Abstract
Background and Aims
Both environmental and genetic effects contribute to phenotypic variation within and among populations. Genetic differentiation of quantitative traits among populations has been shown in many species, yet it can also be accompanied by other genetic changes, such as divergence in phenotypic plasticity and in genetic variance. Sideroxylonal (a formylated phloroglucinol compound or FPC) is an important chemical defence in eucalypts. The effect of environmental variation on its production is a critical gap in our understanding of its genetics and evolution.
Methods
The stability of genetic variation in sideroxylonal was assessed within and among populations of Eucalyptus tricarpa in three replicated provenance/progeny trials. The covariance structure of the data was also modelled to test whether genetic variances were consistent among populations and Fain's test was applied for major gene effects.
Key Results
A significant genotype × environment interaction occurred at the level of population, and was related to temperature range and seasonality in source populations. Within-population genetic variation was not affected by genotype × environment effects or different sampling years. However, within-population genetic variance for sideroxylonal concentration differed significantly among source populations. Regression of family variance on family mean suggested that this trait is subject to major gene effects, which could explain the observed differences in genetic variances among populations.
Conclusions
These results highlight the importance of replicated common-garden experiments for understanding the genetic basis of population differences. Genotype × environment interactions are unlikely to impede evolution or responses to artificial selection on sideroxylonal, but the lack of genetic variation in some populations may be a constraint. The results are broadly consistent with localized selection on foliar defence and illustrate that differentiation in population means, whether due to selection or to drift, can be accompanied by changes in other characteristics, such as plasticity and genetic variance.
Keywords: Additive genetic variance, open-pollinated common-garden experiment, genotype × environment interaction, plasticity, Eucalyptus tricarpa, sideroxylonal, formylated phloroglucinol compounds (FPCs), chemical defence
INTRODUCTION
Geographically structured variation in phenotypic traits can result from genetic and environmental factors. Divergence may be primarily caused by environmental effects, which are mediated by phenotypic plasticity, the differential expression of traits under different environmental conditions (Hoffmann et al., 2005). Alternatively, genetic effects may predominate, diverging by local adaptation or drift (Mitchell-Olds and Schmitt, 2006; Leinonen et al., 2008). Genetic differentiation of quantitative traits among populations has been shown in many species, yet the genetic changes that can accompany divergence have received less attention. These can include changes in phenotypic plasticity, i.e. the ability of an organism to respond to environmental variation, and in genetic architecture, i.e. the genetic factors underlying trait variation. Genetic differentiation among populations can affect not only population means, but also the amount of genetic variation within a population (Widen et al., 2002; Bégin and Roff, 2003; Pressoir and Berthaud, 2004). The plastic response of a trait to environmental variation can also diverge by drift or in response to selection in each source population, creating non-additive effects of genotype and environment, or genotype × environment (G × E) interactions at the level of populations. While there is considerable debate about the role plasticity plays in adaptation to novel environments (de Jong, 2005; West-Eberhard, 2005; Pigliucci et al., 2006), it is generally agreed that reaction norms, which characterize the response of a genotype to a range of environments, can evolve as a result of natural selection (Van Tienderen and Koelewijn, 1994; Lande, 2009). For example, plasticity may be favoured by factors such as heterogeneity and predictability of selection (Lande, 2009). Thus, the partitioning of variation in plasticity within and among populations may reflect the combined action of microevolutionary forces in much the same way as for simple traits.
Studies of G × E interactions are important, not just because of what they can tell us about the causes of geographic differentiation, but also because they have serious implications for the measurement of genetic parameters, both within and among populations. For example, unknown G × E interactions could cause incorrect inferences to be drawn when designing breeding programmes or extrapolating from common-garden results to wild populations (Mitchell-Olds and Rutledge, 1986; Conner et al., 2003). It is common practice to measure genetic variation by rearing genetically different individuals in shared environments. G × E interactions can be studied by manipulating growth conditions to examine the effects of specific environmental variables. A contrasting approach, taken in reciprocal transplant experiments, for example, is to grow plants on sites that differ in many ways, which are often unknown. Replicating genetic experiments at multiple sites rarely identifies the environmental factors responsible for phenotypic differences (Hamann et al., 2000; Costa e Silva et al., 2006), but does provide information on the plasticity or stability of traits and the likelihood that estimates of genetic effects and predicted responses to selection are transferable to new environments (Westcott, 1986; Cooper and DeLacy, 1994). We have taken this approach to investigate the genetic basis and plasticity of among-population and within-population variation in an important plant defence trait.
The ecology and evolutionary outcomes of interactions between plants and animals are directly affected by genetic variation within species (Cipollini et al., 2003; Fornoni et al., 2004) and provide several examples of local adaptation (Sork et al., 1993; Zangerl and Berenbaum, 2003; Stenberg et al., 2006). Community structure and interactions between plants and animals are affected by G × E interactions at multiple scales (Johnson and Agrawal, 2005). G × E interaction effects on defence traits have been observed in response to various environmental factors, including water availability, fertilization, competition and herbivory (Keinänen et al., 1999; Donaldson et al., 2006). Although several studies have detected G × E interaction effects on herbivory and chemical defence at different sites (e.g. Bowers et al., 1992; Stiling and Rossi, 1996; Boege and Dirzo, 2004; Rosner and Hannrup, 2004; Johnson and Agrawal, 2005), the stability of genetic variation (both within and among populations) in foliar defence chemicals across sites is unknown in many systems, especially in long-lived trees (Silfver et al., 2009). Since plant species encounter environments that differ in more than just a few variables, this is a critical gap in our understanding of plant–herbivore interactions, from an applied as well as an evolutionary perspective.
Sideroxylonals are members of the group of potent foliar antifeedant chemicals found in eucalypts that are known as formylated phloroglucinol compounds or FPCs (Lawler et al., 1998; Marsh et al., 2003; Moore and Foley, 2005). This class of compound reduces feeding by both mammalian and at least some insect herbivores (Floyd and Foley, 2001; Andrew et al., 2007). Geographic variation in foliar sideroxylonal concentrations has been identified in Eucalyptus microcorys (Moore et al., 2004b) and has been shown to have a genetic component in E. tricarpa (Andrew et al., 2007). In E. microcorys, high sideroxylonal concentrations in natural populations were related to cool winter temperatures (Moore et al., 2004b). However, no relationship was found between the climates of source populations and the genetic effects on sideroxylonal concentrations in E. tricarpa (Andrew et al., 2007). Given that many eucalypts are distributed across diverse climatic and soil conditions, understanding this discrepancy is an important research goal.
Since the production of sideroxylonal appears not to incur a strong growth cost and variation in concentrations is highly heritable (Andrew et al., 2007), artificial selection for high sideroxylonal concentrations is likely to be successful and confer improved resistance to insect browsing, an important problem in re-establishing trees in farming landscapes in southern Australia (Ohmart and Edwards, 1991; Collett and Neumann, 2002; Floyd et al., 2002). However, genetic improvement of sideroxylonal concentrations would be more difficult if genetic parameters or the rankings of genotypes were to change when this species is grown in different conditions. This is of concern, as significant G × E interactions have been detected for growth and wood traits in studies of other Eucalyptus species grown on multiple sites (Tibbits and Hodge, 1998; Lima et al., 2000; Costa e Silva et al., 2006).
In this study, two aspects of the genetic basis of variation in a foliar defence chemical in Eucalyptus tricarpa were investigated, building on earlier work that connected sideroxylonal concentrations with insect herbivory and demonstrated population differentiation in this trait (Andrew et al., 2007). The study had three aims: (1) to compare the foliar sideroxylonal concentrations of material from different source populations grown in multiple environments; (2) to assess the stability of within-population genetic effects when families are grown at different sites; and (3) to test the homogeneity of quantitative genetic variance among populations.
MATERIALS AND METHODS
Provenance/progeny trial description
Three open-pollinated E. tricarpa provenance/progeny experiments were established by Forests New South Wales and the Victorian Department of Sustainability and the Environment as part of the Australian Low Rainfall Tree Improvement Group (ALRTIG) programme (Harwood et al., 2001). Each experiment was at a different location in south-eastern Australia. One of these, located near Culcairn in New South Wales, was sampled for foliar sideroxylonal chemistry and assessed for insect damage in late 2002 and the resulting data have been presented elsewhere (Andrew et al., 2007). This experiment was sampled again the following year, along with two additional experiments. The sites differed in soil properties observed at the time of planting (D. Stackpole, unpubl. res.; I. Johnson, unpubl. res.) and climatic variables obtained from the BIOCLIM computer program (Centre for Resource and Environmental Studies, Australian National University, Australian Capital Territory, Australia; Table 1). The climates at the three sites as shown in Table 1 are broadly similar: year-to-year variation within sites is probably greater than mean differences between sites. Nevertheless, Lake Tyers is typically less exposed to low winter minima than the other two sites. Each experiment was designed to consist of four complete replicates, with rows and columns as incomplete blocks and families represented by five-tree plots in each replicate. The experiments shared almost all families, which were sampled from 16 populations throughout the range of the species, from central-western Victoria to southern coastal New South Wales.
Table 1.
Site characteristics of E. tricarpa provenance/progeny trials
| Site | Culcairn | Huntly | Lake Tyers |
|---|---|---|---|
| Latitude (S) | 35°43′ | 36°37′ | 37°49′ |
| Longitude (E) | 146°56′ | 144°18′ | 148°06′ |
| Soil type | Loam and sandy loam with clay below 40 cm | Loam with clay below 20 cm | Sandy loam |
| Mean annual temperature (°C) | 15·2 | 14·8 | 14·5 |
| Temperature seasonality (% CV) | 36 | 33 | 23 |
| Annual precipitation (mm) | 613 | 529 | 759 |
| Precipitation seasonality (% CV) | 22 | 23 | 15 |
Climatic variables were obtained using BIOCLIM. Seasonality is measured as the coefficient of variation for the mean monthly temperatures and precipitation.
Sampling
As in a previous study (Andrew et al., 2007), we focused on the progeny of a set of eight populations (Fig. 1) and sampled the first two available progeny from each plot containing the selected families (5–11 per population). Sampling occurred in December 2003 (Huntly), January 2004 (Culcairn) and March 2004 (Lake Tyers) due to constraints on access to the trials. However, care was taken to sample leaves of similar ages at the different sites; new or expanding leaves and senescent leaves were avoided. Approx. 100 g of leaves were stored at −20 °C for later freeze-drying.
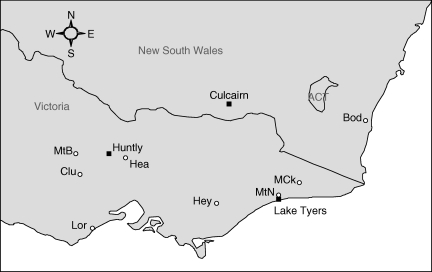
Fig. 1.
Map of south-eastern Australia showing experimental common-garden sites (filled squares) and source population locations (open circles). Population codes are as follows: Bod, Bodalla; Clu, Clunes; Hea, Heathcote; Hey, Heyfield; Lor, Lorne; MtB, Mt Bealiba; MtN, Mt Nowa Nowa; MCk, Martin's Ck.
Chemical analysis
Near infra-red spectroscopy (NIRS) was used to measure sideroxylonals in all 1487 samples, based on laboratory data from a set of calibration samples that included 60 randomly selected trees from each trial site. Laboratory and NIRS measurements of sideroxylonals followed the methods described in Wallis and Foley (2005) and Andrew et al. (2007), respectively. Sideroxylonals A and C were the dominant FPCs in the samples. Trace amounts of sideroxylonal B were detected, but concentrations were too low to be quantified reliably. Sideroxylonals A and C occurred in a constant ratio in the samples, as is seen in other eucalypts (Moore et al., 2004a), and we consider their combined concentration to be an accurate measure of the total FPCs in E. tricarpa. The optimal calibration for sideroxylonals A + C (hereafter referred to as sideroxylonal) was a modified partial least squares model based on 159 samples (excluding spectral outliers) and which used the entire spectrum (408–1092 and 1108–2492 nm). Various combinations of Savitzy–Golay derivative-based spectral smoothing functions provided by the ISI software (Win ISI; Port Matilda, PA, USA) were tested to remove unnecessary spectral signal components and improve the accuracy of the prediction models. The best model employed weighted multiplicative scatter correction and used the second derivative with a gap size of 6 nm, a maximal primary smoothing function and no secondary smoothing function (American Society for Testing and Materials, 1995). This model had an R2 of 0·979 and a standard error of cross-validation of 3·1 mg g−1 of dry matter (dm). The predicted mean and standard deviation were 22·9 and 14·4 mg.g−1 dm, respectively.
NIRS was very successful for predicting foliar sideroxylonal concentrations, which were approximately normally distributed. There were some suggestions of multimodality within populations; however, we considered our statistical analyses valid, as multimodality was not apparent in the residuals.
Statistical analysis
All statistical analysis was performed using GenStat10 (VSN International Ltd, Hemel Hempstead, UK). The experiments were row–column designs, with the nested genetic treatments (population and family within population) applied at the level of the five-tree plot. Replicates were complete blocks; however, only 64 of the approx. 110 plots in each replicate were sampled. Site was always treated as a fixed effect, as we did not consider the sites to be a random sample of the possible environments encountered by the species. Replicate within site was also treated as fixed to limit the number of random terms. Source population was treated as fixed, with families within populations random. Analyses were performed on individual-tree data and plot was included as part of the residual variance for the stratum at which the genetic treatments were applied. Since eucalypts have mixed mating systems, additive genetic variance (VA) was estimated from family variances, assuming a mean relatedness within families of 0·4 and no maternal, dominance or epistatic effects (Williams et al., 2002, p. 103).
Single-site analyses were conducted to compare variance components and fixed effects among sites and thereby assess the stability of the genetic variance estimates. Linear mixed models of data from all three sites were compared to test for G × E interactions and to investigate the sources of variance in chemical concentrations. Mixed models were fitted using residual maximum likelihood (REML) and likelihood ratio tests were performed to assess the significance of random effects (Payne, 2005). Residuals were examined for normality. Wald tests and F statistics were used to assess the significance of fixed effects. Population- or site-specific variances were estimated when likelihood ratio tests demonstrated significantly better fit. If this was not the case, a single variance component was estimated and the significance of the effect tested by dropping it from the model. The details of the linear models tested are given in the Appendix.
Bootstrapping was used as an additional test of the differences in family-level variance among populations. Bootstrapped data sets for each population were created by randomly selecting families (with replacement) from within the population and adding each family to the new data set with new plot identities. Confidence intervals were estimated from 10 000 bootstraps and compared among populations.
The stability of relative sideroxylonal concentrations across years was examined using type A genetic correlations (i.e. correlations among or between pairs of traits, Burdon, 1977; Williams et al., 2002) between the December 2002 and January 2004 data from Culcairn (see Appendix). Additive genetic correlations of sideroxylonal concentrations among sites were estimated using an approach similar to the estimation of more typical, type A genetic correlations, but where the trait of interest is measured on different individuals (Burdon, 1977). These ‘type B’ genetic correlations between sites are expected to be 1·0 in the absence of G × E interactions. The covariances of family effects among sites and variances within sites were estimated using REML and their significance tested using likelihood ratio tests (see Appendix for details). Correlations of population effects across sites were estimated using the predicted population means from the single-site analysis. In addition, Spearman rank correlations of family means were calculated and their confidence intervals estimated by bootstrapping in the R statistical package.
The relationship between the climates of the source populations and a simple measure of their plasticity was explored using univariate regressions. The concentration range for each population was measured as the signed difference in site-specific population mean between the maximum (at Huntly) and the minimum (at Lake Tyers). These values were regressed on each of 19 bioclimatic variables obtained from WORLDCLIM bioclimatic grids (Hijmans et al., 2005) as described previously (Andrew et al., 2007). False discovery rate was used to control for multiple independent tests (Benjamini and Hochberg, 1995).
Fain's test for major gene effects was carried out on family means and variances (Lynch and Walsh, 1998, p. 356). This test is based on the fact that family variance should have a linear relationship with the mean under an infinitesimal model of gene action, whereas genes with major effects are more likely to be homozygous in the families with extreme mean trait values. The presence of a significantly negative quadratic term in a quadratic regression is a conservative test for the presence of genes of major effect. This analysis was conducted on data from each site separately and on the entire data set after controlling for differences in site means.
RESULTS
Heritability at Huntly was somewhat lower than at other sites (Table 2) due primarily to increased σ2e at both the tree and plot level. Although there was substantially lower family variance at Lake Tyers, the measure of evolvability, CVA, was similar among sites, presumably due to mean-variance scaling.
Table 2.
Genetic parameter estimates for foliar sideroxylonal concentration measured at three sites
| Culcairn | Huntly | Lake Tyers | |
|---|---|---|---|
| n* | 505 | 489 | 493 |
| Random model | |||
| σf2 (s.e.)† | 21·41 (5·90) | 21·31 (7·37) | 12·55 (3·5) |
| σplot2 (s.e.)‡ | 6·48 (5·02) | 13·80 (8·19) | 5·90 (2·74) |
| σe2 (s.e.)§ | 62·99 (5·73) | 100·11 (9·12) | 31·99 (2·9) |
| Fixed model replicate (3 d.f.) | |||
| Wald statistic | 10·67 | 8·06 | 4·19 |
| χ2 prob.¶ | 0·014 | 0·045 | 0·242 |
| Fixed model population (7 d.f.) | |||
| Wald statistic | 118·32 | 76·91 | 84·48 |
| χ2 prob. | <0·001 | <0·001 | <0·001 |
| h2 (s.e.)** | 0·59 (0·13) | 0·39 (0·12) | 0·62 (0·14) |
| CVA†† | 48·61 | 48·5 | 37·21 |
* n: total number of trees included in the REML analysis.
† σf2: estimated variance component attributable to families within populations.
‡σplot2: residual plot-level variance component.
§ σe2: residual tree-level variance component.
¶ χ2 probability of Wald statistic and associated degrees of freedom.
** h2: average narrow-sense heritability.
†† CVA: the additive genetic coefficient of variance.
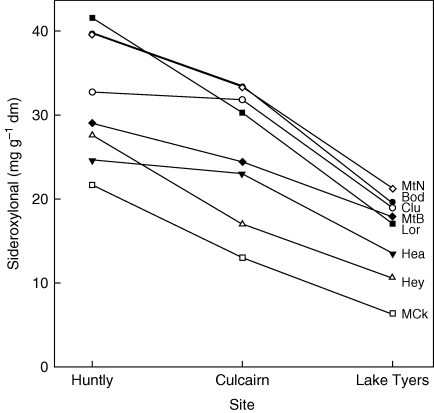
Despite significant site and family effects, the interaction between site and family was not significant (χ2, P = 1·0; Table 3), having been accounted for by the population-level G × E interaction, which was highly significant (Table 3). In general, however, its effect was considerably smaller than the population main effect and crossovers were not prevalent (Table 3, Fig. 2). Population means for foliar sideroxylonal concentration, estimated from single-site analyses as best linear unbiased estimates, were lower at Culcairn than at Huntly, and lowest at Lake Tyers.
Table 3.
Summary of multiple-site analysis
| VC (s.e.) | Probability (identity vs. absent) | Probability (diagonal vs. identity) | |
|---|---|---|---|
| (a) Random model | |||
| σf2 (s.e.) | <0·001 | 0·008 | |
| Bodalla | 19·88 (16·10) | ||
| Clunes | 1·09 (2·00) | ||
| Heathcote | 21·43 (12·21) | ||
| Heyfield | 13·65 (8·54) | ||
| Lorne | 10·76 (9·69) | ||
| Martin's Ck | 46·25 (22·41) | ||
| Mt Bealiba | 0·76 (1·89) | ||
| Mt Nowa Nowa | 8·27 (5·59) | ||
| σfs2 | 1·000 | 0·981 | |
| σplot2 (s.e.) | 6·73 (2·22) | 0·465 | |
| σe2 (s.e.) | <0·001 | ||
| Culcairn | 64·18 (4·87) | ||
| Huntly | 108·18 (7·80) | ||
| Lake Tyers | 31·07 (2·66) | ||
| (b) Fixed model | |||
| Wald statistic | d.f. | χ2 prob | |
| Site | 855·17 | 2 | <0·001 |
| site.replicate | 21·76 | 9 | 0·010 |
| population | 111·95 | 7 | <0·001 |
| site. population | 75·93 | 14 | <0·001 |
Variance components (VC) and tests of fixed effects are given. Likelihood ratio tests were used to test whether variance components should be included (identity vs. absent) and whether each term was best modelled as a single variance component or a separate variance for each level of an appropriate factor (i.e. with a covariance structure based on an identity matrix or a diagonal one). Standard errors are shown in parentheses after each estimate. The interaction between family and site was not significant. Residual variances also varied among sites.
Fig. 2.
Foliar concentrations of sideroxylonal at three trial sites. Lines connect the best linear unbiased estimates (BLUEs) of population means. Abbreviations for source populations are as in Fig. 1; dm = dry matter.
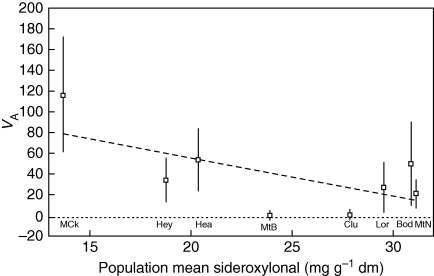
Family variance components differed among populations but not among sites (χ2, P = 0·32), and residual variances differed among sites (Table 3). While large, the bootstrap confidence intervals for Martin's Ck (95 % CI 12·6–203) and Heathcote (95 % CI 15·6–89·3) did not overlap that of Mt Bealiba (95 % CI 7·1 × 10−8–6·09). This was not due to mean-variance scaling, as there was no positive relationship of VA with population means; instead there was an weak negative trend, although only marginally significant (P = 0·079, Fig. 3).
Fig. 3.
Bootstrap mean (with s.d.) of population-specific additive genetic variance (VA, estimated as 2·5σ2f) by population mean. The linear regression line (y = 129 – 3·7x) explained 33 % of the variance in VA and was not significant (P = 0·079). Abbreviations for source populations are as in Fig. 1; dm = dry matter.
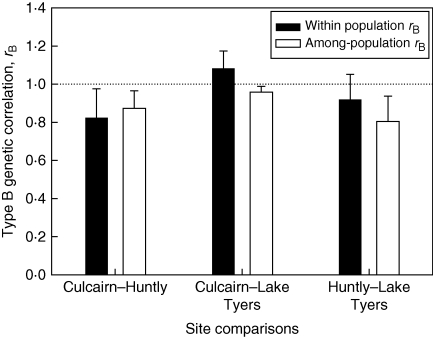
The genetic correlation of foliar sideroxylonal concentrations between years was high (n = 401, rA = 1·02, P < 0·001). Type B genetic correlations among sites at the family-within-population level were strong and highly significant (Fig. 4). They were also close to the upper limit of the parameter, at rB = 1·0. The correlations of population effects among sites were also strongly positive and highly significant (P < 0·001, Fig. 4). However, without correcting for population differences, the rank correlations of family means among sites were strongly positive and significantly different from 1 in each case (Culcairn–Huntly ρ = 0·75, CI = 0·60–0·84; Culcairn–Lake Tyers ρ = 0·85, CI = 0·73–0·91; Huntly–Lake Tyers ρ = 0·079, CI = 0·51–0·82). Pearson correlation coefficients were similar and also significantly less than 1.
Fig. 4.
Genetic correlation (type B) across sites at the family (within-population) and population level for single foliar chemical traits using analysis of plot means. Estimates from each pair of sites are shown, with s.e. Correlations are expected to differ from 1 (indicated by the dashed line) in the presence of genotype × environment interactions.
After correcting for false discovery rate, the difference in foliar sideroxylonal concentrations between Huntly and Lake Tyres was significantly related to five highly correlated temperature-related variables. Of these, the annual temperature range explained the largest proportion of variation in plasticity (Table 4). The univariate regressions suggested that populations with narrower diurnal and annual temperature ranges, higher minimums in the coldest period, lower maximums in the warmest period and less temperature seasonality tended to have a greater difference in sideroxylonal production between Huntly and Lake Tyers. The three populations with the highest means and phenotypic ranges all originate from coastal regions with smaller annual temperature ranges (Bodalla, Lorne and Mt Nowa Nowa; Figs 1 and 2).
Table 4.
Univariate regressions of the difference in mean sideroxylonal concentration between Huntly and Lake Tyers on bioclimatic variables
| Variable | Bioclimatic parameter (P) | Units | Regression slope | R2 | F | P |
|---|---|---|---|---|---|---|
| Annual mean temperature | 1 | °C | 0·29 | 0·10 | 1·81 | 0·23 |
| Mean diurnal temperature range | 2 | °C | −0·33 | 0·81 | 31·47 | 0·0014 |
| Isothermality* | 3 | % | 1·01 | 0·01 | 1·06 | 0·34 |
| Temperature seasonality† | 4 | %CV | −7·2E-03 | 0·84 | 36·92 | 0·0009 |
| Maximum temperature of warmest month | 5 | °C | −0·24 | 0·79 | 27·32 | 0·0020 |
| Minimum temperature of coldest month | 6 | °C | 0·32 | 0·77 | 25·01 | 0·0024 |
| Temperature annual range | 7 | °C | −0·15 | 0·90 | 60·95 | 0·0002 |
| Mean temperature of wettest quarter | 8 | °C | 0·054 | 0·10 | 1·80 | 0·23 |
| Mean temperature of driest quarter | 9 | °C | −0·088 | 0·13 | 2·05 | 0·20 |
| Mean temperature of warmest quarter | 10 | °C | −0·26 | 0·08 | 1·59 | 0·25 |
| Mean temperature of coldest quarter | 11 | °C | 0·32 | 0·67 | 15·25 | 0·0079 |
| Annual precipitation | 12 | mm | 0·013 | 0·04 | 1·29 | 0·30 |
| Precipitation of wettest month | 13 | mm | 0·102 | 0·03 | 1·20 | 0·31 |
| Precipitation of driest month | 14 | mm | 0·087 | −0·13 | 0·21 | 0·66 |
| Precipitation seasonality‡ | 15 | %CV | −0·094 | −0·15 | 0·09 | 0·78 |
| Precipitation of wettest quarter | 16 | mm | 0·047 | 0·07 | 1·50 | 0·27 |
| Precipitation of driest quarter | 17 | mm | 0·034 | −0·09 | 0·41 | 0·54 |
| Precipitation of warmest quarter | 18 | mm | 0·023 | −0·01 | 0·92 | 0·37 |
| Precipitation of coldest quarter | 19 | mm | 0·052 | −0·12 | 0·23 | 0·65 |
| Latitude | ° | 0·60 | −0·05 | 0·69 | 0·44 | |
| Longitude | ° | −2·69 | 0·05 | 1·34 | 0·29 | |
| Altitude | m | −0·033 | 0·55 | 9·51 | 0·022 |
Tests significant at the α = 0·05 level after correction for false discovery rate are shown in bold. Units for regression slopes are sideroxylonal mg g−1 of dry matter per °C, mm or %CV.
* Isothermality measured as (P2/P7) × 100.
† Coefficient of variation based on temperatures measured in K.
‡ Coefficient of variation based on temperatures measured in mm.
Application of Fain's test provided support for the notion that there are genes of major effect influencing foliar sideroxylonals. Variance within families was related to family mean with significant quadratic terms in three of the four tests conducted, in Culcairn (y = 72 – 262x – 166x2, Pquadratic = 0·018, r2adj = 0·22), Huntly (y = 113 – 154x – 214x2, Pquadratic = 0·028, r2adj = 0·08) and overall (y = 80 – 113x – 144x2, Pquadratic = 0·043, r2adj = 0·09). The regression was not significant for the Lake Tyers data set (P = 0·45).
DISCUSSION
Our study of foliar sideroxylonal concentrations in replicated open-pollinated E. tricarpa progeny experiments on multiple sites yielded three important results. (1) Within-population genetic variation is stable across sites and years. (2) Populations differ in their relative responses to each site, as a function of climatic conditions at their source locations. (3) Within-population genetic variances differ among populations and are likely to be influenced by genes of major effect. We will discuss the implications of these results for the genetic improvement of E. tricarpa in tree breeding programmes and the evolution of defence in Eucalyptus, in the broader context of population divergence.
Genetic improvement of E. tricarpa
Our results point to the stability of within-population genetic effects, as demonstrated by the insignificant site × family term and the high genetic correlation of foliar sideroxylonal concentrations between years at Culcairn and among sites. Thus, different genotypes from the same source population are likely to produce consistent responses to changes in environmental conditions, although populations differ in this response. In the only other multiple-site genetic study of herbivore resistance in eucalypts, the rank correlation of family effects on damage to E. grandis by chrysomelid beetles (Henery, 2006) at the two sites was significantly positive but <1 (rB = 0·64, s.e. = 0·17). This implies a small interaction effect between family and site, but is unlikely to have resulted from G × E interactions on phytochemistry because damage by chrysomelids was unrelated to FPC concentrations. Genetic correlations for wood traits across sites range from very low to close to 1 (Tibbits and Hodge, 1998; Hamilton et al., 2009).
While limited to the three sites studied, our finding of low or negligible G × E interactions at the family level has implications both for domestication and for evolutionary research on FPCs. The stable expression of genetic variation is favourable for the genetic improvement of defence in this species. Since the production of sideroxylonal appears not to incur a strong growth cost and variation in concentrations is highly heritable, breeding for high concentrations may be successful (Andrew et al., 2007). In contrast, if genetic parameters or the rankings of genotypes were to change when this species is grown in different conditions, it would be harder to breed for stable increases in concentrations of sideroxylonal across multiple environments. Significant G × E interactions for growth and some wood traits occur in other Eucalyptus species grown on multiple sites (Tibbits and Hodge, 1998; Lima et al., 2000; Costa e Silva et al., 2006; Hamilton et al., 2009). However, our results suggest that the within-population genetic parameters measured at one experiment or plantation site will predict those at a new site reasonably well.
In contrast, there was a significant G × E interaction in foliar sideroxylonal concentrations at the population level, which has not been documented previously for foliar chemical traits in Eucalyptus. A significant population × nutrient availability interaction effect was found on herbivory by common brushtail possums (Trichosurus vulpecula) on E. globulus grown in glasshouses, but the interaction with environment was not significant for FPCs or other chemical traits (O'Reilly-Wapstra et al., 2005). The finding of population × environment interactions for chemical defence indicates that populations differ in how they react to environments and is therefore a key result of the present study.
Although a significant population-level G × E interaction indicated that some populations had more stable levels of defence than others, it should not pose difficulties for artificial selection on sideroxylonal, as changes in rankings across sites were not common and the correlations of predicted population means were strong. The Bodalla and Mt Nowa Nowa progeny were highly ranked at each site. In a previous study of the Culcairn experiment, both of these populations had high growth rates and low levels of both insect damage and Anoplognathus Christmas beetle incidence (Andrew et al., 2007). Likewise, the low level of genetic variance in sideroxylonal concentrations in some populations is unlikely to impede genetic improvement of defence. The Clunes and Mt Bealiba material, which did not display detectable VA despite high average concentrations, may not contribute strongly to ongoing breeding programmes, but Mt Nowa Nowa and Bodalla possessed moderate VA. However, information on long-term survival and growth at multiple sites is required to determine whether these two provenances should be recommended for general use, because both are from the higher rainfall parts of the species' range.
In seeking explanations for this pattern, we found evidence for genes of a major effect influencing sideroxylonal concentration. Major quantitative trait loci for sideroxylonal were also detected in E. nitens (Henery, 2006), suggesting that the genetic architecture of chemical variation may be consistent among different species.
Ecology and evolution of chemical defence
Eucalypts are important foundation species, providing food and habitat for diverse vertebrate, invertebrate and microbial interactors, many of which respond to foliar chemical defences (Lawler et al., 1998; Jones et al., 2002; Marsh et al., 2003; Moore et al., 2005; Andrew et al., 2007). The stability of genetic effects across environments suggests that the evolution of eucalypt phytochemicals can play a role in structuring entire communities (Wimp et al., 2007; Barbour et al., 2009). The evolution of sideroxylonal concentrations appears unlikely to be constrained by G × E interactions; however, the lack of genetic variance for this trait in some populations may prevent adaptation to changing environments.
Populations of E. tricarpa clearly differ in levels of genetic variation in foliar sideroxylonal, independently of mean-variance scaling. Variance structure modelling is increasingly used in the analysis of multisite genetic experiments, as the residual variance may differ among sites (Costa e Silva et al., 2006), and this approach proved useful in detecting differences in family-level variance among source populations. The lack of genetic variance within certain populations suggests that these populations may be less capable of responding to natural or artificial selection on sideroxylonal, although the sampling may have missed some variation present in the population. Differentiation of genetic variances and covariances has been documented previously (Riemenschneider et al., 1994; Widen et al., 2002; Yeaman and Jarvis, 2006), but we are not aware of any similar studies for herbivore defence traits. While the likelihood of differences in the heritability of frost resistance among regions was considered by Tibbits et al. (1998), differences in the heritability or VA of quantitative traits among populations have not been formally demonstrated before in eucalypts. This is surprising because reduced genetic variation would be expected in small populations at the edges of species distributions (Butcher et al., 2009) or those that have experienced prolonged natural selection (Foster et al., 2007). The ranges of eucalypts can encompass considerable environmental variation, over which adaptive divergence can occur (Steane et al., 2006). Our results suggest that the validity of assuming homogeneity of variance should be questioned more frequently, although large sample sizes would be required to detect small differences in variance among populations. The high heritability of sideroxylonal has undoubtedly made the task easier in this study.
Our finding of evidence for gene(s) of major effect is noteworthy because it may help explain the differences in genetic variance among populations. While maternal trees were sampled at a minimum distance of 100 m, which is greater than the extent of positive spatial genetic autocorrelation in the closely related woodland tree, E. melliodora, correlation of sideroxylonal with reproductive traits (e.g. phenology or pollen production) may cause underestimation of genetic variance. In some cases, eucalypts receive pollen from only a subset of the potential pollen donors (Sampson, 1998; Millar et al., 2000; Jones et al., 2008). Although variation in the mating systems of the maternal trees (Borralho and Potts, 1996; Burgess et al., 1996; Butcher and Williams, 2002) may influence differences in variance among populations, it would not explain the apparent lack of genetic variance in some populations. Similarly, the pattern of genetic variance in E. tricarpa does not match that expected if genetic variation is lower near the range limits of species (Aitken and Libby, 1994; Eckstein et al., 2006). Range expansion, genetic drift, introgression, migration and selection may cause differentiation of genetic variances among populations under an infinitesimal model (Yeaman and Jarvis, 2006); these mechanisms are more likely to alter VA if sideroxylonal is influenced by genes of major effect. A combination of genomics and population genetics will be required to ascertain the importance of selection, genes of major effect and/or human disturbance in producing the observed differences in genetic variance among populations. The identification of a major gene affecting FPC concentrations in leaves may offer an unprecedented chance to understand the biosynthesis of this enigmatic class of phytochemicals.
Differences among populations in responses of foliar sideroxylonal concentrations to site environments also accompanied the population divergence in foliar sideroxylonal concentrations that we have documented previously (Andrew et al., 2007). There was a significant G × E interaction at the population level, which was related to the range of temperatures experienced by the source populations. In contrast to patterns of genetic divergence in morphological and life history traits in E. globulus (Dutkowski and Potts, 1999), relationships between bioclimatic variables and physiology or defence in E. tricarpa have been elusive (Andrew et al., 2007). Yet, higher concentrations of sideroxylonals have been found in populations of E. microcorys growing on sites with cold winters (Moore et al., 2004b). Our results suggest that this may be due to environmental as well as genetic effects, highlighting the need for studies in multiple common gardens to understand geographic patterns of phenotypic variation.
While it is unclear whether plasticity in sideroxylonal production is adaptive, or even whether it is active or passive (Pigliucci, 1996; Ghalambor et al., 2007), the link with source population climate suggests that the responses of foliar sideroxylonal concentrations to environmental conditions are ecologically important in this species. Adaptive divergence may be responsible for the differing responses of population means to each site and the lack of genetic variation in plasticity detected within populations, if selection on plasticity varies among source localities. For example, the trend of greater differences in sideroxylonal concentrations between Huntly and Lake Tyers exhibited by families from source populations that experience narrower temperature ranges may result from canalization (evolution of the ability to produce the same phenotype despite environmental perturbations) in inland populations or selection for greater plasticity in coastal populations. Because the inland populations experience greater temperature extremes, there may be greater selective benefit to canalization of foliar sideroxylonal concentration (Buskirk and Steiner, 2009; Lande, 2009).
Eucalypts produce a broad range of secondary chemicals, which can comprise a substantial portion of leaf biomass. FPCs appear to be constitutively expressed (Henery et al., 2008), in contrast to other eucalypt phytochemicals (Rapley et al., 2007) and many defences in other plants that are induced by herbivores or pathogens (e.g. Agrawal et al., 2002; Keeling and Bohlmann, 2006; Björkman et al., 2008). Nevertheless, ontogeny and the timing of phenotypic phase changes may differ between the sites and contribute to the observed population × environment interaction. For this reason, we must be cautious about interpreting the 2-fold difference in mean sideroxylonal concentrations between Huntly and Lake Tyers as suggesting that the strong plasticity of this trait on average has evolved in response to variable environments (Buskirk and Steiner, 2009; Lande, 2009). Strong, positive genetic and phenotypic correlations occurred between seasons, suggesting that the relative levels of defence are stable across years; however, this information was only available for one site. Further study of this species should include longitudinal measurements of growth, herbivory and sideroxylonal concentrations at multiple sites. In order to understand fully the geographic mosaic of genetic variation in foliar sideroxylonal concentrations, the costs and benefits in multiple habitats are of great importance.
In conclusion, our results suggest that geographic divergence in parameters other than population means may be more common than is typically acknowledged. Both environmental effects on phenotypes and genetic variance may differ among populations. This may affect the way in which species respond to rapid environmental changes through adaptive plasticity or evolution.
ACKNOWLEDGEMENTS
We thank Lake Tyers Aboriginal Trust, Forests Victoria, Forests NSW and the Altmeier family for access to the experiments. We thank Andras Keszei and Ben Moore for field assistance, Emlyn Williams for statistical support, and Loren Rieseberg and David Bush for comments on the manuscript. This work was supported by the Rural Industries Research and Development Corporation (RIRDC) (ANU 55A) and the Australian Research Council (LP0667708), and by the RIRDC and Australian Postgraduate Award scholarships to R.L.A.
APPENDIX
G × E interaction and variance component modelling
The generalized mixed model used to analyse sideroxylonal concentrations measured at the three trial sites is:
where X and Zi were incidence matrices for factor levels. b is the list of fixed effects, including site, site.replicate, population and population.site. f, c and p are vectors of the random effects for family.population, family.site and plot, and e is the vector of residual errors. The random effects in the model were assumed to be multivariate normal, with means, variances and covariances:
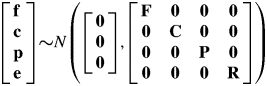 |
where 0 is the null matrix and F, C, P and R are the variance/covariance matrices corresponding to f, c, p and e, respectively. The structures of these matrices were modelled to test for differences among sites in residual and plot variances. For example, the simpler form of the residual variance model was a diagonal matrix, R = Inσ2e, where In is an n × n identity matrix of dimension and n is the number of individuals. This structure was compared with a more complex one,
where i denoted the trial sites, ni were the numbers of trees sampled at each site and ⊕ is the direct sum operation. In this model, the residual variances (σ2e,i for each site, i) were allowed to differ among sites, as described by Costa e Silva et al. (2004). Similarly, the variance due to families within populations was modelled as a single term, σ2f, or as a separate component, σ2f,j for each population, j, with
These models were specified with the VSTRUCTURE directive in GenStat and compared using likelihood ratio tests (Payne, 2005).
Genetic correlation between sampling years
Genetic correlations between trait measurements in December 2002 and January 2004 were estimated using multivariate REML in GenStat (Payne, 2005) to model the covariance among family effects for each year. Replicate and population were included as fixed effects and family was considered random. Unstructured covariance models were estimated for family, plot and residual effects, and the significance of the family-level covariance tested by comparison with a diagonal covariance structure for this term. Additive genetic correlations were estimated from the family variance and covariance components for years 1 and 2 as follows.
Genetic correlations among sites
Genetic correlations were estimated for each pair of sites, s and t, so that each correlation could be tested separately. Type B genetic correlations differ from type A genetic correlations in that traits are measured on different individuals, in this case at different sites. Because of this, the multivariate REML approach was not suitable. Instead, the covariances among sites for family effects were modelled using a covariance structure based on an unstructured matrix as follows.
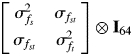 |
where ⊗ denotes the cross-product operation.
Since plots and individuals were not shared across sites, the plot and residual variance structures were again modelled as diagonal matrices, allowing for different variances among sites. The family covariance model was tested by likelihood ratio test with the simpler, diagonal structure,
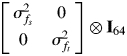 |
Type B genetic correlations were estimated from the family variance and covariance components. For each pair of sites, s and t:
Taylor series expansion was employed to estimate standard errors.
LITERATURE CITED
- Agrawal AA, Conner JK, Johnson MTJ, Wallsgrove R. Ecological genetics of an induced plant defense against herbivores: additive genetic variance and costs of phenotypic plasticity. Evolution. 2002;56:2206–2213. doi: 10.1111/j.0014-3820.2002.tb00145.x. [DOI] [PubMed] [Google Scholar]
- Aitken SN, Libby WJ. Evolution of the pygmy-forest edaphic subspecies of Pinus contorta across an ecological staircase. Evolution. 1994;48:1009–1019. doi: 10.1111/j.1558-5646.1994.tb05289.x. [DOI] [PubMed] [Google Scholar]
- American Society for Testing and Materials. Standard practices for infrared, multivariate, quantitative analysis. Designation: E1655-94. West Conshohocken, PA: American Society for Testing and Materials; 1995. [Google Scholar]
- Andrew RL, Wallis IR, Harwood CE, Henson M, Foley WJ. Heritable variation in the foliar secondary metabolite sideroxylonal in Eucalyptus confers cross-resistance to herbivores. Oecologia. 2007;153:891–901. doi: 10.1007/s00442-007-0784-1. [DOI] [PubMed] [Google Scholar]
- Barbour RC, O'Reilly-Wapstra JM, Little DWD, et al. A geographic mosaic of genetic variation within a foundation tree species and its community-level consequences. Ecology. 2009;90:1762–1772. doi: 10.1890/08-0951.1. [DOI] [PubMed] [Google Scholar]
- Bégin M, Roff DA. The constancy of the G matrix through species divergence and the effects of quantitative genetic constraints on phenotypic evolution: a case study in crickets. Evolution. 2003;57:1107–1120. doi: 10.1111/j.0014-3820.2003.tb00320.x. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Björkman C, Dalin P, Ahrné K. Leaf trichome responses to herbivory in willows: induction, relaxation and costs. New Phytologist. 2008;179:176–184. doi: 10.1111/j.1469-8137.2008.02442.x. [DOI] [PubMed] [Google Scholar]
- Boege K, Dirzo R. Intraspecific variation in growth, defense and herbivory in Dialium guianense (Caesalpiniaceae) mediated by edaphic heterogeneity. Plant Ecology. 2004;175:59–69. [Google Scholar]
- Borralho NMG, Potts BM. Accounting for native stand characteristics in genetic evaluations of open-pollinated progeny from a Eucalyptus globulus base population. New Forests. 1996;11:53–64. [Google Scholar]
- Bowers MD, Collinge SK, Gamble SE, Schmitt J. Effects of genotype, habitat, and seasonal variation on iridoid glycoside content of Plantago lanceolata (Plantaginaceae) and the implications for insect herbivores. Oecologia. 1992;91:201–207. doi: 10.1007/BF00317784. [DOI] [PubMed] [Google Scholar]
- Burdon RD. Genetic correlation as a concept for studying genotype–environment interaction in forest tree breeding. Silvae Genetica. 1977;26:168–175. [Google Scholar]
- Burgess IP, Williams ER, Bell JC, Harwood CE, Owen JV. The effect of outcrossing rate on the growth of selected families of Eucalyptus grandis. Silvae Genetica. 1996;45:97–100. [Google Scholar]
- Buskirk JV, Steiner UK. The fitness costs of developmental canalization and plasticity. Journal of Evolutionary Biology. 2009;22:852–860. doi: 10.1111/j.1420-9101.2009.01685.x. [DOI] [PubMed] [Google Scholar]
- Butcher PA, Williams ER. Variation in outcrossing rates and growth in Eucalyptus camaldulensis from the Petford Region, Queensland; evidence of outbreeding depression. Silvae Genetica. 2002;51:6–12. [Google Scholar]
- Butcher P, McDonald M, Bell J. Congruence between environmental parameters, morphology and genetic structure in Australia's most widely distributed eucalypt, Eucalyptus camaldulensis. Tree Genetics and Genomes. 2009;5:189–210. [Google Scholar]
- Cipollini DF, Busch JW, Stowe KA, Simms EL, Bergelson J. Genetic variation and relationships of constitutive and herbivore-induced glucosinolates, trypsin inhibitors, and herbivore resistance in Brassica rapa. Journal of Chemical Ecology. 2003;29:285–302. doi: 10.1023/a:1022673726325. [DOI] [PubMed] [Google Scholar]
- Collett NG, Neumann FG. Effects of simulated chronic defoliation in summer on growth and survival of blue gum (Eucalyptus globulus Labill.) within young plantations in northern Victoria. Australian Forestry. 2002;65:99–106. [Google Scholar]
- Conner JK, Franks R, Stewart C. Expression of additive genetic variances and covariances for wild radish floral traits: comparison between field and greenhouse environments. Evolution. 2003;57:487–495. doi: 10.1111/j.0014-3820.2003.tb01540.x. [DOI] [PubMed] [Google Scholar]
- Cooper M, DeLacy IH. Relationships among analytical methods used to study genotypic variation and genotype-by-environment interaction in plant breeding multi-environment experiments. Theoretical and Applied Genetics. 1994;88:561–572. doi: 10.1007/BF01240919. [DOI] [PubMed] [Google Scholar]
- Costa e Silva J, Borralho NMG, Potts BM. Additive and non-additive genetic parameters from clonally replicated and seedling progenies of Eucalyptus globulus. Theoretical and Applied Genetics. 2004;108:1113–1119. doi: 10.1007/s00122-003-1524-5. [DOI] [PubMed] [Google Scholar]
- Costa e Silva J, Potts BM, Dutkowski GW. Genotype by environment interaction for growth of Eucalyptus globulus in Australia. Tree Genetics and Genomes. 2006;2:61–75. [Google Scholar]
- Donaldson JR, Kruger EL, Lindroth RL. Competition- and resource-mediated tradeoffs between growth and defensive chemistry in trembling aspen (Populus tremuloides) New Phytologist. 2006;169:561–570. doi: 10.1111/j.1469-8137.2005.01613.x. [DOI] [PubMed] [Google Scholar]
- Dutkowski GW, Potts BM. Geographic patterns of genetic variation in Eucalyptus globulus ssp. globulus and a revised racial classification. Australian Journal of Botany. 1999;47:237–263. [Google Scholar]
- Eckstein RL, O'Neill RA, Danihelka J, Otte A, Kohler W. Genetic structure among and within peripheral and central populations of three endangered floodplain violets. Molecular Ecology. 2006;15:2367–2379. doi: 10.1111/j.1365-294X.2006.02944.x. [DOI] [PubMed] [Google Scholar]
- Floyd RB, Foley WJ. Identifying pest-resistant eucalypts using near-infrared spectroscopy. Canberra, Australia: Rural Industries Research and Development Corporation; 2001. RIRDC Publication No 01/112. [Google Scholar]
- Floyd RB, Farrow RA, Matsuki M. Variation in insect damage and growth in Eucalyptus globulus. Agricultural and Forest Entomology. 2002;4:109–115. [Google Scholar]
- Fornoni J, Valverde PL, Nunez-Farfan J. Quantitative genetics of plant tolerance and resistance against natural enemies of two natural populations of Datura stramonium. Evolutionary Ecology Research. 2004;5:1049–1065. [Google Scholar]
- Foster SA, McKinnon GE, Steane DA, Potts BM, Vaillancourt RE. Parallel evolution of dwarf ecotypes in the forest tree Eucalyptus globulus. New Phytologist. 2007;175:370–380. doi: 10.1111/j.1469-8137.2007.02077.x. [DOI] [PubMed] [Google Scholar]
- Ghalambor CK, McKay JK, Carroll SP, Reznick DN. Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Functional Ecology. 2007;21:394–407. [Google Scholar]
- Hamann A, Koshy MP, Namkoong G, Ying CC. Genotype × environment interactions in Alnus rubra: developing seed zones and seed-transfer guidelines with spatial statistics and GIS. Forest Ecology and Management. 2000;136:107–119. [Google Scholar]
- Hamilton M, Raymond C, Harwood C, Potts B. Genetic variation in Eucalyptus nitens pulpwood and wood shrinkage traits. Tree Genetics and Genomes. 2009;5:307–316. [Google Scholar]
- Harwood CE, Bulman P, Bush D, Mazanec R, Stackpole D. Australian Low Rainfall Tree Improvement Group: compendium of hardwood breeding strategies. Canberra: Rural Industries Research and Development Corporation; 2001. [Google Scholar]
- Henery M. Foliar secondary metabolites in Eucalyptus and their role in resistance to defoliating insects. Canberra, Australia: The Australian National University; 2006. [Google Scholar]
- Henery M, Wallis I, Stone C, Foley W. Methyl jasmonate does not induce changes in Eucalyptus grandis leaves that alter the effect of constitutive defences on larvae of a specialist herbivore. Oecologia. 2008;156:847–859. doi: 10.1007/s00442-008-1042-x. [DOI] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology. 2005;25:1965–1978. [Google Scholar]
- Hoffmann AA, Shirriffs J, Scott M. Relative importance of plastic vs genetic factors in adaptive differentiation: geographical variation for stress resistance in Drosophila melanogaster from eastern Australia. Functional Ecology. 2005;19:222–227. [Google Scholar]
- Johnson MTJ, Agrawal AA. Plant genotype and environment interact to shape a diverse arthropod community on evening primrose (Oenothera biennis) Ecology. 2005;86:874–885. [Google Scholar]
- Jones M, Shepherd M, Henry R, Delves A. Pollen flow in Eucalyptus grandis determined by paternity analysis using microsatellite markers. Tree Genetics and Genomes. 2008;4:37–47. [Google Scholar]
- Jones TH, Potts BM, Vaillancourt RE, Davies NW. Genetic resistance of Eucalyptus globulus to autumn gum moth defoliation and the role of cuticular waxes. Canadian Journal of Forest Research. 2002;32:1961–1969. [Google Scholar]
- de Jong G. Evolution of phenotypic plasticity: patterns of plasticity and the emergence of ecotypes. New Phytologist. 2005;166:101–118. doi: 10.1111/j.1469-8137.2005.01322.x. [DOI] [PubMed] [Google Scholar]
- Keeling CI, Bohlmann J. Genes, enzymes and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. New Phytologist. 2006;170:657–675. doi: 10.1111/j.1469-8137.2006.01716.x. [DOI] [PubMed] [Google Scholar]
- Keinänen M, Julkunen-Tiitto R, Mutikainen P, Walls M, Ovaska J, Vapaavuori E. Trade-offs in phenolic metabolism of silver birch: effects of fertilization, defoliation, and genotype. Ecology. 1999;80:1970–1986. [Google Scholar]
- Lande R. Adaptation to an extraordinary environment by evolution of phenotypic plasticity and genetic assimilation. Journal of Evolutionary Biology. 2009;22:1435–1446. doi: 10.1111/j.1420-9101.2009.01754.x. [DOI] [PubMed] [Google Scholar]
- Lawler IR, Foley WJ, Eschler BM, Pass DM, Handasyde K. Intraspecific variation in Eucalyptus secondary metabolites determines food intake by folivorous marsupials. Oecologia. 1998;116:160–169. doi: 10.1007/s004420050575. [DOI] [PubMed] [Google Scholar]
- Leinonen T, O'Hara RB, Cano JM, Merila J. Comparative studies of quantitative trait and neutral marker divergence: a meta-analysis. Journal of Evolutionary Biology. 2008;21:1–17. doi: 10.1111/j.1420-9101.2007.01445.x. [DOI] [PubMed] [Google Scholar]
- Lima JT, Breese MC, Cahalan CM. Genotype–environment interaction in wood basic density of Eucalyptus clones. Wood Science and Technology. 2000;34:197–206. [Google Scholar]
- Lynch M, Walsh B. Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- Marsh KJ, Foley WJ, Cowling A, Wallis IR. Differential susceptibility to Eucalyptus secondary compounds explains feeding by the common ringtail (Pseudocheirus peregrinus) and common brushtail possum (Trichosurus vulpecula) Journal of Comparative Physiology B: Biochemical, Systemic, and Environmental Physiology. 2003;173:69–78. doi: 10.1007/s00360-002-0318-4. [DOI] [PubMed] [Google Scholar]
- Millar MA, Byrne M, Coates DJ, Stukely MJC, McComb JA. Mating system studies in jarrah, Eucalyptus marginata (Myrtaceae) Australian Journal of Botany. 2000;48:475–479. [Google Scholar]
- Mitchell-Olds T, Rutledge JJ. Quantitative genetics in natural plant populations: a review of the theory. American Naturalist. 1986;127:379–402. [Google Scholar]
- Mitchell-Olds T, Schmitt J. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature. 2006;441:947–952. doi: 10.1038/nature04878. [DOI] [PubMed] [Google Scholar]
- Moore BD, Foley WJ. Tree use by koalas in a chemically complex landscape. Nature. 2005;435:488–490. doi: 10.1038/nature03551. [DOI] [PubMed] [Google Scholar]
- Moore BD, Wallis IR, Pala-Paul J, Brophy JJ, Willis RH, Foley WJ. Antiherbivore chemistry of Eucalyptus – cues and deterrents for marsupial folivores. Journal of Chemical Ecology. 2004a;30:1743–1769. doi: 10.1023/b:joec.0000042399.06553.c6. [DOI] [PubMed] [Google Scholar]
- Moore BD, Wallis IR, Wood JT, Foley WJ. Foliar nutrition, site quality, and temperature influence foliar chemistry of tallowwood (Eucalyptus microcorys) Ecological Monographs. 2004b;74:553–568. [Google Scholar]
- Moore BD, Foley WJ, Wallis IR, Cowling A, Handasyde KA. Eucalyptus foliar chemistry explains selective feeding by koalas. Biology Letters. 2005;1:64–67. doi: 10.1098/rsbl.2004.0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly-Wapstra JM, Potts BM, McArthur C, Davies NW. Effects of nutrient variability on the genetic-based resistance of Eucalyptus globulus to a mammalian herbivore and on plant defensive chemistry. Oecologia. 2005;142:597–605. doi: 10.1007/s00442-004-1769-y. [DOI] [PubMed] [Google Scholar]
- Ohmart CP, Edwards PB. Insect herbivory on eucalyptus. Annual Review of Entomology. 1991;36:637–657. [Google Scholar]
- Payne RW. The guide to Genstat release 8 part 2: statistics. Oxford, UK: VSN International; 2005. [Google Scholar]
- Pigliucci M. How organisms respond to environmental changes: from phenotypes to molecules (and vice versa) Trends in Ecology and Evolution. 1996;11:168–173. doi: 10.1016/0169-5347(96)10008-2. [DOI] [PubMed] [Google Scholar]
- Pigliucci M, Murren CJ, Schlichting CD. Phenotypic plasticity and evolution by genetic assimilation. Journal of Experimental Biology. 2006;209:2362–2367. doi: 10.1242/jeb.02070. [DOI] [PubMed] [Google Scholar]
- Pressoir G, Berthaud J. Population structure and strong divergent selection shape phenotypic diversification in maize landraces. Heredity. 2004;92:95–101. doi: 10.1038/sj.hdy.6800388. [DOI] [PubMed] [Google Scholar]
- Rapley L, Allen G, Potts B, Davies N. Constitutive or induced defences – how does Eucalyptus globulus defend itself from larval feeding? Chemoecology. 2007;17:235–243. [Google Scholar]
- Riemenschneider DE, McMahon BG, Ostry ME. Population-dependent selection strategies needed for 2-year-old black cottonwood clones. Canadian Journal of Forest Research. 1994;24:1704–1710. [Google Scholar]
- Rosner S, Hannrup B. Resin canal traits relevant for constitutive resistance of Norway spruce against bark beetles: environmental and genetic variability. Forest Ecology and Management. 2004;200:77–87. [Google Scholar]
- Sampson JF. Multiple paternity in Eucalyptus rameliana (Myrtaceae) Heredity. 1998;81:349–355. [Google Scholar]
- Silfver T, Roininen H, Oksanen E, Rousi M. Genetic and environmental determinants of silver birch growth and herbivore resistance. Forest Ecology and Management. 2009;257:2145–2149. [Google Scholar]
- Sork VL, Stowe KA, Hochwender C. Evidence for local adaptation in closely adjacent subpopulations of northern red oak (Quercus rubra L.) expressed as resistance to leaf herbivores. American Naturalist. 1993;142:928–936. doi: 10.1086/285581. [DOI] [PubMed] [Google Scholar]
- Steane D, Conod N, Jones R, Vaillancourt R, Potts B. A comparative analysis of population structure of a forest tree, Eucalyptus globulus (Myrtaceae), using microsatellite markers and quantitative traits. Tree Genetics and Genomes. 2006;2:30–38. [Google Scholar]
- Stenberg J, Witzell J, Ericson L. Tall herb herbivory resistance reflects historic exposure to leaf beetles in a boreal archipelago age-gradient. Oecologia. 2006;148:414–425. doi: 10.1007/s00442-006-0390-7. [DOI] [PubMed] [Google Scholar]
- Stiling P, Rossi AM. Complex effects of genotype and environment on insect herbivores and their enemies. Ecology. 1996;77:2212–2218. [Google Scholar]
- Tibbits W, Hodge G. Genetic parameters and breeding value predictions for Eucalyptus nitens wood fiber production traits. Forest Science. 1998;44:587–598. [Google Scholar]
- Van Tienderen PH, Koelewijn HP. Selection on reaction norms, genetic correlations and constraints. Genetics Research. 1994;64:115–125. doi: 10.1017/s0016672300032729. [DOI] [PubMed] [Google Scholar]
- Wallis IR, Foley WJ. The rapid determination of sideroxylonals in Eucalyptus foliage by extraction with sonication followed by HPLC. Phytochemical Analysis. 2005;16:49–54. doi: 10.1002/pca.810. [DOI] [PubMed] [Google Scholar]
- West-Eberhard MJ. Developmental plasticity and the origin of species differences. Proceedings of the National Academy of Sciences, USA. 2005;102:6543–6549. doi: 10.1073/pnas.0501844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westcott B. Some methods of analysing genotype–environment interaction. Heredity. 1986;56:243–253. [Google Scholar]
- Widen B, Andersson S, Rao GY, Widen M. Population divergence of genetic (co)variance matrices in a subdivided plant species, Brassica cretica. Journal of Evolutionary Biology. 2002;15:961–970. [Google Scholar]
- Williams ER, Matheson AC, Harwood CE. Experimental design and analysis for tree improvement. 2nd edn. Collingwood, Australia: CSIRO Publishing; 2002. [Google Scholar]
- Wimp GM, Wooley S, Bangert RK, et al. Plant genetics predicts intra-annual variation in phytochemistry and arthropod community structure. Molecular Ecology. 2007;16:5057–5069. doi: 10.1111/j.1365-294X.2007.03544.x. [DOI] [PubMed] [Google Scholar]
- Yeaman S, Jarvis A. Regional heterogeneity and gene flow maintain variance in a quantitative trait within populations of lodgepole pine. Proceedings of the Royal Society B: Biological Sciences. 2006;273:1587–1593. doi: 10.1098/rspb.2006.3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangerl AR, Berenbaum MR. Phenotype matching in wild parsnip and parsnip webworms: Causes and consequences. Evolution. 2003;57:806–815. doi: 10.1111/j.0014-3820.2003.tb00292.x. [DOI] [PubMed] [Google Scholar]