Abstract
Background
Iron is an essential element for both plant productivity and nutritional quality. Improving plant iron content was attempted through genetic engineering of plants overexpressing ferritins. However, both the roles of these proteins in plant physiology, and the mechanisms involved in the regulation of their expression are largely unknown. Although the structure of ferritins is highly conserved between plants and animals, their cellular localization differs. Furthermore, regulation of ferritin gene expression in response to iron excess occurs at the transcriptional level in plants, in contrast to animals which regulate ferritin expression at the translational level.
Scope
In this review, an overview of our knowledge of bacterial and mammalian ferritin synthesis and functions is presented. Then the following will be reviewed: (a) the specific features of plant ferritins; (b) the regulation of their synthesis during development and in response to various environmental cues; and (c) their function in plant physiology, with special emphasis on the role that both bacterial and plant ferritins play during plant–bacteria interactions. Arabidopsis ferritins are encoded by a small nuclear gene family of four members which are differentially expressed. Recent results obtained by using this model plant enabled progress to be made in our understanding of the regulation of the synthesis and the in planta function of these various ferritins.
Conclusions
Studies on plant ferritin functions and regulation of their synthesis revealed strong links between these proteins and protection against oxidative stress. In contrast, their putative iron-storage function to furnish iron during various development processes is unlikely to be essential. Ferritins, by buffering iron, exert a fine tuning of the quantity of metal required for metabolic purposes, and help plants to cope with adverse situations, the deleterious effects of which would be amplified if no system had evolved to take care of free reactive iron.
Key words: Iron, bacterioferritins, ferritins, oxidative stress, iron storage, seeds, pathogens, nutrition
INTRODUCTION
Metal interactions with organic compounds are numerous in all living organisms. They are essential for many biochemical processes occurring within cells, and concern not only metabolism, but also some regulatory mechanisms of gene expression.
Among metals, iron is of special interest because it is required in most of the cellular redox reactions and it is one of the major metals involved in electron transfer chains. However, its strong reactivity with oxygen makes it a difficult element to handle by aerobic organisms. Indeed, both its insolubility in the form of ferric hydroxides, or its toxicity through the Fenton reaction producing hydroxyl radicals (which are among the most chemically reactive species), have introduced evolutionary constraints in order to enable this metal to be safely utilized by living organisms. The narrow efficient iron concentration required for cellular needs is strictly controlled by biological processes acting both at the transport and the storage levels. In multicellular organisms, transport mechanisms regulate iron traffic from uptake to long distance tissular distribution, and ultimately to subcellular allocation. During developmental processes or in response to environmental challenges, buffering and transitory storage of iron also participates to maintain iron homeostasis. A specific class of iron storage proteins is implicated in this latter function: the ferritins. The main purpose of this paper is to review our current knowledge of plant ferritins, after having summarized first our current knowledge of prokaryote and animal ferritins.
GENERAL FEATURES OF FERRITINS: AN OVERVIEW
Ferritins are a broad superfamily of iron storage proteins, found in all the living kingdom, except in yeast (Andrews et al., 2003; Briat et al., 2006; Arosio et al., 2008). Three sub-classes of these proteins can be defined: (1) haem-free ferritins present both in pro- and eukaryotes; (2) haem-containing bacterioferritins, found only in bacteria; and (3) DNA binding proteins from starved cells (Dps), called miniferritins, present only in prokaryotes (Smith, 2004). Ferritins and bacterioferritins are composed of 24 subunits whereas only 12 identical subunits form the Dps proteins. These subunits assemble in a spherical protein shell defining a central cavity able to accomodate between 2000 and 4000 ferric iron atoms for ferritins and 500 atoms for miniferritins (Andrews et al., 2003; Carrondo, 2003). The oxidation of ferrous iron prior to ferric iron storage within the cavity is catalysed by the ferritin protein shells which possess a binuclear di-iron centre defined as the the ferroxidase centre involved in the oxidation of Fe(II) prior to its accomodation in the Fe(III) mineral core (Arosio et al., 2008). Oxygen and hydrogen peroxide are the major cellular oxidants consumed during this oxidation reaction (Bou-Abdallah et al., 2002; Arosio et al., 2008). Sequestered ferric iron is bio-available in case of cellular needs and is non-reactive with oxygen. Besides their role in iron storage, ferritins and miniferritins are therefore also involved in protection against oxidative stress through their potential detoxification properties of excess iron, dioxygen and, to some extent, hydrogen peroxide (Zhao et al., 2002; Theil et al., 2006; Arosio et al., 2008).
Prokaryote ferritins
Prokaryotes have three sub-classes of proteins able to store up to a few thousands of iron atoms inside a central cavity defined by the protein shell: the miniferritins (Dps), the bacterioferritins (BFR) and the bacterial ferritins (FTN).
Miniferritins, or Dps, are dodecameric ferritins which have been documented in many different bacteria species (Smith, 2004), although most of the knowledge concerning these miniferritins was obtained with Escherichia coli. Escherichia coli Dps belongs to the family of non-specific DNA-binding proteins induced in response to carbon starvation or to H2O2-generated oxidative stress. During exponential growth Dps is degraded by Clp proteases, whereas it is stabilized and its synthesis increased in response to stress (Stephani et al., 2003). Dps participates to protect DNA from various nucleases and oxidative agents by binding to it in a non-specific manner, and consequently it triggers a low level of gene expression. The Dps protein of E. coli is able to oxidize Fe(II) through specific sites containing iron-binding amino acids such as aspartate, glutamate and histidine (Ilari et al., 2000, 2002). It has been characterized as a dodecameric sphere of 205 kDa accommodating 500 Fe(III) atoms in its central cavity.
Bacterioferritin, encoded by the BFR gene in E. coli, consists of 24 identical polypeptides of 18·5 kDa assembled in a hollow sphere of 452 kDa. It is able to accomodate about 1800 Fe(III) atoms in its central cavity. The major peculiarity of bacterioferritins is to contain one haem molecule for two subunits; i.e. 12 haem residues per bacterioferritin molecule–, a methionine residue of each subunit serving as haem ligands (Cheesman et al., 1990, 1993). Although the function of these haem residues is not well characterized, engineered haem-free BFR load about four times more iron in vivo than wild-type BFR (Andrews et al., 1995).
Because BFR, as Dps, preferentially use H2O2 rather than O2 during the Fe(II) oxidation process, they are also considered to play a major role in protection against oxidative stress.
Bacterial ferritin has been largely documented in E. coli. It is a non-haem 24-mers protein of 465 kDa, each 19·5-kDa subunit being encoded by the FTN-A gene, and having a sequence more related to the human H ferritin subunit than to the BFR haem-containing bacterioferritin (Izuhara et al., 1991). It is able to accomodate about 2500 iron atoms in its central cavity through a ferroxidase activity characteristic of H-type ferritins (see the section entitled Mammalian ferritins). As for BFR and Dps, the role of FTN in protecting cells against oxidative damage has been documented (Touati et al., 1995; Touati, 2000). However, such an effect occurs only in a genetic background lacking the Fur transcriptional regulator of bacterial iron homeostasis (Abdul-Tehrani et al., 1999).
Prokaryote maxi-ferritins (FTN and BFR) and mini-ferritins (Dps) have their expression regulated in response to iron or oxidative stress. FTN and BFR gene expression is principally regulated by the Fur–Fe2+ complex (Andrews, 1998; Cartron et al., 2006). In addition, and to a lesser extent than some other genes, BFR mRNA abundance is regulated by the regulatory RNA RyhB. RyhB is a noncoding RNA regulated by the Fur repressor causing the rapid degradation of a number of mRNAs that encode proteins that utilize iron (Masse et al., 2005). Among these proteins, RyhB regulates negatively the synthesis of Fur, therefore creating a negative feedback loop essential for the tight control of iron homeostasis within the cell and impacting indirectly the synthesis of ferritin (Vecerek et al., 2007). Dps proteins are particularly important during oxidative stress or, when protein turnover releases iron (Moore et al., 2005; Reindel et al., 2006). In E. coli, Dps gene expression in response to hydrogen peroxide is coupled to FTN expression during the exponential phase via the transcriptional regulator OxyR, and to BFR expression during the stationary phase via both Sigma S- and the histone-like protein IHF (Altuvia et al., 1994).
Many Bacillus spp., have the peculiarity to possess two mini-ferritin (Dps) genes in their genome but no maxi-ferritin gene. In these bacteria, H2O2-dependent induction of the Dps2 gene expression occurs through modification of the PerR transcription factor by an Fe2+/H2O2 catalysed reaction (Lee and Helmann, 2006). The Dps1 gene is induced by non-specific oxidative stress that is Sigma B-dependent during exponential growth and Sigma B-independent during transitional growth (Antelmann et al., 1997).
Mammalian ferritins
The structure–function relationships of mammalian ferritins have been widely studied (Arosio et al., 2008). Mammalian apoferritins are composed of 24-heteromers, and have a molecular mass of approx. 450 kDa. The central cavity of this hollow sphere can accomodate up to 4500 iron atoms in a safe and bioavailable form, chemically defined as ferrihydrite. Subunits of about 20 kDa each are of two types referred to as H- and L-chains. H- and L-chains share approx. 55% amino acid homology. They are quantitatively differentially abundant in various tissues and, consequently, mammalian apoferritins can be quite variable in their composition depending on the number of L- and H-chains in the protein complex. For example, the heart ferritins are mainly composed of H-chains whereas the liver ferritins contain essentially L-chains. The major difference between the L- and H-chains is the presence in each H-chain of a ferroxidase centre of seven amino acid residues (Glu27, Tyr34, Glu61, Glu62, His65, Glu107 and Glu141in the human ferritin H-chain). The ferroxidase centre enables a rapid oxidation of Fe(II) which binds to it prior to being oxidized into Fe(III), and then incorporated into the mineral core (Levi et al., 1994; Le Brun et al., 1995; Keech et al., 1997). The L-chain has no ferroxidase activity and L-rich ferritins are therefore less efficient in rapid uptake of iron, comparatively to H-rich ferritins. However, long-term storage of iron is privileged in L-rich ferritins since L subunit specific amino acids facing the central cavity of the protein are known to facilitate the mineralization of ferric ions inside this cavity.
In vivo iron loading of ferritin may be more complex than in vitro. Indeed it has recently received support by the finding of a human iron chaperone able to bind ferritin and to facilitate iron incorporation both in vivo and in vitro (Shi et al., 2008). This chaperone corresponds to the poly (rC)-binding protein 1 (PCBP1) which is an ubiquitous mammalian RNA-binding protein found both in the cytosol and nucleus. PCBP1 can bind with high affinity up to three Fe atoms per molecule and is found associated with ferritin only in the presence of ferrous iron.
Iron release from ferritin has been studied in vitro, showing that this metal is stable within the core, and can be released only slowly by strong Fe(III) chelators (Chasteen, 1998). Reducing agents, however, are well known for their ability to promote iron release from ferritin mineral cores (Cassanelli and Moulis, 2001), even without penetrating inside the cavity (Theil et al., 2006). This involves the transfer of electrons through the protein shell to the cavity (Johnson et al., 1999). The hydrophilic channels on the 3-fold axes of the molecule are required for iron release. The melting of the structure around these channels, caused by chaotropic agents or by the specific binding of some peptides that control the pore opening (Liu et al., 2007), increase strongly the reductive release of iron in vitro (Jin et al., 2001). The mechanism of ferritin iron release in vivo is much more difficult to apprehend and it is currently admitted that ferritin iron is mainly released after proteolytic degradation of the protein through different mechanisms (Truty et al., 2001; De Domenico et al., 2006; Kidane et al., 2006). One of them is likely to involve ubiquitination and the proteasome pathway. This proteasome-dependent pathway for ferritin degradation and iron release could be enhanced by oxidative damage (Mehlhase et al., 2005). So far it is still unclear if the reductive processes necessary for ferritin iron release produce pro-oxidant intermediates which are potentially toxic.
Concerning their subcellular localization, although animal ferritins are mostly found within the cytoplasm as soluble proteins, there are few reports of the presence of ferritins inside the nucleus, where they could protect DNA against oxidative stress, or even regulate the transcription of some genes (for a review, see Arosio et al., 2008). Ferritins have also been reported to be located in the mitochondria of some cell types, where they are encoded by an intronless gene (for a review, see Arosio et al., 2008). Animal ferritins are not only intracellular proteins since they can be secreted when synthesized as precursors in worm or insects cells, or even as mature polypeptides in certain mammalian cell types (for a review, see Arosio et al., 2008). The tissue-specific H : L ratio of ferritin subunits is determined by the transcriptional regulation of ferritin genes during development. Mammalian ferritin expression is also regulated at the transcriptional level in response to inducers promoting the antioxidant response. It requires antioxidant-responsive cis elements (AREs) found in the upstream promoter region of ferritin genes, and able to recruit Bach1 and maf proteins as transcription factors (Torti and Torti, 2002; Hintze and Theil, 2005; Theil, 2007). Although of importance, these transcriptional regulations received less attention than the translational control of ferritin mRNA expression in response to iron through the iron responsive element (IRE)/iron response protein (IRP) translational repression system (for a review, see Theil, 2007; Arosio et al., 2008). IREs are specific hairpin structures found in the 5′ UTR of ferritin mRNA which are able to bind with high affinity the repressors IRP1 and IRP2. The IRE-binding activity of the IRPs is regulated by the cellular iron and redox status. Under low iron conditions, the IRE/IRP complex prevents ribosome scanning and translation of ferritin transcripts. Under high iron conditions, IRP1 contains an Fe/S cluster, exhibits aconitase activity and looses its RNA-binding activity. The transition between IRP1 and cytosolic aconitase activity is affected by protein phosphorylation and iron-induced proteasomal degradation (Fillebeen et al., 2003; Clarke et al., 2006; Wang et al., 2007). In contrast to IRP1, IRP2 is not an aconitase. This translational repressor is degraded under high iron conditions via haem interaction, and its S-nitrosylation has been suggested to regulate its stability via the ubiquitin proteasome pathway (Kim and Ponka, 2002; Kim et al., 2004). IRP2 appears to be the major ferritin translational regulator in vivo (Meyron-Holtz et al., 2004; Galy et al., 2005), and the L-ferritin is more tightly regulated by iron at a post-transcriptional level than H-ferritin (Sammarco et al., 2008).
DISTINCTIVE FEATURES OF PLANT FERRITINS
Plant ferritins have a eukaryotic origin, and their amino-acid sequence is well conserved with animal ferritin (Ragland et al., 1990; Andrews et al., 1992). A specific feature, however, has to be noticed at this level. Plant ferritins are synthesized as precursors with a plant-specific N-terminal amino acid extension. This extension is composed of two parts. The shorter one, named extension peptide (EP) belongs to the mature ferritin subunit and is involved in the control of protein stability during iron exchange (Van Wuytswinkel and Briat, 1995; Van Wuytswinkel et al., 1995). Upstream of the EP, a cleavable transit peptide sequence responsible for subcellular targeting of ferritin subunits is observed (Ragland et al., 1990; Proudhon et al., 1989). Indeed, plant ferritins have never been reported to be localized in the cytoplasm. Until recently, they were described as exclusively targeted to non-green plastids, being found accumulated in chloroplasts only in response to stresses (Fobis-Loisy et al., 1995; Briat et al., 1995). In 2004, it was reported that plant ferritins can also be observed in mitochondria, sharing therefore this cellular localization with some animal ferritins (Zancani et al., 2004). However, in contrast to animal ferritins, the gene encoding plant mito-ferritin is not different than others, and the product it encodes could be dual targeted to both mitochondria and plastids. The function of ferritin within plant mitochondria is so far not documented.
As a result of its presence in plastids, a prokaryotic-type environment, the structure of plant ferritin mineral cores is amorphous because of its high P : Fe ratio of 1 : 3, which is different from the ferrihydrite cristal structure found within animal ferritin (Wade et al., 1993; Waldo et al., 1995). The environment of the protein (i.e. richness in P) is more responsible for this difference than the structure of the protein itself since in vitro, in absence of P, plant apoferritin can load iron within its central cavity in the form of pure ferrihydrite (Wade et al., 1993; Waldo et al., 1995).
In addition to their specific NH2-terminal extension plant ferritin subunits have some additional features compared with their animal counterpart. Although the predicted 3-D structure of the pea seed ferritin can be superimposed ±1 Å with mammalian H ferritin (Lobréaux et al., 1992a), there is no distinguishable H- or L-subunits in the plant. Indeed, all the subunits characterized so far contain both a typical H-type ferroxidase centre, and all the amino-acid residues characteristics of an L-type subunit for a better iron nucleation within the cavity (Lobréaux et al., 1992a). Plant ferritin can therefore be considered as an H/L hybrid ferritin. In this respect, it is interesting to notice that in vitro, the ferroxidase activity of purified ferritin is intermediate between recombinant H- or L-human ferritin (Van Wuytswinkel and Briat, 1995).
All eukaryotic ferritins when assembled as 24-mer exhibit channels in the 3-fold and 4-fold symmetry axes of their three dimensional structure. In plant ferritins, both these channels are hydrophilic whereas animal ferritins have their 3-fold channels built up with hydrophilic residues and their 4-fold channels with hydophobic residues. The significance of this difference, if any, is unknown (Lobréaux et al., 1992a).
Finally, a major difference between plant and other eukaryotic ferritins concerns the regulation of their synthesis in response to excess of iron (see below). Whereas this regulation occurs mainly at the translational level through the IRE/IRP system in animals (see the section above; for a review, see Theil, 2007; Arosio et al., 2008), the regulation of plant ferritin gene expression in response to this metal is achieved through a transcriptional control (Lescure et al., 1991). The involvement of an aconitase/IRP switch in the regulation of iron homeostasis in plants has been suggested (Peyret et al., 1995; Navarre et al., 2000). However, no IRE can be predicted within the 5′ UTR of plant ferritin mRNAs, and the iron-mediated expression of the AtFer1 and AtFer3 ferritin genes is unaffected in Arabidopsis thaliana aconitase null mutants (Arnaud et al., 2007). Therefore, the existence of an aconitase/IRP switch in plants to regulate ferritin gene expression is very unlikely to occur.
PLANT FERRITIN SYNTHESIS
Important data documenting the regulation of ferritin synthesis have been obtained with maize (Lobréaux et al., 1992b, 1993; Savino et al., 1997; Petit et al., 2001b), pea (Lobréaux and Briat, 1991) and soybean (Lescure et al., 1991; Wei and Theil, 2000). However, Arabidopsis is now offering the best opportunity to characterize exhaustively the differential expression of the four members of the ferritin gene family present in the genome of this model plant, both during the course of development and in response to environmental signals (Petit et al., 2001a; Ravet et al., 2009).
Developmental regulation of ferritin synthesis
The developmental regulation of ferritins is known to occur from electron microscopy studies in which their presence in various organs and tissues has been reported (for a review, see Seckback, 1982). Ferritin naturally occurs in plant tissues such as cotyledons, root and shoot apices, cells of vessels, in vascular cambium, reproductive cells and senescing cells. However, it is only recently that developmental regulation of plant ferritin has been documented at the molecular level, at defined developmental stages of leaves (Theil and Hase, 1993) and nodules (Ragland and Theil, 1993; Kimata and Theil, 1994), as well as during the life cycle of the pea (Lobréaux and Briat, 1991) and A. thaliana (Petit et al., 2001a) plants.
In pea and A. thaliana, ferritin accumulates in the cotyledons and embryo axis of seeds, while the amount of ferritin in vegetative organs is low (Lobréaux and Briat, 1991; Petit et al., 2001a). Post-transcriptional control at the protein stability level participates to regulate the amount of seed ferritin. Disappearence of ferritin protein from pea seed embryo axes during the first days following germination is accompanied by the appearance of polypeptides of lower molecular weight than the basic ferritin subunit (Lobréaux and Briat, 1991). A mechanism responsible for this specific processing has been proposed, based on iron release experiments performed in vitro (Laulhère et al., 1989, 1990). In vitro release of ferritin iron, when reduced by ascorbate or light, induces conversion of the ferritin subunit from 28 kDa to 26·5 and 25 kDa. As a consequence, pea seed ferritin then has a tendency to aggregate and to become insoluble (Laulhère et al., 1989). Fenton chemistry has been involved in this process since protein cleavage is inhibited by free radical scavengers as well as by iron chelators (Laulhère et al., 1989, 1990). These free radical cleavages occur within the plant-specific NH2-terminus of the pea seed ferritin subunit (Laulhère et al., 1989). These observations have been confirmed by the demonstration that recombinant pea seed ferritin, depleted of its NH2-terminus, is less soluble than recombinant wild-type pea seed ferritin (Van Wuytswinkel et al., 1995). A model explaining the mechanism of ferritin degradation during germination has been proposed (Lobréaux and Briat, 1991). Alternatively, it has been proposed recently that the 28-kDa and 26·5-kDa ferritin subunits from pea seed could be encoded by two different genes, instead of having the smaller subunit produced by processing of the 28-kDa isoform (Li et al., 2009)
Another example of developmental control of ferritin synthesis has been reported in leaves. Indeed the first evidence of such a regulation was suggested because the concentration of ferritin mRNA measured in mature soybean leaves was found to be higher than in young leaves (Ragland et al., 1990). Using the determinate nature of leaf development in maize, a high level of ferritin is only detected in the youngest section of the leaf whereas in the middle of this organ the ferritin concentration reaches its minimum (Theil and Hase, 1993). An unco-ordinated change in ferritin mRNA and ferritin protein concentrations was also reported between the various developmental stages of this leaf development, strengthening the hypothesis that a post-transcriptional control is likely to be an important level of regulation of ferritin synthesis during plant development (Theil and Hase, 1993).
In legumes, iron is also important for nitrogen fixation within the nodules where iron-proteins such as leghaemoglobin and nitrogenase play a key role. Variations in ferritin abundance in soybean nodules depending on their age, on interactions between the Bradyrhizobium strain and the host cultivar, and on mutations affecting the bacterial strain have been reported (Ko et al., 1987). In effective Bradyrhizobium/soybean symbiosis, ferritin has been observed only in young nodules. By contrast, in ineffective nodules produced by Bradyrhhizobium mutants, ferritin is found in nodules of all ages. Hence, ferritin is present in nodules when they are not functional. During the course of soybean nodule ageing, ferritin mRNA concentration remains elevated while the protein concentration decreases 4- to 5-fold (Ragland and Theil, 1993). It again indicates the occurence of a post-transcriptional control of ferritin abundance during a plant-specific developmental programme. During this process of soybean nodule ageing, before complete disappearance of the ferritin protein, breakdown products have been observed (Ragland and Theil, 1993), reminiscent of those observed during ferritin processing following seed germination (Lobréaux and Briat, 1991). Whether reactive oxygen species (ROS)-mediated signalling of ferritin degradation within ageing nodules occurs, as suggested for germinating seeds (Laulhère et al., 1989; Lobréaux and Briat, 1991), remains to be addressed.
In A. thaliana, ferritins are encoded by a small gene family of four members. The analysis of the tissue-specific expression of each of these genes at the transcript level, and kinetically during the life cycle of a plant was therefore straigthtforward to perform (Petit et al., 2001a; Fig. 1). Both AtFer1 and AtFer3 mRNA transcripts are present in rosette leaves, their abundance being higher at later developmental stages. In roots, AtFer1 mRNA has the same abundance at three various time points of the life cycle, whereas AtFer3 mRNA seems to be less abundant than AtFer1. In floral stalks, AtFer1 and AtFer3 transcripts are found in the stem and the leaves, Atfer1 transcript being abundant in this organ compared with AtFer3 which is weakly detected, with no variation between the two time points analysed. Both AtFer1 and AtFer3 mRNA abundances are important in flowers and these transcripts are still detected in siliques. In contrast, they are not observed either in dry siliques or in dry seeds. Both AtFer1 and AtFer3 mRNA are present in tissues during the early stages of germination, after 20 h or 30 h of seed imbibition and in 5-d-old germinating plantlets. The expression pattern of AtFer2 mRNA is markedly different from those observed for AtFer1 and AtFer3 mRNAs since (a) it is abundantly present in mature siliques and dry seeds where no other AtFer transcripts are detected, and (b) conversely it is almost undetectable in roots, rosette or floral stalk leaves, stem, flowers, immature siliques or germinating seeds. The observation that AtFer2 expression is restricted to the seed is consistent with the observation that this gene is activated in response to exogenous ABA application, as was also reported for its ZmFer2 maize orthologue (Lobréaux et al., 1993; Fig. 2). The expression pattern of AtFer4 diverges importantly from what is described above for AtFer1, AtFer3 and AtFer2. Its expression is restricted to the floral stalk and in flowers, with a maximum after pollination.
Fig. 1.
Tissue-specific expression and developmental regulation of ferritin synthesis in Arabidopsis. The expression of the AtFer2 ferritin gene is restricted to mature seeds. In vegetative and reproductive organs, from the germination stage to flowering, AtFer-1, -3 and -4 genes are kinetically and differentially expressed in various tissues and organs (Petit et al., 2001a; Ravet et al., 2009)
Fig. 2.
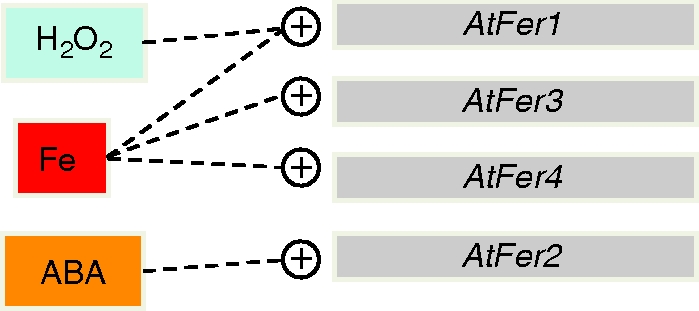
Differential expression of Arabidopsis ferritin genes in response to exogenous signals. Iron excess treatment (Fe) leads to AtFer-1, -3 and -4 gene expression, whereas H2O2 application impacts positively only on the expression of the AtFer1 gene. Consistent with its seed-specific expression, the AtFer2 gene is the only Arabidopsis ferritin gene to be expressed in response to application of exogenous abscisic acid (ABA; Petit et al., 2001a).
At the protein level, the immunodetection of ferritin subunits by western blot of crude protein extracts from leaves and seeds of various ferritin knock out mutants has recently been achieved (Ravet et al., 2009). It makes it possible to conclude that only AtFer1 exists mainly as a 28-kDa mature subunit in leaves, and that it is the more expressed ferritin at the protein level in this organ. It is also deduced from these experiments that AtFer3 and AtFer4 would exist in leaves only as a 26·5-kDa processed subunit. Finally, the only ferritin subunit present in seeds, as a 26·5-kDa subunit, is encoded by the AtFer2 gene, which is consistent with the expression data recorded at the transcript level (Petit et al., 2001a). It is therefore interesting to notice that Arabidopsis ferritin subunits can be observed, as in pea (Laulhere et al., 1989; Lobreaux et al., 1991) and in soybean (Masuda et al., 2001) as two polypeptides of different molecular weight. Ferritin subunits are synthesized as a 32-kDa precursor which contains a unique N-terminal sequence consisting of two domains absent in animal ferritins. The first domain, around 40–50 residues long, is the transit peptide involved in the transport of ferritin subunit precursor into the chloroplast. The second domain is part of the mature protein, and named extension peptide. As indicated above its function could be related to protein stability, and it could be cleaved during the germination process of pea seeds through free radical damages (Lobreaux et al., 1991; Van Wuytswinkel et al., 1995). Another mechanism of ferritin-subunit processing leading to a 26·5-kDa subunit from a mature 28-kDa-subunit has been proposed. Sequencing of purified assembled ferritin from soybean seeds enabled two subunits, H-1 and H-2, to be identified (Masuda et al., 2001). H-2 is an unprocessed 28-kDa subunit, whereas H-1 exhibited a C-terminal truncation of 17 residues leading to the obtention of a 26·5-kDa polypeptide. Cleavage occurs after an arginine residue, absent in H-2 (Fig. 3). Interestingly, in A. thaliana only the FER1 subunit exists in a 28-kDa form, whereas subunits encoded by AtFer2, AtFer3 and AtFer4 genes are only present as a 26·5-kDa polypeptide (Ravet et al., 2009). Alignment of the C-terminal part of the four Arabidopsis ferritin subunits with H-1 and H-2 soybean forms shows that the arginine residue at the cleavage site of H-1 is present in FER2, FER3 and FER4 subunits, but not in FER1 (Fig. 3). This result is consistent with the observation obtained with soybean ferritin, and indicating that a C-terminal cleavage could occur at a conserved arginine residue.
Fig. 3.
Sequence aligment of the C-terminal parts of soybean and Arabidopsis ferritin subunits. Identical and similar residues between at least three proteins are boxed in black and grey, respectively. Amino acids are numbered from the translational start methionine. The arginine in position 234 of Soy H-1 subunit was shown to be the last residue of the processed 26·5-kDa form (Masuda et al., 2001). This residue and the corresponding arginines in Arabidopsis subunits are indicated in red.
Ferritin synthesis in response to iron excess
In maize, the ZmFer1 ferritin gene is activated in response to iron excess (Fobis-Loisy et al., 1995), and its promoter was used to search for iron-dependent cis-regulatory elements (Petit et al., 2001b). The Fe-dependent up-regulation of ZmFer1 expression can be antagonized by antioxidants and PP2A-type phosphatase inhibitors (Savino et al., 1997). The ‘Fe excess’ signal leading to the expression of the ZmFer1 gene could therefore require oxidative stress and phosphorylation/dephosphorylation events to be transduced. Serial deletions and site-directed mutagenesis enabled a 15-bp sequence named IDRS (iron-dependent regulatory sequence) to be characterized within the proximal part of the promoter of the ZmFer1 ferritin gene, necessary for the repression of this gene under Fe-deficiency conditions (Petit et al., 2001b). In Arabidopsis, the AtFer1 gene is the orthologue of the maize ZmFer1 gene, its expression in response to Fe excess also being antagonized by antioxidants and Ser-Thr phosphatase inhibitors (Gaymard et al., 1996; Petit et al., 2001b). Sequence alignments show that the IDRS sequence defined in maize is highly conserved in Arabidopsis. Further analysis has shown that this sequence is functional in planta and, as demonstrated in maize, involved in the transcriptional repression of AtFer1 expression under iron-deficient conditions (Petit et al., 2001b). The IDRS sequence is also required for dark-induced senescence activation of AtFer1 expression but not for age-dependent senescence induction (Tarantino et al., 2003). AtFer1 promoter activity in leaves is observed mainly in the vicinity of the vessels and mutations within the IDRS do not alter this localization. In roots, the AtFer1 promoter activity was restricted to the endoderm. However, IDRS mutagenesis resulted in an extension of the promoter activity to the root cortex and epidermis (Tarantino et al., 2003). These results indicate that, under standard Fe nutrition conditions, the IDRS could be involved in the repression of expression of the AtFer1 gene in the cortex and epidermis cells. In the absence of a functional IDRS, AtFer1 gene repression would not occur any more, leading to an extension of the root expression territories of this gene.
By combining pharmacological and imaging approaches in an Arabidopsis cell culture system, several elements of the signal transduction pathway leading to the increase in AtFer1 transcript level after iron treatment have been clarified (Arnaud et al., 2006). Nitric oxide (NO) quickly accumulates in plastids after excess-iron treatment. This compound acts downstream of iron and upstream of a PP2A-type phosphatase, consistently with a previous report having shown that NO is involved in the transduction pathway between the ‘Fe excess’ signal and AtFer1 promoter de-repression, in an IDRS-, Ser-Thr phosphatase-dependent manner (Murgia et al., 2002). Furthermore, the repressor required for the repression of the AtFer1 gene transcription under low iron conditions is unlikely to be a transcription factor directly bound to the IDRS. It is rather a protein acting upstream in the pathway, which could be ubiquitinated upon iron treatment and subsequently degraded through a 26S proteasome-dependent pathway (Arnaud et al., 2006; Fig. 4).
Fig. 4.
A working model to explain the control of the AtFer1 gene expression in response to iron. (A) Under low-iron conditions, a repressor not directly bound to the AtFer1 promoter would interact with the transcription factor which recognizes the iron-dependent regulatory sequence (IDRS), leading to the repression of AtFer1 gene expression. (B) Under high-iron conditions, an enzymatically produced nitric oxide (NO) burst occurs within the plastids, preceding ubiquitination and proteasome-dependent degradation of the repressor. De-phosphorylation events depending upon a PP2A phosphatase activity would also occur. These events lead to a de-repression of the AtFer1 gene expression (Arnaud et al., 2006). The corresponding ferritin transcript is then translated to give the ferritin precursor polypeptide which is transported to plastids where it is assembled in the 24-mers ferritin protein.
Kinetics of Arabidopsis AtFer3 transcript accumulation in response to Fe overload are very similar to one of the AtFer1 genes, both in roots and shoots. In contrast, the AtFer2gene is not induced by Fe excess and AtFer4 mRNA abundance increases only in leaves with different kinetics than AtFer1 and 3 transcripts (Petit et al., 2001a). Comparison of the promoter sequence of the four Arabidopsis ferritin genes revealed the presence of an IDRS-like element in the same promoter region of the AtFer2, -3 and -4 ferritin genes as the functional IDRS characterized in the AtFer1 promoter. However, it remains to be determined whether or not these IDRS-like are functional. Beside the IDRS, another cis-regulatory element than the IDRS, named FRE (Fe responsive element), has been characterized in the promoter region of a soybean ferritin gene (Wei and Theil, 2000); no such element can be observed in any of the four Arabidopsis ferritin genes (Petit et al., 2001a). No trans-acting factors interacting with these cis-regulatory sequences and involved in plant ferritin gene expression have been characterized so far.
PLANT FERRITIN FUNCTIONS: STORAGE VERSUS STRESS PROTECTION
Despite a wealth of information obtained on ferritin biochemistry or expression in various plant species, the function of this iron-storage protein in plant physiology and development remained largely unknown. Most of the hypotheses formulated regarding ferritin functions in plants were based on correlations between localization of the proteins and responses of their expression to environmental factors and at various developmental stages. Investigating the functions of plant ferritins in planta has remained a difficult task since higher plants possess multiple gene copies coding for these proteins (Fobis-Loisy et al., 1995; Wardrop et al., 1999; Masuda et al., 2001, 2007; Petit et al., 2001; Strozycki et al., 2003; Dong et al., 2007). Furthermore, the high degree of sequence identity between the members of this multigenic family suggests a possible functional redundancy between the different ferritin subunits. Arabidopsis thaliana with its four ferritin genes offered the opportunity to investigate the biological function of plant ferritin through the combination of reverse genetic and physiological approaches. In Arabidopsis, a knock-out mutant in the AtFer1 gene has been isolated and characterized recently. The fer1 knock-out mutant exhibits an increased sensitivity to Erwinia chrysanthemi infection (Dellagi et al., 2005), and a slightly accelerated senescence (Murgia et al., 2007), but no major phenotype alteration is observed, strengthening the hypothesis of a functional redundancy between the four Arabidopsis gene products. New insights into ferritin function in Arabidopsis have been provided through the recent study of multiple knock-out mutants devoid of ferritin in seeds (fer2) or in vegetative and reproductive organs (fer1-3-4). This work has clearly demonstrated that ferritins are not likely to be an essential iron source for plant development, but that they play a significant role in the defence machinery against oxidative stress (Ravet et al., 2009).
Ferritin is not a major player of iron storage in Arabidopsis
During the last decades, ferritin has been considered to be the major iron storage protein in plants, in particular in seeds (Hyde et al., 1963). This hypothesis has been reinforced by studies on seed formation, in particular in pea. An important amount of iron is stored in pea seeds (Lobréaux and Briat, 1991). Immunodetection experiments revealed that ferritin subunits accumulated in embryo during pea seed maturation and remained present in dry seeds (Lobréaux and Briat, 1991; Marentes and Gruzak, 1998). The amount of iron stored inside ferritins is estimated to be 92% of the total seed iron content in the pea embryo axis (Marentes and Gruzak, 1998), leading to the consideration that this protein is the major form of iron storage in seeds. During germination, ferritins are degraded, and the iron released during this process has been postulated to be the main source of iron for the developing plant. In leaves, ferritins accumulated mainly in undifferentiated plastids of young leaves and become almost undetectable in the chloroplasts of the mature leaves (Theil and Hase, 1993). Thus, ferritins were hypothesized to be potential iron donors in the build-up of a functional photosynthetic apparatus where iron is mostly present as Fe–S clusters. In most of these proteins, iron is present as Fe–S clusters. In Arabidopsis mitochondria, the requirement of the frataxin protein as the iron donor for Fe–S cluster biosynthesis has been recently demonstrated (Busi et al., 2006). Since plastids are autonomous for their Fe–S cluster biosynthesis (Balk and Lobréaux, 2005), ferritins were suggested to fulfil the same function in these organelles as frataxin in mitochondria.
Recently, the importance of seed ferritins as the iron donor for Fe-proteins during the germination process has been challenged. Indeed, vacuolar iron remobilization by the NRAMP3 and NRAMP4 tonoplast metal effluxers is essential for germination to take place (Lanquar et al., 2005). Moreover, fer2 plants devoid of ferritin in seeds exhibit no obvious macroscopic phenotype; the seed iron content, the germination rate, and the proper development of the mutant plants are unaffected, even under iron-limiting conditions (Ravet et al., 2009). Such observations are consistent with the calculation that iron contained in ferritins of Arabidopsis seeds represent no more than 5% of the total seed iron (Ravet et al., 2009). To the best of our knowledge, seed iron ferritin content has been documented only for Arabidopsis and pea (Marentes and Gruzak, 1998; Ravet et al., 2009). It would be interesting therefore in the future to have comparative results with other plants.
In view of the elevated abundance of ferritins in young leaves, it was expected that an alteration in these organs would be observed in mutant plants lacking ferritins in vegetative tissues. Surprisingly, no such alteration of young leaves occur in a fer1-3-4 mutant plant devoid of ferritins in vegetative organs, and grown under standard conditions. Moreover, in mature leaves, photosynthesis is not significantly affected by the absence of ferritins; chlorophyll fluorescence parameters and CO2 fixation capacity are similar in wild-type and ferritin-less leaves. It can therefore be concluded that ferritins are not necessary for proper plant development, revealing that they do not constitute a major iron source for plant development. Consistently, chloroplast functionality is not dependent on ferritin presence, suggesting that these proteins do not fulfill the same function in the plastidial Fe–S cluster biosynthesis pathway that frataxin does in mitochondria.
Ferritin: a link between iron and ROS metabolisms
The relationship between iron homeostasis and oxidative stress is clearly established in bacteria, yeast and animals, where the activity of key factors regulating iron homeostasis are modulated by oxidative stress (Rouault and Klausner, 1996; Touati, 2000; Hantke et al., 2001; Kaplan et al., 2006; Theil, 2007; Toledano et al., 2007). In plants, such a link arises from both physiological and molecular evidence. Several studies indicate that iron excess, or mutations leading to an abnormally high level of iron, lead to an increase in ROS production and to the promotion of oxidative stress responses (Pich et al., 1994; Kampfenkel et al., 1995; Caro and Puntarulo, 1996; Pekker et al., 2002). In addition, physiological disorders due to iron toxicity responsible for necrotic spots in leaves have been known for a long time (Ponnamperuma et al., 1955). As described above, ferritin synthesis is activated at the transcriptional level by iron or H2O2 treatments and antagonized by antioxidant molecules. Furthermore, pro-oxidant treatments such as NO or ozone applications, as well as high light intensity, are known to induce ferritin synthesis.
Links between ferritin functions and ROS management have been deduced from the characterization of tobacco transgenic plants over-expressing ferritins. On one hand, plants over-expressing ferritin are less sensitive to methylviologen-promoted oxidative stress. However, on the other hand, ROS detoxifying enzymes are always activated in such plants even when grown under standard non-stressful conditions (Zer et al., 1994; Briat et al., 1999; Deák et al., 1999; Van Wuytswinkel et al., 1999). These data suggest that the control of ferritin synthesis is required for a proper maintenance of the cellular redox status. A clear relationship between iron and ROS has been recently demonstrated by using Arabidopsis ferritin-less mutants (Ravet et al., 2009). The fer2 mutant plant, devoid of seed ferritins, exhibits a comparatively higher sensitivity to methylviologen treatment during germination than to the wild-type plant. Under standard growth conditions, the absence of ferritins in the fer1-3-4 mutant plants leads also to an increased accumulation of ROS in leaves and flowers. However, this mutant can bypass the lack of safe iron storage within ferritins by increasing its ROS detoxifying enzyme activities in order to avoid cellular damages. Thus, despite the energy cost of such a compensation, the development, growth and fertility of the mutant plants lacking ferritins are similar to the those of wild-type plants, except when they are grown under high iron-concentration conditions.
Ferritin requirement for beneficial impact of high iron availability to plants
Ferritins are abundant when plants are grown under high-iron conditions. In such conditions, detoxifying enzymes are strongly induced to avoid deleterious ROS accumulation. This increase of ROS-detoxifying mechanisms in plants devoid of ferritins is not sufficient to bypass the deleterious accumulation of free iron. Consequently, pleiotropic defects both in vegetative and reproductive organs strongly impaired the growth and fertility of the ferritin-less plants (Ravet et al., 2009). Thus, the beneficial effect of elevated iron concentrations in the environment measured by the increased biomass of wild-type plants, is completely abolished when plants are devoid of ferritins. The growth decrease of Arabidopsis fer1-3-4 mutant plants has been shown to result from reduced carbon assimilation (Ravet et al., 2009).
Relationships between photosynthetic activity, ROS production, iron reactivity and ferritins have been evoked. In Arabidopsis, ferritins accumulate strongly in response to photo-inhibition (Murgia et al., 2001). Over-expression of ferritins in transgenic plants has led to divergent conclusions concerning their involvement in the protection against oxidative stress upon light- or cold-mediated photo-inhibition (Murgia et al., 2001; Hegedus et al., 2008). This link between photosynthetic apparatus activity and the protective role of ferritins has been reinforced by the study of ferritin mutants in the green algae Chlamydomonas reinhardtii (Busch et al., 2008). In this organism, ferritins have been shown to be necessary for rapid remodelling of the photosynthetic apparatus and to minimize photo-oxidative stress in response to iron availability. In contrast, in the Arabidopsis fer1-3-4 mutant, chlorophyll fluorescence measurements have shown that PSII efficiency is not affected. Thus, the reduced carbon assimilation observed in this mutant is not a consequence of an alteration of the electron transfer through the photosynthetic apparatus (Ravet et al., 2009), but is more likely originating from a ROS-mediated alteration of Calvin cycle efficiency.
The drastic decrease in fertility of the fer1-3-4 mutant when grown in high-iron condition has been shown to be independent of the absence of ferritins in the rosette. The pleiotropic development defects observed in fer1-3-4 flowers originate from a huge iron overload in this organ, accompanied by a deregulation of many genes related to iron transport and homeostasis in stems and flowers (Ravet et al., 2009). This suggests that modulation of ferritin expression is essential for the plant to establish a balance between availability of iron for metabolism, and sequestration of this metal to avoid subsequent ROS-related damages.
FERRITINS AND PLANT–MICROBE INTERACTIONS
It is well established that part of the innate immunity in vertebrates occurs through withholding iron, which constitutes a powerful antimicrobial mechanism (Ong et al., 2006). Competition for iron nutrition between microbial pathogens and their potential hosts is therefore central for an infection to be successful or not. In plants, the role of iron in pathogenicity has been well studied with E. chrysanthemi infection of saintpaulia or Arabidopsis plants, evidencing the role of high-affinity iron uptake systems mediated by the chrysobactin and achromobactin siderophores (Expert, 2005). Of particular interest is the observation that the expression of Erwininia genes involved in siderophores mediated iron uptake or encoding pectate lyases important for virulence are controlled by Fur, an iron-regulated transcriptional repressor (Franza et al., 2002). It became therefore clear that after penetrating their host, bacteria express their virulence in relation to their iron requirements.
Of interest within the frame of this review are the two following questions. What do we know about: (1) the function of bacterial ferritins in plant–bacteria interactions; and (2) the role of plant ferritin in plant immunity?
Four loci have been identified in E. chrysanthemi (fnA, bfr, dps1 and dps2) encoding, respectively, a ferritin, a haem-containing bacterioferritin and two miniferritins (Boughammoura et al., 2007). bfr and ftnA mutants have been produced (Boughammoura et al., 2008) making it possible to demonstrate that, unlike the bfr mutant, the ftnA mutant has increased sensitivity to iron deficiency and to redox stress conditions. Furthermore, FtnA and Bfr contribute differentially to the virulence of E. chrysanthemi depending on the host. ftnA, bfr, and bfrftnA mutants display a delay in the appearance of symptoms of maceration onto chicory leaves comparatively with wild-type bacteria. In contrast, using saintpaulia as host plants reveals a lag in the appearance of maceration symptoms only for the ftnA mutant. The bfr and bfrftnA mutants are as virulent as the wild-type strain on saintpaulia. More recently, a functional link between bacterioferritin (Bfr), bacterioferredoxin and the Suf protein machinery involved in Fe–S cluster biogenesis and assemby onto apoproteins has been shown to be important for an optimal growth of E. chrysanthemi, and a balanced distribution of iron between essential metalloproteins (Expert et al., 2008).
Attention to the role that plant ferritins could play during plant–pathogen interactions came first from the observation made by Neema et al. (1993), revealing that iron incorporated into plant ferritins drastically decreases in soybean suspension cells challenged with E. chrysanthemi. The role of plant ferritins in defence against pathogens has been strengthened by the observation that transgenic tobacco plants accumulating alfalfa ferritin in their leaves exhibited tolerance to necrotic damage caused by viral (tobacco necrosis virus) or fungal (Alternaria alternata, Botrytis cinerea) attacks (Deák et al., 1999). An increase in the expression of the StF1 potato ferritin gene in response to infection by Phytophthora infestans was also reported, but in that case, the increased ferritin gene expression was not enough to confer resistance to P. infestans (Mata et al., 2001). More recently, a cDNA encoding the AtFer1 ferritin has been pulled out from a cDNA differential screening between A. thaliana plants infected or not with E. chrysanthemi, revealing an up-regulation of this gene in response to a bacterial infection (Dellagi et al., 2005). An A. thaliana atfer1 null mutant has been used to investigate the potential role of the AtFer1 ferritin in an iron-withholding defence mechanism activated in response to bacterial infection. Indeed, spreading of the symptoms is faster and the number of systemic infections is higher in the fer1 mutant line in comparason with the wild-type plants. This result indicates that Atfer1 gene is involved in a basal level of resistance of A. thaliana to E. chrysanthemi (Dellagi et al., 2005). The Atfer1 gene is highly expressed 24 h after inoculation. This up-regulation does not occur after inoculation with an E. chrysanthemi siderophore negative mutant, suggesting a role of siderophores in the Atfer1 gene expression in response to E. chrysanthemi infection. Consistent with this statement, chrysobactin application increases Atfer1 transcript abundance, an effect which is not recorded when the siderophore is loaded with iron. The chrysobactin effect is not specific because AtFer1 up-regulation is also observed by treating plants with desferrioxamine. Neither oxidative stress nor NO are involved in the response to chrysobactin, and the AtFer1 gene expression in response to E. chrysanthemi inoculation is IDRS independent, leaving the mechanism by which siderophores up-regulate AtFer1 Arabidopsis ferritin gene expression an open question (Dellagi et al., 2005).
CONCLUDING REMARKS
Iron is a limiting factor for plant productivity and biomass production (Blain et al., 2007; Cassar et al., 2007; Ravet et al., 2009). However, this beneficial effect of iron on plant biomass and seed production appears dependent on the presence of ferritins. Indeed, growing a ferritin-less mutant under elevated iron conditions revealed major developmental defects associated with iron homeostasis perturbations and oxidative stress (Ravet et al. 2009). In nature, ferritin gene expression is modulated by many environmental factors. Throughout their life cycle, plants often experience these stresses, which are variable in intensity, location and duration, and which can transiently raise intracellular free iron pools, leading to an increased reactivity with oxygen. Thus, the fine-tuning of ferritin gene expression is indicative of the importance of these proteins for the adaptive response of plants to environmental changes. It is very likely that plant ferritins, by buffering iron, exert an appropriate control of the quantity of metal required for metabolic purposes. Therefore, they help plants to cope with adverse situations, the deleterious effects of which would be amplified if no system had evolved to take care of free reactive iron.
LITERATURE CITED
- Abdul-Tehrani H, Hudson AJ, Chang Y-S, et al. Ferritin mutants of Escherichia coli are iron deficient and growth impaired and fur mutants are iron deficient. Journal of Bacteriology. 1999;181:1415–1428. doi: 10.1128/jb.181.5.1415-1428.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altuvia S, Almiron M, Huisman G, Kolter R, Storz G. The Dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Molecular Microbiology. 1994;13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- Andrews SC. Iron storage in bacteria. Advances in Microbiological Physiology. 1998;40:281–351. doi: 10.1016/s0065-2911(08)60134-4. [DOI] [PubMed] [Google Scholar]
- Andrews SC, Arosio P, Bottke H, et al. Structure, function and evolution of ferritins. Journal of Inorganic Biochemistry. 1992;47:161–174. doi: 10.1016/0162-0134(92)84062-r. [DOI] [PubMed] [Google Scholar]
- Andrews SC, Le Brun NE, Barynin V, et al. Site-directed replacement of the coaxial heme ligands of bacterioferritin generates heme-free variants. The Journal of Biological Chemistry. 1995;270:23268–23274. doi: 10.1074/jbc.270.40.23268. [DOI] [PubMed] [Google Scholar]
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiology Reviews. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- Antelmann H, Engelmann S, Schmid R, Sorokin A, Lapidus A, Hecker M. Expression of a pexB-homologous gene is controlled by the alternative sigma factor sigmaB in Bacillus subtilis. Journal of Bacteriology. 1997;179:7251–7256. doi: 10.1128/jb.179.23.7251-7256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud N, Murgia I, Boucherez J, Briat JF, Cellier F, Gaymard F. An iron-induced nitric oxide burst precedes ubiquitin-dependent protein degradation for Arabidopsis AtFer1 ferritin gene expression. The Journal of Biological Chemistry. 2006;281:23579–23588. doi: 10.1074/jbc.M602135200. [DOI] [PubMed] [Google Scholar]
- Arnaud N, Ravet K, Borlotti A, et al. IRE/ IRP1-cytosolic aconitase iron regulatory switch does not operate in plants. The Biochemical Journal. 2007;405:523–531. doi: 10.1042/BJ20061874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arosio P, Ingrassia R, Cavadini P. Ferritins: a family of molecules for iron storage, antioxidation and more. Biochimica et Biophysica Acta. 2008. (in press). doi:10.1016/j.bbagen.2008.09.004. [DOI] [PubMed]
- Balk J, Lobréaux S. Biogenesis of iron-sulfur proteins in plants. Trends in Plant Sciences. 2005;10:324–331. doi: 10.1016/j.tplants.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Blain S, Quéguiner B, Armand L, et al. Effect of natural iron fertilization on carbon sequestration in the Southern Ocean. Nature. 2007;446:1070–1074. doi: 10.1038/nature05700. [DOI] [PubMed] [Google Scholar]
- Bou-Abdallah F, Lewin AC, Le Brun NE, Moore GR, Chasteen ND. Iron detoxification properties of Escherichia coli bacterioferritin: attenuation of oxyradical chemistry. The Journal of Biological Chemistry. 2002;277:37064–37069. doi: 10.1074/jbc.M205712200. [DOI] [PubMed] [Google Scholar]
- Boughammoura A, Franza T, Dellagi A, Roux C, Matzanke-Markstein B, Expert D. Ferritins, bacterial virulence and plant defence. Biometals. 2007;20:347–353. doi: 10.1007/s10534-006-9069-0. [DOI] [PubMed] [Google Scholar]
- Boughammoura A, Matzanke BF, Böttger L, et al. Differential role of ferritins in iron metabolism and virulence of the plant-pathogenic bacterium Erwinia chrysanthemi 3937. Journal of Bacteriology. 2008;190:1518–1530. doi: 10.1128/JB.01640-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat JF, Fobis-Loisy I, Grignon N, et al. Cellular and molecular aspects of iron metabolism in plants. Biology of the Cell. 1995;84:69–81. [Google Scholar]
- Briat JF, Lobréaux S, Grignon N, Vansuyt G. Regulation of plant ferritin synthesis: how and why. Cellular and Molecular Life Sciences. 1999;56:155–166. doi: 10.1007/s000180050014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat JF, Cellier F, Gaymard F. Ferritins and iron accumulation in plant tissues. In: Barton L, Abadia J., editors. Iron nutrition in plants and rhizospheric microorganisms. Berlin: Springer; 2006. pp. 345–361. [Google Scholar]
- Busch A, Rimbauld B, Naumann B., Rensch S, Hippler M. Ferritin is required for rapid remodeling of the photosynthetic apparatus and minimizes photo-oxidative stress in response to iron-availability in Chlamydomonas reinhardtii. The Plant Journal. 2008;55:201–211. doi: 10.1111/j.1365-313X.2008.03490.x. [DOI] [PubMed] [Google Scholar]
- Busi MV, Maliandi MV, Valdez H, et al. Deficiency of Arabidopsis thaliana frataxin alters activity of mitochondrial Fe-S proteins and induces oxidative stress. The Plant Journal. 2006;48:873–882. doi: 10.1111/j.1365-313X.2006.02923.x. [DOI] [PubMed] [Google Scholar]
- Caro A, Puntarulo S. Effect of in vivo iron supplementation on oxygen radical production by soybean roots. Biochimica et Biophysica Acta. 1996;1291:245–251. doi: 10.1016/s0304-4165(96)00071-2. [DOI] [PubMed] [Google Scholar]
- Carrondo MA. Ferritins, iron uptake and storage from the bacterio-ferritin viewpoint. EMBO Journal. 2003;22:1959–1968. doi: 10.1093/emboj/cdg215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. Feo-transport of ferrous iron into bacteria. Biometals. 2006;19:143–157. doi: 10.1007/s10534-006-0003-2. [DOI] [PubMed] [Google Scholar]
- Cassanelli S, Moulis J. Sulfide is an efficient iron releasing agent for mammalian ferritins. Biochimica et Biophysica Acta. 2001;1547:74–182. doi: 10.1016/s0167-4838(01)00182-0. [DOI] [PubMed] [Google Scholar]
- Cassar N, Bender ML, Barnett BA, et al. The Southern Ocean biological response to aeolian iron deposition. Science. 2007;317:1067–1070. doi: 10.1126/science.1144602. [DOI] [PubMed] [Google Scholar]
- Chasteen ND. Ferritin: uptake, storage and release of iron. Metal Ions in Biological Systems. 1998;35:479–514. [PubMed] [Google Scholar]
- Cheesman MR, Thomson AJ, Greenwood C, Moore GR, Kadir F. Bis-methionine axial ligation of haem in bacterioferritin from Pseudomonas aeruginosa. Nature. 1990;346:771–773. doi: 10.1038/346771a0. [DOI] [PubMed] [Google Scholar]
- Cheesman MR, Le Brun NE, Kadir FHA, et al. Haem and non-haem iron sites in Escherichia coli bacterioferritin: spectroscopic and model building studies. The Biochemical Journal. 1993;292:47–56. doi: 10.1042/bj2920047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SL, Vasanthakumar A, Anderson SA, et al. Iron-responsive degradation of iron-regulatory protein 1 does not require the Fe-S cluster. EMBO Journal. 2006;25:544–553. doi: 10.1038/sj.emboj.7600954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deák M, Horváth GV, Davletova S, et al. Plants ectopically expressing the iron-binding protein, ferritin, are tolerant to oxidative damage and pathogens. Nature Biotechnology. 1999;17:192–196. doi: 10.1038/6198. [DOI] [PubMed] [Google Scholar]
- De Domenico I, Vaughn MB, Li L, et al. Ferroportin-mediated mobilization of ferritin iron precedes ferritin degradation by the proteasome. EMBO Journal. 2006;25:5396–5404. doi: 10.1038/sj.emboj.7601409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellagi A, Rigault M, Segond D, et al. Siderophore-mediated upregulation of Arabidopsis ferritin expression in response to Erwinia chrysanthemi infection. The Plant Journal. 2005;43:262–272. doi: 10.1111/j.1365-313X.2005.02451.x. [DOI] [PubMed] [Google Scholar]
- Dong X, Sun Q, Wei D, et al. A novel ferritin gene, SferH-5, reveals heterogeneity of the 26·5-kDa subunit of soybean (Glycine max) seed ferritin. FEBS Letters. 2007;581:5796–5802. doi: 10.1016/j.febslet.2007.11.049. [DOI] [PubMed] [Google Scholar]
- Expert D. Genetic regulation of iron in Erwinia chrysanthemi as pertains to bacterial virulence. In: Barton L, Abadia J., editors. Iron nutrition in plants and rhizospheric microorganisms. Berlin: Springer; 2005. pp. 215–227. [Google Scholar]
- Expert D, Boughammoura A, Franza T. Siderophore-controlled iron assimilation in the enterobacterium Erwinia chrysanthemi: evidence for the involvement of bacterioferritin and the suf iron-sulfur cluster assembly machinery. The Journal of Biological Chemistry. 2008;283:36564–36572. doi: 10.1074/jbc.M807749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillebeen C, Chahine D, Caltagirone A, Segal P, Pantopoulos K. A phosphomimetic mutation at Ser-138 renders iron regulatory protein 1 sensitive to iron-dependent degradation. Molecular and Cellular Biology. 2003;19:6973–6981. doi: 10.1128/MCB.23.19.6973-6981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fobis-Loisy I, Loridon K, Lobréaux S, Lebrun M, Briat JF. Structure and differential expression in response to iron and abscisic acid of two maize ferritin genes. European Journal of Biochemistry. 1995;231:609–619. doi: 10.1111/j.1432-1033.1995.tb20739.x. [DOI] [PubMed] [Google Scholar]
- Franza T, Michaud-Soret I, Piquerel P, Expert D. Coupling of iron assimilation and pectinolysis in Erwinia chrysanthemi 3937. Molecular Plant–Microbe Interactions. 2002;15:1181–1191. doi: 10.1094/MPMI.2002.15.11.1181. [DOI] [PubMed] [Google Scholar]
- Galy B, Ferring D, Minana B, et al. Altered body iron distribution and microcytosis in mice deficient in iron regulatory protein 2 (IRP2) Blood. 2005;106:2580–2589. doi: 10.1182/blood-2005-04-1365. [DOI] [PubMed] [Google Scholar]
- Gaymard F, Boucherez J, Briat JF. Characterization of a ferritin mRNA from Arabidopsis thaliana accumulated in response to iron through an oxidative pathway independent of abscisic acid. The Biochemical Journal. 1996;318:67–73. doi: 10.1042/bj3180067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K. Iron and metal regulation in bacteria. Current Opinion in Microbiology. 2001;4:172–177. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- Hegedus A, Janda T, Horváth GV, Dudits D. Accumulation of overproduced ferritin in the chloroplast provides protection against photoinhibition induced by low temperature in tobacco plants. Journal of Plant Physiology. 2008;165:1647–1651. doi: 10.1016/j.jplph.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Hintze KJ, Theil EC. DNA and mRNA elements with complementary responses to hemin, antioxidant inducers, and iron control ferritin-L expression. Proceedings of the National Academy of Sciences of the USA. 2005;102:15048–15052. doi: 10.1073/pnas.0505148102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde BB, Hodge AJ, Kahn A, Birnstiel ML. Studies on phytoferritin. I. Identification and localization. Journal of Ultrastructure Research. 1963;59:248–258. doi: 10.1016/s0022-5320(63)80005-2. [DOI] [PubMed] [Google Scholar]
- Ilari A, Ceci P, Ferrari D, Rossi GL, Chiancone E. Iron incorporation into Escherichia coli Dps gives rise to a ferritin-like microcrystalline core. The Journal of Biological Chemistry. 2002;277:37619–37623. doi: 10.1074/jbc.M206186200. [DOI] [PubMed] [Google Scholar]
- Ilari A, Stefanini S, Chiancone E, Tsernoglou D. The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site. Nature Structural Biology. 2000;7:38–43. doi: 10.1038/71236. [DOI] [PubMed] [Google Scholar]
- Izuhara M, Takamune K, Takata R. Cloning and sequencing of an Escherichia coli K12 gene which encodes a polypeptide having similarity to the human ferritin H subunit. Molecular General Genetics. 1991;225:510–513. doi: 10.1007/BF00261694. [DOI] [PubMed] [Google Scholar]
- Jin W, Takagi H, Pancorbo B, Theil EC. ‘Opening’ the ferritin pore for iron release by mutation of conserved amino acids at interhelix and loop sites. Biochemistry. 2001;40:7525–7532. doi: 10.1021/bi002509c. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Norcross DC, Arosio P, Frankel RB, Watt GD. Redox reactivity of animal apoferritins and apoheteropolymers assembled from recombinant heavy and light human chain ferritins. Biochemistry. 1999;38:4089–4096. doi: 10.1021/bi982690d. [DOI] [PubMed] [Google Scholar]
- Kampfenkel K, Van Montagu M, Inze D. Effects of iron excess on Nicotiana plumbaginifolia plants (implications to oxidative stress) Plant Physiology. 1995;107:725–735. doi: 10.1104/pp.107.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J, McVey Ward D, Crisp RJ, Philpott CC. Iron-dependent metabolic remodeling in S. cerevisiae. Biochimica et Biophysica Acta. 2006;1763:646–651. doi: 10.1016/j.bbamcr.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Keech AM, Le Brun NE, Wilson MT, Andrews SC, Moore GR, Thomson AJ. Spectroscopic studies of cobalt (II) binding to Escherichia coli bacterioferritin. The Journal of Biological Chemistry. 1997;272:422–429. doi: 10.1074/jbc.272.1.422. [DOI] [PubMed] [Google Scholar]
- Kidane TZ, Sauble E, Linder MC. Release of iron from ferritin requires lysosomal activity. American Journal of Physiology: Cell Physiology. 2006;291:C445–C455. doi: 10.1152/ajpcell.00505.2005. [DOI] [PubMed] [Google Scholar]
- Kim S, Ponka P. Nitric oxide-mediated modulation of iron regulatory proteins: implication for cellular iron homeostasis. Blood Cells Molecular Diseases. 2002;29:400–410. doi: 10.1006/bcmd.2002.0579. [DOI] [PubMed] [Google Scholar]
- Kim S, Wing SS, Ponka P. S-nitrosylation of IRP2 regulates its stability via the ubiquitin-proteasome pathway. Molecular and Cellular Biology. 2004;24:330–337. doi: 10.1128/MCB.24.1.330-337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Theil EC. Posttranscriptional regulation of ferritin during nodule development in soybean. Plant Physiology. 1994;104:263–270. doi: 10.1104/pp.104.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko MP, Huang PY, Huang JS, Barker KR. The occurrence of phytoferritin and its relationship to effectiveness of soybean nodules. Plant Physiology. 1987;83:299–305. doi: 10.1104/pp.83.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanquar V, Lelievre F, Bolte S, et al. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO Journal. 2005;24:4041–4051. doi: 10.1038/sj.emboj.7600864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laulhère JP, Labouré AM, Briat JF. Mechanism of the transition from plant ferritin to phytosiderin. The Journal of Biological Chemistry. 1989;264:3629–3635. [PubMed] [Google Scholar]
- Laulhère JP, Labouré AM, Briat JF. Photoreduction and incorporation of iron into ferritins. The Biochemical Journal. 1990;269:79–84. doi: 10.1042/bj2690079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Brun NE, Andrews SC, Guest JR, Harrison PM, Moore GR, Thomson AJ. Identification of the ferroxidase centre of Escherichia coli bacterioferritin. The Biochemical Journal. 1995;312:385–392. [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- Lescure AM, Proudhon D, Pesey H, Ragland M, Theil EC, Briat JF. Ferritin gene transcription is regulated by iron in soybean cell cultures. Proceedings of the National Academy of Sciences of the USA. 1991;88:8222–8226. doi: 10.1073/pnas.88.18.8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi S, Santambrogio P, Cozzi A, et al. The role of the L-chain in ferritin iron incorporation: studies of homo and heteropolymers. Journal of Molecular Biology. 1994;238:649–654. doi: 10.1006/jmbi.1994.1325. [DOI] [PubMed] [Google Scholar]
- Li C, Hu X, Zhao G. Two different H-type subunits from pea seed (Pisum sativum) ferritin that are responsible for fast Fe(II) oxidation. Biochimie. 2009;91:230–239. doi: 10.1016/j.biochi.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Liu XS, Patterson LD, Miller MJ, Theil EC. Peptides selected for the protein nanocage pores change the rate of iron recovery from the ferritin mineral. The Journal of Biological Chemistry. 2007;282:31821–31825. doi: 10.1074/jbc.C700153200. [DOI] [PubMed] [Google Scholar]
- Lobréaux S, Briat JF. Ferritin accumulation and degradation in different organs of pea (Pisum sativum) during development. The Biochemical Journal. 1991;274:601–606. doi: 10.1042/bj2740601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobréaux S, Hardy T, Briat JF. Abscisic acid is involved in the iron-induced synthesis of maize ferritin. EMBO Journal. 1993;12:651–657. doi: 10.1002/j.1460-2075.1993.tb05698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobréaux S, Yewdall S, Briat JF, Harrison PM. Amino acid sequence and predicted three-dimensional structure of pea seed ferritin. The Biochemical Journal. 1992a;288:931–939. doi: 10.1042/bj2880931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobréaux S, Massenet O, Briat JF. Iron induces ferritin synthesis in maize plantlets. Plant Molecular Biology. 1992b;19:563–575. doi: 10.1007/BF00026783. [DOI] [PubMed] [Google Scholar]
- Marentes E, Grusak MA. Iron transport and storage within the seed coat and embryo of developing seeds of pea (Pisum sativum L.) Seed Science Research. 1998;8:575–582. [Google Scholar]
- Masse E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. Journal of Bacteriology. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T, Goto F, Yoshihara T. A novel plant ferritin subunit from soybean that is related to a mechanism in iron release. The Journal of Biological Chemistry. 2001;276:19575–19579. doi: 10.1074/jbc.M011399200. [DOI] [PubMed] [Google Scholar]
- Masuda T, Goto F, Yoshihara T, et al. Construction of homo- and heteropolymers of plant ferritin subunits using an in vitro protein expression system. Protein Expression and Purification. 2007;56:237–246. doi: 10.1016/j.pep.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Mata CG, Lamattina L, Cassia RO. Involvement of iron and ferritin in the potato-Phytophthora infestans interaction. European Journal of Plant Pathology. 2001;107:557–562. [Google Scholar]
- Mehlhase J, Sandig G, Pantopoulos K, Grune T. Oxidation-induced ferritin turnover in microglial cells: role of proteasome. Free Radicalals in Biology and Medicine. 2005;38:276–285. doi: 10.1016/j.freeradbiomed.2004.10.025. [DOI] [PubMed] [Google Scholar]
- Meyron-Holtz EG, Ghosh MC, Iwai K, et al. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO Journal. 2004;23:386–395. doi: 10.1038/sj.emboj.7600041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CM, Gaballa A, Hui M, Ye RW, Helmann JD. Genetic and physiological responses of Bacillus subtilis to metal ion stress. Molecular Microbiology. 2005;57:27–40. doi: 10.1111/j.1365-2958.2005.04642.x. [DOI] [PubMed] [Google Scholar]
- Murgia I, Briat JF, Tarantino D, Soave C. Plant ferritin accumulates in response to photoinhibition but its ectopic overexpression does not protect against photoinhibition. Plant Physiology and Biochemistry. 2001;39:1–10. [Google Scholar]
- Murgia I, Delledonne M, Soave C. Nitric oxide mediates iron-induced ferritin accumulation in Arabidopsis. The Plant Journal. 2002;30:521–528. doi: 10.1046/j.1365-313x.2002.01312.x. [DOI] [PubMed] [Google Scholar]
- Murgia I, Vazzola V, Tarantino D, et al. Knock-out of ferritin AtFer1 causes earlier onset of age-dependent leaf senescence in Arabidopsis. Plant Physiology and Biochemistry. 2007;45:898–907. doi: 10.1016/j.plaphy.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Navarre DA, Wendehenne D, Durner J, Noad R, Klessig DF. Nitric oxide modulates the activity of tobacco aconitase. Plant Physiology. 2000;122:573–582. doi: 10.1104/pp.122.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neema C, Laulhère JP, Expert D. Iron deficiency induced by chrysobactin in Saintpaulia leaves inoculated with Erwinia chrysanthemi. Plant Physiology. 1993;102:967–973. doi: 10.1104/pp.102.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong ST, Ho JZ, Ho B, Ding JL. Iron-withholding strategy in innate immunity. Immunobiology. 2006;211:295–314. doi: 10.1016/j.imbio.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Pekker I, Tel-Or E, Mittler R. Reactive oxygen intermediates and glutathione regulate the expression of cytosolic ascorbate peroxidase during iron-mediated oxidative stress in bean. Plant Molecular Biology. 2002;49:429–438. doi: 10.1023/a:1015554616358. [DOI] [PubMed] [Google Scholar]
- Petit JM, Briat JF, Lobréaux S. Structure and differential expression of the four members of the Arabidopsis thaliana ferritin gene family. The Biochemical Journal. 2001a;359:575–582. doi: 10.1042/0264-6021:3590575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit JM, Van Wuytswinkel O, Briat JF, Lobréaux S. Characterization of an iron-dependent regulatory sequence involved in the transcriptional control of AtFer1 and ZmFer1 plant ferritin genes by iron. The Journal of Biological Chemistry. 2001b;276:5584–5590. doi: 10.1074/jbc.M005903200. [DOI] [PubMed] [Google Scholar]
- Peyret P, Perez P, Alric M. Structure, genomic organization, and expression of the Arabidopsis thaliana aconitase gene: plant aconitase shows significant homology with mammalian iron-responsive element-binding protein. The Journal of Biological Chemistry. 1995;270:8131–8137. doi: 10.1074/jbc.270.14.8131. [DOI] [PubMed] [Google Scholar]
- Pich A, Scholz G, Stephan UW. Iron dependent changes of heavy metals, nicotianamine and citrate in different plant organs and in the xylem exudate of two tomato genotypes: nicotianamine as possible copper translocator. Plant and Soil. 1994;165:189–196. [Google Scholar]
- Ponnamperuma FN, Bradfield R, Peech M. Physiological disease of rice attributable to iron toxicity. Nature. 1955;175:275–279. [Google Scholar]
- Proudhon P, Briat JF, Lescure AM. Iron induction of ferritin synthesis in soyabean cell suspensions. Plant Physiology. 1989;90:586–590. doi: 10.1104/pp.90.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland M, Theil EC. Ferritin (mRNA, protein) and iron concentrations during soybean nodule development. Plant Molecular Biology. 1993;21:555–560. doi: 10.1007/BF00028813. [DOI] [PubMed] [Google Scholar]
- Ragland M, Briat JF, Gagnon J, Laulhère JP, Massenet O, Theil EC. Evidence for conservation of ferritin sequences among plants and animals and for a transit peptide in soybean. The Journal of Biological Chemistry. 1990;265:18339–18344. [PubMed] [Google Scholar]
- Ravet K, Touraine B, Boucherez J, Briat JF, Gaymard F, Cellier F. Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. The Plant Journal. 2009;57:400–412. doi: 10.1111/j.1365-313X.2008.03698.x. [DOI] [PubMed] [Google Scholar]
- Reindel S, Schmidt CL, Anemuller S, Matzanke BF. Expression and regulation pattern of ferritin-like DpsA in the archaeon Halobacterium salinarum. Biometals. 2006;19:19–29. doi: 10.1007/s10534-005-3682-1. [DOI] [PubMed] [Google Scholar]
- Rouault TA, Klausner R. Iron-sulfur clusters as biosensors of oxidants and iron. Trends in Biochemical Sciences. 1996;21:174–177. [PubMed] [Google Scholar]
- Sammarco MC, Ditch S, Banerjee A, Grabczyk E. Ferritin L and H subunits are differentially regulated on a post-transcriptional level. The Journal of Biological Chemistry. 2008;283:4578–4587. doi: 10.1074/jbc.M703456200. [DOI] [PubMed] [Google Scholar]
- Savino G, Briat JF, Lobréaux S. Inhibition of the iron-induced ZmFer1 maize ferritin gene expression by antioxidants and serine/threonine phosphatase inhibitors. The Journal of Biological Chemistry. 1997;272:33319–33326. doi: 10.1074/jbc.272.52.33319. [DOI] [PubMed] [Google Scholar]
- Seckback JJ. Ferreting out the secrets of plant ferritin: a review. Journal of Plant Nutrition. 1982;5:369–394. [Google Scholar]
- Shi H, Bencze KZ, Stemmler TL, Philpott CC. A cytosolic iron chaperone that delivers iron to ferritin. Science. 2008;320:1207–1210. doi: 10.1126/science.1157643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL. The physiological role of ferritin-like compounds in bacteria. Critical Reviews in Microbiology. 2004;30:173–185. doi: 10.1080/10408410490435151. [DOI] [PubMed] [Google Scholar]
- Stephani K, Weichart D, Hengge R. Dynamic control of Dps protein levels by ClpXP and ClpAP proteases in Escherichia coli. Molecular Microbiology. 2003;49:1605–1614. doi: 10.1046/j.1365-2958.2003.03644.x. [DOI] [PubMed] [Google Scholar]
- Strozycki PM, Skapska A, Szczesniak K, Sobieszczuk E, Briat JF, Legocki AB. Differential expression and evolutionary analysis of the three ferritin genes in the legume plant Lupinus luteus. Physiologia Plantarum. 2003;118:380–389. [Google Scholar]
- Tarantino D, Petit JM, Lobréaux S, Briat JF, Soave C, Murgia I. Differential involvement of the IDRS cis-element in the developmental and environmental regulation of the AtFer1 ferritin gene from Arabidopsis. Planta. 2003;217:709–716. doi: 10.1007/s00425-003-1038-z. [DOI] [PubMed] [Google Scholar]
- Theil EC. Coordinating responses to iron and oxygen stress with DNA and mRNA promoters: the ferritin story. Biometals. 2007;20:513–521. doi: 10.1007/s10534-006-9063-6. [DOI] [PubMed] [Google Scholar]
- Theil EC, Hase T. Plant and microbial ferritins. In: Barton LL, Hemings B, editors. Iron chelation in plants and soil microorganisms. New York, NY: Academic Press; 1993. pp. 133–156. [Google Scholar]
- Theil EC, Matzapetakis M, Liu X. Ferritins: iron/oxygen biominerals in protein nanocages. Journal of Biological Inorganic Chemistry. 2006;11:803–810. doi: 10.1007/s00775-006-0125-6. [DOI] [PubMed] [Google Scholar]
- Toledano MB, Kumar C, Le Moan N, Spector D, Tacnet F. The system biology of thiol redox system in Escherichia coli and yeast: differential functions in oxidative stress, iron metabolism and DNA synthesis. FEBS Letters. 2007;581:3598–3607. doi: 10.1016/j.febslet.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Torti FM, Torti SV. Regulation of ferritin genes and protein. Blood. 2002;99:3505–3516. doi: 10.1182/blood.v99.10.3505. [DOI] [PubMed] [Google Scholar]
- Touati D. Iron and oxidative stress in bacteria. Archives in Biochemistry and Biophysics. 2000;373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in fur mutants of Escherichia coli: protective role of superoxide dismutase. Journal of Bacteriology. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truty J, Malpe R, Linder MC. Iron prevents ferritin turnover in hepatic cells. The Journal of Biological Chemistry. 2001;276:48775–48780. doi: 10.1074/jbc.M105392200. [DOI] [PubMed] [Google Scholar]
- Van Wuytswinkel O, Briat JF. Conformational changes and in vitro core formation modifications induced by site directed mutagenesis of the specific amino terminus (EP) of pea seed ferritin. The Biochemical Journal. 1995;305:959–965. doi: 10.1042/bj3050959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wuytswinkel O, Savino G, Briat JF. Purification and characterization of recombinant pea seed ferritins expressed in Escherichia coli: influence of amino terminus deletions on protein solubility and in vitro core formation. The Biochemical Journal. 1995;305:253–261. doi: 10.1042/bj3050253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wuytswinkel O, Vansuyt G, Grignon N, Fourcroy P, Briat JF. Iron homeostasis alteration in transgenic tobacco overexpressing ferritin. The Plant Journal. 1999;17:93–97. doi: 10.1046/j.1365-313x.1999.00349.x. [DOI] [PubMed] [Google Scholar]
- Vecerek B, Moll I, Bläsi U. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. The EMBO Journal. 2007;26:965–975. doi: 10.1038/sj.emboj.7601553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade VJ, Treffry A, Laulhère JP, et al. Structure and composition of ferritin cores from pea seed. Biochimica et Biophysica Acta. 1993;1161:91–96. doi: 10.1016/0167-4838(93)90201-2. [DOI] [PubMed] [Google Scholar]
- Waldo GS, Wright E, Wang ZH, Briat JF, Theil EC, Sayers DE. Formation of the ferritin iron mineral occurs in plastids: an X-ray absorption spectroscopy study. Plant Physiology. 1995;109:797–802. doi: 10.1104/pp.109.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Fillebeen C, Chen G, Biederbick A, Lill R, Pantopoulos K. Iron-dependent degradation of apo-IRP1 by the ubiquitin-proteasome pathway. Molecular and Cellular Biology. 2007;7:2423–2430. doi: 10.1128/MCB.01111-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardrop AJ, Wicks RE, Entsch B. Occurrence and expression of members of the ferritin gene family in cowpeas. The Biochemical Journal. 1999;337:523–530. [PMC free article] [PubMed] [Google Scholar]
- Wei J, Theil EC. Identification and characterization of the iron regulatory element in the ferritin gene of a plant (soybean) The Journal of Biological Chemistry. 2000;275:17488–17493. doi: 10.1074/jbc.M910334199. [DOI] [PubMed] [Google Scholar]
- Zancani M, Peresson C, Biroccio A, et al. Evidence for the presence of ferritin in plant mitochondria. European Journal of Biochemistry. 2004;271:3657–3664. doi: 10.1111/j.1432-1033.2004.04300.x. [DOI] [PubMed] [Google Scholar]
- Zer H, Peleg I, Chevion M. The protective effect of desferrioxamine on paraquat-treated pea (Pisum sativum) Physiologia Plantarum. 1994;92:437–442. [Google Scholar]
- Zhao G, Ceci P, Ilari A, et al. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells: a ferritin-like DNA-binding protein of Escherichia coli. The Journal of Biological Chemistry. 2002;277:27689–27696. doi: 10.1074/jbc.M202094200. [DOI] [PubMed] [Google Scholar]





