Abstract
Addictive drugs including opioids activate signal transduction pathways that regulate gene expression in the brain. However, changes in CNS gene expression following morphine exposure are poorly understood. We determined changes in gene expression following short- and long-term morphine treatment in the hypothalamus and pituitary using genome-wide DNA microarray analysis and confirmed those alterations in gene expression by real-time reverse transcriptase polymerase chain reaction (RT-PCR) analysis. In the hypothalamus, short-term morphine administration up-regulated (at least 2-fold) 39 genes and down-regulated six genes. Long-term morphine treatment up-regulated 35 genes and down-regulated 51 genes. In the pituitary, short-term morphine administration up-regulated 110 genes and down-regulated 29 genes. Long-term morphine treatment up-regulated 85 genes and down-regulated 37 pituitary genes. Microarray analysis uncovered several genes involved in food intake (neuropeptide Y, agouti-related protein, and cocaine and amphetamine-regulated transcript) whose expression was strongly altered by morphine exposure in either the hypothalamus or pituitary. Subsequent RT-PCR analysis confirmed similar regulation in expression of these genes in the hypothalamus and pituitary. Finally, we found functional correlation between morphine-induced alterations in food intake and regulation of genes involved in this process. Changes in genes related to food intake may uncover new pathways related to some of the physiological effects of opioids.
Keywords: microarray, RT-PCR, opiates, gene regulation, pituitary, hypothalamus, drug addiction, food intake
Introduction
Drug addiction is a chronic relapsing disorder that results from gradual adaptations of the brain to repeated drug exposure. The current understanding of this complex phenomenon is that neurons responding to natural reinforcers, such as food, sex, and social interactions are abnormally stimulated leading to strong dysregulation of brain reward pathways (Koob and Le Moal, 1997) and aberrant learning processes (Robbins and Everitt, 1999). Addiction has many phases, including initiation and maintenance of drug consumption, withdrawal episodes, protracted abstinence, and relapse. The brain circuits of addiction involve reward pathways, in association with stress, obsessive-compulsive, habit-forming systems and molecular adaptations to chronic drug use (Gerrits et al., 2003). Many neurotransmitter systems are recruited during this process, the most widely studied is the dopaminergic system (Volkow et al., 2004, Zhang et al., 2004) and the endogenous opioid system (Gerrits et al., 2003, Kreek et al., 2004).
While the clinical phenomena of drug addiction are more understood today, less is known about the underlying genetic mechanisms that determine susceptibility to addiction and the molecular consequences of drug exposure. Addictive drugs like morphine and cocaine activate signal transduction pathways that regulate brain gene expression and such regulation may be modulated by the presence of certain transcription factors present in individual neurons including cAMP response element binding protein (CREB) and the Fos and Jun families of immediate early genes (Chao and Nestler, 2004). The characterization of drug-induced changes in gene expression shows promise for better understanding of drug addiction following exposure to drugs of abuse such as morphine, cocaine and ethanol.
Opioid use is associated with modifications in neural physiology in which altered gene expression is detected even after a single administration and can persist for a long time following cessation of drug exposure (Koob et al., 2004). Although much research has focused on opiate use altering various neurotransmitter systems, other studies have found that opiates cause changes in neuroendocrine/neuropeptide systems, such as the endogenous opioid peptides (β-endorphin, enkephalins, dynorphins), corticotrophin-releasing hormone (CRH), and anti-opiate peptides, such as orphanin FQ/nociceptin and neuropeptide AF and FF in animals or humans exposed to drugs (Garcia de Yebenes and Pelletier, 1993, Spangler et al., 1996, Zhou et al., 1996, Rodriguez de Fonseca et al., 1997).
In humans, most drug users do not become drug-dependent (Koob and Le Moal, 1997). Koob and Le Moal (Koob and Le Moal, 1997) postulated that the organism tries to maintain homeostasis when challenged by exogenous drugs such as opiates, but is eventually unable to adapt to the environmental challenges. As the organism has a new, albeit pathological, set-point, tolerance to the drug occurs, and if drug intake ceases, withdrawal symptoms occur. Thus, the individual becomes addicted to the drug of abuse and is transformed from a state of voluntary drug intake to a state where drug craving, drug seeking and compulsive drug consumption occur (Koob et al., 2004). Chronic drug exposure resulting in “the addicted brain” is likely to lead to a different alteration in gene expression compared to short-term drug exposure. These altered genes and proteins can act in concert as the “switch” that moves the individual into a state of compulsive drug seeking and abuse (Leshner, 1997). In order to understand this “switch to addiction”, we investigated gene expression patterns and behavior in response to both short-term and long-term opiate exposure using an in vivo mouse model.
Microarray technology has become a valuable tool to evaluate the expression of many genes simultaneously (Lockhart and Winzeler, 2000). It can also be used to uncover new genes involved in pharmacological and addictive properties of opiates as well as to uncover genes not previously known to be involved in opiate addiction. In order to understand opiate-induced regulation of the neuroendocrine system, we studied morphine-induced gene expression changes in both the hypothalamus and pituitary. We performed DNA microarray analysis on these two central nervous system (CNS) regions, as these are the major sites of hormonal biosynthesis. Gene expression changes were confirmed by real-time reverse transcriptase polymerase chain reaction (RT-PCR) and peptide analyses and correlated with behavioral studies in mice.
Experimental Procedures
Mice
For all experiments, we used 12–16 week-old male C57BL/6 mice (Taconic, Hudson, NY). The animals were housed in groups of five under a 12 h light/dark cycle with food and water available ad libitum. All animal protocols were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at The Charles Drew University.
Drug treatments
Mice were implanted subcutaneously on the dorsal aspect of the neck with 25 mg morphine pellets [generously provided by the National Institute on Drug Abuse (NIDA), Rockville, MD, USA], with one pellet implanted for six h (short-term treatment) or four days (long-term treatment) under isoflurane anesthesia (Attane, Minrad INC, Bethlehem, PA, USA). Control animals were implanted with a matching placebo pellet (NIDA) for the same duration as morphine-treated animals. Treatment with morphine pellets for at least three days leads to high levels of morphine dependence (Roy et al., 2005). On the other hand, our pilot studies have shown that short-term morphine (morphine pellet implantation for six h) does not lead to signs of abstinence withdrawal. Thus, we used a similar treatment for our short-term studies.
Abstinence withdrawal in mice exposed to short-term and long-term morphine
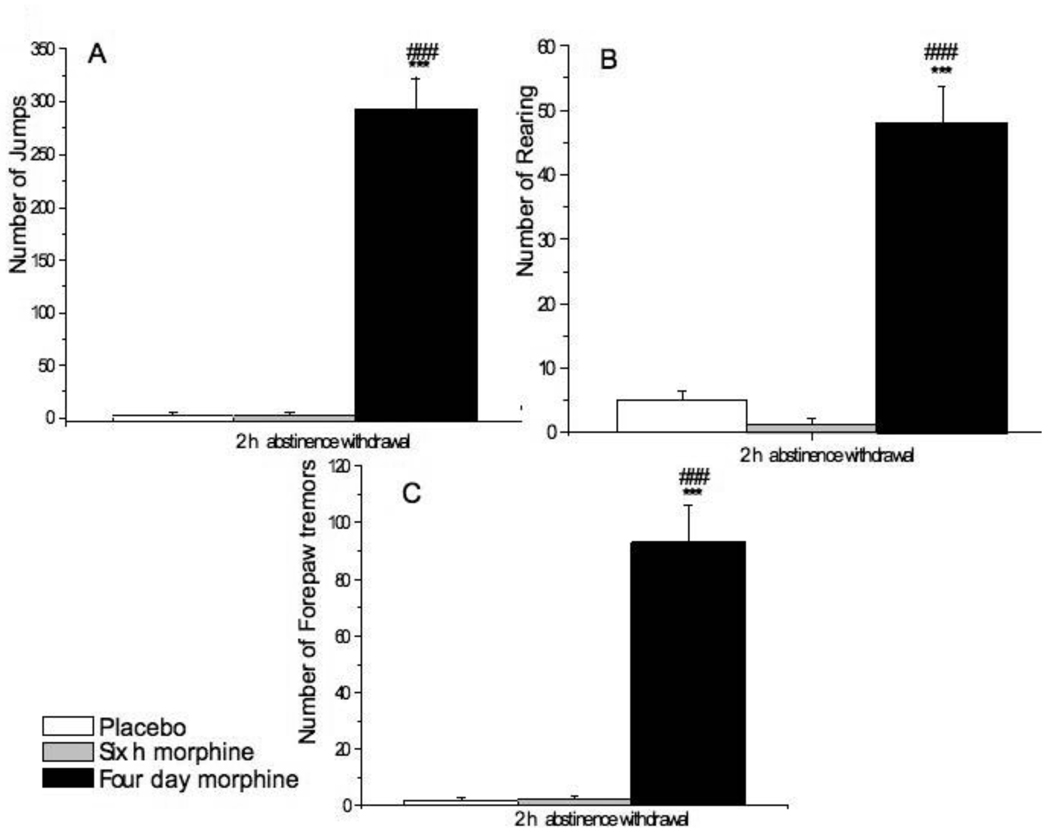
Mice were implanted subcutaneously with a placebo pellet for six h (n=3) or four days (n=4) or a morphine pellet for 6 h (n=6) or 4 days (n=5), the pellets were removed (during the light cycle) and opiate abstinence withdrawal was determined by counting the number (± SEM) of jumps, rearing and forepaw tremors over the next two h. The two placebo groups were pooled for analysis since none of these mice showed any signs of withdrawal.
RNA extraction
Mice were sacrificed by decapitation and the brain and pituitary were removed. The whole hypothalamus was removed en bloc by sharp dissection on the ventral side of the brain (Baker et al., 1983). The boundaries of the block were: anterior, just posterior to the optic chiasm; lateral, the choroidal fissures; and posterior, the anterior portion of the mammillary bodies. The tissue was removed by a horizontal cut 3 mm from the ventral surface of the brain. The pituitary and hypothalamus were rapidly placed in 1.0 ml RNAlater solution (Ambion, Austin, TX) and stored at −80°C to prevent RNA degradation. RNA was isolated from pooled tissue samples from three to five mice (to reduce inter-animal variability) that received the same treatment using Trizol reagent (Invitrogen, San Diego, CA, USA) according to the instructions of the manufacturer. After chloroform extraction, RNA was precipitated with ethanol and further purified with RNeasy Micro Kit (Qiagen, Valencia, CA, USA) according to the instructions provided by the manufacturer. The concentration of total RNA was measured by ultraviolet (UV) spectrophotometry at 260/280 nm, and RNA quality was assessed by electrophoresis on a 1% agarose gel. Only the samples with a 260/280 ratio ≥ 1.9 and no signs of degradation on agarose electrophoresis were used for analysis.
Microarray analysis
Microarray analysis was performed as described previously (Lockhart and Winzeler, 2000) with a few modifications. Briefly, total RNA (3–5 µg per array isolated from pools of tissue from three to five mice) was converted to cDNA, amplified, and labeled using Amino Allyl MessageAmp aRNA (Ambion, Austin, TX) according to the manufacturer protocol. The amplified RNA (aRNA) from three independent pools of sample was then coupled to fluorescent dye esters (Cy3 and Cy5). Size exclusion column purification steps preceded and followed the dye-coupling reaction.
For hybridizations, we used the “dye–swap” design for technical replicates in which duplicate microarrays were performed per RNA sample but the Cy dyes were “swapped”, that is, one of the duplicates contained control (placebo) and morphine-treated samples labeled with Cy3 and Cy5, respectively (called polarity P+), and the other contained control and treatment samples that were reversely labeled with Cy5 and Cy3, respectively (called polarity P−). The differentially expressed genes (DEG) were determined based on each gene’s average differential expression of the dye swap pair (P+ and P−). The arrays were prehybridized in 40 µl of 5× SSC with 0.1% SDS and 1% BSA at 42° C for 30 min. The prehybridization solution was removed, and arrays were hybridized for 16 h at 42°C in 5× SSC buffer containing Cy3/Cy5 labeled targets, 25% formamide, 0.1% SDS, 1 µg Cot-1 DNA, and 1 µg poly A RNA. After washing in molecular biology-grade water for 5 times, followed by one wash in isopropanol, hybridized microarrays were scanned for Cy3 and Cy5-labelled probes using a GenePix 4100A confocal scanner (Axon Instruments, Union City, CA) at 532 and 635 nm, respectively. Photomultiplier tube (PMT) voltages were adjusted at the time of data acquisition to balance Cy5 and Cy3 signals over the whole intensity range using GenePix image balance histograms.
DNA microarrays were made at the SCCPRR Gene Array Facility (Department of Pharmacology, University of Washington) using the Programs for Genomic Applications (PGA) mouse 70mer oligo library generated at Massachusetts General Hospital (Boston, MA). The 19,554 oligos on this array were designed using the OligoPicker software (available at http://pga.mgh.harvard.edu/oligopicker) (Wang and Seed, 2003). Oligos were selected using a stringent set of filters to ensure specific representation of all protein-coding sequences in the National Center for Biotechnology Information (NCBI) GenPept database. The program designs oligos that have a Tm within a 10° C range, to ensure hybridization under similar conditions. Also, OligoPicker avoids sequences that are likely to cross-hybridize with other genes or to self-anneal (form primer-dimers). The sequence specificity of each oligo was tested with a global BLAST score and all oligos with a BLAST score above a threshold (Altschul et al., 1990) were discarded. Additional information about the library, including sequences of oligos, is available on the MGH website: https://dnacore.mgh.harvard.edu/microarray/index.shtml.
There were two independent pooled RNA samples each from hypothalamus and pituitary. Dye-swap duplicate microarrays were performed for each RNA, therefore, four microarrays were performed for short-term treatment, four microarrays for long-term treatment for a total of eight microarrays on hypothalamus and eight microarrays on pituitary RNA.
Data processing and analyses
The GenePix Pro 6.0 software (Silicon Genetics, Redwood City, CA) was used to acquire and analyze the microarray images that included spot finding, identification and quantification of fluorescent signal intensities. The GenePix result files (".gpr" file), including signal, background, standard deviation, pixel statistics and quality parameters for both Cy3 and Cy5 channels were stored in a local microarray database.
Multiple individual “.gpr” files were imported into GeneSpring® (Agilent Technologies, Santa Clara, CA) to create a single experiment. After the “.gpr” files were imported into GeneSpring®, all slides were normalized using a LOWESS (Locally Weighted Regression and Smoothing) function to address intensity-dependent dye biases. Once all the slides are entered and normalized, the slides corresponding to each treatment group (including dye-swaps) were merged, the background-subtracted data for each spot log-transformed, spots not above the background threshold were removed, and the data visualized and analyzed for statistical significance. From the experiments created in GeneSpring®, we generated Excel spreadsheet files, which includes the normalized ratio of treatment to control, the treatment average for each spot, the control average for each spot, the T-test p value of treatment and control values for each spot, and the annotation (Plate ID, Genbank, LocusLink, Gene Name, Gene Symbol, and Gene Description) for each spot.
The data (“.gpr” files generated by GenePix) were filtered on the basis of signal levels and spot quality using Acuity software version 4 (Molecular Devices, Sunnyvale, CA). First arrays were normalized by median-centering the logarithmic ratios so that the median ratio of all genes that passed through the filters was equal to 1 (global normalization). Local background values were subtracted from spot intensities to obtain signal values. Data were included if the signal-to-background ratio was ≥ 2, the signal intensity was >500 fluorescence units above background, the spot diameter was between 50 and 180 microns, and at least 70% of the pixels had fluorescence intensities that were two standard deviations above background. Cy5/Cy3 ratios were calculated for each spot and logarithmically transformed (log 2). A ratio with absolute value equal to 1 was taken as the threshold value to consider a sequence differentially expressed in the two populations (a ratio of 1 in log base 2 equals an expression ratio difference with a factor of 2).
The statistical significance (p value) for each gene between two classes for each experiment design was determined with the Student’s t test. A significant differential expression was defined as 2-fold compared to controls, as others have used (Chen et al., 2007). The Acuity and Gene Expression Investigation Suite (GENESIS developed by Alexander Sturn, Graz University of Technology, Austria) (Sturn et al., 2002) were used for the generation of hierarchical clusters of the filtered data. Average linkage hierarchical clustering of samples was based on a Pearson correlation similarity metric using genes selected by statistical significance (p ≤0.05). Genes were included for which there was 100% data present. The final complete microarray data (unprocessed “.gpr” files and normalized data representing the ratio of morphine/placebo), used to generate hierarchical cluster figures have been deposited in the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) of the NCBI and are accessible through GEO Series accession number GSE9525.
Quantitative real-time RT-PCR
We confirmed gene expression changes by two-step real-time RT- PCR for selected genes with significantly changed expression detected on our arrays. RNA was extracted from pooled tissue (3 animals each) from two independent groups of mice. Each group included three to five animals that were separate from those used in the array study. In the first step, we reverse transcribed the mRNA to cDNA using QuantiTect Reverse Transcription kit (Qiagen, Valencia, CA) according to the instructions of the manufacturer. In the second step, we performed real-time PCR using QuantiTect SYBR Green RT-PCR kit (Qiagen, Valencia, CA) according to the instructions of the manufacturer. Three hundred ng per reaction of cDNA was used and the total volume per reaction was 50 µl. Primer sequences for each gene of interest were provided by Primer Bank database (http://pga.mgh.harvard.edu/primerbank/) and synthesized by GenoMechanix (Gainesville, FL).
The PCR reactions were performed in the iCycler (Bio-Rad Laboratories, Hercules, CA) using a PCR program of an activation step for 15 min at 95°C, followed by 35 cycles of amplification, with each cycle consisting of a denaturation step at 94°C for 15 s, followed by an annealing step at 60°C for 30 seconds and an extension step at 72°C for 30 s. We also performed a melting curve analysis (temperature range 65–95°C) to check for the formation of primer-dimers and production of nonspecific products. Each reaction was performed in triplicate and threshold cycles (CT) were calculated using the second derivative of the reaction. The CT of each gene was normalized against that of GAPDH, which showed no regulation by morphine on our microarrays. Fold changes were determined using the −ΔΔCT method. Controls without RNA were performed to ensure that amplification of products was specific and was not due to non-specific contamination.
Food intake, body weight and hypothalamic neuropeptide Y (NPY) and agouti-related peptide (AgRP) expression in morphine-treated, placebo-treated or pair-fed mice
Mice were divided into three groups and treated for four days with morphine or placebo pellets or placebo pellets (day 0) pair-fed to match food intake on the prior day of morphine pellet-mice. Pair-feeding occurred on days 1–4. Animals were weighed daily and the weight change from baseline was calculated. The amount of food consumed per mouse was also determined daily. Baseline food intake and weight were measured for two days prior to pellet implantation in placebo- and pair-fed animals and for one day in morphine-treated animals. Hypothalamic NPY and AgRP expression was determined by real-time RT-PCR using the conditions and primers above and corrected for expression of 18S RNA.
Measurement of hypothalamic peptides related to food intake in morphine-treated and placebo-treated mice
Mice were divided into two groups and implanted with a morphine or placebo pellet for four days. Mice were sacrificed, brains were removed and hypothalamus of each mouse was carefully dissected out, extracted in RIPA buffer (Upstate, Lake Placid, NY, USA) containing a protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA) and passed through a Strata C-18E (55 µm, 70A) Sep-Column (Phoenix Pharmaceuticals, Inc, Burlingame, CA, USA). The eluant was lyophilized and reconstituted in the assay buffer supplied in the enzyme immunoassay kit. Hypothalamic levels of NPY, AgRP (82–131 amide), CART (61–102) and α-MSH were determined using enzyme immunoassay kits (Phoenix Pharmaceuticals, Belmont, CA). The NPY assay cross-reacts with NPY 3–36 at 14.3% and does not cross-react with peptide YY or other tested peptides. The AgRP assay does not cross-react with α-MSH, orexins or other tested peptides. The CART assay cross-reacts with CART (55–102) at 100% and does not cross-react with α-MSH, orexins, AgRP, NPY or other tested peptides. The α-MSH assay cross-reacts with desacetyl-α-MSH at 79% and does not cross-react with CART, ACTH, AgRP, NPY or other tested peptides. The inter-assay variation was 5–10% and the intra-assay variation was <15% for all kits. Peptide levels were expressed per µg of protein as determined by the Bradford assay (Bradford, 1988).
Statistical analyses
Data were expressed as the mean ± SEM. Repeated measure analysis of variance with Newman-Keuls’s post-hoc testing used to compare treatments in the behavioral and abstinence studies, except Tukey-Kramer post-hoc testing was used in the experiments on food intake and body weight following morphine pellets. Statistical significance for correlations between real-time PCR and microarray data used the Fisher’s z test. Significance level for all experiments was set at p< 0.05.
RESULTS
Abstinence withdrawal in mice implanted with morphine for six h or four days
To assess the degree of dependence on morphine in mice exposed to morphine for four days as compared to six h, we removed the morphine or placebo pellets and monitored the mice for signs of abstinence withdrawal over the next two h (Fig. 1). Jumps (A), rearing (B) and forepaw tremors (C) were frequent in mice treated with morphine pellets for 4-days. In contrast, no withdrawal signs were seen in mice implanted with morphine for six h. These results indicate that long-term, but not short-term morphine administration leads to opiate dependency. Thus, we used these two different forms of morphine treatment to study the effect of short-term and long-term morphine treatment on gene expression.
Fig. 1.
Four day, but not six h morphine pellet administration leads to opioid dependence as determined by abstinence withdrawal. Mice were implanted with a placebo [6 h (n=3) or 4 days (n=4)] or morphine [6 h (n=6) or 4 days (n=5)] pellet. The pellets were removed after 6 h or 4 days and mice were observed for signs of opiate withdrawal that was determined by counting the number (± SEM) of jumps (A), rearing (B) and forepaw tremors (C) over the next two h following pellet removal. The 6 h and 4 day placebo groups were pooled. *** P<0.001 vs placebo pellet, ### P<0.001 vs 6 h morphine pellet.
Microarray analysis
We performed DNA microarray analysis in the hypothalamus and pituitary of mice treated with short-term or long-term morphine in order to identify genes that are regulated by acute and/or chronic morphine treatment. To uncover such genes, we employed two common data mining strategies. First, we ranked the data according to quantitative level of gene expression change and identified genes with significant up-regulation or down-regulation of 2-fold or more (Table 1–Table 3, Supplemental Fig. 1– 4). Second, we used unbiased hierarchical clustering to identify groups of genes with patterns of gene expression changes that correlated with particular morphine treatment and/or tissue type. The hierarchical clustering feature sorts the data, bringing genes and experiments with similar profiles together in a visual form.
Table 1.
Regulation of gene expression in hypothalamus and pituitary.
| Short-term (6 hour) | Long-term (4 day) | |||
|---|---|---|---|---|
| Organ | ↑ | ↓ | ↑ | ↓ |
| Hypothalamus | ||||
| ≥2 fold | 39 | 6 | 35 | 51 |
| 1.5–1.9 fold | 113 | 46 | 38 | 185 |
| Pituitary | ||||
| ≥2 fold | 110 | 29 | 85 | 37 |
| 1.5–1.9 fold | 256 | 117 | 283 | 200 |
A total number of 19,554 oligos were present on each slide. ↑ up-regulation, ↓ down-regulation. The table lists the genes that were significantly regulated (p≤ 0.05).
Table 3.
Pituitary genes with highest fold regulation by morphine.
| Name | GeneBank # | 6 h | P value | 4 d | P value |
|---|---|---|---|---|---|
| Carbonic anhydrase 2 | NM_009801 | 4.8 | 0.0008 | 1.2 | 0.04 |
| CD79B antigen | NM_008339 | 6.4 | 0.001 | 1.8 | 0.004 |
| Cytochrome P450, family 27, subfamily a, polypeptide 1 |
AK004977 | 1.5 | 0.03 | 4.5 | 0.02 |
| Heterogeneous nuclear ribonucleoprotein D-like |
NM_016690 | 3.0 | 0.001 | 1.3 | 0.005 |
| ORF; chromosome 24p3 gene, partial cds |
L47696 | 4.5 | 0.007 | 2.1 | 0.02 |
| Phosphatidylserine synthase 2 | NM_013782 | 3.1 | 0.02 | 1.8 | 0.04 |
| RIKEN cDNA 1110036H21 gene | NM_025404 | 5.0 | 0.0007 | 3.3 | 0.03 |
| RIKEN cDNA 2610042L04 gene | AK011748 | 4.2 | 0.007 | 2.0 | 0.02 |
| RIKEN cDNA 4930435C18 gene | AK015320 | 4.4 | 0.01 | 1.3 | 0.007 |
| Chemokine (C-C motif) ligand 12 | NM_011331 | −3.3 | 0.006 | −1.5 | 0.003 |
| Myeloid cell leukemia sequence 1 | NM_008562 | 3.2 | 0.001 | −1.4 | 0.006 |
| Neoplastic progression 3 | Z31362 | 4.0 | 0.002 | −1.5 | 0.008 |
| X transporter protein 2 | AF075264 | 3.1 | 0.0008 | −1.3 | 0.01 |
| DNA Segment, Chr 6, human D12S2489E |
NM_033078 | −1.6 | 0.001 | 3.0 | 0.03 |
The table lists the genes that were significantly regulated.
Regulation of hypothalamic gene expression by morphine
We identified hypothalamic genes whose expression was changed by short- and long-term morphine treatment. We found 39 transcripts were significantly up-regulated at least two-fold by short-term morphine treatment as compared to the placebo group (Table 1). Of those, six transcripts were genes with unknown function, three transcripts were related to immunological processes [lymphatic vessel endothelial HA receptor-1 (Xlkd1), cytotoxic T lymphocyte-associated protein 2 alpha (Ctla2a) and thymopoietin (tmpo)], two are RAS related [RAB11B, member RAS oncogene family (Rab11b) and RAS, dexamethasone-induced 1 (Rasd1)]. In contrast, only six transcripts were down-regulated (two-fold or more) in the hypothalamus. Of the six down-regulated genes, two were involved in immunological processes [interferon-stimulated protein (Isg20) and T-cell replacing factor (Tcrf) (Kinashi et al., 1986, Grander et al., 1998)], one was a regulator of mitochondrial network [ganglioside-induced differentiation-associated-protein 1 (Gdap1) (Niemann et al., 2005)], one was involved in apoptotic processes [apoptosis-associated speck-like protein containing a CARD (Pycard) (Stehlik et al., 2003)], one was part of odorant receptor [odorant receptor M4 (Olfr63) gene (Lewcock and Reed, 2004)] and one was a gene with unknown function (RIKEN cDNA 2310051N18 gene).
After long-term morphine treatment, the pattern of gene regulation changed more dramatically as compared to the six-hour morphine-treated group. We found 35 transcripts were up-regulated at least 2-fold and 51 transcripts were down-regulated following the long-term morphine exposure (Table 1).
Regulation of pituitary gene expression by morphine
At 6 h after morphine pellet implantation, we found extensive gene regulation changes in the pituitary. Over 100 genes were up-regulated (2-fold or more) and similar numbers were down-regulated (Table 1).
At four-day morphine exposure, up-regulated genes predominated over down-regulated genes in the pituitary. In general, the expression levels of more genes in the pituitary were altered than in the hypothalamus (Table 1).
Comparison of short-term versus long-term morphine-induced regulation of gene expression in the hypothalamus and pituitary
In the hypothalamus (Supplemental Fig. 1), we found several genes that were differentially regulated by short-term versus long-term morphine treatment. Six genes were down-regulated by short-term, yet up-regulated by long-term morphine treatment. This category included several heat shock proteins [1∝ (Hspca), 1β (Hspcb) and 8 (Hspa8)] and pro-opiomelanocortin-alpha (Pomc1) and carboxypeptidase E (Cpe), CREBBP/EP300 inhibitory protein 1 (Cri1). Other hypothalamic genes were up-regulated following short-term and down-regulated following long-term treatment. This category included several mitochondrial ribosomal proteins (L12, L23 and L52) (Mrpl1, Mrpl23 and Mrpl2), NADH dehydrogenase (ubiquinone) (Ndufa2), and cadherin 3 (Cdh3).
In the pituitary (Supplemental Fig. 2), we found a group of differentially regulated genes in which several genes were up-regulated by both short- and long-term morphine treatment and some genes were up-regulated by short-term treatment but down-regulated upon long-term treatment. There was also a group of genes that was down-regulated with short-term but up-regulated with long-term morphine treatment. For example, ubiquinone (Ndufa1) was up-regulated by both short-term and long-term morphine treatment, while heat-shock protein 8 (Hspa8) and Cpe were up-regulated by short-term but down-regulated by long-term treatment. Interestingly, this differential regulation of Cpe in the pituitary was the converse of the pattern observed in the hypothalamus in which Cpe was down-regulated by short-term but up-regulated by long-term morphine treatment.
Comparison of morphine-induced regulation of gene expression between hypothalamus and pituitary
In order to extract additional informative patterns of gene expression, we performed hierarchical clustering in which we compared between the pituitary and hypothalamus genes expression following short-term (Supplemental Fig. 3) and long-term (Supplemental Fig. 4) morphine treatment. Gene clustering allowed us to visualize the expression of different genes that follow the same pattern of regulation.
Analyzing data generated by GeneSpring software, we observed that short-term morphine treatment induced up-regulation of 79 transcripts and down-regulation of 31 transcripts in both tissues. The up-regulated genes in both tissues included kinases (phosphatidylinositol-4-phosphate 5-kinase (Pip5k), FMS-like tyrosine kinase 1 (Flt1), CDC like kinase 4 (Clk4) and casein kinase 1 (Csnk1)), phosphatases [protein tyrosine phosphatase (Ptp) and protein phosphatase 2 (Ppp2r2a)], cell cycle mediators [cyclin-dependent kinase inhibitor 1A (Cdcn1a), retinoblastoma binding protein 7 (Rbbp7)]. The cluster shown in Supplemental Fig. 3 illustrates groups of genes that were up-regulated or down-regulated by short-term morphine treatment in both the hypothalamus and pituitary. The up-regulated genes include: RAS-related pathway genes [ras homolog gene family (Rhoa) and RAB, member of RAS oncogene family (Rab4a)], a zinc finger protein, two mitochondrial ATP synthase subunits (Atp5j2 and Atp5e), and two RIKEN transcript with unknown function. The down-regulated genes in both tissues included metallothionein 1 (Mt1), lipocalin 2 (Lcn2), and retinoblastoma binding protein 7 (Rbbp7). Following long-term morphine treatment, we found nine transcripts in which the hypothalamic gene was up-regulated and the pituitary gene was down-regulated. These include heat shock protein 8 (Hspa8) and 5 (Hspa5), secretogranin II (Scg2), Cpe and lipocalin 2. Conversely, we found 18 transcripts in which hypothalamic genes were down-regulated and the pituitary genes were up-regulated. These include a proteasome subunit (Psma2) and several transcripts with unknown yet function (RIKEN genes) (Supplemental Fig. 4).
Morphine-regulated genes involved in food intake
Our microarray analysis uncovered several genes regulated by morphine that play a role in food intake, a novel finding which demonstrates the unique power of the microarray approach. These genes showed a high degree of regulation in the mouse hypothalamus and/or pituitary after short-term or long-term morphine administration (Table 4). Both neuropeptide Y (NPY) and agouti-related protein (AgRP) were strongly up-regulated following long-term morphine treatment in the hypothalamus. Pomc1 was decreased after 4 d treatment in the hypothalamus, but not in the pituitary. Other genes [leptin receptor (Lepr), cocaine and amphetamine regulated transcript (CART), adiponutrin (Adpn) and peptide YY (Pyy)] involved in obesity pathway were selectively regulated in both the pituitary and hypothalamus (Table 4).
Table 4.
| Gene | GenBank # | Hypo- 6 h (fold change) |
P value | Pituitary- 6 h (fold change) |
P value | Hypo- 4 d (fold change) |
P value | Pituitary- -4 d (fold change) |
P value |
|---|---|---|---|---|---|---|---|---|---|
| Npy | NM_023456 | 1.2 | 0.01 | 1.1 | 0.09 | 2.6 | 0.002 | 1.5 | 0.02 |
| Npyr Y1 | D63819 | −1.2 | 0.4 | 1.1 | 0.05 | 1.2 | 0.05 | 1.1 | 0.08 |
| AgRP | NM_007427 | 1.6 | 0.001 | N/A | N/A | 2.4 | 0.02 | 1.5 | 0.02 |
| CPE | NM_013494 | 1.4 | 0.2 | 1.2 | 0.04 | −1.3 | 0.03 | 2.1 | 0.04 |
| Lepr | AF039456 | 1.2 | 0.09 | −1.1 | 0.5 | 1.8 | 0.03 | −2.9 | 0.5 |
| Peptide YY | BC010821 | 1.1 | 0.07 | −1.2 | 0.1 | 1.2 | 0.06 | 1.1 | 0.3 |
| Adiponutrin | NM_054088 | 1.1 | 0.008 | −1.7 | 0.08 | −4.0 | 0.02 | −1.9 | 0.01 |
| Pomc1 | NM_01_10929 | 1.1 | 0.4 | 1.3 | 0.1 | −1.5 | 0.01 | 1.3 | 0.02 |
| CART | NM_013732 | 1.1 | 0.6 | 2.2 | 0.002 | −1.1 | 0.09 | 1.2 | 0.3 |
Genes involved in the food intake pathway as elucidated by microarray analysis. Hypo-, hypothalamus, Npy neuropeptide Y, Npyr neuropeptide Y receptor, AgRP agouti-related protein, Lepr leptin receptor, isoform Re, CART cocaine and amphetamine regulated transcript, CPE carboxypeptidase E. The table lists the genes that were significantly regulated.
Confirmation of regulated genes by real-time RT-PCR
We were able to validate by real-time RT-PCR (Table 5) the morphine regulation of several significantly regulated genes found by microarray analysis. Examining the nine genes listed in Table 4, validation of the microarray results was reflected in the high correlation (R2=0.71, p=0.002) of mRNA expression as measured by microarray and that measured by real-time RT-PCR.
Table 5.
Real-time RT-PCR results compared with microarray data (expressed as percent of control.
| Hypothalamus 4 d | Ratio of expression following 4 d of morphine treatment compared to placebo treatment |
Ratio of expression following 4 d of morphine treatment compared to placebo treatment |
|
|---|---|---|---|
| Gene | GenBank # | Microarray | Real-time RT-PCR |
| Unknown (RIKEN cDNA 5730513H21 gene) |
AK017769 | 0.67 | 0.42 ± 0.2 |
| Unknown (RIKEN cDNA 3110001K13 gene) |
AK013955 | 0.44 | 0.78 ± 0.01 |
| AgRP | NM_007427 | 2.4 | 3.4 ± 0.01 |
| Npy | NM_023456 | 2.6 | 4.3 ± 0.1 |
| proSAAS | NM_013892 | 0.94 | 1.1 ± 0.1 |
| Pituitary 4 d | |||
| Gene | GenBank | Microarray | Real-time RT-PCR |
| ACP homolog | 0.38 | 0.64 ± 0.02 | |
| (RIKEN cDNA 2610003B19 gene) |
AK008788 | ||
| AgRP | NM_007427 | 1.5 | 3.6 ± 0.2 |
| Npy | NM_023456 | 1.5 | 4.3 ± 0.2 |
| proSAAS | NM_013892 | 1.3 | 1.2 ± 0.04 |
ACP Acyl Carrier Protein, AgRP agouti-related protein, Npy neuropeptide Y. The table lists the genes that were significantly regulated (p≤ 0.05). Data is expressed as mean ± SEM. RT-PCR results are from 2 separate pools of mRNA each from 3 animals.
Morphine treatment reduced food intake and body weight
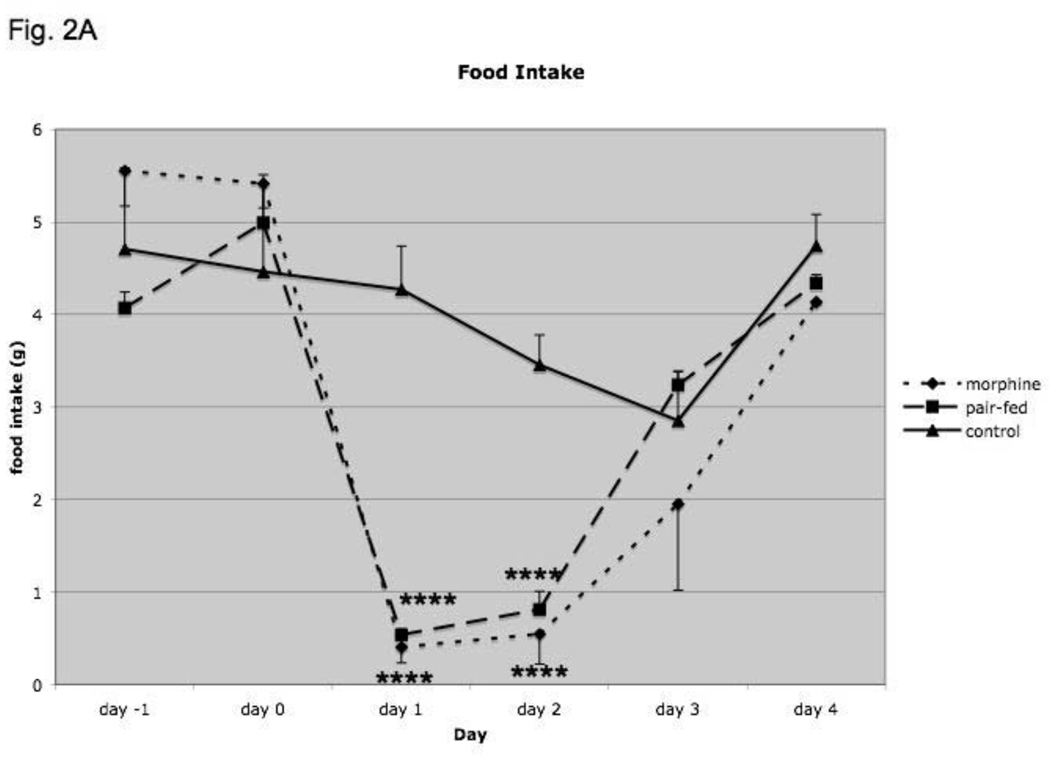
To determine if there was a physiologic response that may be associated with morphine-induced regulation of genes involved in food intake, we measured changes in food intake and body weight in mice treated with morphine for 4 days (Fig. 2A and B). We also determined whether the changes are due to morphine treatment or alterations in food intake; thus, we examined the effect of pair-feeding on these parameters by using mice exposed to placebo pellets that were pair-fed to match food intake on the prior day of morphine pellet-mice. For food intake, there was a significant effect of treatment (F2,29 = 3.95; p<0.05), a significant effect of time (day with regard to placebo or morphine pellet implantation; F5,117 = 25.4; p<0.000001) and a significant interaction (F10,117 = 5.24; p<0.000005), indicating that food intake was significantly altered following morphine as compared to placebo implantation (Fig. 2A). Post-hoc analysis revealed a significant reduction (p<0.05) in food intake in morphine-treated and pair-fed mice as compared to placebo group for days 1 and 2. Food intake in pair-fed mice was not significantly different from morphine-treated mice, as expected.
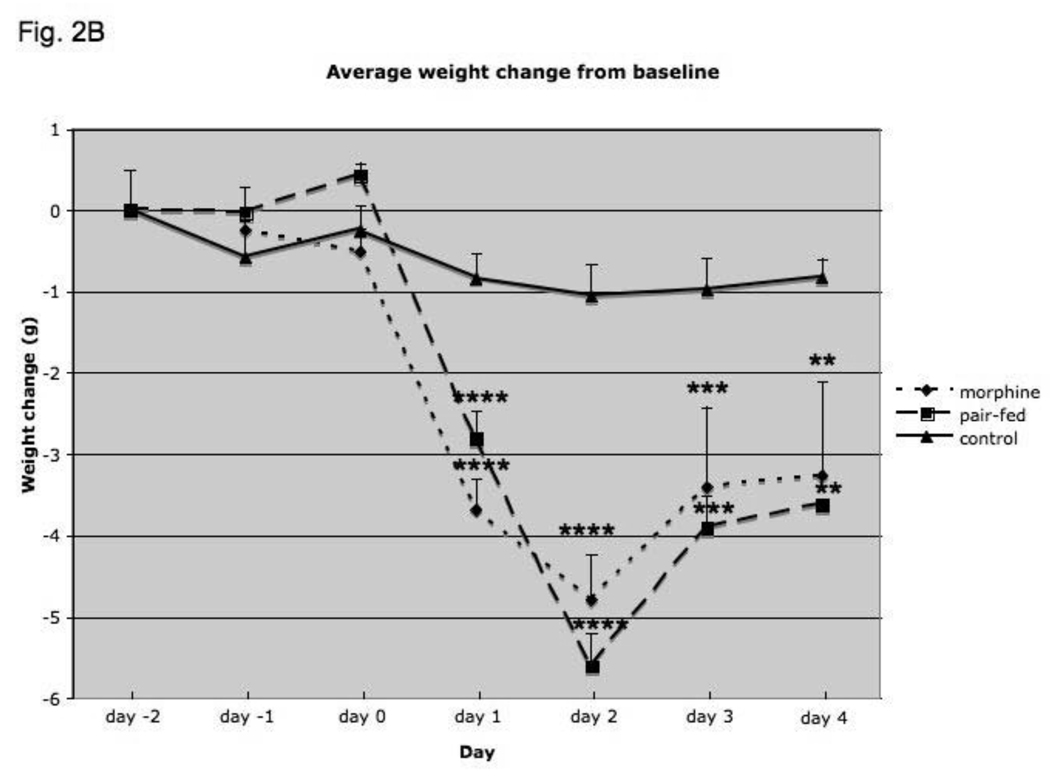
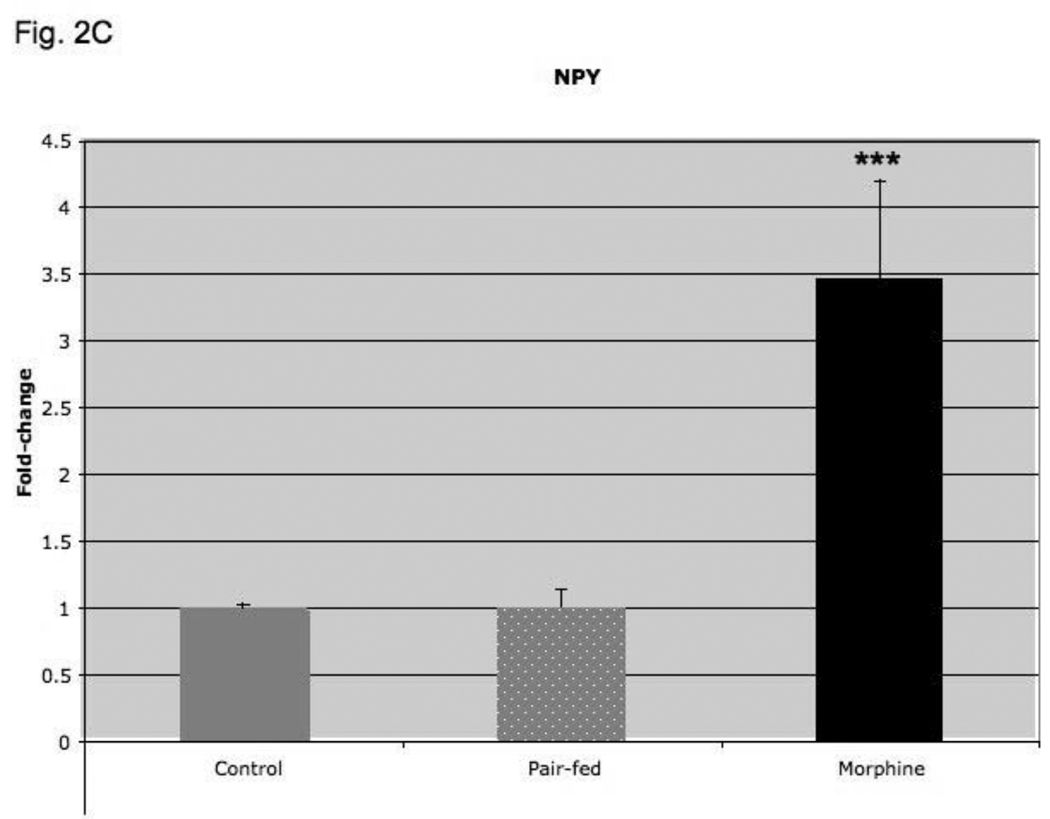
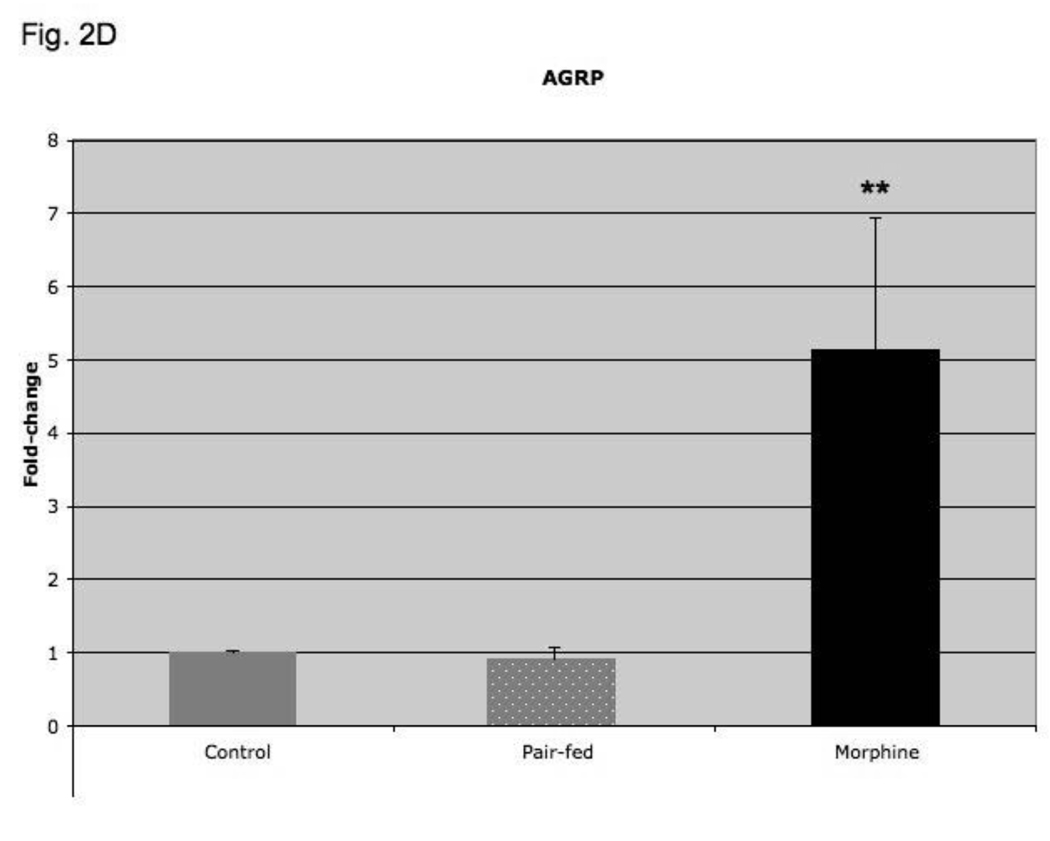
Fig. 2.
Food intake (A), average weight change from baseline (B), NPY expression (C) and AgRP (D) expression in mice treated for four days with morphine or placebo pellets or placebo pellets pair-fed to match food intake on the prior day of morphine pellet-mice. Pair-feeding occurred on days 1–4 and morphine mice are shifted by one day so that their food intake is aligned with that of pair-fed mice. Animals were weighed daily and weight change from baseline was calculated. The amount of food consumed per mouse was determined daily. Baseline food intake and weight were measured for two days prior to pellet implantation in placebo- and pair-fed animals and for one day in morphine-treated animals. Hypothalamic NPY and AgRP expression was determined by real-time RT-PCR and corrected for expression of 18S RNA. Food intake, N=8–11 animals, body weight, N=10–12 animals, Gene expression, N=7–14 animals. *, P< 0.05, **, P< 0.01, ***, P< 0.001, ****, P< 0.0001.
For body weight, there was a significant effect of treatment (F2,31 = 17.2; p<0.0001), a significant effect of time (day of treatment with placebo, pair-fed or morphine) (F5, 137 = 67.0; p<0.000001) and a significant interaction between treatment and time (F10, 137 = 12.1; p<0.000001), indicating that body weight was significantly altered following morphine-treated and pair-fed mice as compared to placebo implantation (Fig. 2B). Post-hoc analysis revealed a significant decrease (p<0.05) in body weight by morphine as compared to placebo-treated control group for days 1–4. Body weight in pair-fed mice was not significantly different from morphine-treated mice. Preliminary observations revealed that locomotion activity was increased following morphine implantation but returned to normal by 24 h (data not shown).
Four-day morphine treatment but not pair-feeding increased hypothalamic NPY and AgRP expression
In order to determine if four-day morphine treatment regulated hypothalamic NPY and AgRP expression per se, or whether their regulation is as a consequence of the decreased food intake and weight loss observed in morphine-treated as compared to their placebo-treated mice, we performed pair-feeding experiments. As shown in Fig. 2C, morphine treatment increased hypothalamic NPY expression 3-fold (p < 0.001; compared to placebo-treated mice), but pair-feeding was without effect (p=NS; versus placebo-treated group). Similarly, morphine treatment increased hypothalamic AgRP expression 5-fold (p < 0.01 compared to placebo-treated group), but pair-feeding as compared to placebo treatment was without effect (Fig. 2D; p=NS).
Four-day morphine treatment increased hypothalamic NPY, CART and α-MSH peptide levels
In order to determine if four-day morphine treatment regulated the peptide products from the hypothalamic genes involved in food intake, we used specific peptide RIAs. Hypothalamic levels of NPY, CART and α-MSH were significantly elevated in morphine treated mice compared to mice receiving placebo pellets (Table 6). AgRP peptide level was not significantly affected by morphine treatment.
Table 6.
Levels of food-related peptides in the hypothalamus following 4 d of morphine treatment
| Peptide | Morphine treatment protein) |
(pg/µg | Placebo treatment (pg/µg protein) |
P value |
|---|---|---|---|---|
| Npy | 12.2 ± 3.3 | 3.9 ± 0.9 | 0.02 | |
| AgRP | 4.6 ± 0.9 | 2.9 ±0.6 | 0.13 | |
| CART | 3.4 ± 1.1 | 1.0 ± 0.1 | 0.04 | |
| α-MSH | 3.3 ± 0.3 | 2.3 ± 0.2 | 0.008 |
AgRP, agouti-related peptide; α-MSH, α-melanocyte-stimulating hormone, CART, cocaine and amphetamine regulated transcript; Npy, neuropeptide Y. Mean ± SEM is depicted. N=6 for morphine-treated mice and N=7 for placebo-treated mice.
DISCUSSION
Opioids alter a series of physiological processes including nociceptive information, respiration, gastrointestinal motility, carbohydrate metabolism, reproduction and food intake; many of these processes may be regulated by changes in endogenous neuropeptides (Morley, 1981, Vuong et al., 2009 [Epub ahead of print]). The molecular, cellular and physiological mechanisms that mediate the transition from occasional drug use to the loss of control that, in part, defines addiction are not fully understood. Using short-term (6 h) and a long-term (4 day) morphine exposure in mice, we attempted to model an “unaddicted” brain that would then transition to an “addicted” brain and used microarray chips to uncover known and unknown genes involved in this transition. We focused on the hypothalamus and the pituitary as these tissues are the sites of neuropeptide biosynthesis. In both the hypothalamus and the pituitary, we found multiple genes regulated by both short-term and long-term morphine exposure. We found three types of genes regulated by morphine: (1) genes known and expected to be regulated by opioids, (2) genes not known to be regulated by opioids and (3) previously unknown genes. The known genes include POMC (Garcia de Yebenes and Pelletier, 1993) and heat-shock proteins (Ammon-Treiber et al., 2004a). POMC expression is likely due to auto-regulation by endogenous opiates, while heat-shock proteins are likely up-regulated as a protective effect against opioid-induced neurotoxicity. Genes in categories not previously shown to be regulated by opioids, included kinases and phosphatases, cell cycle mediators and genes involved in immunological processes. We have also uncovered several previously uncharacterized genes (RIKENs) whose altered expression may be important for development of opioid addiction.
We posit that there may be genes involved in the transition from the unaddicted brain to the addicted brain that are regulated differently after long-term compared to short-term morphine exposure. We found several genes that were regulated in opposite directions at 6 h compared to 4 d morphine treatment. Several genes were up-regulated at 6 h and down-regulated at 4 d including hypothalamic fibronectin 1 (Fn1), pituitary myeloid cell leukemia sequence 1 (Mcl1), pituitary neoplastic progression 3 (Npn3), pituitary X transporter protein 2 (Slc6a18), pituitary DNA Segment (D11Wsu68e), whereas another group of genes was down-regulated at 6 h and up-regulated at 4 d: Chr 6 (Ing4), human D12S2489E and pituitary leptin receptor, isoform Re (Lepr). Further studies on the role of these morphine-regulated genes in the processes related to opioid addiction are needed.
Several recent studies using different morphine-exposure paradigms and examining different brain regions have analyzed genes regulated by morphine using microarray technology (Loguinov et al., 2001, Ammon et al., 2003, Ammon-Treiber and Hollt, 2005, McClung et al., 2005, Grice et al., 2007, Korostynski et al., 2007). In the first published study, Loguinov et al. showed that 45 genes/ESTs were down-regulated in the mouse striatum and/or the spinal cord after a single morphine injection, whereas only nine genes/ESTs up-regulated, without major differences between the two regions (Loguinov et al., 2001).
In a study in rat frontal cortex using a 10-day treatment schedule with escalating morphine doses leading to morphine tolerance, 14 of 8000 genes were found to be induced after the last dose and only one gene was found to be reduced (Ammon et al., 2003). The majority of the morphine-induced genes coded for heat shock proteins (hsp70, hsp27, hsp40, hsp105, GRP78). In our study, we also found substantial regulation of heat shock proteins by morphine. Ammon-Treiber et al. (Ammon-Treiber et al., 2004b) identified additional genes following naloxone-precipitated morphine withdrawal and found 36 ESTs were induced greater than two-fold by withdrawal. Seven of the genes were transcription factors.
In the locus ceruleus (LC), using DNA microarray analysis, chronic morphine treatment regulated tyrosine hydroxylase, prodynorphin, and galanin in both mouse and rat but different gene expression changes occurred in the ventral tegmental area (VTA) (McClung et al., 2005). Microarray analysis of the nucleus accumbens of C57BL/6 and DBA/2J mice identified axon guidance genes, particularly the semaphorins, as showing altered expression in the presence of morphine, and plasticity genes as showing altered expression across strains (Grice et al., 2007). Single and repeated morphine administration induced numerous novel regulation (e.g. Olig2 and Camk1g) and a number of transcripts with strain-specific changes in expression (e.g. Hspa1a and Fzd2) in the striatum of selected inbred mouse strains (129P3/J, DBA/2J, C57BL/6J and SWR/J) (Korostynski et al., 2007).
Additionally, Spijker et al. (Spijker et al., 2004) performed a real-time RT-PCR-based approach to identify the temporal expression profile of a set of 159 genes expressed in the nucleus accumbens (NAc) of rats during a 14-day exposure to morphine and during the subsequent 3 weeks of abstinence. Surprisingly, only a few genes examined in this study displayed a relative change greater than 2-fold.
More recently, Befort et al. (Befort et al., 2008) identified 25 genes in the lateral hypothalamus whose expression was altered by an escalating dose morphine regimen in wild-type but not mu-receptor null mice. Six of these genes were confirmed by RT-PCR. These genes could be involved in the altered reward processes seen in addiction.
Thus, although microarray analysis following morphine administration has been studied before, our study is unique in that we a) examined the effect in neuropeptide-rich regions of the hypothalamus and pituitary and b) we specifically uncovered genes that were preferentially regulated after long-term morphine administration compared to short-term administration, c) we observed regulation of potentially important genes related to food intake. Our results are similar to the previous reports, in that we found regulation of several heat shock proteins and kinases by morphine. Our results are limited, however, in that we studied gene regulation in the whole hypothalamus and pituitary. Therefore, further studies are needed to examine gene regulation in distinct hypothalamic nuclei to reveal the importance of these nuclei in morphine-regulated food intake and body weight homeostasis. Also, experiments using receptor-selective drugs are necessary to characterize the importance of different opioid receptors in these actions of morphine.
One of the most noteworthy findings revealed by this microarray analysis was the uncovering of morphine-induced alterations in appetite-regulating genes. We found that long-term morphine treatment increased hypothalamic and pituitary NPY and AgRP expression as well as the pituitary CART expression, whereas, the same treatment decreased pituitary NPY1 receptor and hypothalamic peptide YY expression. Additionally, POMC expression which following post-translational processing leads to α-MSH was down-regulated by 4-day morphine treatment. Both AgRP and NPY increase food intake, while PYY, CART and α-MSH decrease food intake (Schwartz, 2001, Pedrazzini, 2004, Chen et al., 2006). Furthermore, carboxypeptidase E (CPE), a prohormone-processing exopeptidase and a prohormone sorting receptor for the regulated secretory pathway (Cawley et al., 2004), were up-regulated in the pituitary and down-regulated in the hypothalamus following 4-day morphine treatment. CPE-null mice were obese, hyperphagic and have decreased metabolic rate (Cawley et al., 2004). In contrast, 6-h morphine exposure decreased the pituitary leptin receptor and hypothalamic and pituitary adiponutrin expression. Thus, morphine treatment has complex effects on the regulation of potent stimulators and inhibitors of food intake that also depends on the duration of morphine treatment.
Due to translation or post-translational regulation, it cannot be assumed that the hypothalamic peptide levels related to food intake were changed along with mRNA levels. Therefore, we measured NPY, α-MSH, CART and AgRP peptide levels in the hypothalamus of 4-day morphine-treated mice and found that all peptides were significantly increased, except AgRP levels which did not reach a statistically significant level (Table 6). The lack of significance in the AgRP measurement may be due to the large variance found. While hypothalamic NPY exhibited an increase in both mRNA and peptide levels, CART and POMC/α-MSH exhibited slight down-regulation of mRNA levels with up-regulation of peptide levels. We interpret these findings as regulation by morphine of the prohormone convertases of which PC1/3 and PC2 are the most important for processing prohormones to active peptides (Seidah and Chretien, 1999). We recently showed that short-term (24-h) morphine exposure down-regulated hypothalamic PC1/3 and PC2 and longer-term (7-days) morphine exposure up-regulated these processing enzymes (Espinosa et al., 2008). In our current 4-day morphine experiment, potential up-regulation of hypothalamic PC1/3 and PC2 could lead to more α-MSH [generated from POMC by PC1/3 and PC2 (Zhou et al., 1993)], NPY [generated from pro-NPY primarily by PC1/3 (Brakch et al., 1997) and PC5/6 (Stein et al., 2006)], CART55–102 [generated from pro-CART primarily by PC1/3 (Dey et al., 2003)] and AGRP83–132 [generated from pro-AgRP primarily by PC1/3 (Creemers et al., 2006)]. Furthermore, translational regulation, alteration of exopeptidases, such as CPE, as well as regulation of peptide catabolism may also influence peptide levels.
The increase in CART peptide found in the hypothalamus of 4-day morphine treated mice may be accompanied by increase in other pro-CART derived peptides possessing biological activity (Dylag et al., 2006). Similarly, the increase in α-MSH may be accompanied by an increase in other POMC-derived peptides, including ACTH and β-endorphin (Eipper and Mains, 1980).
Examining the data in Table 4– 6, we found that the orexigenic effects of NPY and AgRP may be balanced by the anorexigenic effects of α-MSH and CART. However, changes in peptide levels in the hypothalamus are more likely to be important in food intake than changes in the pituitary peptides, which although they may be released into the circulation, may not be involved in food intake. A similar caveat is that changes in expression levels of genes or peptides found in the whole hypothalamus may not be localized in neurons involved in food intake.
Some of the changes in gene expression may be a direct result of morphine, while other changes may be compensatory and related to the decreased food intake. In order to determine if morphine-regulation of these genes caused decreased food intake and weight loss or whether their regulation of NPY and AGR expression is an effect of the decreased food intake and weight loss, we performed pair-feeding experiments which did not show changes in hypothalamic NPY and AgRP on day 4, indicating that morphine itself regulated hypothalamic NPY and AgRP. In contrast to our findings that 4-day pair-feeding in mice (with reduced, but not complete food restriction) led to no change in hypothalamic NPY and AgRP expression, Li et al. (Li et al., 2002) found that 48-h fasted rats had a 1.3 fold increase in hypothalamic NPY expression. AgRP expression was not noted.
The effects of opiates on food intake is controversial and includes reports of both hyperphagia (Grandison and Guidotti, 1977, Cooper and Sanger, 1984, Morley et al., 1984) as well as anorexia (Frenk and Rogers, 1979, Marks-Kaufman and Kanarek, 1980, Sanger and McCarthy, 1980, Cooper, 1981, Leshem, 1981, Kunihara et al., 1983, Wolgin and Benson, 1991) depending on the dose and dosing regimen [reviewed in (Levine et al., 1985)]. Morphine has also been shown to have a triphasic effect on feeding (Leshem, 1981, Leshem, 1988). Following injection of 15 mg/kg, morphine suppressed intake during the first h, enhanced intake during the next 3 h, and then suppressed intake again for up to 24 h. These studies were all performed prior to the discovery of many of the appetite-regulating pathways uncovered in the last 20 years. We used a morphine pellet regimen and showed that decreases in food intake and body weight ensued following morphine pellet implantation. Decreased food intake was maximal on the first day of pellet implantation and returned towards that of placebo-implanted control mice, but continued to be below that of the control group on subsequent days. Weight loss was maximal on day 2. The effect was not due to anesthesia or surgery as placebo-implanted mice had only a minimal weight loss. Of note, morphine-treated mice had less weight loss than pair-fed animals, indicating that morphine may facilitate an adaptive behavior to decrease weight loss in the face of decreased food intake, possibly by decreasing metabolic rate. Preliminary observations by our group (Marquez et al., 2006) as well as behavioral studies in this report suggest that hyperlocomotion followed morphine exposure consistent with prior reports (Kotlinska et al., 2007), so the decreased food intake was not due to catalepsy, which is observed in rats exposed to morphine.
There are several caveats related to performing and interpreting microarray experiments (Soverchia et al., 2005). These include collecting samples at different times of the day, stress in animal handling, non-uniformity in dissecting the complete tissue, degradation of mRNA, non-linear amplification and variation between animals and between experiments. We have taken care to mitigate these potential problems by avoiding stress in animal handling, collecting our samples at the same time of day, taking care to prevent mRNA degradation, ensuring linear amplification, pooling animals to reduce inter-animal variability, and performing hybridization on multiple occasions.
We used quantitative real-time RT-PCR to confirm our microarray results. There was a high (R2=0.71) correlation between microarray and real-time RT-PCR results. In general, real-time RT-PCR gave higher fold changes than microarray experiments following morphine treatment. Differences between real-time RT-PCR and microarray experiments can occur for several reasons, including different probes used for the microarray and real-time RT-PCR experiments (which can capture differential expression in splice variants), differences in the methods for normalization of expression data and possible false-positive expression changes.
In conclusion, we used microarray experiments to uncover genes regulated by short-term and long-term morphine exposure. Several patterns of genes were exposed including genes involved in food intake. Our results on hypothalamic genes involved in food intake can be further investigated using microdissection of specific hypothalamic nuclei or in situ hybridization/immunohistochemistry, in order to determine specific localization of this regulation. Our findings demonstrate the advantages of the microarray approach to understand mechanisms involved in the process of addiction especially by examining differential changes in expression of genes following short-term and long-term morphine exposure.
Supplementary Material
Table 2.
Hypothalamic genes with highest fold regulation by morphine.
| Name | GeneBank # | 6 h | P value | 4 d | P value | |
|---|---|---|---|---|---|---|
| 3-hydroxy-3-methylglutaryl- Coenzyme A synthase 2 |
AK004865 | 2.7 | 0.03 | 1.7 | 0.03 | |
| Neuropeptide Y | NM_023456 | 1.2 | 0.01 | 2.6 | 0.002 | |
| Sulfotransferase family 1A, phenol-preferring, member 1 |
BC005413 | 1.5 | 0.001 | 2.1 | 0.0002 | |
| Agouti-related protein | NM_007427 | 1.6 | 0.001 | 2.4 | 0.02 | |
| Apoptosis-associated speck-like protein containing a CARD |
NM_023258 | −2.2 | 0.005 | −1.5 | 0.03 | |
| Fibronectin 1 | BC004724 | 2.7 | 0.0002 | −1.4 | 0.03 |
Acknowledgements
This work was supported by NIH grants R01 DA14659 and K21 DA00276 and a Charles E. Culpeper Foundation Research Award to T.C.F., RCMI Program grant G21 RR03026, an NIH (R24DA017298) Minority Institution Drug Abuse Research Program (MIDARP) Program and a Center of Clinical Research Excellence grant (U54 RR14616) and the Drew/UCLA Cooperative Reproductive Science Research Centers at Minority Institutions (U54HD41748) to Charles R. Drew University of Medicine & Sciences. K.L. was supported by NIH grant R01 DA16682.
We thank the SCCPRR Gene Array Center (Department of Pharmacology, University of Washington) for provide us with the microarrays and performing the hybridizations
Abbreviations
- Adpn
adiponutrin
- AgRP
agouti-related peptide
- CART
cocaine and amphetamine regulated transcript
- Cdcn1a
cyclin-dependent kinase inhibitor 1A
- Clk4
CDC like kinase 4
- CNS
central nervous system
- Cpe
carboxypeptidase E
- CRH
corticotrophin-releasing hormone
- Cri1
CREBBP/EP300 inhibitory protein 1
- Csnk1
casein kinase 1
- Ctla2a
cytotoxic T lymphocyte-associated protein 2 alpha
- DEG
differentially expressed genes
- Flt1
FMS-like tyrosine kinase 1
- GEO
Gene Expression Omnibus
- LC
locus ceruleus
- Lcn2
lipocalin 2
- Lepr
leptin receptor
- LOWESS
Locally Weighted Regression and Smoothing
- Mt1
metallothionein 1
- NAc
nucleus accumbens
- NCBI
National Center for Biotechnology Information
- NIDA
National Institute on Drug Abuse
- NPY
neuropeptide Y
- Pip5k
phosphatidylinositol-4-phosphate 5-kinase
- PMT
Photomultiplier tube
- Pomc1
pro-opiomelanocortin-alpha
- Ppp2r2a
protein phosphatase 2
- Ptp
protein tyrosine phosphatase
- Pyy
peptide YY
- Rasd1
RAS, dexamethasone-induced 1
- Rbbp7
retinoblastoma binding protein 7
- Rhoa
ras homolog gene family
- RT-PCR
reverse transcriptase polymerase chain reaction
- Scg2
secretogranin II
- SEM
standard error of the mean
- Tcrf
T-cell replacing factor
- tmpo
thymopoietin
- VTA
ventral tegmental area
- Xlkd1
lymphatic vessel endothelial HA receptor-1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: There is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ammon S, Mayer P, Riechert U, Tischmeyer H, Hollt V. Microarray analysis of genes expressed in the frontal cortex of rats chronically treated with morphine and after naloxone precipitated withdrawal. Brain Res Mol Brain Res. 2003;112:113–125. doi: 10.1016/s0169-328x(03)00057-3. [DOI] [PubMed] [Google Scholar]
- Ammon-Treiber S, Grecksch G, Stumm R, Riechert U, Tischmeyer H, Reichenauer A, Hollt V. Rapid, transient, and dose-dependent expression of hsp70 messenger RNA in the rat brain after morphine treatment. Cell Stress Chaperones. 2004a;9:182–197. doi: 10.1379/CSC-42.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammon-Treiber S, Hollt V. Morphine-induced changes of gene expression in the brain. Addict Biol. 2005;10:81–89. doi: 10.1080/13556210412331308994. [DOI] [PubMed] [Google Scholar]
- Ammon-Treiber S, Tischmeyer H, Riechert U, Hollt V. Gene expression of transcription factors in the rat brain after morphine withdrawal. Neurochem Res. 2004b;29:1267–1273. doi: 10.1023/b:nere.0000023613.44988.9d. [DOI] [PubMed] [Google Scholar]
- Baker H, Joh TH, Ruggiero DA, Reis DJ. Variations in number of dopamine neurons and tyrosine hydroxylase activity in hypothalamus of two mouse strains. J Neurosci. 1983;3:832–843. doi: 10.1523/JNEUROSCI.03-04-00832.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Befort K, Filliol D, Darcq E, Ghate A, Matifas A, Lardenois A, Muller J, Thibault C, Dembele D, Poch O, Kieffer BL. Gene expression is altered in the lateral hypothalamus upon activation of the mu opioid receptor. Ann N Y Acad Sci. 2008;1129:175–184. doi: 10.1196/annals.1417.028. [DOI] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1988;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brakch N, Rist B, Beck-Sickinger AG, Goenaga J, Wittek R, Burger E, Brunner HR, Grouzmann E. Role of prohormone convertases in pro-neuropeptide Y processing: coexpression and in vitro kinetic investigations. Biochemistry (Mosc) 1997;36:16309–16320. doi: 10.1021/bi9714767. [DOI] [PubMed] [Google Scholar]
- Cawley NX, Zhou J, Hill JM, Abebe D, Romboz S, Yanik T, Rodriguiz RM, Wetsel WC, Loh YP. The carboxypeptidase E knockout mouse exhibits endocrinological and behavioral deficits. Endocrinology. 2004;145:5807–5819. doi: 10.1210/en.2004-0847. [DOI] [PubMed] [Google Scholar]
- Chao J, Nestler EJ. Molecular neurobiology of drug addiction. Annu Rev Med. 2004;55:113–132. doi: 10.1146/annurev.med.55.091902.103730. [DOI] [PubMed] [Google Scholar]
- Chen G, Bridenbaugh EA, Akintola AD, Catania JM, Vaidya VS, Bonventre JV, Dearman AC, Sampson HW, Zawieja DC, Burghardt RC, Parrish AR. Increased susceptibility of aging kidney to ischemic injury: identification of candidate genes changed during aging, but corrected by caloric restriction. Am J Physiol Renal Physiol. 2007 doi: 10.1152/ajprenal.00138.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Celik A, Georgeson KE, Harmon CM, Yang Y. Molecular basis of melanocortin-4 receptor for AGRP inverse agonism. Regul Pept. 2006;136:40–49. doi: 10.1016/j.regpep.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Cooper SJ. Behaviourally-specific hyperdipsia in the non-deprived rat following acute morphine treatment. Neuropharmacology. 1981;20:469–471. doi: 10.1016/0028-3908(81)90179-9. [DOI] [PubMed] [Google Scholar]
- Cooper SJ, Sanger DJ. Endorphinergic mechanisms in food, salt and water intake: an overview. Appetite. 1984;5:1–6. doi: 10.1016/s0195-6663(84)80043-4. [DOI] [PubMed] [Google Scholar]
- Creemers JW, Pritchard LE, Gyte A, Le Rouzic P, Meulemans S, Wardlaw SL, Zhu X, Steiner DF, Davies N, Armstrong D, Lawrence CB, Luckman SM, Schmitz CA, Davies RA, Brennand JC, White A. Agouti-related protein is posttranslationally cleaved by proprotein convertase 1 to generate agouti-related protein (AGRP)83-132: interaction between AGRP83-132 and melanocortin receptors cannot be influenced by syndecan-3. Endocrinology. 2006;147:1621–1631. doi: 10.1210/en.2005-1373. [DOI] [PubMed] [Google Scholar]
- Dey A, Xhu X, Carroll R, Turck CW, Stein J, Steiner DF. Biological processing of the cocaine and amphetamine-regulated transcript precursors by prohormone convertases, PC2 and PC1/3. J Biol Chem. 2003;278:15007–15014. doi: 10.1074/jbc.M212128200. [DOI] [PubMed] [Google Scholar]
- Dylag T, Kotlinska J, Rafalski P, Pachuta A, Silberring J. The activity of CART peptide fragments. Peptides. 2006;27:1926–1933. doi: 10.1016/j.peptides.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Eipper BA, Mains RE. Structure and biosynthesis of pro-adrenocorticotropin/endorphin and related peptides. Endocr Rev. 1980;1:1–27. doi: 10.1210/edrv-1-1-1. [DOI] [PubMed] [Google Scholar]
- Espinosa VP, Liu Y, Ferrini M, Anghel A, Nie Y, Tripathi PV, Porche R, Jansen E, Stuart RC, Nillni EA, Lutfy K, Friedman TC. Differential regulation of prohormone convertase 1/3, prohormone convertase 2 and phosphorylated cyclic-AMP-response element binding protein by short-term and long-term morphine treatment: implications for understanding the "switch" to opiate addiction. Neuroscience. 2008;156:788–799. doi: 10.1016/j.neuroscience.2008.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenk H, Rogers GH. The suppressant effects of naloxone on food and water intake in the rat. Behav Neural Biol. 1979;26:23–40. doi: 10.1016/s0163-1047(79)92855-3. [DOI] [PubMed] [Google Scholar]
- Garcia de Yebenes E, Pelletier G. Opioid regulation of proopiomelanocortin (POMC) gene expression in the rat brain as studied by in situ hybridization. Neuropeptides. 1993;25:91–94. doi: 10.1016/0143-4179(93)90087-q. [DOI] [PubMed] [Google Scholar]
- Gerrits MA, Lesscher HB, van Ree JM. Drug dependence and the endogenous opioid system. Eur Neuropsychopharmacol. 2003;13:424–434. doi: 10.1016/j.euroneuro.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Grander D, Sangfelt O, Skoog L, Hansson J. In vivo induction of the interferon-stimulated protein 2'5'-oligoadenylate synthetase in tumor and peripheral blood cells during IFN-alpha treatment of metastatic melanoma. Journal of interferon & cytokine research. 1998;18:691–695. doi: 10.1089/jir.1998.18.691. [DOI] [PubMed] [Google Scholar]
- Grandison L, Guidotti A. Stimulation of food intake by muscimol and beta endorphin. Neuropharmacology. 1977;16:533–536. doi: 10.1016/0028-3908(77)90019-3. [DOI] [PubMed] [Google Scholar]
- Grice DE, Reenila I, Mannisto PT, Brooks AI, Smith GG, Golden GT, Buxbaum JD, Berrettini WH. Transcriptional profiling of C57 and DBA strains of mice in the absence and presence of morphine. BMC Genomics. 2007;8:76. doi: 10.1186/1471-2164-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinashi T, Harada N, Severinson E, Tanabe T, Sideras P, Konishi M, Azuma C, Tominaga A, Bergstedt-Lindqvist S, Takahashi M, et al. Cloning of complementary DNA encoding T-cell replacing factor and identity with B-cell growth factor II. Nature. 1986;324:70–73. doi: 10.1038/324070a0. [DOI] [PubMed] [Google Scholar]
- Koob GF, Ahmed SH, Boutrel B, Chen SA, Kenny PJ, Markou A, O'Dell LE, Parsons LH, Sanna PP. Neurobiological mechanisms in the transition from drug use to drug dependence. Neurosci Biobehav Rev. 2004;27:739–749. doi: 10.1016/j.neubiorev.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Koob GF, Le Moal M. Drug abuse: hedonic homeostatic dysregulation. Science. 1997;278:52–58. doi: 10.1126/science.278.5335.52. [DOI] [PubMed] [Google Scholar]
- Korostynski M, Piechota M, Kaminska D, Solecki W, Przewlocki R. Morphine effects on striatal transcriptome in mice. Genome Biol. 2007;8:R128. doi: 10.1186/gb-2007-8-6-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlinska J, Pachuta A, Dylag T, Silberring J. The role of neuropeptide FF (NPFF) in the expression of sensitization to hyperlocomotor effect of morphine and ethanol. Neuropeptides. 2007;41:51–58. doi: 10.1016/j.npep.2006.09.048. [DOI] [PubMed] [Google Scholar]
- Kreek MJ, Schlussman SD, Bart G, Laforge KS, Butelman ER. Evolving perspectives on neurobiological research on the addictions: celebration of the 30th anniversary of NIDA. Neuropharmacology. 2004;47 Suppl 1:324–344. doi: 10.1016/j.neuropharm.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Kunihara M, Kanbayashi M, Ohshima T. Opposite effects of morphine on feeding and drinking in rats relative to administration time. Jpn J Pharmacol. 1983;33:829–835. doi: 10.1254/jjp.33.829. [DOI] [PubMed] [Google Scholar]
- Leshem M. Morphine-induced anorexia in lateral hypothalamic rats. Psychopharmacology (Berl) 1981;75:48–53. doi: 10.1007/BF00433501. [DOI] [PubMed] [Google Scholar]
- Leshem M. Morphine induces delayed anorexia in rats. Psychopharmacology (Berl) 1988;94:254–258. doi: 10.1007/BF00176855. [DOI] [PubMed] [Google Scholar]
- Leshner AI. Addiction is a brain disease, and it matters. Science. 1997;278:45–47. doi: 10.1126/science.278.5335.45. [DOI] [PubMed] [Google Scholar]
- Levine AS, Morley JE, Gosnell BA, Billington CJ, Bartness TJ. Opioids and consummatory behavior. Brain Res Bull. 1985;14:663–672. doi: 10.1016/0361-9230(85)90116-9. [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Proc Natl Acad Sci U S A. 2004;101:1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Lescure PA, Misek DE, Lai YM, Chai BX, Kuick R, Thompson RC, Demo RM, Kurnit DM, Michailidis G, Hanash SM, Gantz I. Food deprivation-induced expression of minoxidil sulfotransferase in the hypothalamus uncovered by microarray analysis. J Biol Chem. 2002;277:9069–9076. doi: 10.1074/jbc.M110467200. [DOI] [PubMed] [Google Scholar]
- Lockhart DJ, Winzeler EA. Genomics, gene expression and DNA arrays. Nature. 2000;405:827–836. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- Loguinov AV, Anderson LM, Crosby GJ, Yukhananov RY. Gene expression following acute morphine administration. Physiol Genomics. 2001;6:169–181. doi: 10.1152/physiolgenomics.2001.6.3.169. [DOI] [PubMed] [Google Scholar]
- Marks-Kaufman R, Kanarek RB. Morphine selectively influences macronutrient intake in the rat. Pharmacol Biochem Behav. 1980;12:427–430. doi: 10.1016/0091-3057(80)90048-9. [DOI] [PubMed] [Google Scholar]
- Marquez P, Baliram R, Gajawada N, Friedman TC, Lutfy K. Differential involvement of enkephalins in analgesic tolerance, locomotor sensitization, and conditioned place preference induced by morphine. Behav Neurosci. 2006;120:10–15. doi: 10.1037/0735-7044.120.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA, Nestler EJ, Zachariou V. Regulation of gene expression by chronic morphine and morphine withdrawal in the locus ceruleus and ventral tegmental area. J Neurosci. 2005;25:6005–6015. doi: 10.1523/JNEUROSCI.0062-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JE. The endocrinology of the opiates and opioid peptides. Metabolism. 1981;30:195–209. doi: 10.1016/0026-0495(81)90172-4. [DOI] [PubMed] [Google Scholar]
- Morley JE, Levine AS, Gosnell BA, Billington CJ. Which opioid receptor mechanism modulates feeding? Appetite. 1984;5:61–68. doi: 10.1016/s0195-6663(84)80051-3. [DOI] [PubMed] [Google Scholar]
- Niemann A, Ruegg M, La Padula V, Schenone A, Suter U. Ganglioside-induced differentiation associated protein 1 is a regulator of the mitochondrial network: new implications for Charcot-Marie-Tooth disease. The Journal of cell biology. 2005;170:1067–1078. doi: 10.1083/jcb.200507087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrazzini T. Importance of NPY Y1 receptor-mediated pathways: assessment using NPY Y1 receptor knockouts. Neuropeptides. 2004;38:267–275. doi: 10.1016/j.npep.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Robbins TW, Everitt BJ. Drug addiction: bad habits add up. Nature. 1999;398:567–570. doi: 10.1038/19208. [DOI] [PubMed] [Google Scholar]
- Rodriguez de Fonseca F, Carrera M, Navarro M, Koob GF, Weiss F. Activation of corticotropin-releasing factor in the limbic system during cannabinoid withdrawal. Science. 1997;276:2050–2054. doi: 10.1126/science.276.5321.2050. [DOI] [PubMed] [Google Scholar]
- Roy S, Guo X, Kelschenbach J, Liu Y, Loh HH. In vivo activation of a mutant mu-opioid receptor by naltrexone produces a potent analgesic effect but no tolerance: role of mu-receptor activation and delta-receptor blockade in morphine tolerance. J Neurosci. 2005;25:3229–3233. doi: 10.1523/JNEUROSCI.0332-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger DJ, McCarthy PS. Differential effects of morphine on food and water intake in food deprived and freely-feeding rats. Psychopharmacology (Berl) 1980;72:103–106. doi: 10.1007/BF00433813. [DOI] [PubMed] [Google Scholar]
- Schwartz MW. Brain pathways controlling food intake and body weight. Exp Biol Med (Maywood) 2001;226:978–981. doi: 10.1177/153537020122601103. [DOI] [PubMed] [Google Scholar]
- Seidah NG, Chretien M. Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res. 1999;848:45–62. doi: 10.1016/s0006-8993(99)01909-5. [DOI] [PubMed] [Google Scholar]
- Soverchia L, Ubaldi M, Leonardi-Essmann F, Ciccocioppo R, Hardiman G. Microarrays--the challenge of preparing brain tissue samples. Addict Biol. 2005;10:5–13. doi: 10.1080/13556210412331327803. [DOI] [PubMed] [Google Scholar]
- Spangler R, Zhou Y, Maggos CE, Zlobin A, Ho A, Kreek MJ. Dopamine antagonist and "binge' cocaine effects on rat opioid and dopamine transporter mRNAs. Neuroreport. 1996;7:2196–2200. doi: 10.1097/00001756-199609020-00028. [DOI] [PubMed] [Google Scholar]
- Spijker S, Houtzager SW, De Gunst MC, De Boer WP, Schoffelmeer AN, Smit AB. Morphine exposure and abstinence define specific stages of gene expression in the rat nucleus accumbens. FASEB J. 2004;18:848–850. doi: 10.1096/fj.03-0612fje. [DOI] [PubMed] [Google Scholar]
- Stehlik C, Lee SH, Dorfleutner A, Stassinopoulos A, Sagara J, Reed JC. Apoptosis-associated speck-like protein containing a caspase recruitment domain is a regulator of procaspase-1 activation. Journal of immunology (Baltimore, Md. 2003;171:6154–6163. doi: 10.4049/jimmunol.171.11.6154. [DOI] [PubMed] [Google Scholar]
- Stein J, Shah R, Steiner DF, Dey A. RNAi-mediated silencing of prohormone convertase (PC) 5/6 expression leads to impairment in processing of cocaine- and amphetamine-regulated transcript (CART) precursor. Biochem J. 2006;400:209–215. doi: 10.1042/BJ20060506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Fowler JS, Wang GJ, Swanson JM. Dopamine in drug abuse and addiction: results from imaging studies and treatment implications. Mol Psychiatry. 2004;9:557–569. doi: 10.1038/sj.mp.4001507. [DOI] [PubMed] [Google Scholar]
- Vuong C, Van Uum SH, O'Dell LE, Lutfy K, Friedman TC. The effects of opioids and opioid analogs on animal and human endocrine systems. Endocr Rev. 2009 doi: 10.1210/er.2009-0009. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Seed B. Selection of oligonucleotide probes for protein coding sequences. Bioinformatics. 2003;19:796–802. doi: 10.1093/bioinformatics/btg086. [DOI] [PubMed] [Google Scholar]
- Wolgin DL, Benson HD. Role of associative and nonassociative mechanisms in tolerance to morphine "anorexia". Pharmacol Biochem Behav. 1991;39:279–286. doi: 10.1016/0091-3057(91)90180-a. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lou D, Jiao H, Zhang D, Wang X, Xia Y, Zhang J, Xu M. Cocaine-induced intracellular signaling and gene expression are oppositely regulated by the dopamine D1 and D3 receptors. J Neurosci. 2004;24:3344–3354. doi: 10.1523/JNEUROSCI.0060-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Bloomquist BT, Mains RE. The prohormone convertases PC1 and PC2 mediate distinct endoproteolytic cleavages in a strict temporal order during proopiomelanocortin biosynthetic processing. J Biol Chem. 1993;268:1763–1769. [PubMed] [Google Scholar]
- Zhou Y, Spangler R, LaForge KS, Maggos CE, Ho A, Kreek MJ. Corticotropin-releasing factor and type 1 corticotropin-releasing factor receptor messenger RNAs in rat brain and pituitary during "binge"-pattern cocaine administration and chronic withdrawal. J Pharmacol Exp Ther. 1996;279:351–358. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.