Abstract
Background
Point mutations in the mitofusin 2 (MFN2) gene has been identified exclusively in Charcot-Marie-Tooth type 2 (CMT2), and in a single family with intermediate CMT. MFN2 point mutations are probably the most common cause of CMT2.
Methods
Two-hundred and thirty-two consecutive unselected and unrelated CMT families with available DNA from all regions in Norway were included. We screened for point mutations in the MFN2 gene.
Results
We identified four known and three novel point mutations in 8 unrelated CMT families. The novel point mutations were not found in 100 healthy controls. This corresponds to 3.4% (8/232) of CMT families have point mutations in the MFN2 gene. The phenotypes were compatible with CMT1 in two families, CMT2 in four families, intermediate CMT in one family and distal Hereditary Motor Neuropathy (dHMN) in one family. This corresponds to 2.3% of CMT1, 5.5% of CMT2, 12.5% of intermediate CMT and 6.7% of dHMN families have a point mutation in the MFN2 gene. Point mutations in the MFN2 gene is likely to be the fourth most common cause to CMT after duplication of the peripheral myelin protein 22 (PMP22) gene, and point mutations in the Connexin32 (Cx32) and myelin protein zero (MPZ) genes.
Conclusions
The identified known and novel point mutations in the MFN2 gene expand the clinical spectrum from CMT2 and intermediate CMT to also include possibly CMT1 and the dHMN phenotypes. Thus, genetic analyses of the MFN2 gene should not be restricted to persons with CMT2.
Background
Charcot-Marie-Tooth (CMT) disease is a heterogeneous group of inherited peripheral neuropathies with an estimated prevalence of 1 in 2,500[1]. At present 40 different genes has been identified to cause inherited peripheral neuropathies[2]. CMT is subdivided into type 1 and 2, depending on whether the motor conduction velocity (MCV) is less or above 38 m/s[3,4]. A third form has intermediate MCV[5]. Intermediate CMT is defined as MCVs between 25 and 45 m/s. Distal Hereditary Motor Neuropathy (dHMN) is the spinal form of CMT[6].
Twelve different genes have been described to cause CMT2. The majority of the mutations are reported in the mitofusin 2 (MFN2) gene[2]. This gene is responsible for 18-23% of those with CMT2, and is probably the most common cause of CMT2 [7-9]. A MFN2 mutation has also been described in a single family with intermediate CMT[9].
Mitofusins are evolutionary conserved GTPases of the mitochondrial outer membrane, and has an essential role for the controlled fusion of the mitochondria membrane[10]. The MFN2 protein spans the mitochondrial outer membrane with a large N-terminal and a relatively short C-terminal exposed to the cytosol[11]. The MFN2 mutation so far has almost exclusively been missense mutations that affect the N- or the C-terminal of the MFN2 protein[2,9].
We analyzed 232 Norwegian CMT families for mutations in the MFN2 gene.
Methods
Patients
Two-hundred and thirty-two consecutive unselected and unrelated CMT families with available DNA from all regions in Norway were included. The total Norwegian population is 4.8 millions people. Based on clinical and neurophysiological data, 86 families had CMT1 (47.3%), 73 families had CMT2 (40.1%), 8 families had intermediate CMT (4.4%) and 15 families had dHMN (8.2%). The neurophysiological phenotype was unknown in 50 families.
Clinical interview, genetic and neurological examination
Probands and their relatives had a semi-structured clinical interview, a genetic and a neurological examination by geneticist and neurologist GJB. The diagnosis CMT was based on clinical information and exclusion of acquired causes of neuropathy such as endocrine, immunologic, infectious, metabolic, nutritional, toxic drugs and substances, and connective tissue and paraneoplastic disorders. Other hereditary disorders with neuropathy such as Friedreich's ataxia, hereditary spastic paraplegia, myotonic dystrophy etc. were also excluded. All findings were recorded in a neuropathy scheme that has been used and evaluated in Norway [12-14]. This scheme had some supplementary items added by GJB. Cranial nerves, muscle weakness, reflexes and sensation was scored according to the Neuropathy Impairment Score (NIS)[15]. Muscle weakness includes neck flexion, respiratory muscles, and 9 muscle groups in each upper extremity (UE) and 8 muscle groups in each lower extremity (LE). The muscle strength is scored linearly, i.e. normal strength is scored 0 and paralysis is scored 4, while 25%, 50% and 75% muscle weakness are scored 1, 2 and 3.
Neurophysiology
Standard techniques with surface electrodes were used for antidromic sensory conduction and orthodromic motor conduction. Needle electrodes were used, if the surface electrodes failed to record conduction velocities. Electromyography (EMG) was performed.
Mutation analysis
Genomic DNA was extracted from white blood cells using FlexiGene DNA Kit (QIAGEN). The coding exons were derived from the published sequences in National Center for Biotechnology Information (NP_055689 and NM_014874, MFN2). Primers for PCR amplification of exons and intronic splice junctions were designed with Primer3[16]. Fifty ng of patient genomic DNA was used for the analysis. Each region was amplified in 35 cycles. The sequencing was carried out using the PCR primers, internal sequencing primers when needed, and BigDye Terminator kit version 1.1. Sequencing was performed using Applied Biosystems ABI3130x1 Genetic analyzer and aligned with the Sequencher programme (Gene Codes Corporation, Ann Arbor, MI). The numbering of nucleotides is according to the open reading frame of the cDNA sequences[17]. In families in which a de novo point mutation was suspected, paternity was confirmed using 17 STR markers (CSF1PO, D3S1358, D5S818, D7S820, D8S1179, D12S392 D13S317, D16S539, D17S906, D18S51, D21S11, FGA, Penta D, Penta E, TH01, TPOX, and vWA) and Amelogenin. One-hundred persons without neuropathy were screened as controls for sequence variations. For further details see Additional file 1.
We analyzed for mutations consecutively in the genes; peripheral myelin protein 22 (PMP22), Connexin32 (Cx32), myelin protein zero (MPZ), small integral membrane protein of lysosome/late endosome (SIMPLE), early growth response 2 (EGR2), and MFN2. The PMP22 duplication was investigated by Real-time quantitative PCR[18]. Coding exons were derived from published sequences in GenBank (Cx32 (GJB1): NM_000166, EGR2: NM_000399, MFN2: BC017061, MPZ: NM_000530, PMP22: NM_000304 and SIMPLE_NM_004862, AB034747. Primers for PCR amplification of exons and intronic splice junctions were designed with Primer3[16].
Ethics
The study was approved by the ethical committees. Participation was based on informed consent.
Results
Molecular genetic analysis
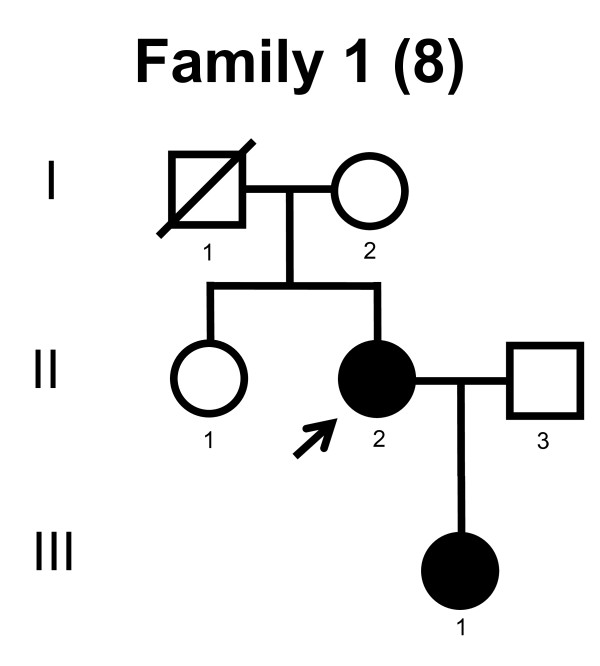
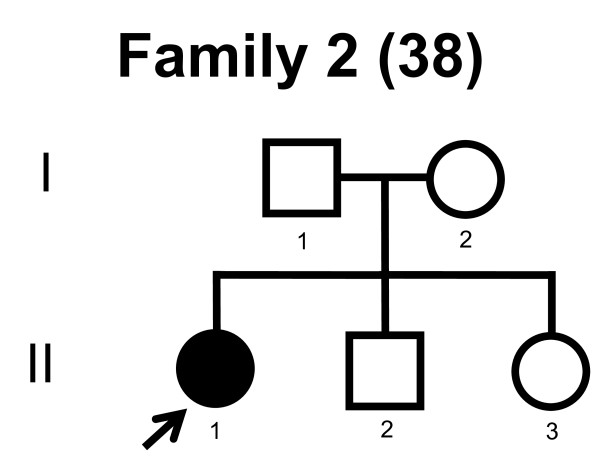
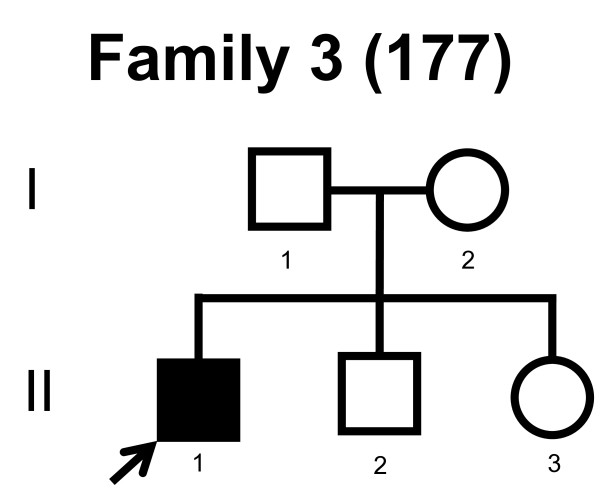
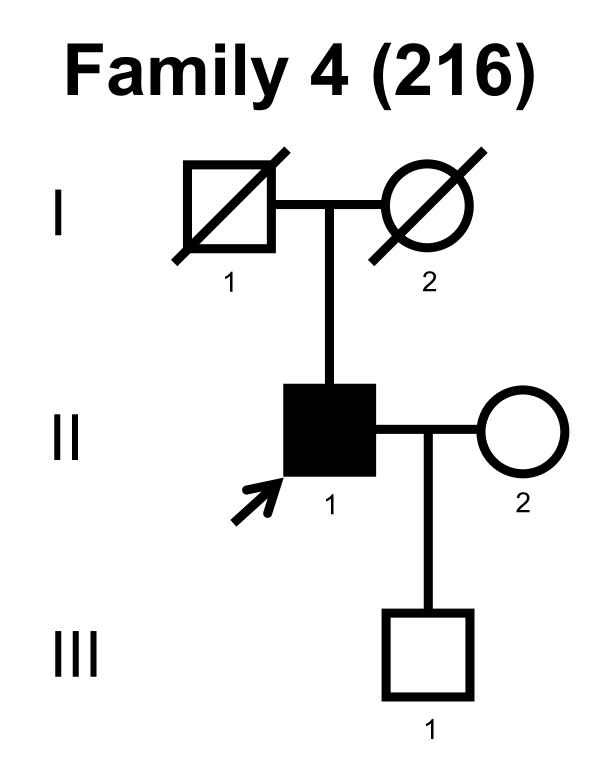
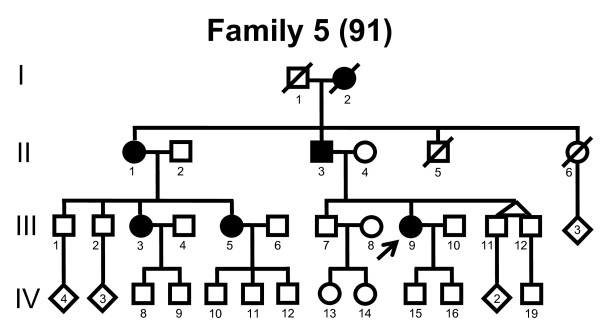
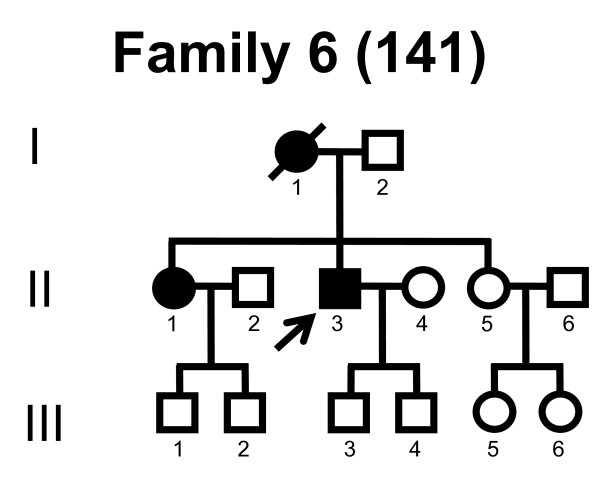
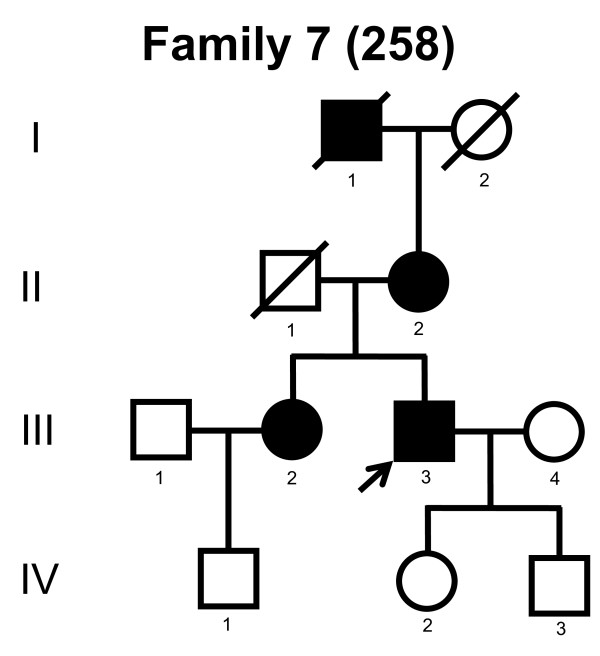
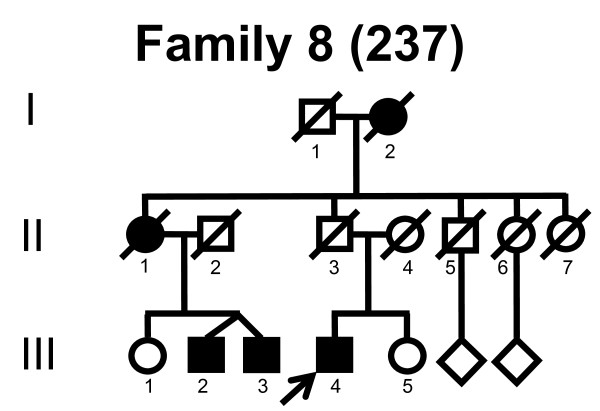
We screened the entire coding region and the intron/exon boundaries of the MFN2 gene. A total of 232 unrelated Norwegian families with CMT were included. We identified 7 point mutations in 8 CMT families (Table 1). Four point mutations were known, and three point mutations were novel. All our point mutations were missense mutation that caused an amino acid change. The point mutations were located in exons 4, 14, 15 and 18. The novel point mutations c.1709A>G and c.2119C>T were found in the probands, while c.2146G>A was found in the proband and his affected first cousin. The pedigrees, family 1, 2, 3, 4, 5, 6, 7 and 8 are shown in figure 1, 2, 3, 4, 5, 6, 7 and 8, respectively. Five pedigrees had a segregation pattern compatible with autosomal dominant inheritance (Figures 1, 5, 6, 7 and 8). In four probands the MFN2 point mutation occurred de novo. Paternity was confirmed in one pedigree (Figure 2). HLA haplotyping was compatible with paternity in another family (Figure 1), while DNA was unavailable in two fathers (Figure 3 and 4). None of the novel point mutations was found in 200 control chromosomes of healthy Norwegians. However, we found a c.1569C>T polymorphism in eight persons, that did not change the amino acid sequence (Ser523Ser). We found no duplication in PMP22 gene, and no point mutations in the PMP22, Cx32, MPZ, SIMPLE or EGR2 genes in the affected with MFN2 missense mutations.
Table 1.
Gender, age at onset, initial symptoms of Charcot-Marie-Tooth disease, phenotype and point mutations in the eight unrelated Norwegian families with mutation in the MFN2 gene.
| Family no. | Family member | Gender | Age at onset (yrs) |
Initial symptoms | Phenotype | Point mutation |
Exon | Amino acid Change |
|---|---|---|---|---|---|---|---|---|
| 1 | II-2 | ♀ | 4 | Weakness in legs | CMT1 | c.280C>T | 4 | Arg94Trp |
| III-1 | ♀ | 4 | Weakness in legs and pes cavus | |||||
| 2 | II-1 | ♀ | 4 | Stumble and dorsal motion weakness in toes | CMT2 | c.281G>A | 4 | Arg94Gln |
| 3 | II-1 | ♂ | 2 | Foot deformity and stumbled | CMT1 | c.1403G>A | 14 | Arg468His |
| 4 | II-1 | ♂ | 63 | Weakness in the right foot, and difficulties walking | dHMN | c.1709A>G | 15 | Asn570Ser |
| 5 | III-9 | ♀ | 10 | Weakness in legs and pes cavus | CMT2 | c.2113G>A | 18 | Val705Ile |
| 6 | II-3 | ♂ | 47 | Paresthesia in feet | CMT2 | c.2113G>A | 18 | Val705Ile |
| 7 | III-3 | ♂ | 44 | Muscular pain during exercise in left leg | CMT2 | c.2119C>T | 18 | Arg707Trp |
| 8 | III-2 | ♂ | 23 | Recurrent ankle sprains, weakness and reduced balance | Intermediate CMT | c.2146G>A | 18 | Ala716Thr |
| III-4 | ♂ | 50 | Paresthesia in foot |
Relatives of probands and novel point mutations are shown in italics.
Figure 1.
Pedigree of family 1 with point mutation in the MFN2 gene.
Figure 2.
Pedigree of family 2 with point mutation in the MFN2 gene.
Figure 3.
Pedigree of family 3 with point mutation in the MFN2 gene.
Figure 4.
Pedigree of family 4 with point mutation in the MFN2 gene.
Figure 5.
Pedigree of family 5 with point mutation in the MFN2 gene.
Figure 6.
Pedigree of family 6 with point mutation in the MFN2 gene.
Figure 7.
Pedigree of family 7 with point mutation in the MFN2 gene.
Figure 8.
Pedigree of family 8 with point mutation in the MFN2 gene.
Clinical and neurophysiological findings
Table 2 shows a summary of the neurophysiology and molecular genetic findings in 232 unrelated Norwegian CMT families. We identified a MFN2 point mutation in 3.4% (8/232) of the CMT families. The phenotype was compatible with CMT1 in two probands, CMT2 in four probands, intermediate CMT in one proband, and distal Hereditary Motor Neuropathy (dHMN) in one proband. This corresponds to 2.3% of CMT1, 5.5% of CMT2, 12.5% of intermediate CMT and 6.7% of dHMN families have a point mutation in the MFN2 gene.
Table 2.
Summary of neurophysiology and MFN2 mutations in 232 CMT families.
| Phenotype | Neurophysiology % (n) |
MFN2 mutations % (frequency) |
|---|---|---|
| CMT1 | 37.1 (86) | 2.3 (2/86)) |
| CMT2 | 31.5 (73) | 5.5 (4/73) |
| Intermediate CMT | 3.4 (8) | 12.5 (1/8) |
| dHMN | 6.5 (15) | 6.7 (1/15) |
| CMT neurophysiology unknown | 21.6 (50) | 0 (0/50) |
| Total | 100 (232) | 3.4 (8/232) |
The mean age at onset was 3.0 years in the CMT1 probands and 26.3 years in the CMT2 probands, while the probands with intermediate CMT and dHMN had late onset (Table 1). The initial symptoms were muscle weakness, difficulties walking and frequent falls. Pes cavus, hammertoes and muscle wasting in feet and legs was frequently observed (Additional file 2, table S3). The sensory modality touch and pain were mildly affected. Vibration and proprioception in the toes, Achilles and patellar reflexes and Romberg was frequently affected. Muscle weakness in the lower limbs varied considerable (Additional file 3, table S4). One proband was wheelchair bound, while her daughter had severe difficulties walking (Family 1). Additional features such as tremor in arm/leg and kyphoscoliosis were sometimes present (Additional file 2, table S3).
The proband in family 4 had a complex history prior to weakness of the right foot and walking difficulties at age 63 years. At age 57 years he had a L4 compression fracture causing spinal stenosis and intermittent claudicatio, until he had a laminectomy, facetectomy and discectomy two years later. A 1/2 year later he experienced lumbar pain and paresthesia in the thighs and had a new laminectomy at age 63 years. One year prior he had bilateral arthroplasty due to osteoarthrosis. He was operated for an abdominal aortic aneurism at age 65 years and four years later he had radiotherapy for a prostate cancer.
Neurophysiological data were available from eight probands and one relative (Additional file 3, table S4). The conduction velocities (CV) were reduced in the probands with the CMT1 phenotypes (Family 1 and 3). The slight reduction in peroneal motor nerve CV within four years could be compatible with demyelination, but we interpreted it as CMT2, see discussion for details (Family 2). The proband in family 4 had normal motor and sensory CV and chronic denervation compatible with dHMN. The CV were slightly reduced but >38 m/s in the probands with CMT2 phenotypes (Family 5-7). The range of the CV in family 8 is compatible with intermediate CMT phenotype. The compound motor action potential (CMAP) and sensory nerve action potential (SNAP) was generally reduced or could not be elicited.
Discussion
We screened 232 unselected and unrelated Norwegian CMT families with available DNA. A total of 7 MFN2 missense mutations were identified in 8 families (Table 1). The literature describes three nonsense mutations and one deletion, but missense mutations in far the most common types of mutations in the MFN2 gene[2,9,19].
We identified 4 de novo mutations in the MFN2 gene, paternity could not be confirmed in two families (family 3 and 4) due to lack of paternal DNA. Our 25-50% (2-4/8) frequency of de novo mutations in an population sample corresponds well with the 34% and possibly 55% frequency found in a major collaboration on MFN2 mutations[2]. Two of our de novo missense mutation occurred in codon 94. The literature describes 5 other families with de novo mutations in codon 94 with confirmed paternity[9,20]. These mutations have also been described in families with different ethnicity[2]. This strongly suggest that codon 94 is a hot spot for mutations. The de novo c.1403G>A mutation has previously been described in three CMT families, but it was also found in a healthy control[2,21,22]. The c.2113G>A was found in two unrelated Norwegian families, and has also been described by others[2,21]. The c.1709A>G in exon 15 was a novel missense mutations and a de novo mutation found in the proband. Mutations in exon 15 have not previously been described. The other novel missense mutations were found in probands from CMT families with several affected, but we only had DNA from a single first cousin whom also carried the mutation. We excluded duplication of PMP22 and point mutation in PMP22, Cx32, MPZ, SIMPLE and EGR2 genes in the affected with MFN2 missense mutations.
Previous mutations in the MFN2 gene have all been assigned to a CMT2 phenotype, except from a single family that was assigned intermediate CMT[2,7-9,19-21]. Additional symptoms such as ataxia, optic neuritis, optic atrophy, pyramidal signs, scoliosis and tremor have also been described[2,23-25].
Missense mutations in codon 94, i.e. amino acid change Arg94Trp and Arg94Gln cause intermediate CMT, CMT2 or CMT2 with additional features[9]. The proband in the family with intermediate CMT had motor CV <38 m/s in all tested motor nerves including median nerve and should therefore be classified CMT1. However, his affected father had motor CV >38 m/s, and the family was classified intermediate CMT. The missense mutation Arg94Trp occurred de novo in our family 1. Since we had no neurophysiological data on the probands affected daughter, we classified the family CMT1 rather than the more infrequent intermediate CMT. The missense mutation in codon 468, i.e amino acid change Arg468His was found in a proband with CMT2 and her father with Parkinson disease and distal neuropathy indicating co-segregation as well as in 1 of 260 control chromosomes[21]. A patient with a severe CMT carried two mutations, the Arg468His in MFN2 and Gln95stop in ganglioside-induced differentiation-associated protein 1 (GDAP1)[22]. Two family members carried exclusively the missense mutation in the MFN2 that caused the amino acid change Arg468His. One had a mild axonal neuropathy at age 56 years, while the other was unaffected at age 18 years[22]. Three other family members with the Gln95stop in GDAP1 were unaffected. The proband in our family 3 with the Arg468His amino acid change had reduced motor CV, while 1 of 250 control chromosomes had the missense mutation carried by an unaffected woman age 25 years.
We analyzed all types of CMT families and found MFN2 missense mutation in CMT1, CMT2, intermediate CMT and dHMN patients.
The CV in the median motor nerve is used as reference for classifying CMT1, CMT2 and intermediate CMT [3-5]. dHMN is defined with normal motor and sensory CV and chronic denervation on EMG[6]. We do not have CV of the median motor nerve in the probands with CMT1. However, they all had reduced CV in motor nerves compatible with CMT1. The CV is equal in the ulnar and the median motor nerve. Thus, the proband with CV <38 m/s in the ulnar motor nerve was classified CMT1 (Family 1). We interpreted the marked reduction in motor CV of the tibial nerve as demyelination (Family 3). The slight reduction in motor CV of the peroneal nerve in Family 2 was interpreted as CMT2, as the CV cut off for the normal value is less in the peroneal than the median nerve, and an extrapolation with this proportional factor would increase the CV from 35.2 to 42.1 m/s, while a similar extrapolation in family 3 would increase the CV from 15.7 to 18.8 m/s (Additional file 2, table S3). Our neurophysiological data is far from optimal. The 232 consecutive unselected and unrelated CMT families with available DNA were from all regions in Norway, and were included into the study irrespective of the perfection of neurophysiological parameters, if the clinical or other parameters suggested it was CMT. An International study of mutations in the MFN2, included 323 unrelated probands of whom 44 unrelated probands had an unknown neurophysiology[9]. The lack of complete neurophysiological data makes the neurophysiological classification a challenge, along with the fact that CMAP were reduced in several recordings. Severely reduced CMAP and low CV in lower limbs suggest CMT2. However, the neurophysiological classification on CMT is based on CV alone not including CMAP or other neurophysiological features. For that reason we focused on CV in our interpretation. This provides a challenge in family 2, 3, 4 and 5 since neurophysiological data is only available from the lower limbs. Family 4 is diagnosed as dHMN based on a motor and a sensory CVs were normal, but the amplitudes of the sensory potentials were not measured which makes it difficult to judge whether the sensory nerves were involved or not.
The International study on MFN2 mentioned above included 323 unrelated probands, of whom 249 (77%) probands were diagnosed as CMT2, 20 (6%) had intermediate CMT, six (2%) had CMT1, three (1%) had dHMN and one (<1%) had hereditary sensory and autonomic neuropathy (HSAN, while 44 (14%) had an unknown neurophysiological phenotype. MFN2 mutations were found in 28 probands with CMT2 and in one family with intermediate CMT. We found the MFN2 mutation in 2 of 86 (2.3%) unrelated CMT1 families and in 1 of 15 (6.7%) unrelated dHMN families. These low frequencies of MFN2 mutations in CMT1 and dHMN made it unlikely for the International study to obtain MFN2 mutations in these CMT subtypes. Unfortunately, we do not have CV from the median nerve in the two families classified as CMT1. Thus, further studies are needed to confirm that MFN2 mutations can cause CMT1.
Clinically the age at onset was early in the CMT1 families (Family 1 and 3), while it was late in CMT2, intermediate and dHMN (Family 4-8), with one exception (Family 2). Additional symptoms such as ataxia, kyphoscoliosis and tremor were found three families (Table 2). The phenotype genotype correlation can be questioned in the proband with dHMN, since the classification was based on single motor and sensory nerve and he had multiple other disorders that might have caused his motor and sensory symptoms (for details see result section). However, since the motor and sensory CVs were normal, we characterized the proband as dHMN. The identified missense mutation c.1709A>G in exon 15 might be a polymorphism, although we did not find the mutation among 200 control chromosomes.
Point mutations in the MFN2 gene is likely to be the fourth most common cause to CMT after the duplication of PMP22 gene, and point mutations in the Cx32 and MPZ genes. We found that 2.3% of CMT1, 5.5% of CMT2, 12.5% of intermediate CMT and 6.7% of dHMN families have a point mutation in the MFN2 gene. We analyzed for mutations in five other common genes that may cause CMT1, CMT2, intermediate CMT or dHMN. However the exclusion analyses were not exhaustive since mutations in at least 40 genes can cause CMT. We suggest that genetic analyses of MFN2 should not be restricted exclusively to those with a CMT2 phenotype.
Conclusions
Today only people with CMT2 receive testing for the MFN2 gene, as the literature provides no evidence for testing other subtypes of CMT. Our main finding indicates that MFN2 mutations might be found in all subtypes of CMT. Thus, MFN2 gene testing should not be restricted to CMT2.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
GJB acquired the material, conceived the study, participated in the design of the study and drafted the manuscript. JCS, AL and HH carried out the molecular genetic studies and the sequence alignment. MBR conceived the study, participated in the design of the study and drafted the manuscript. All authors read and approved the final manuscript.
Authors' informations
GJB is a neurologist and geneticist. JCS, AL and HH are cand. scient. MBR is professor of neurology, PhD and DMSci. The authors main interest is genetics in neurological disorders with special emphasis on CMT.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Appendix 1. Primer sequences used for amplifying and sequencing the coding regions of the MFN2 gene.
Table S3. Clinical characteristics of the patients with point mutations in the MFN2 gene.
Table S4. Neurophysiology in patients with Charcot-Marie-Tooth disease caused by point mutations in the MFN2 gene.
Contributor Information
Geir J Braathen, Email: g.j.braathen@medisin.uio.no.
Jette C Sand, Email: jettesand@hotmail.com.
Ana Lobato, Email: alobatopascual@gmail.com.
Helle Høyer, Email: Helle.Hoyer@sthf.no.
Michael B Russell, Email: m.b.russell@medisin.uio.no.
Acknowledgements
The study was financially supported by University of Oslo, Faculty Division Akershus University Hospital, South-Eastern Norway Regional Health Authority, Akershus University Hospital HF and Nansen Foundation.
References
- Skre H. Genetic and clinical aspects of Charcot-Marie-Tooth's disease. Clin Genet. 1974;6:98–118. doi: 10.1111/j.1399-0004.1974.tb00638.x. [DOI] [PubMed] [Google Scholar]
- The Mutation Database of Inherited Peripheral Neuropathies. http://www.molgen.ua.ac.be/CMTMutations/default.cfm
- Dyck PJ, Lambert EH. Lower motor and primary sensory neuron diseases with peroneal muscular atrophy. I. Neurologic, genetic and electrophysiologic findings in hereditary polyneuropathies. Arch Neurol. 1968;18:603–618. doi: 10.1001/archneur.1968.00470360025002. [DOI] [PubMed] [Google Scholar]
- Harding AE, Thomas PK. The clinical features of hereditary motor and sensory neuropathy types I and II. Brain. 1980;103:259–280. doi: 10.1093/brain/103.2.259. [DOI] [PubMed] [Google Scholar]
- Nicholson G, Nash J. Intermediate nerve conduction velocities define X-linked Charcot-Marie-Tooth neuropathy families. Neurology. 1993;43:2558–2564. doi: 10.1212/wnl.43.12.2558. [DOI] [PubMed] [Google Scholar]
- Harding AE. In: Peripheral Neuropathy. 4. Dyck PJ, Thomas PK, editor. Philadelphia: Elsevier, Saunders; 2005. Inherited Neuronal Atrophy and degeneration predominantly of lower motor neurons; pp. 1603–1621. [Google Scholar]
- Kijima K, Numakura C, Izumino H. Mitochondrial GTPase mitofusin 2 mutation in Charcot-Marie-Tooth neuropathy type 2A. Hum Genet. 2005;116:23–27. doi: 10.1007/s00439-004-1199-2. [DOI] [PubMed] [Google Scholar]
- Lawson VH, Graham BV, Flanigan KM. Clinical and electrophysiologic features of CMT2A with mutations in the mitofusin 2 gene. Neurology. 2005;65:197–204. doi: 10.1212/01.wnl.0000168898.76071.70. [DOI] [PubMed] [Google Scholar]
- Verhoeven K, Claeys KG, Züchner S. MFN2 mutation distribution and genotype/phenotype correlation in Charcot-Marie-Tooth type 2. Brain. 2006;129:2093–2102. doi: 10.1093/brain/awl126. [DOI] [PubMed] [Google Scholar]
- Santel A. Get the balance right: mitofusins roles in health and disease. Biochim Biophys Acta. 2006;1763:490–499. doi: 10.1016/j.bbamcr.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Rojo M, Legros F, Chateau D. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J Cell Sci. 2002;115:1663–1674. doi: 10.1242/jcs.115.8.1663. [DOI] [PubMed] [Google Scholar]
- Skre H. Hereditary ataxias and their associated traits. A clinical, genetic, and epidemiological study in Western Norway. Bergen and Oslo: University of Bergen and University of Oslo (Thesis); 1976. pp. 1–288. [Google Scholar]
- Skre H. Neurological signs in a normal population. Acta Neurol Scand. 1972;48:575–606. doi: 10.1111/j.1600-0404.1972.tb07577.x. [DOI] [PubMed] [Google Scholar]
- Skre H. Application of a quantitative scoring system in the investigation of some hereditary neurological disorders. Clin Genet. 1974;5:163–172. doi: 10.1111/j.1399-0004.1974.tb01678.x. [DOI] [PubMed] [Google Scholar]
- Dyck PJ, Turner DW, Davies JL. Electronic case-report forms of symptoms and impairments of peripheral neuropathy. Can J Neurol Sci. 2002;29:258–266. doi: 10.1017/s0317167100002043. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S, Misener S, editor. Totowa, NJ: Humana Press; 2000. Primer3 on the WWW for general users and for biologist programmers; pp. 365–386.http://fokker.wi.mit.edu/primer3 [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
- Aarskog NK, Vedeler CA. Real-time quantitative polymerase chain reaction. A new method that detects both the peripheral myelin protein 22 duplication in Charcot-Marie-Tooth type 1A disease and the peripheral myelin protein 22 deletion in hereditary neuropathy with liability to pressure palsies. Hum Genet. 2000;107:494–498. doi: 10.1007/s004390000399. [DOI] [PubMed] [Google Scholar]
- Züchner S, Mersiyanova IV, Muglia M. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- Neusch C, Senderek J, Eggermann T. Mitofusin 2 gene mutation (R94Q) causing severe early onset axonal polyneuropathy (CMT2A) Eur J Neurol. 2007;14:575–577. doi: 10.1111/j.1468-1331.2006.01688.x. [DOI] [PubMed] [Google Scholar]
- Engelfried K, Vorgerd M, Hagedorm M. Charcot-Marie-Tooth neuropathy type 2A:novel mutations in the mitofusin 2 gene (MFN2) BMC Med Genet. 2006;7:53. doi: 10.1186/1471-2350-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchs I, Casasnovas C, De Jorge L. A novel mutation in GDAP1 and a change in MFN2 gene in a family with a severe form of Charcot-Marie-Tooth. Eur J Hum Genet. 2008;16(suppl 2):S88. [Google Scholar]
- Zhu D, Kennerson MI, Walizada G. Charcot-Marie-Tooth with pyramidal signs is genetically heterogeneous:families with and without MFN2 mutations. Neurology. 2005;65:496–497. doi: 10.1212/01.wnl.0000171345.62270.29. [DOI] [PubMed] [Google Scholar]
- Chung KW, Kim SB, Park KD. Early onset severe and late-onset mild Charcot-Marie-Tooth disease with mitofusin 2 (MFN2) mutations. Brain. 2006;129:2103–2118. doi: 10.1093/brain/awl174. [DOI] [PubMed] [Google Scholar]
- Züchner S, De Jonghe P, Jordanova A. Axonal neuropathy with optic atrophy (HMSN VI) is caused by mutations in mitofusin 2. Ann Neurol. 2006;59:276–281. doi: 10.1002/ana.20797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 1. Primer sequences used for amplifying and sequencing the coding regions of the MFN2 gene.
Table S3. Clinical characteristics of the patients with point mutations in the MFN2 gene.
Table S4. Neurophysiology in patients with Charcot-Marie-Tooth disease caused by point mutations in the MFN2 gene.