eos and various truncated forms of Foxp3. The N-terminal 198 amino acid fragment of Foxp3 was found to be required for interaction with Eos (Fig. S1D). We further mapped the Eos-interacting domain in Foxp3 that is sufficient to mediate its interaction with Eos using the mammalian two-hybrid assay (14). We thus generated different fragments of Foxp3 fused to a Gal4-DNA binding domain, and Eos fused to the Gal4-activation domain. An interaction between the two fusion proteins should lead to reconstitution of Gal4 transcriptional activity, driving the expression of a luciferase reporter gene under the control of multiple Gal4-binding sites (pG5-Luc). It was found that a 93-amino acid fragment of Foxp3 (106–198) was necessary and sufficient for binding to Eos (Fig. 1F). This was further confirmed in a co-immunoprecipitation experiment using lysates of HEK293 cells co-transfected with constructs encoding tagged Eos and different fragments of Foxp3 (Fig. 1G). To further map the minimal interaction domain between Foxp3 and Eos, similar coimmunoprecipitation experiments in HEK 293T cells were carried out, leading to the identification of a 51 amino acid (148–198) fragment of Foxp3 that is necessary and sufficient to bind to the C-terminal region (281–586 amino acids) of Eos (Fig. 1H,I). Since Eos is associated with the N-terminus of Foxp3, which has been shown to be critical for repression of some genes, we considered the possibility that Eos itself might mediate the transcriptional repression activity of Foxp3.
It has been shown that Foxp3 directly binds to the IL-2 promoter and suppresses IL-2 expression (in addition to other effector cytokines) in both T cell lines and primary T cells (4). Since Foxp3 directly interacts with Eos, we investigated whether Eos might be involved in IL-2 suppression by Foxp3 in T cells. We first generated mouse primary CD4+ T cells in which Eos was efficiently knocked down by a lentivirus-mediated RNAi (pFup3.1-siEos carrying a GFP marker) while primary CD4+ cells treated with pFUP3.1-siRL (control siRNA, RL:Renilla Luciferase) served as the experimental control. Two different si-Eos targeting sequences (located in the Eos coding region or 3′UTR) were tested, and a similar knock-down efficiency was achieved (see material and method for detail)(15). As shown in Fig. 2A, Eos expression was knocked down more than 95% upon si-Eos treatment compared to that of si-RL. Also, the scrambled Eos siRNA control group gave a similar result as the RL siRNA group (data not shown). Forty-eight hours after lentivirus transduction, the modified T cells were re-transduced with a biscistronic retroviral vector expressing Foxp3 (1–429), ΔFoxp3 (with 148–198 deletion) and RFP or the RFP alone as a control. For the rescue experiment, the modified T cells were additionally re-transduced with a retroviral vector (carrying a Thy1.1 marker) expressing Eos with one base pair mutation at the 3′ UTR region (siRNA targeted region, RV-Eos). After an additional 72 hours, the GFP+RFP+ or GFP+RFP+Thy1.1+ T cells were sorted and stimulated with anti-CD3 in the presence of APCs to stimulate IL-2 production. As expected, the expression of the full-length Foxp3 almost completely blocked IL-2 production (Fig. 2B), and Eos protein level remains unchanged upon overexpression of Foxp3 (Fig. S3). The deletion of the 51-amino acid fragment required for Eos interaction completely abrogated the ability of Foxp3 to repress IL-2 production. Importantly, knockdown of Eos reversed Foxp3-mediated suppression of IL-2 production (Fig. 2B), which could be rescued by re-transduction with a retroviral vector expressing Eos with one base pair mutation at the siRNA targeted region. The 148–198 deletion of Foxp3 (ΔFoxp3) abrogates recruitment of Eos/CtBP1 complexes without disrupting its association with NFAT or AML1/Runx1(Fig. S4), further demonstrating that this domain of Foxp3 specifically mediates the Foxp3-Eos interaction. Taken together, these results indicated that the Foxp3-mediated IL-2 gene suppression requires Eos and this 51aa Eos-interacting domain of Foxp3.
Fig. 2.
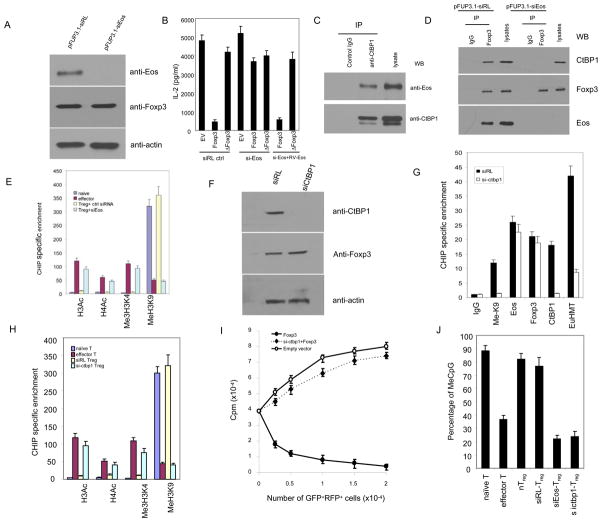
Foxp3 recruits Eos/CtBP co-repressor complexes to the core promoter region of IL-2 and epigenetically modify the promoter activity in Treg cells. (A) Knockdown of Eos by lentivirus-mediated RNA interference in Tregs. CD4+CD25high Treg cells were sorted from Balb/c mice and transduced with indicated lentivirus. 72h post-transduction, the CD4+GFP+ cells were sorted out and subject to Western blot and blot with anti-Eos(top), anti-Foxp3 (middle), and anti-actin, respectively. The target sequence for si-Eos was listed in the supplementary table 1. (B) Knockdown of Eos could reverse Foxp3-mediated IL-2 expression in T cells. CD4+ T cell were transduced with a retroviral vector (carrying RFP as an internal marker) expressing nothing (EV), full length Foxp3 or Foxp3 with the 148–198 Eos binding fragment deleted (ΔFoxp3). Cells were then retransduced with either the control siRNA expressing lentiviral vector or lentiviral vector (carrying GFP as an internal marker) expressing Eos siRNA (targeting to the 3′ UTR of Eos) in the presence or absence of a retroviral vector (carrying Thy1.1+ as an internal marker) expressing mouse Eos gene with one base pair mutation of the siRNA targeted region. 72h post-transduction, the RFP+GFP+ or RFP+GFP+Thy1.1+ cells were sorted out, and re-stimulated with anti-CD3/CD8, IL-2 production was assessed by ELISA. (C) Endogenous interaction between Eos and CtBP1 in Treg cells. Treg cells were lysed, subjected to immunoprecipitation (IP) with anti-CtBP1 antibody or control IgG. The immunoprecipitates were resolved on SDS-PAGE followed by Western blot analysis (WB) with anti-Eos (top) and anti-CtBP1 (Santa Cruz, bottom) antibodies, respectively. (D) Effect of Eos knockdown on the association between Foxp3 and CtBP1 in Treg cells. Eos knockdown in Treg cells were carried out with the same protocol as the panel A and B. Cell lysates were prepared from sorted siRL or si-Eos introduced Treg cells, followed by immunoprecipitation with anti-Foxp3 (eBioscience) or control IgG, and then analyzed by Western blotting with anti-CtBP1 (top), anti-Foxp3 (middle) and anti-Eos (bottom), respectively. (E) Effect of knockdown of Eos on indicated histone modifications at the Foxp3-binding region of the IL-2 promoter in Treg cells. The indicated cell population was sorter purified, followed by CHIP analysis (see material and method section for detail). (F) Knockdown of CtBP1 expression by RNA interference in Tregs. Cell lysates were prepared from sorted siRL(control) or si-CtBP1 transduced Treg cells, followed by Western blotting with anti-CtBP1(top), and anti-actin (bottom), respectively. The targeted sequence for si-CtBP1 is listed in the supplementary table 1. (G) CtBP1 siRNA decreases CtBP complex occupancy and H3 Lys9 methylation at the IL-2 promoter in Treg cells. siRL or si-CtBP1 treated Treg cells were sorted out and subjected to CHIP analysis. (H) Effect of knocking down CtBP1 on histone H3 and H4 modifications in the Foxp3-binding region at the IL-2 promoter in Treg cells. si-CtBP1 Treg cells were prepared (the same as panel F and G), and quantitative ChIP analysis was performed to determine the modification of histone at the IL-2 promoter using real-time PCR. (I) Effects of knockdown of CtBP1 on the suppressive activity of Treg cells in vitro. GFP+RFP+ cells from DO11.10/RAG2−/− CD4+ cells transduced with MIGR1-Foxp3-RFP/pFup3.1-siRL-GFP (closed circle), MIGR1-Foxp3-RFP/pFUP3.1-si-CtBP1-GFP (closed rhombus) or MIGR1-RFP/pFUP3.1-siRL-GFP (open circle) were cultured with 2×104 freshly isolated DO11.10 CD4+ in the presence of OVA peptide and 4×104 APCs (irradiated CD3- cells). [3H]-thymidine incorporation for the last 8 hrs of cell culture was measured as an indicator of cell proliferation and was expressed as the mean ± s.d. for triplicate cultures. (J) Effect of Eos and CtBP1 knockdown on the DNA methylation status at the core region of the IL-2 promoter in Treg cells. The results shown are the means ± s.d. of three independent experiments. All primers for the CHIP analysis are listed in the supplementary table 1.
Eos is known to interact with CtBP1(13), which has been shown to recruit co-repressor complexes to modify chromatin structures and silence gene expression(16–18). We hypothesized that Eos might serve as a co-repressor in Treg to recruit CtBP1 and its associated negative regulators of gene expression to Foxp3 in order to attenuate transcription at promoters of genes targeted for silencing by Foxp3, such as IL-2. To test this hypothesis, we determined the association between Eos and CtBP1 in Treg cells and assessed IL-2 promoter epigenetic status in Treg cells upon Eos knockdown. As shown in Fig. 2C, Eos indeed interacts with CtBP1 in Treg. Moreover, Eos not only co-immunoprecipitated with Foxp3, but also CtBP1, while knock-down of Eos blocks CtBP1 incorporation into Foxp3 suppressive complexes (Fig. 2D). These data suggest that CtBP1 is involved in the Foxp3/Eos suppression complex. It has been reported that CtBP1 exerts transcription suppression by affecting histone methylation and acetylation(16). In order to ascertain whether such was the case in Treg, chromatin immunoprecipitation (ChIP) analysis was applied to detect the histone modifications at the IL-2 promoter region in Treg cells by examining histone methylation (H3K4, H3K9) and acetylation (H3Ac, H4Ac). As shown in Fig. 2E, in vitro-primed effector CD4+ cells, which transcribe the IL-2 gene more rapidly than naïve cells upon activation(19), exhibited a 10 to 40-fold increase in histone trimethylation (Me3H3K4) and acetylation (H3Ac and H4Ac), but a concurrent decrease in histone methylation (MeH3K9) at the IL-2 promoter (Fig. 2E). However, Treg cells exhibited little or no change in histone methylation and acetylation at the IL-2 promoter. Indeed, upon knockdown of Eos, histone trimethylation (Me3H3K4) and acetylation (H3Ac and H4Ac) were significantly increased at the IL-2 promoter (Fig. 2E), but not at the CD3ε promoter that does not contain a Foxp3 binding site (data not shown). The epigenetic modification of histones at the IL-2 promoter is inferred to be due to abrogation of the Foxp3 interaction with CtBP1 in siEos Treg cells (Fig. 2D) since Foxp3 itself still binds to IL-2 promoter even after siEos knockdown (Fig. S5). It has been shown that CtBP1 associates with a co-repressor complex including EuHMT(G9a), which is responsible for histone 3 lysine 9 (H3K9) methylation in other cell types (20, 21). We thus used RNAi to determine whether CtBP1 is associated with the methylation of H3K9 at the IL-2 promoter in Treg cells. As shown in Fig. 2F, CtBP1 expression in si-CtBP1 Treg cells was significantly decreased. These cells were then analyzed by ChIP for histone H3K9 methylation, CtBP1 and EuHMT at the IL-2 promoter, and siRL transduced Treg cells were used as control (Fig. 2G). After inhibition of CtBP1 expression by RNAi, we observed a marked decrease in CtBP1 and EuHMT occupancy at the IL-2 promoter while there was no significant change to Foxp3/Eos occupancy. These changes were accompanied by a significant decrease in H3K9 methylation (MeK9, Fig. 2G). In addition, we observed that Histone H3 and H4 acetylation were enhanced upon si-CtBP1 treatment (Fig. 2H). At the functional level, knockdown of CtBP1 in Treg abrogated their suppressive activity (Fig. 2I), although without affecting Treg cells proliferation itself (data not shown). Similar epigenetic histone modifications were also observed at the IFN-γ promoter in Treg cells (fig. S6).
Another epigenetic modification that regulates IL-2 gene expression is methylation of genomic DNA at CpG dinucleotides in the promoter. We thus determined whether knockdown of Eos might influence the level of DNA methylation at the IL-2 locus. Given the documented correlation between methylation status of the −68 CpG dinucleotide in the IL-2 promoter region and IL-2 transcription (22, 23), we examined the DNA methylation status of the CpG islands flanking the −68 region within the IL-2 promoter using a methylation-sensitive PCR assay in Treg cells. As shown in Fig. 2J, knockdown of Eos or CtBP1 led to demethylated CpG dinucleotides at the IL-2 promoter upon CD3 and CD28 treatment. Taken together, these findings lend further support to the notion that Eos/CtBP represses the IL-2 promoter through epigenetic modification mechanisms in Treg.
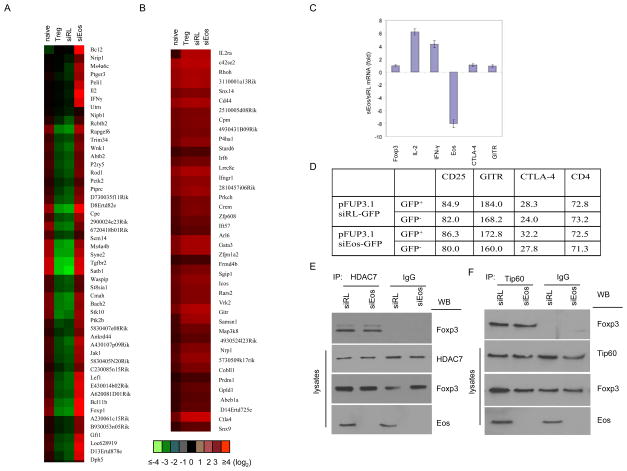
While the data presented thus far strongly supports a central role for Eos in mediating Foxp3-dependent gene silencing of well characterized index genes (ie IL-2) in Treg, we sought to determine whether Eos played a broader role in Treg gene silencing and also whether that role was selective for gene silencing vs gene activation. Using the Foxp3 dependent gene set from natural Treg defined by Rudensky and colleagues (6) as a basis for comparison, we analyzed changes in global gene expression patterns from Eos-siRNA vs. control RL-siRNA transduced Treg. This analysis revealed that the majority (>70%) of the si-Eos affected genes were Foxp3-dependent (fig. S7). Among them, 90% of the genes known to be suppressed by Foxp3 were no longer down-regulated upon Eos knockdown in Treg (Fig 3A). In striking contrast, expression of the vast majority (>95%) of known Foxp3-dependent up-regulated genes from Rudensky and colleagues (6) were unaltered by Eos knockdown (Fig. 3B). Consistent with the microarray data, qRT-PCR (Fig. 3C) and cell surface staining (Fig. 3D) further supported the notion that the expression of some Foxp3 up-regulated genes such as CD25, cytotoxic T-lymphocyte-associated antigen-4 (CTLA-4) and glucocorticoid-induced TNF-receptor-family-related protein (GITR) remains unchanged. The expression of some known Foxp3-interacting proteins such as Tip60 and HDAC7 was not significantly changed upon si-Eos treatment, and the interaction between Foxp3 and HDAC7/Tip60 is Eos-independent (Fig. 3E, F), although HDAC7 could associate with Eos suppression complexes in Treg cells (Fig. S8). Thus, global gene expression analysis, analysis of specific index genes and analysis of selected cell membrane proteins all demonstrate that Eos and its co-repressor are specifically involved in Foxp3-mediated gene silencing, but not gene activation, in Treg cells.
Fig. 3.
Gene expression differences between siEos versus siRL-transduced Treg cells. Shown are the genes of up- (A) and down- (B) regulated transcripts in siEos vs. siRL-transduced Treg cells. Naïve T cell and nTreg cell gene expression profiling was carried out in parallel. siRL transduced Treg cell gene expression signature was comparable with non-transduced nTreg. Foxp3 dependent up- or down- regulated genes were compared with the databases reported by Zheng et al [Nature. 445,936 (2007)]. Genes with expression increased or decreased by two-fold or more in Treg vs. naïve T cells were considered up-or down-regulated, respectively, by Foxp3. (C) Knockdown of Eos enhances IL-2 and IFN-γ gene expression in Treg cells. Si-Eos and siRL Treg cells were sorted out and subjected to q-RT PCR assay using indicated gene specific primers. The primer sequences were listed in the supplementary table 1. (D) Mean fluorescence intensity for various cell surface molecules on CD25+CD4+ cells transduced with pFUP3.1siRL-GFPor pFUP3.1-siEos-GFP (see methods section for the antibodies). (E, F) The association between Foxp3 and Tip60 or HDAC7 in Treg cells remains unchanged upon si-Eos treatment. Cell lysates were prepared from sorted siRL or si-Eos transduced Treg cells, followed by immunoprecipitation with (E) anti-HDAC7, (F) anti-Tip60 (Santa Cruz) or control IgG, then analyzed by Western blotting with anti-Foxp3. The whole cell lysates were blotted with (E) anti-HDAC7, (F) anti-Tip60, anti-Foxp3 and anti-Eos as indicated.
The selective effect of Eos knockdown on Foxp3-repressed genes without altering expression of Foxp3-activated genes prompted us to determine the specific role of Foxp3 repressed genes on Treg function. We thus examined the effects of Eos knockdown on the function of Treg in vitro. Sorted Treg, transduced with the same siEos lentivirus as above, were subjected to an in vitro suppression assay. While both fresh wt-Treg and si-RL transduced Treg were capable of suppressing the proliferation of normal T cells, Treg with Eos knockdown lost their suppressive activity (Fig. S9), which could be rescued by a functional Eos construct (human Eos, hEos) that bears a mutation in the siRNA targeted region (data not shown). Notably, si-Eos Treg cells themselves could proliferate to some extent upon stimulation with anti-CD3 and CD28 compared to the wt or si-RL Treg cells (Fig. S10). To further confirm that Eos is involved specifically in Foxp3 mediated suppression, we examined the effects of Eos knockdown on the suppressive activity of Foxp3-transduced naïve CD4+ T cells invitro. It has been reported that ectopic expression of Foxp3 confers suppressive activity on naïve T cells (24). Recombination-activating gene (RAG)-deficient (Rag2−/−) DO11.10 CD4+ T cells, which express ovalbumin (OVA) peptide-specific transgenic TCRs, were used for these studies. In the absence of OVA, almost all T cells from Rag2−/− DO11.10 mice remained in a naïve state. As shown in Fig. 4A, transgenic CD4+ T cells transduced with MIGR1-Foxp3-RFP suppressed the proliferation of freshly prepared non-transduced transgenic T cells upon stimulation with specific OVA peptides, whereas those transduced with the control MIGR1-RFP or MIGR1-ΔFoxp3-RFP mutant (with 148–198 deletion, which is unable to interact with Eos) did not. Also, we found that lentivirus-mediated siRNA knockdown of Eos reduced the suppressive activity of Foxp3-expressing antigen-specific CD4+ T cells, which could be rescued by re-transduction with RV-Eos. If the Foxp3-Eos interaction is crucial in Foxp3-mediated suppression, we reasoned that expression of a ΔFoxp3/hEos fusion protein (ΔFoxp3 fused with human Eos that does not contain the target sequences for our Eos siRNA) should be able to rescue the defective suppression phenotype of MIGR1- ΔFoxp3-RFP transduced T cells. Indeed, when a chimeric construct was used, it exerted suppression activity comparable to the wt Foxp3 (Fig. 4A). Moreover, si-Eos treatment could no longer block the chimeric Foxp3/hEos-mediated suppression (Fig. 4A). As expected, neither Eos alone nor Eos co-transfected with ΔFoxp3 could rescue the Eos-knockdown phenotype (data not shown). Foxp3 protein levels remained unchanged upon siEos treatment (Fig. S11). Taken together, these results suggested that the association between Eos and Foxp3 is required for Foxp3-mediated suppressive activity of Treg cells.
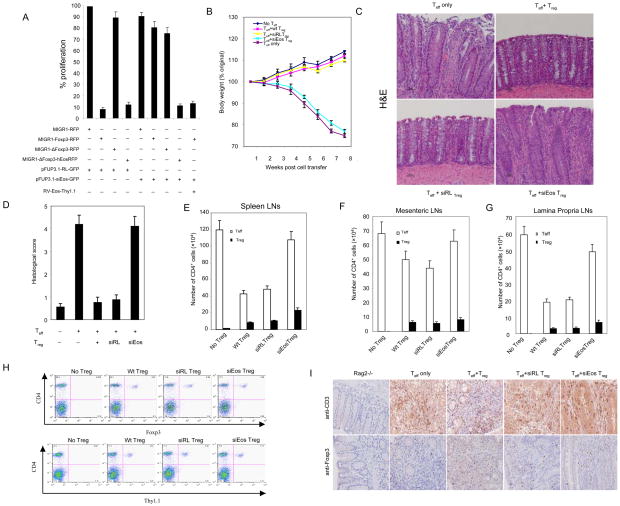
Fig. 4.
Effects of knockdown of Eos on the suppressive activity of Treg cells invitro and in vivo. (A) Knockdown of Eos by siRNA reduces the suppressive activity of Foxp3-transduced naïve T cells. Sorted GFP+RFP+ or GFP+RFP+Thy1.1+ T cells from DO11.10/RAG-2−/− CD4+ cells transduced with the indicated viruses were co-cultured with 2×104 freshly isolated and CFSE labeled DO11.10 CD4+ (Thy1.2+, ratio 1:1) in the presence of OVA peptide and 4×104 APCs (irradiated CD3- cells) for 80h, followed by surface marker, Foxp3 and Ki67 staining. To quantify both frequency and absolute cell number, all cells under each condition were collected. Cell proliferation was determined by CFSE dilution and upregulation of Ki67. The proliferation index was calculated: the absolute number of Ki67+ CFSE diluting Thy1.2+CD4+Foxp3- T responder cells (co-cultured with the indicated Treg cells at 1:1 ratio) were divided by the number of proliferating T responder alone. The results were expressed as the mean ± s.d. for triplicate cultures. (B) Percentage weight change after transfer of the indicated cells demonstrates that co-transfer of siEos transduced Treg cannot suppress weight loss induced by transfer of CD4+CD25− CD62Lhigh Teff into Rag2−/− mice. (C) Representative photomicrography of the distal colon of Rag2−/− mice following T cells transfer. CD4+CD25− CD62Lhigh cells isolated from WT BALB/c were able to induce colitis in BALB/c Rag2−/− recipients (top left). Co-transfer of WT CD4+CD25+ Treg cells (top right) or siRL-transduced Treg (bottom left) was able to prevent colitis. However, siEos-transduced Treg had no effect on colitis (bottom right). Data represent 9–12 mice per group in each panel. (D) Colonic histology scores of experimental mice. Colons were removed from mice 8 weeks after T cells reconstitution and fixed in 10% neutral buffered formalin. Five-micrometer paraffin-embedded sections were cut and stained with haematoxylin and eosin (H&E). Pathology of the colon was evaluated by routine microscopic examination, then scored blindly using a semi-quantitative scale of zero to five (see method section for detail). 9–12 mice were used in each group. (E–G) Analysis of spleen (E), mensenteric LN (F), and lamina propria LN (G) Teff (CD4+Foxp3−) and Treg (CD4+Foxp3+) cell numbers. Foxp3 expression was determined by intracellular staining (eBioscience). (H) Comparable Treg cells are found in Eos deficient and control Treg recipient mice. FACS analysis of spleen and draining mesenteric lymph nodes of mice 8 weeks post-adopt transfer T cells in the model of IBD. Sorted Thy1.1+ Treg cells transduced with siRL or siEos were cotransfered with or without CD4+CD25−CD62Lhi T cells marked as Thy1.1− into Rag2−/− mice. 8 weeks post adoptive transfer, pooled spleen and lymph nodes were stained with APC-CD4 together with FITC-Foxp3 or Thy1.1. Data shown are a representative staining of three independent experiments. (I) Immunohistochemistry (IHC) staining of CD3+ or Foxp3+ T cells in colon tissue. IHC staining of paraffin sections of colon was performed by a peroxidase technique, using the Vectastain Elite ABC kit (Vector Laboratories, Burlingame,CA) and diaminobenzidine substrate kit (Vector). CD3+ T cells were visualized using goat anti-CD3 polyclonal antibodies (Santa Cruz, top panel), and Foxp3+ cells were visualized with rat anti-Foxp3 clone FJK-16 antibody (eBioscience, bottom panel). Haematoxylin serves as a counterstaining (magnification, x 20). Substitution of irrelevant antibodies of the same isotype was used as a control (not shown). The IHC analysis was performed on five mice per group.
Lastly, we determined the role of Eos in the function of Treg cells in an in vivo colitis model (inflammation bowel disease, IBD) in which severe T helper type-1 (Th1)-mediated inflammation within the colon is induced following the transfer of naïve CD4+CD25−CD62Lhi T cells in the absence of Treg into immunodeficient recipients (25). The co-transfer of CD4+CD25+ Treg cells is sufficient to prevent the development of disease (26). We used this IBD model to test the function of si-Eos transduced Treg in vivo. Purified wild type BALB/c CD4+CD25−CD62Lhi Thy1.2 T cells, either alone or in the presence of wild type, si-RL or si-Eos transduced Thy1.1 Treg, were adoptively transferred into Rag2−/− mice. Disease progression was monitored by weight loss and colitis assessed by histological analysis of colon tissue sections (27). As shown in Fig. 4B–D, wild-type or si-RL modified Treg cells were able to effectively suppress the development of disease as judged by both body weight loss (Fig. 4B) and histological analysis of tissue isolated from the colon (Fig. 4C, D). However, transfer of si-Eos Treg cells in the presence of CD4+CD25−CD62Lhi T effector cells failed to prevent the disease development (Fig. 4B–D). In vivo tracking of transferred cells demonstrated that the colitis that developed in mice receiving CD4+CD25−CD62Lhi T cells and si-Eos transduced Treg was not due to impaired accumulation or homing Foxp3 Treg cells to the mesenteric lymph nodes or spleen (Fig. 4E–I). Given that Eos knockdown de-represses effector cytokine genes in Treg, we used the in vivo colitis model to determine whether Treg developed effector function in the absence of Eos. Indeed, transfer of higher doses of pure si-Eos Treg cells (5×105) into Rag2−/− mice produced colitis (Fig. S12–14), which was significantly less severe than that caused by transfer of similar numbers of Teff. At lower doses of transferred cells (2×105), Teff still induced colitis whereas si-Eos Treg did not (Fig. S13). Thus, we conclude that Eos knockdown alone partially converts Treg into functional T effector cells. This finding is consistent with our finding that the inhibitory functions associated with Foxp3 activated genes in Treg are unaffected by Eos knockdown. Taken together, these results demonstrate that Eos is required for the regulatory activity of Treg in vivo.
Collectively, our findings demonstrate that Foxp3 suppresses gene expression through its interaction with Eos and define Eos as a key missing link in this process. Unlike other Foxp subfamily members, the mechanisms of Foxp3 mediated suppression have been undefined since Foxp3 lacks an intrinsic motif for recruiting negative regulators of gene transcription, which were known to be crucial for Foxp1, 2 and 4 function (28–30). We postulate that the unique requirement for a distinct molecule, Eos, to coordinate gene repression by Foxp3 reflects the evolution of a greater complexity and plasticity of gene regulation in Treg programming than those mediated by other Foxp members in other cell types. The N-terminal region (200 aa) of Foxp3 has been reported to contain a potential “repressor” domain that is necessary and sufficient to repress gene transcription in several previous studies (4, 8–10, 29). Tip60 and HDCA7 have been reported to interact with the N-terminal region. However, knockdown of Tip60 only partially diminishes the suppressive activity of Foxp3(28), and here we found that knockdown of HDAC7 did not affect the expression of Foxp3-dependent genes such as IL-2 and IFN-γ (Fig. S15). These results suggested that another molecule was critical to this process. Our data showed that knockdown of Eos could completely abolish the suppressive activity in Treg in vitro and in vivo, although the association between Tip60/HDAC7 and Foxp3 was not affected upon Eos knockdown (Fig. 3E, F). In fact, the function of Tip60 instead appears to be involved in Foxp3 post-translational modification and enhancement of binding of Foxp3 to the promoters of its target genes(31).
Foxp3 has also been shown to interact with the transcription factor NFAT and AML1/Runx1 to orchestrate the cellular and molecular program involved in Treg function(1). A recent study has shown that NFAT interacts with C-terminal region of Foxp3, and facilitates Foxp3-dependent gene activation and suppression(4). AML1/Runx1 has been reported to bind to the IL-2 promoter constitutively, and might facilitate the assembly of transcriptional activator complexes including NFAT to activate IL-2 gene expression upon T cell activation. In Treg cells, AML1/Runx1 physically binds to Foxp3 at its N-terminal region between the leucine zipper domain and the forkhead box. Disrupting the interaction between Foxp3 and AML1/Runx1 impaired the Foxp3-dependent suppression of IL-2 gene expression and attenuated the suppressive activity without affecting the association between Foxp3 and NFAT(8). However, deletion of the N-terminal region of Foxp3 totally abrogated its suppressive function even though this Foxp3 mutant could still bind to Runx1 and NFAT(4). Here, we have identified Eos, a member of the Ikaros transcription factor family, as another important partner with Foxp3, capable of associating with its N-terminal 51 amino acid region (148-198aa). Furthermore, ΔFoxp3 (with 148-198aa deletion), although retaining the capacity to bind Runx1 and NFAT, could no longer suppress IL-2 promoter activity and lacked suppressive function. Thus, Eos is required for Foxp3-mediated gene silencing. We further demonstrated that Eos directly interacts with Foxp3 in Treg. Eos appears to serve as a co-repressor to recruit CtBP1 associated repression complexes to the promoter region of Foxp3 target genes, leading to epigenetic modification of the target genes’ chromatin structure to achieve the goal of gene silencing. In contrast, Eos does not affect Foxp3-dependent gene activation in Treg. Various genes previous reported to be up-regulated in Treg such as CTLA-4 and GITR did not exhibit significant changes in gene expression upon knockdown of Eos. However, the same Eos knockdown reversed the transcriptional repression of genes that are targeted by Foxp3, such as IL-2 and IFN-γ. These results suggest that Eos plays an essential and selective role in Treg function through the recruitment of CtBP1 and its associated negative transcriptional regulators, thereby allowing selective gene silencing in Treg. The abrogation of functional Treg activity upon Eos knockdown demonstrates that, apart from genes activated in Treg, gene silencing is critical to their function. Our findings raise the possibility that the interaction between Foxp3 and Eos along with its associated co-repressor complexes might be a potential therapeutic target for controlling physiological and pathological immune responses. This may be particularly true in the setting of cancer or chronic infections, where excessive Treg activity seems to play an important role. Further dissection of the subset of genes silenced by Eos/Foxp3 complexes will likely lead to the identification of downstream molecules involved in the regulation of Treg function.
METHODS
Mice
Rag2−/− DO11.10 and Rag2−/− Balb/c mice were initially provided by H. Levisky. They were subsequently obtained from our own breeding colony housed at Johns Hopkins School of Medicine. Foxp3 GFP mice were kindly provided by A. Rudensky. All animal experiments were performed in specific-pathogen-free, Helicobacter-free facilities in the Johns Hopkins Animal Resource Center following national, state and institutional guidelines. Animal protocols were approved by the Johns Hopkins Animal Care and Use Committee.
RNA interference and In vitro suppression assays for Treg cells
Sorted CD4+CD25high Treg cells (defined as the high 2% fraction of CD4+ T cells) were transduced with lentivirus-mediated siRNA against Eos, with CMV driven-GFP as a marker for transduction efficiency. siRNA against Renilla luciferase (RL) was used as a control. At 16 h after transduction, 20U ml−1 human recombinant IL-2 (eBioscience) was added to the culture. At 40 h after transduction, GFP+ Treg cells were sorted for western blot, immunoprecipitation, and/or suppression assay as indicated. CD4+CD25− T cells were similarly sorted and used as responders. CD3− cells were irradiated and used as APCs. A total of 10,000 Treg cells were mixed with 20,000 APCs with or without 10,000 responder cells. Cells were stimulated with 100ng/ml anti-CD3 (HIT3a, BD Pharmingen) for 4 days. [3H] Thymidine incorporation for the last 8 h of cell culture was measured as an indicator of cell proliferation and is expressed as the mean ± s.d. for triplicate cultures.
Retroviral transduction
For co-transduction experiments, cells were first transduced with pFUP3.1-siEos-GFP or pFup3.1-siRL-GFP and kept for additional 8h at 37°C with 5%CO2 and were subsequently transduced with pMIG-Foxp3-RFP. Gene transduction into CD4+CD25− conventional T cells and CD4+CD25+ Treg cells was performed by stimulating cells with plate-bound anti-CD3 (10μg ml−1) and soluble anti-CD28 (1μg ml−1) with 60U ml−1 human recombinant IL-2 for 16 h. Activated T cells were transduced with viral supernatants supplemented with 60U ml−1 IL-2 and 8 μg ml−1 polybrene, followed by centrifugation for 1h at 2,500 rpm. Cells were cultured at 37°C with 5% CO2 for an additional 24 h and sorted out for ELISA analysis.
CHIP
CHIP analysis was carried out according to the manufacturer’s protocol (Upstate/Millipore, Massachusetts, USA). Cells (1–5×106) were fixed with 1% formaldehyde, and chromatin was precleared by incubation with Protein A/G agarose/salmon sperm DNA (Upstate/Millipore). Subsequently, chromatin was immunoprecipitated by overnight incubation at 4°C with 4μg antibodies (rabbit isotype, Santa Cruz; anti-acetyl-histone H3, H4, anti-trimethyl-K4 histone H3 and anti-methyl-K9 histone H3, Upstate/Millipore, USA) followed by incubation with protein A/G agarose/salmon sperm DNA for 1h. Precipitates were defixed and DNA was purified. The amount of immunoprecipitated DNA was quantified by real-time PCR with the ABI PRISM 7500 Sequence Detection System (Applied Biosystems) using SYBR Green and the primer pair was listed in the supplementary table 1.
Methylation-sensitive PCR (MSP) assay
MSP assay was performed by using the Promoter methylation PCR kit (Panomics, Redwood City, CA) following the manufacturer’s instruction. Briefly, genomic DNA from T cells was prepared using the DNeasy Tissue Kit (Qiagen). Genomic DNA was digested with MseI (NEB). The methylated DNA fragments were isolated by a spin column affinity purification using MeCP2, and were amplified by PCR. The following primer was used for PCR analysis of methylated DNA: forward 5′-CCAGAGAGTCATCAGAAGAGGAA-3′ and reverse 5′-AATAATATGGGGGTGTCACGA-3′. This primer amplified a 109 bp region including the −68 CpG islands with −157 to −49 of IL-2 promoter.
RNA, complementary DNA and quantitative real-time PCR
T-cell RNA was isolated from whole cells using the Qiagen miniRNA extraction kit following the manufacturer’s instructions. RNA was quantified spectrophotometrically, and complementary DNA was reverse-transcribed using the cDNA archival kit (Applied Biosystem) following the manufacturer’s instruction. The cDNA samples were distributed on plates at 20ng/well and run in triplicate. PCR reactions were set up in 25-μl volumes using TaqMan Universal PCR Master Mix (Applied Biosystem) on an ABI Prism 7500 Sequence Detection System. Quantification of relative mRNA expression was determined by the comparative CT (critical threshold) method where the amount of target mRNA, normalized to endogenous β-actin or 18s rRNA expression, is determined by the formula 2−ΔCT. Primer sequences were listed in the supplementary table 1.
Immunoprecipitation and western blotting
Immunoprecipitation and western blotting were performed as described previously(15). Immunoprecipitation experiments were carried out by using Pierce crosslink IP kit and clearn-Blot IP detection system (Thermo Scientific). The peptide SSGDSSLEKDSL (corresponding to amino acides[aa] 58 to 69 of mouse Eos was used to make specific antibody against Eos in rabbits (QCB/Biosource, Hopkinton, MA).
GST pull-down
pCDNA3-Foxp3 or pCDNA3-Eos were labeled by incorporation of S35-labelled methionine during in vitro translation (TNT coupled reticulocyte lysate system, Promega Corporation). Five microliters of S35-labelled protein was incubated with 3μg of the affinity purified GST protein (with GSTrap column, Amersham) as indicated in the presence of 0.2%BSA in BC100 (20mM Tris pH7.9, 100mM NaCl, 10%glycerol, 0.2mM EDTA, 0.1% Triton X-100) on a rotator overnight at 4°C. The proteins were pulled down using GST beads, followed by five times washes with BC100 before elution with 50μl of BC100 plus 20mM reduced glutathione for 10 mins with gentle rotation. Eluted materials were resolved on SDS-PAGE. The presence of S35-labelled protein was detected by autoradiography.
Colitis induction and histological assessment
Naïve CD4+CD25−CD62Lhigh T cells were isolated from BALB/c and injected via tail vein (i.v.) into BALB/c RAG2−/− immunodeficient recipients (1×106/mouse). BALB/c wild-type CD4+CD25+ Treg or siRL/siEos transduced Treg cells (2×105) were co-injected i.v. where indicated. Mice were monitored weekly for wasting disease and mice losing more than 20% of its starting body weight or showing severe signs of disease were sacrificed. Colons were removed from mice 8 weeks after T cells reconstitution and fixed in 10% formalin. Five-micrometer paraffin-embedded sections were cut and stained with haematoxylin and eosin (H&E). CD3+ T cells were visualized using goat anti-CD3 polyclonal antibodies (Santa Cruz) and diaminobenzidine chromagen with haematoxylin as a counterstain. Foxp3+ cells were visualized with rat anti-Foxp3 clone FJK-16 antibody (eBioscience). Pathology of colon tissue was scored in a blinded fashion, on a scale of 0–5 where a grade of 0 was given when there were no changes observed. Changes associated with other grades were as follows(32): grade 1, minimal scattered mucosal inflammatory cell infiltrates, with or without minimal epithelial hyperplasia; grade 2, mild scattered to diffuse inflammatory cell infiltrates, sometimes extending into the submusoca and associated with erosions, with mild to moderate epithelial hyperplasia and mild to moderate mucin depletion from goblet cells; grade 3, moderate inflammatory cell infiltrates that were sometimes transmural, with moderate to severe epithelial hyperplasia and mucin depletion; grade 4, marked inflammatory cell infiltrates that were often transmural and associated with crypt abscesses and occasional ulceration, with marked epithelial hyperplasia, mucin depletion; and grade 5, marked transmural inflammation with severe ulceration and loss of intestinal glands.
Isolation of lamina propria leukocytes (LPL) and FACS analysis
Mice (pooled from 4–6 mice per group) were euthanized and the colons excised and placed in ice-cold PBS. After extensive washing of the colonic lumen with PBS, colons were minced into 0.3–0.5cm pieces, and repeatedly incubated in Ca- and Mg-free HBSS containing 10% FCS and 5mM EDTA to release intestinal epithelial cells. The remaining tissue was further incubated with a digestion cocktail [Liberase and DNaseI (Roche)] at 37°C for 1h, and the LPL were then layered on discontinuous Percoll gradient (Amersham Biosciences). After centrifugation for 30 min at 2000 rpm at room temperature, the LPL population was recovered at the 40%/75% interphase, washed twice in HBSS/BSA, and counted by trypan blue staining. Cells were stained first with surface staining antibodies and then fixed/permeablizied, followed by intracellular cytokine staining (eBioscience). Flow cytometric analysis was performed on FACSCalibur (BD Bioscience) and data was analyzed using FlowJo software (Tree Star Inc.). The antibodies used were CD62L, CD25, GITR, CTLA-4, Thy1.1, Thy1.2 (all from BD Biosciences) and CD4, Foxp3 (from eBioscience).
Microarrays
Total RNA was isolated from sorted cell populations using TriZol reagent (Invitrogen) followed by RNeasy mini kit with DNase on-column digestion (Qiagen). RNA was quantified with NanoDrop ND-1000 followed by quality assessment with 2100 Bioanalyzer (Agilent Technologies) according to manufacturer’s protocol. Sample amplification and labeling procedures were carried outby using Low RNA Input Fluorescent Linear Amplification Kit (Agilent Technologies) with minor modifications. Briefly, 0.4 ug total RNA was reverse-transcribed into cDNA by MMLV-RT using an oligo dT primer (System Biosciences) that incorporates a T7 promoter sequence. The cDNA is then used as a template for in vitro transcription in the presence of T7 RNA polymerase and Cyanine-3 labeled CTPs. The labeled cRNA is purified using RNeasy mini kit (Qiagen). RNA spike-in controls (Agilent Technologies) are added to RNA samples before amplification and labeling according to manufacturer’s protocol.0.825 ug of each Cy3-labeled samples was used for hybridization on Agilent 4x 44K whole mouse genome microarray which contains 41,174 unique probes (G4122F) at 65°C for 17 hours in a hybridization oven with rotation. After hybridization, microarrays are washed and dried according to Agilent microarray processing protocol using stabilization and drying solution. Microarrays were scanned using an Agilent Scanner controlled by Agilent Scan Control 7.0 software. Data were extracted with Agilent Feature Extraction 9.5.3.1 software. Agilent FE processed signal intensities were imported into GeneSpring GX 9.0.1 (Agilent Technologies). Normalization was done with all intensities higher than 5, log2 transformation and cross-array quartile normalization. Only transcripts with both an adjusted P value < 0.05 and a fold-change of > 2.0 were considered differentially expressed.
Supplementary Material
Supplementary Table S1: Primer/Oligo Sequences
Jurkat T cells were cotransfected with expression plasmids HA-tagged or Flag-Eos together with MigR1-Foxp3, HA-Ikaros or Myc-Helios as indicated, then immunoprecipitated with anti-HA or Flag M2, followed by Western blotting with either (a) anti-Foxp3 (eBioscience); (b) anti-HA (sigma); or (c) anti-Myc (sigma), respectively. (d)The N-terminal portion of Foxp3 is required for the physical interaction with Eos. HA-tagged Eos was co-transfected with Flag-tagged full-length Foxp3, ΔC-Foxp3 (2-320aa), or ΔN-Foxp3 (199-432aa) into HEK293T cells and immunoprecipitated with anti-Flag antibody, followed by western blot with anti-HA (top) or anti-Flag (middle) antibodies. The expression of Eos in cell lysates was detected by immunoblotting with an anti-HA antibody (bottom).
Sorted GFP+Foxp3+ (from GFP-Foxp3 mice) Treg cells were subjected to co-immunoprecipitation (A) and CHIP assay (B), separately. Treg cells were lysed and immunoprecipitated with anti-Eos antibodies or control IgG. The immunoprecipitates were resolved on SDS-PAGE followed by Western blot analysis with anti-Foxp3 (top) and anti-Ikaros (bottom) antibodies, respectively. For CHIP assay, sorted Treg cells were treated with mock or anti-CD3 and anti-CD28 for 4hrs and subjected to Ikaros CHIP to examine Ikaros occupancy of the IL-2 promoter region. The primers were listed in the supplementary table 1.
Sorted GFP+RFP+ cells from DO11.10/RAG2−/− CD4+ cells transduced with the indicated viruses were subjected to SDS-PAGE, followed by Western blotting with anti-Eos (Top), anti-Foxp3 (eBioscience; Middle) and reprobed with anti-actin antibody (Santa Cruz, Bottom).
Jurkat T cells were cotransfected with expression plasmids Myc-tagged ΔFoxp3 together with HA-NFAT, HA-AML1 or HA-CtBP1 as indicated, then immunoprecipitated with anti-Myc (Sigma), followed by Western blotting with either (a) anti-NFAT; (b) anti-AML1; or (c) anti-CtBP1 (Santa Cruz), respectively.
Sorted si-RL or si-Eos Treg cells were treated with mock or anti-CD3 and anti-CD28 for 4hrs and subjected to Foxp3 CHIP to examine Foxp3 occupancy of the IL-2 promoter region.
Quantitative ChIP analysis was performed to determine the modification of histone H3 and H4 at the IFN-γ promoter using real-time PCR. The results shown are the means ± s.d. of three independent experiments.
Shown are the numbers of up- and down-regulated transcripts in siEos vs. siRL transduced Treg cells. Naïve T cells and nTreg cells gene expression profiling was carried out in parallel. siRL transduced Treg cells gene expression signature was comparable with the untransduced nTregs. Foxp3 dependent up- or down- regulated genes were compared with the databases reported by Zheng et al [Nature. 445,936 (2007)]. In the siEos transduced Treg cells, 62 genes were up-regulated in siEos Treg cells relative to the siRL transduced Treg cells. Of these genes, ~70% were previously identified as Foxp3-dependent down-regulated genes (51). The majority of genes (40) known to be up-regulated by Foxp3 remains unchanged in both groups. Genes with expression increased or decreased by two-fold or more in Treg vs. naïve T cells were considered up-regulated or down-regulated by Foxp3.
Treg cells were lysed, subjected to immunoprecipitation (IP) with anti-Eos antibody or control IgG. The immunoprecipitates were resolved on SDS-PAGE followed by Western blot analysis (WB) with anti-HDAC7 (top) and anti-Eos (Santa Cruz, bottom) antibodies, respectively.
CFSE labeled Thy1.1+ T responder cells were co-cultured with or without wt or siEos Treg cells (Thy1.2+) for 80 hrs, followed by surface marker staining. Proliferation was assessed by CFSE dilution.
Modified Tregs sorted from lentivirus transduced Treg cells were stimulated with indicated reagents for 72h. [3H]-thymidine incorporation for the last 8 hrs of cell culture was measured as an indicator of cell proliferation and was expressed as the mean ± s.d. for triplicate cultures.
Sorted GFP+RFP+ cells from DO11.10/RAG2−/− CD4+ cells transduced with the indicated viruses were subjected to SDS-PAGE, followed by Western blotting with anti-Foxp3 (eBioscience; Top) and reprobed with anti-actin (Sigma, Bottom).
Higher dose of siEos transduced Treg cells only could cause weight loss in the Rag2−/− mice.
Rag2−/− mice received higher dose (A) or lower dose (B) of si-Eos Treg cells as indicated. After 6–8 weeks adoptive transfer, histological analysis of mice was performed as described in Figure 4. Data represent 6–8 mice per group in each panel.
CD4+CD25−CD62Lhigh cells isolated from Wt BALB/c were able to induce colitis in BALB/c Rag2−/− recipients (panel a,e). Higher dose of siEos-Treg cells (1×106) only could induce lesion upon adoptive transfer into Rag2−/− recipients (panel b,f). The same dose of wt Treg or siRL Treg cells did not cause observed colitis disease (panel c,d and g,h). The histological sections from each group were stained with H&E (panel a to d), or Alcian blue (panel e to h). Data represent 6–8 mice per group.
CD4+CD25high T cells were isolated from BALB/c mice and transduced with indicated lentivirus. 72h post-transduction, GFP+CD4+ T cells were sorted and subjected to Western blot (A) and q-RT PCR assay (B). Si-Eos group was carried out as experimental control. The primer sequences were listed in the supplementary table 1.
Acknowledgments
We would like to thank members of the Pardoll laboratory and G. Zhou for helpful discussion, KJ Rhee for pathology assistance, and A. Rudensky (University of Washington), Y. Shi (Harvard Medical School), M. Crossley (University of Sydney, Australia) and S. Smale (University of California at Los Angeles) for plasmids, H Levisky (Johns Hopkins University) for providing Rag2−/− BALB/c and DO11.10 breeding colony. In addition, we would like to thank CV Dang and JD Powell for helpful suggestions. This work was supported by grants from the US National Institutes of Health, the Janey Fund and Seraph Foundation, and gifts from Bill and Betty Topecer and Dorothy Needle.
References
- 1.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Cell. 2008 May 30;133:775. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Maloy KJ, Powrie F. Nat Immunol. 2001 Sep;2:816. doi: 10.1038/ni0901-816. [DOI] [PubMed] [Google Scholar]
- 3.Shevach EM. Annu Rev Immunol. 2000;18:423. doi: 10.1146/annurev.immunol.18.1.423. [DOI] [PubMed] [Google Scholar]
- 4.Wu Y, et al. Cell. 2006 Jul 28;126:375. [Google Scholar]
- 5.Marson A, et al. Nature. 2007 Feb 22;445:931. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Y, Rudensky AY. Nat Immunol. 2007 May;8:457. doi: 10.1038/ni1455. [DOI] [PubMed] [Google Scholar]
- 7.Hill JA, et al. Immunity. 2007 Nov;27:786. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Ono M, et al. Nature. 2007 Apr 5;446:685. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- 9.Li B, et al. Proc Natl Acad Sci U S A. 2007 Mar 13;104:4571. doi: 10.1073/pnas.0700298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bettelli E, Dastrange M, Oukka M. Proc Natl Acad Sci U S A. 2005 Apr 5;102:5138. doi: 10.1073/pnas.0501675102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang CT, et al. Immunity. 2004 Oct;21:503. doi: 10.1016/j.immuni.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 12.Fisson S, et al. J Exp Med. 2003 Sep 1;198:737. doi: 10.1084/jem.20030686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perdomo J, Crossley M. Eur J Biochem. 2002 Dec;269:5885. doi: 10.1046/j.1432-1033.2002.03313.x. [DOI] [PubMed] [Google Scholar]
- 14.Fearon ER, et al. Proc Natl Acad Sci U S A. 1992 Sep 1;89:7958. doi: 10.1073/pnas.89.17.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan F, et al. Nature. 2007 Jan 25;445:433. [Google Scholar]
- 16.Shi Y, et al. Nature. 2003 Apr 17;422:735. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 17.Koipally J, Georgopoulos K. J Biol Chem. 2002 Aug 2;277:27697. doi: 10.1074/jbc.M201694200. [DOI] [PubMed] [Google Scholar]
- 18.Hu R, et al. Mol Cell Biol. 2007 Jun;27:4018. doi: 10.1128/MCB.01839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sojka DK, Bruniquel D, Schwartz RH, Singh NJ. J Immunol. 2004 May 15;172:6136. doi: 10.4049/jimmunol.172.10.6136. [DOI] [PubMed] [Google Scholar]
- 20.Heo K, et al. J Biol Chem. 2007 May 25;282:15476. doi: 10.1074/jbc.M610270200. [DOI] [PubMed] [Google Scholar]
- 21.Tachibana M, et al. Genes Dev. 2002 Jul 15;16:1779. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murayama A, et al. Embo J. 2006 Mar 8;25:1081. doi: 10.1038/sj.emboj.7601012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruniquel D, Schwartz RH. Nat Immunol. 2003 Mar;4:235. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 24.Hori S, Nomura T, Sakaguchi S. Science. 2003 Feb 14;299:1057. [Google Scholar]
- 25.Powrie F, Leach MW, Mauze S, Caddle LB, Coffman RL. Int Immunol. 1993 Nov;5:1461. doi: 10.1093/intimm/5.11.1461. [DOI] [PubMed] [Google Scholar]
- 26.Izcue A, Coombes JL, Powrie F. Immunol Rev. 2006 Aug;212:256. doi: 10.1111/j.0105-2896.2006.00423.x. [DOI] [PubMed] [Google Scholar]
- 27.Collison LW, et al. Nature. 2007 Nov 22;450:566. doi: 10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- 28.Li B, Greene MI. Cell Cycle. 2007 Jun 15;6:1432. [PubMed] [Google Scholar]
- 29.Lopes JE, et al. J Immunol. 2006 Sep 1;177:3133. doi: 10.4049/jimmunol.177.5.3133. [DOI] [PubMed] [Google Scholar]
- 30.Schubert LA, Jeffery E, Zhang Y, Ramsdell F, Ziegler SF. J Biol Chem. 2001 Oct 5;276:37672. doi: 10.1074/jbc.M104521200. [DOI] [PubMed] [Google Scholar]
- 31.Samanta A, et al. Proc Natl Acad Sci U S A. 2008 Sep 16;105:14023. doi: 10.1073/pnas.0806726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leach MW, Bean AG, Mauze S, Coffman RL, Powrie F. Am J Pathol. 1996 May;148:1503. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: Primer/Oligo Sequences
Jurkat T cells were cotransfected with expression plasmids HA-tagged or Flag-Eos together with MigR1-Foxp3, HA-Ikaros or Myc-Helios as indicated, then immunoprecipitated with anti-HA or Flag M2, followed by Western blotting with either (a) anti-Foxp3 (eBioscience); (b) anti-HA (sigma); or (c) anti-Myc (sigma), respectively. (d)The N-terminal portion of Foxp3 is required for the physical interaction with Eos. HA-tagged Eos was co-transfected with Flag-tagged full-length Foxp3, ΔC-Foxp3 (2-320aa), or ΔN-Foxp3 (199-432aa) into HEK293T cells and immunoprecipitated with anti-Flag antibody, followed by western blot with anti-HA (top) or anti-Flag (middle) antibodies. The expression of Eos in cell lysates was detected by immunoblotting with an anti-HA antibody (bottom).
Sorted GFP+Foxp3+ (from GFP-Foxp3 mice) Treg cells were subjected to co-immunoprecipitation (A) and CHIP assay (B), separately. Treg cells were lysed and immunoprecipitated with anti-Eos antibodies or control IgG. The immunoprecipitates were resolved on SDS-PAGE followed by Western blot analysis with anti-Foxp3 (top) and anti-Ikaros (bottom) antibodies, respectively. For CHIP assay, sorted Treg cells were treated with mock or anti-CD3 and anti-CD28 for 4hrs and subjected to Ikaros CHIP to examine Ikaros occupancy of the IL-2 promoter region. The primers were listed in the supplementary table 1.
Sorted GFP+RFP+ cells from DO11.10/RAG2−/− CD4+ cells transduced with the indicated viruses were subjected to SDS-PAGE, followed by Western blotting with anti-Eos (Top), anti-Foxp3 (eBioscience; Middle) and reprobed with anti-actin antibody (Santa Cruz, Bottom).
Jurkat T cells were cotransfected with expression plasmids Myc-tagged ΔFoxp3 together with HA-NFAT, HA-AML1 or HA-CtBP1 as indicated, then immunoprecipitated with anti-Myc (Sigma), followed by Western blotting with either (a) anti-NFAT; (b) anti-AML1; or (c) anti-CtBP1 (Santa Cruz), respectively.
Sorted si-RL or si-Eos Treg cells were treated with mock or anti-CD3 and anti-CD28 for 4hrs and subjected to Foxp3 CHIP to examine Foxp3 occupancy of the IL-2 promoter region.
Quantitative ChIP analysis was performed to determine the modification of histone H3 and H4 at the IFN-γ promoter using real-time PCR. The results shown are the means ± s.d. of three independent experiments.
Shown are the numbers of up- and down-regulated transcripts in siEos vs. siRL transduced Treg cells. Naïve T cells and nTreg cells gene expression profiling was carried out in parallel. siRL transduced Treg cells gene expression signature was comparable with the untransduced nTregs. Foxp3 dependent up- or down- regulated genes were compared with the databases reported by Zheng et al [Nature. 445,936 (2007)]. In the siEos transduced Treg cells, 62 genes were up-regulated in siEos Treg cells relative to the siRL transduced Treg cells. Of these genes, ~70% were previously identified as Foxp3-dependent down-regulated genes (51). The majority of genes (40) known to be up-regulated by Foxp3 remains unchanged in both groups. Genes with expression increased or decreased by two-fold or more in Treg vs. naïve T cells were considered up-regulated or down-regulated by Foxp3.
Treg cells were lysed, subjected to immunoprecipitation (IP) with anti-Eos antibody or control IgG. The immunoprecipitates were resolved on SDS-PAGE followed by Western blot analysis (WB) with anti-HDAC7 (top) and anti-Eos (Santa Cruz, bottom) antibodies, respectively.
CFSE labeled Thy1.1+ T responder cells were co-cultured with or without wt or siEos Treg cells (Thy1.2+) for 80 hrs, followed by surface marker staining. Proliferation was assessed by CFSE dilution.
Modified Tregs sorted from lentivirus transduced Treg cells were stimulated with indicated reagents for 72h. [3H]-thymidine incorporation for the last 8 hrs of cell culture was measured as an indicator of cell proliferation and was expressed as the mean ± s.d. for triplicate cultures.
Sorted GFP+RFP+ cells from DO11.10/RAG2−/− CD4+ cells transduced with the indicated viruses were subjected to SDS-PAGE, followed by Western blotting with anti-Foxp3 (eBioscience; Top) and reprobed with anti-actin (Sigma, Bottom).
Higher dose of siEos transduced Treg cells only could cause weight loss in the Rag2−/− mice.
Rag2−/− mice received higher dose (A) or lower dose (B) of si-Eos Treg cells as indicated. After 6–8 weeks adoptive transfer, histological analysis of mice was performed as described in Figure 4. Data represent 6–8 mice per group in each panel.
CD4+CD25−CD62Lhigh cells isolated from Wt BALB/c were able to induce colitis in BALB/c Rag2−/− recipients (panel a,e). Higher dose of siEos-Treg cells (1×106) only could induce lesion upon adoptive transfer into Rag2−/− recipients (panel b,f). The same dose of wt Treg or siRL Treg cells did not cause observed colitis disease (panel c,d and g,h). The histological sections from each group were stained with H&E (panel a to d), or Alcian blue (panel e to h). Data represent 6–8 mice per group.
CD4+CD25high T cells were isolated from BALB/c mice and transduced with indicated lentivirus. 72h post-transduction, GFP+CD4+ T cells were sorted and subjected to Western blot (A) and q-RT PCR assay (B). Si-Eos group was carried out as experimental control. The primer sequences were listed in the supplementary table 1.