Abstract
Background and Aims
Despite extensive study of polyploidy, its origin, and ecogeographical differences between polyploids and their diploid progenitors, few studies have addressed ploidy-level structure and patterns of ecogeographical differentiation at various spatial scales using detailed sampling procedures. The pattern of coexistence of polyploids in the geophyte Allium oleraceum at the landscape and locality scale and their ecology were studied.
Methods
Flow cytometry and root-tip squashes were used to identify the ploidy level of 4347 plants from 325 populations sampled from the Czech Republic using a stratified random sampling procedure. Ecological differentiation among ploidy levels was tested by comparing sets of environmental variables recorded at each locality.
Key Results
Across the entire sampling area, pentaploids (2n = 5x = 40) predominated, while hexaploids (2n = 6x = 48) and tetraploids (2n = 4x = 32) were less frequent. The distribution of tetra- and hexaploids was partially sympatric (in the eastern part) to parapatric (in the western part of the Czech Republic) whereas pentaploids were sympatric with other cytotypes. Plants of different ploidy levels were found to be ecologically differentiated and the ruderal character of cytotypes increased in the direction 4x → 5x → 6x with the largest realized niche differences between tetra- and hexaploids. Most populations contained only one ploidy level (77 %), 22 % had two (all possible combinations) and 1 % were composed of three ploidy levels. The majority of 4x + 5x and 5x + 6x mixed populations occurred in sympatry with uniform populations of the participating cytotypes in sites with ecologically heterogeneous or marginal environment, suggesting secondary contact between cytotypes. Some mixed 4x + 6x populations dominated by tetraploids being sympatric and intermixed with uniform 4x populations might represent primary zones of cytotype contact. Almost no mixed accessions were observed on the fine spatial scale in mixed populations.
Conclusions
The results provide evidence for adaptive differences among ploidy levels, which may contribute to their complex distribution pattern. The prevalence of asexual reproduction, limited dispersal and equilibrium-disrupting processes may support local coexistence of cytotypes.
Keywords: Allium oleraceum, contact zones, Czech Republic, ecological differentiation, distribution, DNA ploidy level, ploidy mixture, polyploidy, spatial scales
INTRODUCTION
Polyploidy is a highly dynamic process that plays a major role in the evolution and speciation of angiosperms, and a significant role in the evolutionary history of other eukaryotes (Grant, 1981; Thompson and Lumaret, 1992; Wendel, 2000; Soltis et al., 2003). Previous studies have estimated the proportion of polyploids within angiosperms at about 50 % (Müntzing, 1936; Grant, 1981). Later studies suggested that at least 70 % of angiosperms are of polyploid origin (Goldblatt, 1980; Masterson, 1994). Recent genomic studies suggest that perhaps all eukaryotes possess genomes with gene redundancy, much of which is the result of past genome duplication. Most plants have probably undergone polyploidization followed by diploidization through genomic rearrangements, gene silencing and gene divergence (Wendel, 2000; Soltis et al., 2003). Molecular data support the recurrent origin of many polyploids (e.g. Soltis and Soltis, 1993, 1999, 2000; Segraves et al., 1999; Soltis et al., 2003; Guo et al., 2005). Different genotypes resulting from independent polyploidization events come into contact and afford the opportunity for recombination and production of new genotypes. Recurrent formation and gene flow between progenitors and polyploids enrich the gene pool of polyploids (Soltis and Soltis, 1995).
One of the prerequisites for polyploid research is knowledge of the geographical distribution of cytotypes. Distribution data can offer insight into the extent of reproductive isolation between cytotypes and the mechanisms responsible for their spatial separation (Baack, 2004; Suda et al., 2007a, b; Kao, 2008). Such data may serve as a foundation for answering questions about polyploid origins and exploring the history of contemporary distribution patterns using molecular techniques (van Dijk and Bakx-Schotman, 1997; Segraves et al., 1999).
Two scenarios have been proposed to explain differences in patterns of cytotype distribution. According to the adaptive evolutionary scenario, novel genetic combinations can produce novel characters that endow polyploids with new responses to environmental conditions (Levin, 1983, 2002; Soltis and Soltis, 1993; Otto and Whitton, 2000; Soltis et al., 2003). Consequently, polyploid populations often expand to wider or different ranges of habitats relative to those of their progenitors (e.g. Rothera and Davy, 1986; Bayer and Stebbins, 1987; Lumaret et al., 1987; Jay et al., 1991; Petit and Thompson, 1999; Petit et al., 1999; Levin, 2002; Johnson et al., 2003; Soltis et al., 2003). Ecological sorting along environmental gradients usually results in spatial separation of polyploids and their ancestors, a phenomenon observed in many plant species (Stebbins, 1950; Lewis, 1980). Such spatial relationships between cytotypes may be of several types, ranging from sympatry to parapatry or even allopatry (Levin, 2002). Accordingly, the coexistence of sympatric cytotypes requires niche differentiation and highly localized spatial patterns of habitat differentiation between cytotypes (Thompson and Lumaret, 1992). In the case of parapatry, contact zones (= ecotones) are maintained by divergent selection pressures along some gradient, often with selection against parental types in non-native environments (Barton and Hewitt, 1985; Fritsche and Kaltz, 2000).
Although ecological sorting along abiotic and biotic environmental gradients has been considered as the main mechanism underlying spatial separation between polyploids and their diploid ancestors and between cytotypes within polyploid complexes (Endler, 1977; Levin, 2002), alternative, environmentally independent explanations (‘non-adaptive scenarios’) also exist. Spatial separation between cytotypes may be directed by frequency-dependent mating success that results from low fitness of hybrids formed from between-cytotype matings and which gradually leads to the elimination of the minority cytotype from the population (‘minority cytotype exclusion model’; Levin, 1975; Fowler and Levin, 1984; Ramsey and Schemske, 1998). As a consequence, this model predicts that most populations should be cytologically uniform and that cases of multiple coexisting cytotypes represent transient situations following frequent generation or immigration of an alternative cytotype (Kao, 2007). Although this process was originally considered for primary hybrid zones (sensu Petit et al., 1999), an analogous process can occur across zones of secondary contacts between cytotypes when dispersal leads to mixed cytotype populations (Dorken and Pannell, 2007).
Differences in present-day distribution among cytotypes may also reflect history, i.e. the place of origin or past environmental heterogeneity. Widespread cytotypes may have been superior colonizers of areas which became available upon amelioration of the climate after the last ice age (Pleistocene) or due to human activities such as deforestation and agricultural use (Levin, 1983; Stebbins, 1985; Gornall and Wentworth, 1993; Xie-Kui et al., 2008). Alternatively, such distribution patterns may be explained non-adaptively through the position of past cytotype refuges relative to the sites which became available for colonization by single cytotypes (van Dijk et al., 1992; van Dijk and Bakx-Schotman, 1997; Štěpánková, 2001; Mandáková and Münzbergová, 2006).
The increasing number of diploid–polyploid contact zones studied, accelerated by the introduction of flow cytometric techniques, allowing more samples to be analysed than previously possible by classical chromosome counting, has revealed that mixed cytotype populations are much more frequent than anticipated (e.g. Keeler, 1990, 1998; Burton and Husband, 1999; Suda, 2002; Suda et al., 2004, 2007a; Halverson et al., 2008; Kao, 2008). This influx of data is challenging and altering our knowledge of the establishment, persistence and distribution of polyploids (Kron et al., 2007; Suda et al., 2007b). More recent models investigating the role of several mechanisms that increase the probability of polyploids evading minority cytotype exclusion have shown (Felber, 1991; Rodriguez, 1996; Felber and Bever, 1997) that when diploids produce relatively high frequencies of unreduced gametes or when cytotypes differ in fitness, fecundity, longevity and level of self-compatibility (but see Mable, 2004), polyploids can become established and maintained in the populations. Husband (2004) showed in computer simulations evaluating the effect of triploids on autotetraploid evolution in Chamerion angustifolium that partially fit triploids can increase the likelihood of diploid–tetraploid coexistence, and in some cases they can facilitate tetraploid fixation. In addition, the evolution of assortative mating – attained by a variety of factors such as divergence in flowering time or differences in pollinators (Fowler and Levin, 1984; van Dijk and Bijlsma, 1994; Segraves and Thompson, 1999), iteroparity, parthenogenesis (Bierzychudek, 1985; Yamauchi et al., 2004; Kao, 2007, 2008) or short-distance pollen and seed dispersal (Li et al., 2004; Baack, 2005) – might suffice to allow coexistence of cytotypes.
Numerous studies have examined ploidy-level structure and patterns of ecogeographical differentiation at various spatial scales in established polyploid complexes (Chmielewski and Semple, 1983; Stutz and Sanderson, 1983; Rothera and Davy, 1986; Lumaret et al., 1987; Keeler, 1990; Brochmann and Elven, 1992; Hroudová and Zákravský, 1993; Burton and Husband, 1999; McArthur and Sanderson, 1999; Hardy et al., 2000; Suda, 2002; Weiss et al., 2002; Johnson et al., 2003; Stuessy et al., 2004; Baack, 2004, 2005; Suda et al., 2004, 2007a; Mandáková and Münzbergová, 2006; Halverson et al., 2008; Kao, 2008; Mráz et al., 2008; Španiel et al., 2008; Xie-Kui et al., 2008; Kolář et al., 2009), but many of these studies were based on rough measures of the environment and small population samples. In consequence, ecogeographical patterns observed vary widely when multiple data are compared (Mable, 2003). There is a need for a quantitative approach (Johnson et al., 2003; Halverson et al., 2008), and this may be particularly important for detecting less obvious cases of habitat differentiations, as in ploidy variation within species (Lewis, 1980).
Allium oleraceum (Alliaceae), a bulbous geophyte distributed in most of Europe (Stearn, 1980; Hultén and Fries, 1986), is a polyploid complex comprising four documented ploidy levels ranging from triploids to hexaploids (2n = 24, 32, 40, 48; see Table 1). Low generative reproduction is a common feature of all the ploidy levels, but there is significant asexual reproduction by means of daughter bulbs and vegetative bulbils within the inflorescence (Duchoslav, 2000; Karpavičienė, 2002; Åström and Hæggström, 2004). A review of published A. oleraceum chromosome counts across Europe shows (Table 1) that, except for four instances, papers report only a single ploidy level per population with tetraploids and pentaploids being the most frequently reported cytotypes in Europe. Fialová (1996) and Karpavičienė (2007), using few population samples, observed the occasional co-occurrence of penta- and hexaploids and tetra- and pentaploids in the Czech Republic and Lithuania, respectively. The occurrence of three of four known ploidy levels and evidence for the unusual co-occurrence of penta- and hexaploids in the Czech Republic make this area very suitable for exploring patterns of ecogeographical distribution and the frequency of coexistence of different cytotypes. Understanding the factors responsible for the distribution of the cytotypes in this region may offer excellent opportunities to gain deeper knowledge of the ecological and evolutionary significance of chromosome number variation within this polyploid complex. There are two additional reasons for choosing this species as a model. First, A. oleraceum is relatively common throughout the whole of central Europe, so studies can be based on extensive sampling, and distribution patterns can therefore be insensitive to random fluctuations (see also results on another common species, Pilosella officinarum – Mráz et al., 2008). Secondly, the limited possibility of seed reproduction and, for that reason, low dispersion ability over larger distances, is another advantage of this species, as it conserves distribution patterns.
Table 1.
Summary of the ploidy levels found in Allium oleraceum in Europe according to the literature and present records
* Occurrence of cytotype-mixed populations was reported in the source.
Here a large sampling study of A. oleraceum populations and their environment at two spatial scales in the Czech Republic (central Europe) was performed. The following questions were addressed. (1) What are the frequencies and distribution patterns of plants of different ploidy levels at the landscape and local scales? (2) Are there any ecological differences between ploidy levels allowing for interpretation of the observed distribution pattern as a result of environmentally dependent selection? (3) Do populations with cytotype mixtures exist? And if so, (4) can their composition and ecogeographical distribution allow the inference of their mode of origin (primary or secondary hybrid zones or contact zones without inter-cytotype gene flow)? (5) Are such mixed populations dominated by a single cytotype, which might suggest the ‘minority cytotype exclusion’ effect?
MATERIALS AND METHODS
Study species
Allium oleraceum L. is a bulbous geophyte occurring in most of Europe (Meusel et al., 1965). It is distributed throughout western, central and eastern Europe and in southern Scandinavia. In the Czech Republic, the species is quite common and its distribution is concentrated between 300 and 500 m a.s.l. (Duchoslav, 2001a). It grows in a wide range of natural and human-influenced habitats, ranging from rocky grounds and dry grasslands through field margins and road ditches to scrub and deciduous forests (Duchoslav, 2001a, b, 2009; Karpavičienė, 2004; Hæggström and Åström, 2005).
The plant has one to four leaves. They are linear to filiform, fistular in the lower part and sheathing the bottom half of the scape. The terminal bulb in non-flowering plants and the major offset bulb in flowering plants replace the parent bulb at the end of the growing season. The plants rarely form non-dormant daughter bulbs. Sexually mature plants produce a lax umbel with a few hermaphroditic protandrous flowers (0–20) and many bulbils (10–60) at the top of the scape. Each flower can produce up to six seeds (Stearn, 1980), but in practice seed production varies greatly and seedling establishment is low (Duchoslav, 2000; Karpavičienė, 2002; Åström and Hæggström, 2004).
The origin of the A. oleraceum polyploid complex is still unclear. Levan (1938) considered A. oleraceum to be an autopolyploid form of diploid Allium paniculatum that arose by somatic doubling of the chromosomes, although he did not rule out fusion of unreduced gametes as an alternative. An autopolyploid origin of the species was proposed by Pastor (1982) and Fialová (1996). Vosa (1976), using the C-banding technique, stated that tetraploid plants of A. oleraceum are of allopolyploid origin. Careful analysis of Levan's paper shows that the ‘synthetic’ A. oleraceum obtained therein by crossing plants from two distant diploid populations [one being ‘A. paniculatum’ from the botanical garden in Cluj-Napoca (Romania) and the other A. podolicum from the botanical garden in Stockholm] may actually be an interspecific hybrid formed by unreduced gametes. It is not clear whether the Romanian plant was a true specimen of A. paniculatum or another member of this group – there are at least two other native species in Romania, namely A. fuscum and A. fussii (Brullo et al., 1996). Also, the second parent is not certain, and the name suggests a Ukrainian origin. Russian and Ukrainian authors (Omelchuk-Myakushko, 1979; Dobrotchaeva et al., 1999; Seregin, 2005) distinguish three species within this group (A. paniculatum, A. podolicum and A. praescissum) occurring in Ukraine.
Study area and sampling procedure
The present research was carried out in the Czech Republic (78 865 km2), which is covered by a heterogeneous cultural landscape of arable fields, broadleaved and coniferous forests, and human settlements. Its western part (the Bohemian massif) has a Palaeogene relief of rolling plains, hills and plateaus surrounded by the densely forested Hercynian mountains. The younger areas comprise river canyons and areas with Tertiary volcanism. Mostly acidic Variscan regions were later covered by Permo-Carboniferous and Mesozoic sediments. Base-rich bedrocks are concentrated in the lower altitudes. The eastern part of the Czech Republic is the flat northernmost projection of the Pannonnian basin and is surrounded by the western slopes of the Carpathian mountains. Relief here is of Tertiary age. The bedrock is more diverse than in the west, being mostly of Mesozoic and Tertiary origin. Calcium-rich substrates occur from the lowlands to the mountains. The vegetation cover has a more fine-grained distribution in comparison with the Bohemian massif (Ložek, 1988).
Samples of A. oleraceum were collected throughout the Czech Republic during early spring from 2001 to 2004. A stratified random sampling procedure was used to sample populations and subsamples within populations. The Czech Republic was divided according to a road atlas (Hlaváček, 2000) into 144 quadrats, each with an area of approx. 6·0 × 9·3 km. Within each quadrat, we randomly searched for two populations and within each sampled population all plants in at least five randomly placed subsamples were sampled, each with an area of approx. 30 × 30 cm. To take into account habitat diversity and variation in population density (Duchoslav, 2001a), additional rules concerning the sampling procedures were defined. (1) In each quadrat one population from a natural habitat and one from an human-impacted one (see ‘Habitat and population characteristics’ below for explanations) were sampled (if available). (2) The minimum distance between two populations from the same type of habitat was specified as 10 km. (3) The minimum distance between sampled subsamples was (if available) specified as 1 m. The standard sampling procedure (i.e. number of populations per quadrat and number of subsamples per population) was modified in some cases to reflect population size, population density at the landscape level and habitat variation within quadrats (number of sampled populations per quadrat: mean 2·27, s.d. 1·15, minimum 1, maximum 8). Samples were transported to and planted in the garden of the Palacký University in Olomouc, Czech Republic. In total, 325 populations and 4481 plants of A. oleraceum were collected in the field (number of sampled plants per population: mean 13, s.d. 4, minimum 3, and maximum 32). The early spring was chosen as the sampling time because at that time of year even populations consisting of non-flowering plants are easily recognizable in the field. Later in the year, when other plants grow as well, it is difficult to find A. oleraceum, and vegetative plants usually disappear during June (Duchoslav, 2009).
Habitat and population characteristics
Initially, the habitat of each population was investigated in the field. The following set of primary variables (see Appendix for a survey) was recorded at each locality. (1) Habitat type was assessed in the field according to EUNIS habitat classification (Davies et al., 2004). Because of the low frequency of some habitats in the sample, they were translated here into one of seven common habitat types (rock; steppe; mesic & wet grassland; semi-natural forest; ruderal scrub; planted Robinia pseudacacia forest; arable field & field margins). Correspondence between this and EUNIS habitat classifications is explained in the Appendix. (2) Light conditions were assessed in the field according to the visually estimated proportion of full sunlight reaching the ground during late spring (1 = strong shade, 2 = half-shade, 3 = low shade, 4 = full insolation). (3) Populations were classified into two categories according to their distance to the nearest arable field (‘Presence of arable land’; 0 = distance to the nearest field > 20 m, 1 = distance to the nearest field ≤ 20 m). (4) Populations were classified into two categories according to the degree of anthropogenic impact [‘Habitat naturalness’; 0 = vegetation strongly influenced or created by humans, typically with higher proportions of ruderal or alien species (‘human-impacted’), 1 = natural and semi-natural vegetation without strong anthropogenic influence (‘natural’)]. Examples of ‘human-impacted’ vegetation represent natural forests with ruderal or alien species, woody vegetation outside forest, intensively managed or disturbed grasslands, etc.
A secondary data set of geographical characteristics of the sites was obtained from tourist maps and from a digitized database of climatic parameters as follows: (1) altitude was estimated using 1 : 50 000 tourist maps (SHOCart, Inc., Zádveřice, Czech Republic), and (2) each locality was classified into one of three categories according to prevailing climatic conditions (‘Climatic region’; C = cold region, SW = slightly warm region, W = warm region; Quitt, 1971).
Soil samples (topsoil, 5–10 cm) were taken from the sites during field sampling. Soil samples were passed through a 2-mm sieve. Soil pH was measured in water suspension potentiometrically. The oxidizable carbon concentration (C) was determined by oxidation with potassium dichromate in sulphuric acid. Oxidative mixture redundancy was determined via volumetry with Mohr's salt (Zbíral, 1995). Organic nitrogen concentrations (N) were determined after mineralization with sulphuric acid, conversion to ammonium ions and subsequent distillation with water vapour (Zbíral, 1997) on a Kjeltec System Instrument (TECATOR; FOSS, Inc., Hillerød, Denmark). Phosphorus pentoxide concentrations (PO43−) were determined after extraction in Mehlich II solution (Mehlich, 1978) using a DR 2000 spectrophotometer. Determination of metallic cation (Ca, Mg, K) concentrations in soil samples were made after extraction in Mehlich II solution using an AVANTA atomic absorption spectrometer.
The size of each population was assessed visually on an ordinal scale (less than 50, 51–500, more than 500 individuals) and population area was estimated in square metres. The spatial pattern of individuals within each population (‘Morphological pattern’) was described according to the prevailing type observed in situ (i.e. either separate individuals or clumping).
Chromosome counts
Chromosome counts were obtained from somatic mitotic cells in root-tip cuttings of pot-cultivated plants. Plants for chromosome counts were raised in a greenhouse. After 2 weeks, the root tips were excised and pre-treated with a 0·5 % solution of colchicine at room temperature for 3 h, fixed in a cold mixture of ethanol and acetic acid (3 : 1) overnight and then stored at 4 °C in 70 % ethanol until use. The fixed root tips were hydrolysed in cold 5 m HCl, stained with Feulgen and squashed in 45 % acetic acid (Lillie, 1951). Chromosomes were counted using an Olympus CX-31 light microscope.
Estimation of DNA ploidy levels
DNA ploidy levels (Suda et al., 2006) were measured from most of the surviving plants (n = 4347, i.e. 97 % of all sampled plants) using flow cytometry. Leaf tissue of analysed Allium plant(s) with an appropriate volume of the internal reference standard (Triticum aestivum ‘Saxana’) were chopped with a new razor blade in a Petri dish containing 1 mL LB01 buffer (Doležel et al., 1989). The suspension was filtered through a 42-μm nylon mesh and the samples were stained with DAPI (final concentration 2 µg mL−1). The relative fluorescence intensity of stained nuclei was analysed using a Partec PAS instrument (Partec GmbH, Münster, Germany) equipped with an HBO-100 mercury arc lamp. Histograms of fluorescence intensity were registered over 512 channels. In each sample, 1000–2000 nuclei of both the standard and the test plant G1 peaks were analysed. The gain of the instrument was adjusted so that the G1 peak of wheat was approximately on channel 50. The ploidy level of each sample was determined by the position of its G1 peak relative to the G1 peak of an internal standard. Known tetra-, penta- and hexaploid plants with known chromosome counts were used for the specification of internal standard-sample position. The ratios between the positions of sample and internal reference standard peaks were 2·4–2·6, 2·8–3·0 and 3·3–3·4 for tetraploids, pentaploids and hexaploids, respectively. DAPI staining yielded histograms with coefficients of variance (CV) below 5 % for both the standard and the sample in the majority of DNA-ploidy measurements (mean standard CV = 4·48 %, s.d. = 0·94; mean sample CV = 4·67 %, s.d. = 0·82). In total, 99·9 % of the surviving plants were successfully analysed by flow cytometry. Chromosome numbers for samples that could not be analysed by flow cytometry were ascertained cytologically.
Statistical analyses
Univariate statistical analyses of variation in the environmental and population parameters of ploidy levels were performed using NCSS 2001 software (Hintze, 2001). Two data sets were prepared from the original data matrix. The first set included only cytotype-uniform populations (n = 250) while the second data set comprised all (uniform and mixed) populations (n = 325). The first data set was used for an analysis of morphological patterns without accounting for the effects of different ploidy levels in mixed populations. The second data set was used in the rest of the analyses. In the case of mixed populations and univariate analyses, the environmental data were duplicated for each respective ploidy level. Either a paired t-test or an F-test (randomized blocks; ANOVA) evaluated whether one ploidy level was consistently dominant in the mixed cytotype sites. Contingency tables were used for the analyses of qualitative environmental variables; ANOVA and the Kruskal–Wallis test were used for the analyses of quantitative and ordinal data, respectively (Zar, 1996). The Bonferroni correction of α for multiple tests (Gotelli and Ellison, 2004) was applied in the case of environmental variables.
Environmental variables were subsequently subjected to multivariate data analysis. Due to different types of descriptors (nominal, ordinal, quantitative), the primary data matrix was replaced by a secondary data matrix with the Gower general coefficient of similarity for combined data (Legendre and Legendre, 1998) using MVSP 3·12 software (Kovach Computing Service; http://www.kovcomp.co.uk/). The secondary matrix was subjected to principal coordinate analysis (PCoA; Legendre and Legendre, 1998) in MVSP 3·12. PCoA results were then subjected to constrained PCoA (db-RDA; Legendre and Anderson, 1999) where the independent X matrix contained ploidy-level identifiers and the dependent Y matrix consisted of the principal coordinates. Calculations were done in the program CANOCO 4·5 (ter Braak and Šmilauer, 2002) according to Lepš and Šmilauer (2003). First, differences in habitat conditions among different cytotypes were tested with a Monte Carlo permutation test (999 permutations). Fuzzy coding of independent variables was used to accommodate the existence of mixed populations and variable representation of respective ploidy levels within mixed populations. Differences in habitat conditions between pairs of ploidy levels were tested by partial db-RDA with respective pairs as explanatory variables and the third ploidy level as a covariable. Secondly, differences in habitat conditions were tested among groups of populations classified by ploidy-level composition with a Monte Carlo permutation test (999 permutations). In total, six groups were established, which included uniform (4x, 5x, 6x) and mixed (4x + 5x, 4x + 6x, 5x + 6x) populations. Groups of populations with mixed 4x + 5x + 6x cytotypes were excluded from the analysis due to the small number of these populations. Because the overall test was significant (see Table 6), we next tested for differences in habitat conditions between a priori selected pairs of groups on a reduced set of data matrices consisting of only populations of the respective groups. The Bonferroni correction of α (at α = 0·05) for multiple tests was applied.
Table 6.
Survey of environmental variables that are best correlated with occurrence of ploidy levels in constrained principal coordinates analysis applied to environmental variables recorded in populations of Allium oleraceum in the Czech Republic
| 4x (+) vs. 5x (−) | r | 4x (+) vs. 6x (−) | r | 5x (+) vs. 6x (−) | r |
|---|---|---|---|---|---|
| Forest | 0·68 | Habitat naturalness | 0·65 | Habitat naturalness | 0·57 |
| Habitat naturalness | 0·65 | Forest | 0·64 | Soil Ca2+ | 0·54 |
| Colder climate | −0·28 | Light conditions | −0·55 | Colder climate | −0·49 |
| Light conditions | −0·42 | Soil Ca2+ | 0·26 | pH | 0·42 |
| Arable field & field margin | −0·57 | Arable field & field margin | −0·58 | Arable field & field margin | −0·52 |
| Presence of arable land | −0·66 | Presence of arable land | −0·75 | Presence of arable land | −0·67 |
Within each analysis, variables showing the highest positive or negative correlations with the first canonical axis are reported together with their sign and correlation coefficient. The sign of correlation coefficients corresponds to the position of the respective ploidy level along the first canonical axis within each analysis (i.e. 4x vs. 5x; 4x vs. 6x; 5x vs. 6x). For explanations of variables see Methods.
The niche breadth of each ploidy level was expressed by the Gower general coefficient of dissimilarity (1 − G, where G is the Gower similarity coefficient). The mean and its 95 % bootstrap confidence interval (from 200 bootstrap samples) were calculated for all dissimilarity coefficients among populations in the presence of the respective ploidy level.
RESULTS
Cytotype composition in the Czech Republic
Chromosome numbers were estimated for two individuals from each of 16 populations (32 individuals in total), confirming the presence of tetra- (2n = 4x = 32), penta- (2n = 5x = 40) and hexaploid (2n = 6x = 48) cytotypes (see Supplementary data, available online). DNA tetraploid, pentaploid and hexaploid cytotypes were also observed within the study area. Neither other ploidy levels nor aneuploid counts were found. Of the 325 populations, tetraploids occurred in 18 %, pentaploids in 65 % and hexaploids in 41 %.
Within the area sampled, populations consisted of one, two or three ploidy levels (Table 2). Most of the populations (77 %) contained only one ploidy level and 22 % contained two. Populations that contained three ploidy levels were extremely rare (1 %). Among the populations consisting of a single ploidy level, 57 % contained pentaploids, 29 % contained hexaploids and 14 % contained tetraploids. Among the populations comprising two ploidy levels, 71 % contained pentaploids and hexaploids, while combinations of tetraploids and hexaploids had the lowest frequency (Table 2).
Table 2.
Ploidy-level composition of 325 populations of Allium oleraceum from the Czech Republic
| Number of ploidy levels per population |
Populations containing one ploidy level |
Populations containing two ploidy levels |
||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4x | 5x | 6x | 4x + 5x | 4x + 6x | 5x + 6x |
| 0·77 | 0·22 | 0·01 | 0·14 | 0·57 | 0·29 | 0·21 | 0·08 | 0·71 |
Populations were grouped and relative frequencies calculated according to the number of ploidy levels per population (one, two or three). Populations were further subdivided according to specific cytotypes present.
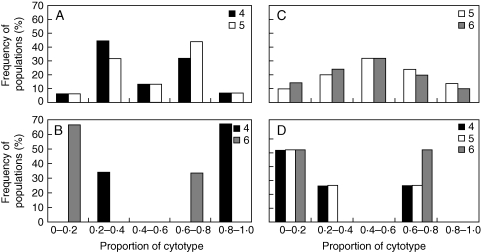
There was strong heterogeneity in the relative frequency of ploidy levels among populations containing cytotype mixtures (Fig. 1). Hence, no single ploidy level was consistently dominant in mixed populations containing various cytotype combinations (Table 3). Within 4x + 5x mixed populations, usually one ploidy level tended to be predominant, and the other was in the minority. The distribution of tetraploids and hexaploids within mixed 4x + 6x populations showed an antimodal pattern with either tetraploids or hexaploids predominating. By contrast, the distributions of pentaploids and hexaploids within 5x + 6x mixed populations were similar, and the two ploidy levels were almost uniformly distributed with a tendency toward evenly mixed populations. The distribution of ploidy levels in mixed 4x + 5x + 6x populations was characterized by the weak dominance of some ploidy levels and rare occurrences of others (Fig. 1).
Fig. 1.
Ploidy-level frequencies in Allium oleraceum mixed populations of tetra- and pentaploids (A), tetra- and hexaploids (B), penta- and hexaploids (C), and tetra-, penta- and hexaploids (D) in the Czech Republic.
Table 3.
Ploidy-level relative frequencies in mixed populations of Allium oleraceum in the Czech Republic
| Ploidy-level combination | n | Mean frequency of tetraploids ± s.e. | Mean frequency of pentaploids ± s.e. | Mean frequency of hexaploids ± s.e. | Paired t (F*) | P |
|---|---|---|---|---|---|---|
| 4x + 5x | 15 | 0·46 ± 0·06 | 0·54 ± 0·06 | – | −0·02 | 0·98 |
| 4x + 6x | 6 | 0·70 ± 0·13 | – | 0·30 ± 0·13 | 1·03 | 0·35 |
| 5x + 6x | 50 | – | 0·53 ± 0·03 | 0·47 ± 0·03 | 1·17 | 0·25 |
| 4x + 5x + 6x | 4 | 0·32 ± 0·14 | 0·28 ± 0·12 | 0·40 ± 0·14 | 0·01* | 0·99 |
The mean frequency of tetraploid, pentaploid and hexaploids is given for populations containing two ploidy levels and populations containing three ploidy levels from the entire sampling area.
Either a paired t-test on the number of 4x vs. 5x, 4x vs. 6x and 5x vs. 6x individuals or GLM ANOVA (F-test) on the number of 4x vs. 5x vs. 6x individuals evaluated whether one ploidy level was consistently dominant in the mixed sites.
Small-scale spatial pattern of ploidy levels within mixed populations
The small-scale spatial pattern of ploidy levels was rather uniform, and ploidy levels formed mostly homogeneous stands at a fine spatial scale (30 × 30 cm). In total, only 5·0 % of subsamples within mixed populations contained two or more ploidy levels. Populations of tetraploids mixed with any other ploidy level showed a higher (but not significantly different: χ2 = 0·61, d.f. = 2, P = 0·74) proportion of mixed subsamples (4x + 5x: 3·1 %; 4x + 6x: 3·2 %) than mixed populations of penta- and hexaploids (1·7 %).
Population parameters
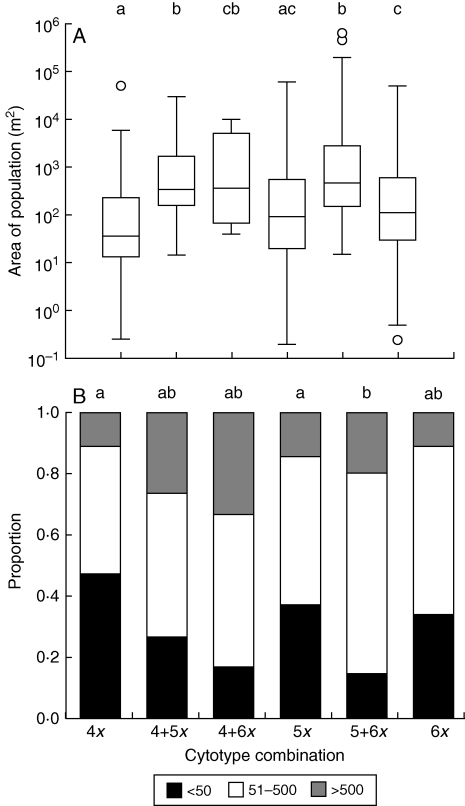
Population size and area differed significantly with respect to the cytotype composition of populations (size: chi-square test, χ2 = 35·00, d.f. = 5, P < 0·001; area: Kruskal–Wallis test, χ2 = 17·83, d.f. = 5, P = 0·003). Populations of uniform Ploidy level showed a tendency towards smaller population sizes and areas than mixed populations. Both parameters increased slightly with increasing population ploidy level (Fig. 2). An increasing tendency to form clumps of individuals was observed with increasing population ploidy level (4x: 34·3 %, 5x: 53·2 %, 6x: 71·2 %; chi-square test, χ2 = 14·00, d.f. = 2, P = 0·001).
Fig. 2.
Box plot of the area of the population (A) and frequency diagram of population size (B; <50, 51–500, >500 individuals, as indicated) in single- and mixed-ploidy-level populations of Allium oleraceum in the Czech Republic. Mixed populations of 4x + 5x + 6x cytotypes were excluded from the analyses due to small sample size. Note the log-scale of the y-axis in (A). Significant differences in medians between pairs of populations with different ploidy-level combinations (A; Dunn's test with P < 0·05) and in frequencies of population size categories between pairs of populations with different ploidy-level combinations (Cross-tabulation; with P < 0·05) are marked by different letters in rows above the diagrams.
Ploidy-level distribution in the Czech Republic
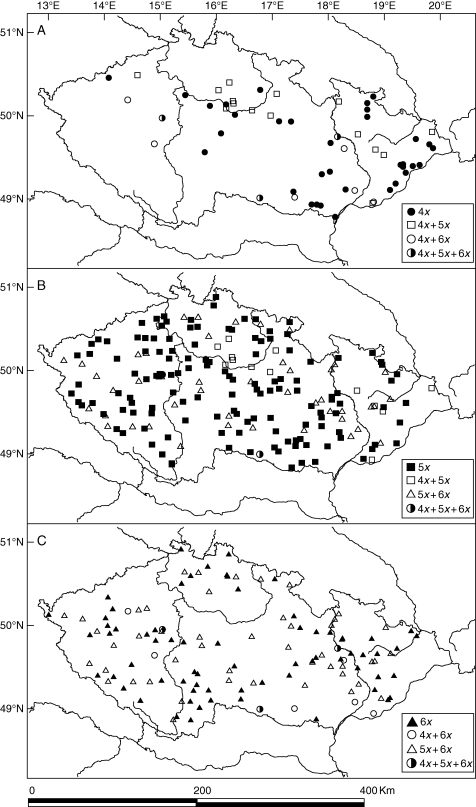
The distribution of particular ploidy levels in the Czech Republic is shown in Fig. 3. It is clear from this that ploidy levels differ in their distribution. Uniform pentaploid populations occur regularly throughout the entire study area. By contrast, uniform tetra- and hexaploid populations occur in narrower ranges and are partially sympatric in the eastern part but rather parapatric in the western part of the Czech Republic. Some small, single-cytotype areas were observed: for example, those of hexaploids in the western part and those of tetraploids in the eastern part of the Czech Republic. Except for mixed populations consisting of tetra- and hexaploids, the distribution of mixed populations coincides with areas of sympatric occurrence of uniform populations of the respective ploidy levels. Mixed populations of tetra- and hexaploids are mostly located in broad contact zones between uniform tetra- and hexaploid populations. Mixed populations of three ploidy levels occur rarely in areas of sympatric occurrence of all ploidy levels.
Fig. 3.
Geographical distribution of uniform and mixed-ploidy-level populations of Allium oleraceum in the Czech Republic. (A) Uniform 4x and mixed 4x + 5x, 4x + 6x and 4x + 5x + 6x populations; (B) uniform 5x and mixed 4x + 5x, 5x + 6x and 4x + 5x + 6x populations; (C) uniform 6x and mixed 4x + 6x, 5x + 6x and 4x + 5x + 6x populations.
Ecological differentiation among ploidy levels – univariate analyses
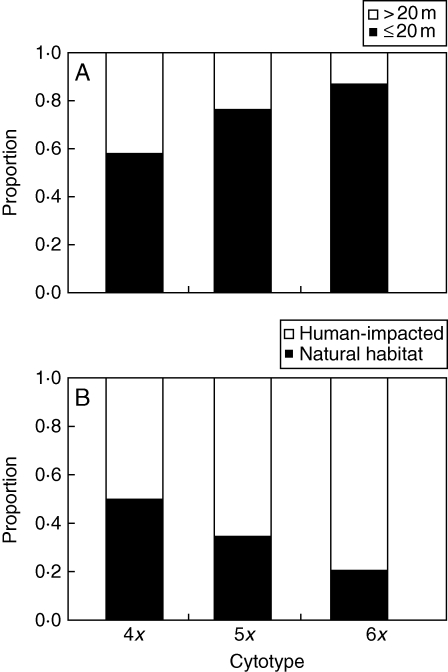
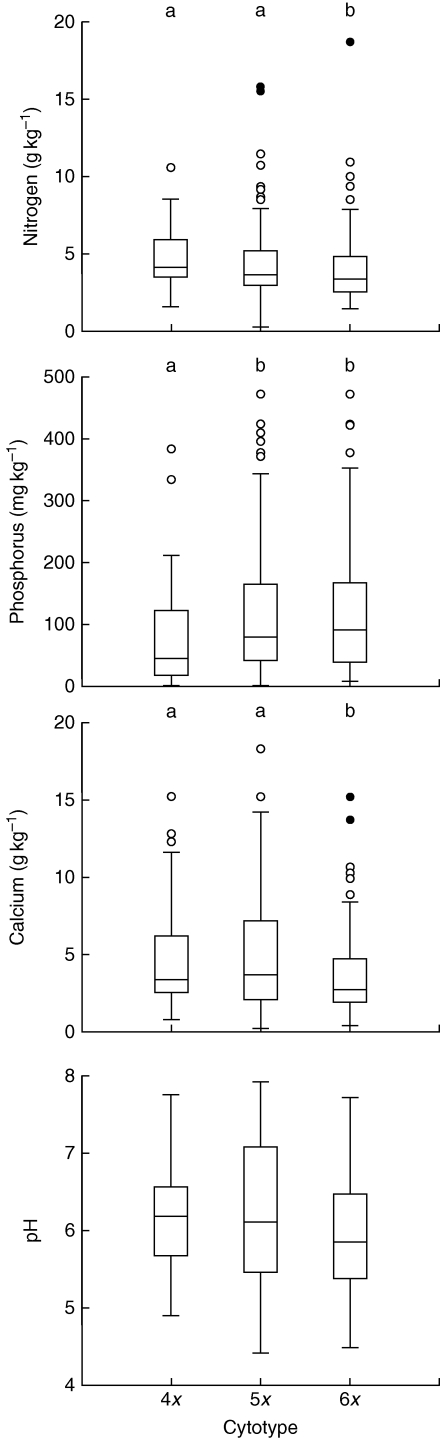
No significant differences in relative frequencies of tetra-, penta- and hexaploids in different habitat types were found (Table 4), although tetraploids tended to occur more frequently in deciduous forests (oak–hornbeam and hardwood floodplain forests) and less frequently in field and field margins than plants of other ploidy levels. However, penta- and hexaploids were increasingly frequent in localities in close contact with arable land (Armitage test for trend in proportions, Z-value = 4·33, P < 0·001) and in human-impacted vegetation (Z-value = 4·34, P < 0·001; Table 4, Fig. 4). Whereas tetraploids were evenly distributed between natural and human-impacted vegetation, hexaploids mostly (80·4 %) occurred in human-impacted vegetation: field margins, road verges, in ruderal scrub, eutrophicated forests, etc. Tetra- and pentaploids occurred on soils with a higher content of nitrogen and calcium than hexaploids. Both ploidy levels also occurred on slightly less acidic soils than hexaploids, but the difference was not significant. By contrast, penta- and hexaploids occurred on soils with a higher content of phosphorus than tetraploids (Fig. 5). There was no difference in other soil properties (C, Mg2+, K+), light conditions and altitude among ploidy levels (Table 4).
Table 4.
Summary of the associations between ploidy levels and selected environmental variables in populations of Allium oleraceum in the Czech Republic
| Variable | Test | d.f. | Test statistics | P |
|---|---|---|---|---|
| Habitat type | CT | 12 | 14·32 | 0·286 |
| Presence of arable land | CT | 2 | 18·96 | ≪0·001 |
| Habitat naturalness | CT | 2 | 20·39 | ≪0·001 |
| Climatic region | CT | 4 | 46·94 | ≪0·001 |
| Altitude | KW | 2 | 3·40 | 0·183 |
| Light conditions | KW | 2 | 1·47 | 0·478 |
| Soil C* | ANOVA | 2 | 2·12 | 0·122 |
| Soil N | KW | 2 | 6·68 | 0·035 |
| Soil PO43− | KW | 2 | 12·48 | 0·002 |
| Soil pH | KW | 2 | 5·49 | 0·064 |
| Soil Ca2+ | KW | 2 | 8·43 | 0·014 |
| Soil Mg2+* | ANOVA | 2 | 0·92 | 0·631 |
| Soil K+* | ANOVA | 2 | 0·68 | 0·534 |
Differences were tested either by one-way ANOVA, Kruskal–Wallis test (KW) or contingency tables (CT).
*Data were log (x + 1) transformed before analysis. P-values in bold are significant after Bonferroni correction (P < 0·004).
Fig. 4.
(A) Relative frequencies of ploidy levels in relation to the distance of their populations from the nearest arable field (‘Presence of arable land’; distance to the nearest field >20 m or ≤20 m, as indicated). (B) Relative frequencies of ploidy levels in relation to the degree of anthropogenic impact applied on their populations (‘Habitat naturalness’; human-impacted or natural habitat, as indicated).
Fig. 5.
Box plots of selected chemical soil properties at sites with occurrence of different ploidy levels of Allium oleraceum. Significant differences in medians between ploidy levels (Dunn's test; P < 0·05) are marked by different letters in rows above the box plots. For overall tests see Table 4.
Analysis of the cytotype frequencies in different climatic regions showed that tetraploids, in contrast to the other ploidy levels, were not concentrated in any climatic region. Pentaploids and hexaploids, however, were concentrated in areas with mesic climatic conditions (Table 4).
Ecological differentiation among ploidy levels – multivariate analyses
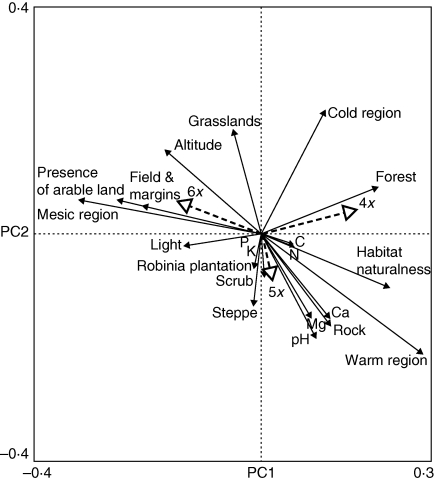
Although all three ploidy levels have overlapping ecological niches, overall and paired tests showed that the ploidy levels are ecologically differentiated, with the largest realized niche differences between tetra- and hexaploids (Tables 5 and 6; Fig. 6). In general, hexaploids occurred predominantly in human-impacted open habitats (usually at field margins and in fields) in mesic climatic conditions at higher altitudes, whereas tetraploids were confined to more natural habitats, both exposed and shaded along the whole altitudinal gradient. Pentaploids were confined to warm and mesic regions in the lower and middle altitudes, but were found in a wide range of habitats on soils with higher levels of minerals and higher soil pH. The niche breadth of tetra- and pentaploids was similar and higher than that of hexaploids [Gower dissimilarity coefficient: mean (95 % bootstrap confidence interval) – 4x: 0·361 (0·354–0·367); 5x: 0·359 (0·358–0·361); 6x: 0·320 (0·319–0·321)].
Table 5.
Summary of the analyses using constrained principal coordinates analysis applied to environmental variables recorded in populations of Allium oleraceum in the Czech Republic: (a) effects of ploidy levels; (b) effects of groups of populations classified by observed ploidy-level combinations
| Model | Trace (1st axis) | F | P |
|---|---|---|---|
| (a) Ploidy levels | |||
| Overall model | 0·047 | 7·22 | 0·001 |
| 4x vs. 5x | 0·033 | 5·70 | 0·001 |
| 4x vs. 6x | 0·082 | 9·06 | 0·001 |
| 5x vs. 6x | 0·016 | 3·99 | 0·001 |
| (b) Groups | |||
| Overall model | 0·063 | 3·93 | 0·001 |
| 4x vs. 5x | 0·030 | 5·18 | 0·001 |
| 4x vs. 6x | 0·084 | 9·21 | 0·001 |
| 5x vs. 6x | 0·016 | 4·00 | 0·002 |
| 4x vs. 4x + 5x | 0·010 | 1·76 | 0·071 |
| 4x vs. 4x + 6x | 0·018 | 1·96 | 0·039 |
| 5x vs. 4x + 5x | 0·016 | 3·12 | 0·003 |
| 5x vs. 5x + 6x | 0·007 | 1·75 | 0·065 |
| 6x vs. 4x + 6x | 0·009 | 0·95 | 0·473 |
| 6x vs. 5x + 6x | 0·008 | 1·98 | 0·033 |
All effects were tested by Monte Carlo permutation tests using 999 random permutations. See Methods for details.
P-values in bold (except for overall models) are significant after Bonferroni correction [P < 0·017 in the section (a)] or are significant at P = 0·01 in the section (b).
Fig. 6.
The first and the second axis of the constrained principal coordinate analysis testing environmental differences among ploidy levels of Allium oleraceum. Vectors of the environmental variables were used as supplementary data to help interpret the ordination. See Methods for analysis settings.
Significant differences in habitat conditions were found even among groups of populations classified according to ploidy-level composition (Table 5b). Pure 4x, 5x and 6x populations differed from each other with regard to environmental conditions. By contrast, the environments of mixed-ploidy-level populations did not differ from those of uniform populations of the respective cytotypes, pointing to the heterogeneity of the environments where cytotypes co-occur. This is in agreement with the finding that mixed ploidy populations occur more frequently (24·3 %) at sites with two or more adjoining habitats than uniform populations (16·9 %).
DISCUSSION
The results of this study indicate that pentaploids are the most common cytotype in the Czech Republic, and surprisingly, that hexaploids are more frequent than tetraploids there (Table 2). Other ploidy levels or aneuploid plants were not detected in this area. This is in good agreement with previous reports of chromosome numbers in the Czech Republic (Table 1), especially with the local study of Fialová (1996) who investigated 206 individuals from 24 A. oleraceum populations in the eastern part of the Czech Republic and found mostly pentaploids and hexaploids, with only rare occurrences of tetraploids and no aneuploid plants. The absence of mature aneuploid plants, particularly within those populations containing pentaploids, does not mean that they could not be formed. Fialová (1996) observed a few aneuploid seedlings from 5x maternal plants. It is probable that aneuploid offspring are either non-viable or are eventually out-competed due to their reduced fitness.
Screening results also clearly indicate that ploidy-level distribution patterns are much more complex and finely grained than previously thought (see Table 1 and Fig. 3). Widespread sympatry of cytotypes of A. oleraceum that result in the contact zones, where at least two ploidy levels are intermixed, contrasts with the majority of diploid–polyploid contact zones studied. They are thought to have resulted from secondary contacts, the distribution of cytotypes is mostly parapatric, and only a few inter-cytotype hybrids are normally recorded (Soltis and Soltis, 1993; Petit et al., 1999; Segraves et al., 1999). The present study also found more mixed-ploidy-level A. oleraceum populations (23 %) than had previously been reported (8·3 %, Fialová, 1996; 12 %, Karpavičienė, 2007). In fact, co-occurrences of all possible combinations of 4x, 5x and 6x ploidy levels were observed, and among these 4x + 6x and 4x + 5x + 6x populations had not previously been identified in Europe.
High frequency of mixed-ploidy-level populations has been recently observed in several other plant species (Burton and Husband, 1999; Suda, 2002; Keeler, 2004; Suda et al., 2007a; Halverson et al., 2008; Kao, 2008). However, it is impossible to identify unambiguously the sole unifying mechanism that explains the existence of those populations. Likewise, the complex distribution patterns of the three ploidy levels of A. oleraceum observed do not permit an easy explanation. Several non-exclusive mechanisms that could explain the observed geographical patterns of the three cytotypes are discussed below.
Ecological differentiation among ploidy levels
Although their ecological preferences partially overlap, both univariate and multivariate analyses indicate that A. oleraceum of different ploidy levels show ecological differentiation. This difference in ecological requirements of the cytotypes is thus consistent with the generally accepted fact that polyploidization can produce novel characters which lead to niche shifts (reviewed by Ehrendorfer, 1980; Lewis, 1980; Levin, 1983, 2002; Soltis et al., 2003), although we acknowledge that the differences observed may have emerged later via selection (Petit et al., 1999). Higher ploidy levels also sometimes confer a wider ecological amplitude (e.g. Hancock and Bringhurst, 1981; Rothera and Davy, 1986; Thompson and Lumaret, 1992; Burton and Husband, 1999). Brochmann and Elven (1992) showed in a detailed study of three diploid and 13 polyploid (4x–16x) species of Draba that ecological amplitude, heterozygosity and biochemical diversity all increased significantly with increasing ploidy level. This is not the case in A. oleraceum here: the niche breadth of tetra- and pentaploids is similar, being higher than that of hexaploids. This is in agreement with the markedly lower biochemical diversity of hexaploids than either tetra- or pentaploids noted by Staňková (2005) in an electrophoretic enzyme study of 30 Czech populations of A. oleraceum. Similar observations, in which a broader ecological niche was found for lower ploidy levels, were recently reported from a diploid–hexaploid complex of Senecio carniolicus (Schönswetter et al., 2007) and diploid–tetraploid complexes of Centaurea stoebe (Španiel et al., 2008) and Santolina pectinata (Rivero-Guerra, 2008).
Ploidy-level distributions on regional and local scales: is the adaptive scenario the most plausible explanation?
Hypotheses concerning ecological diversification among cytotypes predict trends towards parapatry or allopatry of cytotypes at larger spatial scales if the fitness of cytotypes is a function of the environment, which itself changes with geographical scale (Engen et al., 2002; Johnson et al., 2003), or towards partial or complete sympatry but ecological isolation between cytotypes if the environmental factors are mosaic in structure (Thompson and Lumaret, 1992; Levin, 2002). The character of ecogeographical differentiation observed among the A. oleraceum ploidy levels is more probably consistent with the latter model because the most important environmental factors contributing to the ecological differentiation among the cytotypes (e.g. habitat naturalness; presence of arable land) have a rather mosaic pattern in the central European landscape. Furthermore, the coarse-grained spatial pattern of certain environmental factors that cause lower landscape heterogeneity in some Czech regions, especially at higher altitudes (Petřík and Wild, 2006), may also explain (1) the existence of a few small single-cytotype areas, especially those of hexaploids; and (2) the extremely rare occurrence of tetraploids in the large upland areas of central and western parts of the Czech Republic (Bohemia) in contrast to their common occurrence in climatically similar or even harsher upland areas in the eastern part of the Czech Republic (eastern Moravia), despite their broad ecological niche (see Fig. 3). The contrasting distributions of tetraploids probably reflect differences in environmental conditions and historical processes that influence floristic composition in these regions. Rather acidic and mineral-poorer soils dominate in the Bohemian upland; a mosaic of mineral-poor and mineral-rich soils occur in eastern Moravia (Demek, 1987; Ložek, 1988). In fact, the distribution of tetraploids in Bohemia roughly corresponds to the occurrence of mineral-rich soil substrates at altitudes under approx. 350(400) m (Ložek, 1988; Sádlo, 2007), and this may also partly explain the absence of hexaploids from large areas of the Bohemian lowlands despite the vast expanses of arable land. Mráz et al. (2008) recorded similar cytotype distribution patterns in Pilosella officinarum in Bohemia, albeit with rare penta- and hexaploids confined to warm, low-elevation regions and common tetraploids prevailing in the whole of Bohemia.
At the population level, detected differences in ecological niches among cytotypes of A. oleraceum seem to support the observation of predominantly monotypic populations, i.e. a situation when plants invading a population of the other cytotype would do poorly in the unsuitable habitat (Baack, 2004). The present data demonstrate that a majority of 4x + 5x and 5x + 6x mixed populations do not show distinct geographical patterns, and are sympatric and intermixed with single-ploidy-level populations. The existence of such mixed populations could be best explained as a stochastic event based on mutually independent dispersion of cytotypes through the landscape followed either by: (1) their successful establishment and persistence at sites that represent less typical or marginal (unfavourable) environments for either one or both cytotypes but where they can both persist due to partial overlaps in realized ecological niches; or (2) local secondary contacts between cytotype-different, but uniform, populations at borders between different habitats. The present analyses support the idea that environmental conditions at mixed-population sites are marginal to or intermediate (‘heterogeneous’) between the environmental conditions of the respective uniform cytotype population sites. Similar patterns, i.e. the coexistence of cytotypes in sites possessing either sufficient environmental heterogeneity or ecologically marginal environment, were observed by Lumaret et al. (1987), Keeler (1990), and Husband and Schemske (1998) in mixed-cytotype populations of Dactylis glomerata, Andropogon gerardii and Chamaerion angustifolium, respectively.
The hypothesis of secondary contacts between ploidy levels is further supported by the larger population sizes and areas of mixed populations relative to the uniform populations (Fig. 2). The present results suggest that mixed-ploidy populations occur somewhat more frequently at sites with two or more adjoining habitats than do uniform populations, i.e. the larger area of mixed-cytotype populations also entails higher environmental heterogeneity (Legendre and Legendre, 1998; Koenig, 1999). As no single cytotype was strongly dominant in the majority of mixed 4x + 5x and 5x + 6x populations, which cytotype dominates at a site could therefore be due either to specific responses of the cytotypes to local environmental conditions or to random factors such as chance colonization (Kliber and Eckert, 2005; Kao, 2008). Cytotype uniformity of small populations can be explained through events such as colonization of a new site by a small number of individuals, probably representing a clone (founder effect), or through processes such as habitat change when large populations are strongly reduced in size (bottleneck effect and/or drift).
By contrast, here some mixed 4x + 5x and 5x + 6x populations were observed at sites with homogeneous environment typical of one of the participating cytotypes. This suggests that niche differentiation is insufficient to explain the existence of these mixtures, although we cannot exclude that such differentiation occurs on a very fine spatial scale. Furthermore, ecological differentiation fails to explain the markedly contrasting cytotype frequencies in a majority of mixed populations of tetra- and hexaploids, because, in most of these populations, tetraploids predominate even if habitat conditions appear less suitable for them than for hexaploids on the basis of multivariate analysis.
We therefore consider ecological differentiation to be an important but not sole driving force behind the distribution patterns observed. Rather, our non-exclusive explanations take into account (1) the generation of cytotype mixtures as a result of the interploidy crosses or the emergence of a higher polyploid within a lower polyploid population, (2) predominant vegetative reproduction and localized dispersal that retard the effect of exclusion of emerging or invading cytotype and (3) human impact influencing distribution of cytotypes.
First, we suggest that in situ de novo production of hexaploids in some tetraploid populations seems to occur in A. oleraceum through the union of reduced and unreduced gametes, as evidenced by the fact that (1) some mixed 4x + 6x populations are intermixed with uniform 4x populations outside the hexaploid range, (2) the cytotype structure of the majority of 4x + 6x populations is characterized by strong dominance of tetraploids and rare occurrence of hexaploids, and (3) the tetra- and hexaploids have identical multilocus isozyme phenotypes within the two 4x + 6x populations (Staňková, 2005). However, the origin of hexaploid plants in tetraploid populations seems to be an extremely rare event given that no previous study on the cytology of A. oleraceum (see Table 1) reported mixed 4x + 6x populations, despite the frequent reports of tetraploid populations. In turn, the origin of 4x + 5x and 5x + 6x mixed populations is difficult to explain on the basis of a primary origin of a novel ploidy level (Ramsey and Schemske, 1998) because of the absence of one ‘compatible’ donor ploidy level, i.e. 4x in 5x + 6x or 6x in 4x + 5x mixed-ploidy populations. Indeed, we cannot exclude the possibility that our research simply failed to detect the minority (4x or 6x, respectively) cytotype, but this is unlikely because (1) the within-population screening was quite extensive, and, as stated above, (2) the ‘missing’ donor ploidy usually did not occur in the surroundings. Hybridization between the 4x and 6x plants would yield pentaploids, but only three mixed populations of tetra-, penta- and hexaploids were found. This suggests limited gene flow between 4x and 6x cytotypes due to extremely rare production of flowers in hexaploids (Ohryzek, 2007) and rather indicates secondary contacts between cytotypes.
Second, asexual reproduction via aerial bulbils and daughter bulbs strongly predominates over sexual reproduction in A. oleraceum (Duchoslav, 2000; Karpavičienė, 2002; Åström and Hæggström, 2004; Ohryzek, 2007). Hence, even a single plant of one cytotype emerging within or invading a uniform population of another cytotype has the potential to persist as it can maintain itself and spread through asexual reproduction (Kao, 2007). This feature is analogous to apomictic seed formation, which allows plants of the other cytotype to produce their exact copies and thus to escape reproductive costs due to their minority status. Yamauchi et al. (2004) showed that if asexuality dominates over sexual reproduction, the outcome of mixed-ploidy occurrences are probably determined by direct competition between the cytotypes. We therefore suggest that existence of some mixed populations is a result of (recent) cytotype invading a uniform population of another cytotype and that eventually one cytotype suppress the other(s) in the ensuing period. Longevity of A. oleraceum individuals and their ability to propagate via daughter bulbs even under suboptimal ecological conditions hindering the production seeds and bulbils, for example in deep shade (Duchoslav, 2009), may retard competitive exclusion of one cytotype.
Additionally, direct competition among cytotypes can be reduced if cytotypes show local pollen/propagule dispersal leading to local spatial segregation of cytotypes (Li et al., 2004; Baack, 2005). The present data show that A. oleraceum forms predominantly cytotype-homogeneous patchy stands within mixed populations and that local cytotype homogeneity tends to increase with ploidy level. This pattern may be explainable simply by limited dispersion (Hardy and Vekemans, 2001; Meirmans et al., 2003). A. oleraceum is functionally similar to A. vineale, for which Ronsheim (1994) found that dispersion distances did not differ between bulbils and seeds and that most propagules fall within 30 cm of their mother plants. She also observed, however, that a very small number of seeds dispersed far from mother plants (> 1 m). This can, in the case of co-occurring cytotypes, result in a mixture with established patches of the other cytotype. Seed recruitment may, however, be inhibited by competition for safe sites from clonal bulbils and daughter bulbs (Abrahamson, 1980; Eriksson, 1997; Kliber and Eckert, 2005). This would also decrease the probability of invasion by an ‘alien’ cytotype. Much scarcer seed production by higher ploidy levels (the average production of seeds increases from zero seeds per plant in hexaploids to one and two seeds per plant in penta- and tetraploids, respectively; M. Fialová and M. Duchoslav, pers. obs.) and prevailing vegetative reproduction could explain not only increased local cytotype homogeneity within mixed populations of higher ploidy levels but also the increased clumping of higher ploidy levels presented here. Local dispersal and clumping of A. oleraceum cytotypes can thus effectively separate the cytotypes and thereby decrease reproductive interference and inter-cytotype competition (Baack, 2005). Recently, Kolář et al. (2009) proposed that founder effects together with limited seed distribution capacity leading to clumping is a plausible explanation for the existence of some mixed-ploidy populations of Knautia arvensis agg. Undoubtedly, data regarding the fine-scale spatial and environmental distribution of cytotypes within mixed-ploidy-level populations is needed to assess the relative importance of various coexistence mechanisms.
Third, the present-day complex distribution patterns of ploidy levels and high proportion of mixed-ploidy populations may also reflect human impact (Balfourier et al., 2000; Perný et al., 2008), i.e. the weedy character of A. oleraceum (Duchoslav, 2001a). The spread of the species was indeed influenced by agricultural practices and transport of crops and hay. Its frequent present-day occurrence along roadsides and field margins is evidence of its past abundance in arable land before the implementation of subsoil ploughing and the use of selective herbicides (Håkansson, 1963; Willmans, 1985; Duchoslav, 2001a). In the face of these equilibrium-disrupting processes, the abilities of niche differentiation and competitive exclusion to limit co-occurrence of cytotypes may be weakened.
General pattern of ploidy-level distribution in Europe and hypothetical origins of polyploidy
The pattern of ploidy-level distribution observed by us in the Czech Republic contrasts with that seen on the European scale. Because published karyological data on A. oleraceum are based on chromosome counting in small numbers of plants, such an approach is able to detect the most frequent cytotypes but fails to detect rare ones (Burton and Husband, 1999; Halverson et al., 2008). It can therefore evaluate frequencies and large-scale spatial patterns of detected cytotypes inaccurately. Hence, it is highly possible that the current understanding of cytotype distribution patterns in Europe is inaccurate.
The origin of A. oleraceum is still not fully understood. Polyploid A. oleraceum could be of autopolyploid and/or of allopolyploid origin. We consider Levan's (1938) experimental results, i.e. the creation of tetraploids in one step from diploids, to provide a highly probable explanation for the origin of A. oleraceum. The A. paniculatum group is an extremely complicated set of species of the section Codonoprasum that occurs from the westernmost parts of Macaronesia, northern Africa (de Wilde-Duyfjets, 1976) and the Iberian peninsula (Pastor and Valdes, 1983) through the whole Mediterranean area (Zahariady, 1975; Stearn, 1980; Jauzein and Tison, 1999, 2001; Brullo et al., 1996, 2001) to Iran (Wendelbo, 1971) and south-western Siberia (Frizen, 1988). This group could represent the hypothetical parents. Furthermore, they are not only diploid, as were used by Levan, but also triploid, tetraploid and pentaploid. Thus, the high variation in ploidy levels of A. oleraceum from triploid to hexaploid might be the result of independent crosses between different members of this complex. At present, this hypothesis is supported by the occurrence of two types of hexaploids found by us: one type is represented by populations that differ from tetra- and pentaploids in their ecological requirements, and the second hexaploid type comprises rare individuals admixed in populations of tetraploid plants that probably originated from fusion of reduced and unreduced gametes.
No triploid plants were detected during our detailed screening within the Czech Republic, and only extremely rare records of triploids are known, all of which are from the northern edge of the ranges of the supposed diploid progenitors (Vakhtina, 1984; Krahulcová, 2003). Triploids probably result from the hybridization of reduced and unreduced gametes of diploid progenitors, but they may arise from pollinations between ancestor diploid and tetraploid A. oleraceum (Type I hybrids; Petit et al., 1999). Currently, it is not possible to differentiate between these two hypotheses.
Both tetra- and pentaploids are widely distributed and probably sympatric (Karpavičienė, 2007; present study) throughout Europe, even in northern areas that were covered by glaciers during the last glacial maximum (Huntley & Birks, 1983). By contrast, hexaploids are presently known just in the Czech Republic, Austria and Spain (Table 1). Pastor (1982) mentioned that higher cytotypes (5x, 6x) probably arose in tetraploid populations through the production of (un)reduced gametes and interploidy crosses, and this mode is partially (6x) supported by the present data. However, alternative origins of the higher cytotypes cannot yet be ruled out, including backcrosses of triploids or tetraploids with parental taxa and eventual polyploidization, and even polyphyletic crosses (see above). Apart from the origin of ploidy levels, the wide present-day distribution of tetra- and pentaploids probably reflects their superior colonization abilities, as evidenced by the breadth of their ecological niches, which were found to be greater than that of hexaploids. Alternatively, the common occurrence of hexaploids in the Czech Republic may represent evidence of a recent range expansion of a newly established hexaploid type in anthropogenic habitats.
Conclusions
A detailed investigation of the distribution of different ploidy levels of A. oleraceum showed a distribution pattern much more complex than could be deduced from published chromosome counts. Individual cytotypes differ in their ecological requirements at the regional scale and this contributes to the distribution patterns observed. Local cytotype coexistence is, however, widespread. It is therefore considered that high frequency of mixed-ploidy populations is a result of both the cumulative effects of various isolating mechanisms, including niche differentiation, localized dispersal and prevailingly asexual propagation, and equilibrium-disrupting processes, i.e. agricultural practices. Distributional data support the existence of both primary and secondary zones of cytotype contact.
The extensive distribution range of A. oleraceum over most parts of Europe, as compared with the narrower distribution of its supposed diploid (and perhaps also polyploid) progenitors (species of the A. paniculatum group) in southern Europe, is consistent with the idea that polyploids are more ecologically tolerant and therefore are able to colonize harsher environments than their diploid progenitor(s). We suggest the recurrent formation of particular cytotypes and consider their polyphyletic origin as highly probable. A logical extension of this research would be to determine whether the present results could be extrapolated outside central Europe. The growing number of chromosome counts reported from individual regions suggests an increasingly complex cytogeographical pattern.
SUPPLEMENTARY DATA
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. Ohryzek, H. Staňková, M. Fialová, M. Jandová, A. Jírová, J. Duchoslavová and R. Slípková for their help with fieldwork and maintenance of samples in the experimental garden, and to M. Dančák, B. Trávníček, R. J. Vašut, A. Jírová, M. Fialová, Z. Skála and Z. Hradílek for sampling of some populations. Our thanks also go to J. S. Heslop-Harrison and two anonymous reviewers for their comments and advice on the manuscript. Conn Breen and Fred Rooks kindly revised the English text. This work was supported by the Grant Agency of the Czech Republic (grant numbers 206/01/P097, 206/04/P115 and 206/09/1126 to M.D., and 206/07/0706 to F.K.).
APPENDIX
Survey of environmental and population variables used in the study. Codes of habitats corresponding to the EUNIS habitat classification (Davies et al., 2004), which are included within each habitat type used in the study, are given in parentheses.
| Environmental/population variable | Explanation [units] |
|---|---|
| Habitat type | rock (H2, H3); steppe (E1, E5·2); mesic & wet grassland (E2, E3); semi-natural forest (G1·2, G1·6, G1·7, G1·8, G1.A, G3·4, F3 p.p.); ruderal scrub (G5, FA, F3 p.p.); planted Robinia pseudacacia forest (G1.C); arable field & field margins (E5·1, H5·6, I, J4) |
| Presence of arable land | 0 – distance to the nearest field >20 m, otherwise 1 |
| Habitat naturalness | 0 = human-impacted habitat, 1 = natural habitat |
| Light conditions | 1 = strong shade, 2 = half-shade, 3 = low shade, 4 = full insolation |
| Chemical soil parameters | pH of water extract, PO43− [mg kg−1], Mg2+ [g kg−1], K+ [g kg−1], Ca2+[g kg−1], N [g kg−1], C [%] |
| Altitude | [m a. s. l.] |
| Climatic region | C = cold region, SW = slightly warm region, W = warm region |
| Population size | <50, 51–500, >500 individuals |
| Area of population | [m2] |
| Morphological pattern | individuals, clusters |
LITERATURE CITED
- Abrahamson WG. Demography and vegetative reproduction. In: Solbrig OT, editor. Demography and evolution in plant populations. Oxford: Blackwell; 1980. pp. 89–106. [Google Scholar]
- Agapova ND. Numeri chromosomatum Magnoliophytorum florae URSS. Aceraceae – Menyantaceae. Leningrad: Nauka; 1990. [Google Scholar]
- Arohonka T. Kromosomilukumäärityksiä nauvon seilin saaren putkilokasveista. Turun Yliopiston Biologian-Laitoksen Julkaisuja. 1982;3:1–12. [Google Scholar]
- Åström H, Hæggström C. Generative reproduction in Allium oleraceum (Alliaceae) Annales Botanici Fennici. 2004;41:1–14. [Google Scholar]
- Baack EJ. Cytotype segregation on regional and microgeographic scales in snow buttercups (Ranunculus adoneus: Ranunculaceae) American Journal of Botany. 2004;91:1783–1788. doi: 10.3732/ajb.91.11.1783. [DOI] [PubMed] [Google Scholar]
- Baack EJ. To succeed globally, disperse locally: effects of local pollen and seed dispersal on tetraploid establishment. Heredity. 2005;94:538–546. doi: 10.1038/sj.hdy.6800656. [DOI] [PubMed] [Google Scholar]
- Balfourier F, Imbert C, Charmet G. Evidence for phylogeographic structure in Lolium species related to the spread of agriculture in Europe. A cpDNA study. Theoretical and Applied Genetics. 2000;101:131–138. [Google Scholar]
- Baranyi M, Greilhuber J. Genome size in Allium: quest of reproducible data. Annals of Botany. 1999;83:687–695. [Google Scholar]
- Barton NH, Hewitt GM. Analysis of hybrid zones. Annual Review of Ecology and Systematics. 1985;16:113–148. [Google Scholar]
- Bayer RJ, Stebbins GL. Chromosome numbers, patterns of distribution, and apomixis in Antennaria (Asteraceae: Inuleae) Systematic Botany. 1987;12:305–319. [Google Scholar]
- Bierzychudek P. Patterns in plant parthenogenesis. Experientia. 1985;41:1255–1264. doi: 10.1007/978-3-0348-6273-8_9. [DOI] [PubMed] [Google Scholar]
- ter Braak CJF, Šmilauer P. CANOCO reference manual and CanoDraw for Windows User's guide: software for canonical community ordination (version 4.5) Ithaca, NY: Microcomputer Power; 2002. [Google Scholar]
- Brochmann C, Elven R. Ecological and genetic consequences of polyploidy in arctic Draba (Brassicaceae) Evolutionary Trends in Plants. 1992;6:111–124. [Google Scholar]
- Brullo S, Guglielmo A, Pavone P, Scelsi F, Terrasi MC. Cytotaxonomic consideration of Allium fuscum Waldst. et Kit. (Liliaceae), a critical species of the European flora. Folia Geobotanica et Phytotaxonomica. 1996;31:465–472. [Google Scholar]
- Brullo S, Guglielmo A, Pavone P, Salmeri C. Osservazioni tassonomiche e cariologiche sulle specie del ciclo di Allium paniculatum L. in Italia. Informatore Botanico Italiano. 2001;33:500–506. [Google Scholar]
- Burton TL, Husband BC. Population cytotype structure in the polyploid Galax urceolata (Diapensiaceae) Heredity. 1999;82:381–390. doi: 10.1038/sj.hdy.6884910. [DOI] [PubMed] [Google Scholar]
- Capineri R, D'amato G, Marchi P. Numeri cromosomici per la Flora Italiana. 534–583. Informatore Botanico Italiano. 1978;10:421–465. [Google Scholar]
- Chmielewski JG, Semple JC. The cytogeography of Aster lanceolatus. 3. Cytoecology in southern Ontario. Canadian Journal of Botany. 1983;61:1879–1886. [Google Scholar]
- Davies CE, Moss D, Hill MO. EUNIS habitat classification revised 2004. Copenhagen: European Environment Agency & Paris: European Topic Centre on Nature Protection and Biodiversity; 2004. [Google Scholar]
- Demek J. Hory a nížiny. Prague: Academia; 1987. [Google Scholar]
- De Wilde-Duyfjets BEE. A revision of the genus Allium (Liliaceae) in Africa. Mededelingen landbouwhogeschool Wageningen. 1976 76/11. [Google Scholar]
- van Dijk P, Bakx-Schotman T. Chloroplast DNA phylogeography and cytotype geography in autopolyploid Plantago media. Molecular Ecology. 1997;6:345–352. [Google Scholar]
- van Dijk PK, Bijlsma R. Simulations of flowering time displacement between two cytotypes that form inviable hybrids. Heredity. 1994;72:522–535. [Google Scholar]
- van Dijk P, Hartog M, van Delden W. Single cytotype areas in autopolyploid Plantago media L. Biological Journal of the Linnean Society. 1992;46:315–331. [Google Scholar]
- Dobeš C, Vitek E. Documented chromosome number checklist of Austrian vascular plants. 2000. Wien: Verlag des Naturhistorischen Museums Wien. [Google Scholar]
- Dobrotchaeva DN, Kotov MI, Prokudin JN. Opredetitel' vyssich rastenij Ukrainy. Kiev: Institute of Botany; 1999. [Google Scholar]
- Doležel J, Binarova P, Lucretti S. Analysis of nuclear DNA content in plant cells by flow cytometry. Biologia Plantarum. 1989;31:113–120. [Google Scholar]
- Dorken ME, Pannell JR. The maintenance of hybrid zones across a disturbance gradient. Heredity. 2007;99:89–101. doi: 10.1038/sj.hdy.6800969. [DOI] [PubMed] [Google Scholar]
- Duchoslav M. Comparative ecology of Allium oleraceum and A. vineale. 2000 PhD thesis, Department of Botany, Faculty of Science, Palacký University, Olomouc, Czech Republic. [Google Scholar]
- Duchoslav M. Allium oleraceum and A. vineale in the Czech Republic: distribution and habitat differentiation. Preslia. 2001a;73:173–184. [Google Scholar]
- Duchoslav M. Small-scale spatial pattern of two common European geophytes Allium oleraceum and A. vineale in contrasting habitats. Biologia. 2001b;56:57–62. [Google Scholar]
- Duchoslav M. Effects of contrasting habitats on the phenology, seasonal growth, and dry-mass allocation pattern of two bulbous geophytes (Alliaceae) with partly different geographic ranges. Polish Journal of Ecology. 2009;57:15–32. [Google Scholar]
- Ehrendorfer F. Polyploidy and distribution. In: Lewis WH, editor. Polyploidy – biological relevance. New York: Plenum; 1980. pp. 45–60. [Google Scholar]
- Endler JA. Geographic variation, speciation and clines. Princeton, NJ: Princeton University Press; 1977. [PubMed] [Google Scholar]
- Engen S, Lande R, Saether BE. The spatial role of population fluctuation and quasi-extinction risk. American Naturalist. 2002;160:439–451. doi: 10.1086/342072. [DOI] [PubMed] [Google Scholar]
- Eriksson O. Clonal life histories and the evolution of seed recruitment. In: De Kroon H, van Groenendael J, editors. The ecology and evolution of clonal plants. Leiden: Backhuys Publishers; 1997. pp. 211–226. [Google Scholar]
- Felber F. Establishment of a tetraploid cytotype in a diploid population: effect of relative fitness of the cytotypes. Journal of Evolutionary Biology. 1991;4:195–207. [Google Scholar]
- Felber F, Bever JD. Effect of triploid fitness on the coexistence of diploids and tetraploids. Biological Journal of the Linnean Society. 1997;60:95–106. [Google Scholar]
- Fernandez Casas J, Pon-Sorolla A, Susanna A. Números cromosomáticos de plantas occidentales, 64–69. Anales del Jardin Botanico de Madrid. 1980;37:199–201. [Google Scholar]
- Fialová R. Polyploidní komplexy u rodu Allium. 1996 PhD thesis, Department of Botany, Faculty of Science, Palacký University, Olomouc, Czech Republic. [Google Scholar]
- Fowler N, Levin D. Ecological contrasts on the establishment of a novel polyploid in competition with its diploid progenitor. American Naturalist. 1984;124:703–711. [Google Scholar]
- Fritsche F, Kaltz O. Is the Prunella (Lamiaceae) hybrid zone structured by an environmental gradient? Evidence from a reciprocal transplant experiment. American Journal of Botany. 2000;87:995–1003. [PubMed] [Google Scholar]
- Frizen NV. Lukovyje Sibiri. Novosibirsk: Nauka; 1988. [Google Scholar]
- Geitler L, Tschermak-Woess L. Cytologie der Wildbestände von Allium carinatum und Allium oleraceum bein Lunz. Naturwissenschaften. 1946;33:27. [Google Scholar]
- Goldblatt P. Polyploidy in Angiosperms: monocotyledons. In: Lewis W, editor. Polyploidy, biological relevance. New York: Plenum; 1980. [Google Scholar]
- Gornall RJ, Wentworth JE. Variation in the chromosome number of Parnassia palustris L. in the British Isles. New Phytologist. 1993;123:383–388. [Google Scholar]
- Gotelli NJ, Ellison AM. A primer of ecological statistics. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- Grant V. Plant speciation. 2nd edn. New York: Columbia University Press; 1981. [Google Scholar]
- Guo YP, Saukel J, Mettermayr R, Ehrendorfer F. AFLP analyses demonstrate genetic divergence, hybridisation, and multiple polyploidisation in the evolution of Achillea (Asteraceae-Anthemideae) New Phytologist. 2005;166:273–289. doi: 10.1111/j.1469-8137.2005.01315.x. [DOI] [PubMed] [Google Scholar]
- Hæggström CA, Åström H. Allium oleraceum (Alliaceae) in Finland: distribution, habitats and accompanying vascular plant species. Memoranda Societas pro Fauna et Flora Fennica. 2005;81:1–18. [Google Scholar]
- Håkansson S. Allium vineale L. as a weed. Plant Husbandry. 1963;19:1–208. [Google Scholar]
- Halkka L. Chromosome counts of Finish vascular plants. Annales Botanici Fennici. 1985;22:315–317. [Google Scholar]
- Halverson K, Heard SB, Nason JD, Stireman JO. Origins, distribution, and local co-occurrence of polyploid cytotypes in Solidago altissima (Asteraceae) American Journal of Botany. 2008;95:50–58. doi: 10.3732/ajb.95.1.50. [DOI] [PubMed] [Google Scholar]
- Hancock JF, Bringhurst RS. Evolution in California populations of diploid and octoploid Fragaria (Rosaceae): a comparison. American Journal of Botany. 1981;68:1–5. [Google Scholar]
- Hardy OJ, Vekemans X. Patterns of allozyme variation in diploid and tetraploid Centaurea jacea at different spatial scales. Evolution. 2001;55:943–954. doi: 10.1554/0014-3820(2001)055[0943:poavid]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Hardy OJ, Vanderhoeven S, de Loose M, Meerts P. Ecological, morphological and allozymic differentiation between diploid and tetraploid knapweeds (Centaurea jacea) from a contact zone in the Belgian Ardennes. New Phytologist. 2000;146:281–290. doi: 10.1046/j.1469-8137.2000.00631.x. [DOI] [PubMed] [Google Scholar]
- Hintze J. NCSS 2001 – Number Cruncher Statistical System. Kaysville, UT: NCSS; 2001. [Google Scholar]
- Hlaváček R. Česká republika: autoatlas. Praha: Kartografie; 2000. [Google Scholar]
- Hroudová Z, Zákravský P. Ecology of two cytotypes of Butomus umbellatus. III. Distribution and habitat differentiation in the Czech and Slovak Republics. Folia Geobotanica et Phytotaxonomica. 1993;28:425–435. [Google Scholar]
- Hultén E, Fries M. Atlas of north European vascular plants north of the Tropic of Cancer I. Köningstein: Koeltz; 1986. [Google Scholar]
- Huntley B, Birks HJB. An atlas of past and present pollen maps for Europe. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Husband BC. The role of triploid hybrids in the evolutionary dynamics of mixed-ploidy populations. Biological Journal of the Linnean Society. 2004;82:537–546. [Google Scholar]
- Husband BC, Schemske DW. Cytotype distribution at a diploid–tetraploid contact zone in Chamerion (Epilobium) angustifolium (Onagraceae) American Journal of Botany. 1998;85:1688–1694. [PubMed] [Google Scholar]
- Jauzein P, Tison JM. Hypothèses sur les liens entre Allium oleraceum L., Allium oporinanthum Brullo, Pavone & Salmeri et Allium savii Parl. Journal de Botanique de la Societé de Botanique de la France. 1999;11:55–58. [Google Scholar]
- Jauzein P, Tison JM. Étude analytique du genre Allium L., sous-genre Codonoprasum (Reichenb.) Zahar., section Codonoprasum Rechenb., en France. Journal de Botanique de la Societé Botanique de la France. 2001;15:29–50. [Google Scholar]
- Jay M, Reynaud J, Blaise S, Cartier D. Evolution and differentiation of Lotus corniculatus/Lotus alpinus populations from French south-western Aps. III. Conclusions. Evolutionary Trends in Plants. 1991;5:157–160. [Google Scholar]
- Johnson MTJ, Husband BC, Burton TL. Habitat differentiation between diploid and tetraploid Galax urceolata (Diapensiaceae) International Journal of Plant Sciences. 2003;164:703–710. [Google Scholar]
- Kao RH. Asexuality and coexistence of cytotypes. New Phytologist. 2007;175:764–772. doi: 10.1111/j.1469-8137.2007.02145.x. [DOI] [PubMed] [Google Scholar]
- Kao RH. Origins and widespread distribution of co-existing polyploids in Arnica cordifolia (Asteraceae) Annals of Botany. 2008;101:145–152. doi: 10.1093/aob/mcm271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpavičienė B. Allium oleraceum populations: ecological attachment and reproduction. Botanica Lithuanica. 2002;8:103–110. [Google Scholar]
- Karpavičienė B. Allium genties rūšių paplitimas Lietuvoje. Botanica Lithuanica. 2004:19–30. Suppl. 6. [Google Scholar]
- Karpavičienė B. Chromosome numbers of Allium from Lithuania. Annales Botanici Fennici. 2007;44:345–352. [Google Scholar]
- Keeler KH. Distribution of polyploid variation in big bluestem (Andropogon gerardii, Poaceae) across the tallgrass prairie region. Genome. 1990;33:95–100. [Google Scholar]
- Keeler KH. Population biology of intraspecific polyploidy in grasses. In: Cheplick GP, editor. Population biology of grasses. Cambridge: Cambridge University Press; 1998. pp. 183–206. [Google Scholar]
- Keeler KH. Impact of intraspecific polyploidy in Andropogon gerardii (Poaceae) populations. American Midland Naturalist. 2004;152:63–74. [Google Scholar]
- Kish RY. Genus Allium L. (Alliaceae) in transcarpathian flora. Subgen. Allium and Amerallium Traub. Ukrainski Botanicheskii Zhurnal. 2001;58:693–700. (in Ukrainian) [Google Scholar]
- Kliber A, Eckert CG. Interaction between founder effect and selection during biological invasion in an aquatic plant. Evolution. 2005;59:1900–1913. [PubMed] [Google Scholar]
- Koenig WD. Spatial autocorrelation of ecological phenomena. Trends in Ecology and Evolution. 1999;14:22–26. doi: 10.1016/s0169-5347(98)01533-x. [DOI] [PubMed] [Google Scholar]
- Kolář F, Štech M, Trávníček P, Rauchová J, Urfus T, Vít P, Kubešová M, Suda J. Towards resolving the Knautia arvensis agg. (Dipsacaceae) puzzle: primary and secondary contact zones and ploidy segregation at landscape and microgeographic scales. Annals of Botany. 2009;103:963–974. doi: 10.1093/aob/mcp016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahulcová A. Chromosome numbers in selected monocotyledons (Czech Republic, Hungary, and Slovakia) Preslia. 2003;75:97–113. [Google Scholar]
- Kron P, Suda J, Husband BC. Applications of flow cytometry to evolutionary and population biology. Annual Review of Ecology, Evolution, and Systematics. 2007;38:847–876. [Google Scholar]
- Laane MM, Lie T. Fremstilling av kromosompreparater med enkle metoder. Blyttia. 1985;43:7–15. [Google Scholar]
- Legendre P, Anderson MJ. Distance-based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecological Monographs. 1999;69:1–24. [Google Scholar]
- Legendre P, Legendre L. Numerical ecology. Amsterdam: Elsevier Science; 1998. [Google Scholar]
- Lepš J, Šmilauer P. Multivariate analysis of ecological data using CANOCO. Cambridge: Cambridge University Press; 2003. [Google Scholar]
- Levan A. Cytological studies in the Allium paniculatum group. Hereditas. 1938;23:317–370. [Google Scholar]
- Levin DA. Minority cytotype exclusion in local plant populations. Taxon. 1975;24:35–43. [Google Scholar]
- Levin DA. Polyploidy and novelty in flowering plants. American Naturalist. 1983;122:1–25. [Google Scholar]
- Levin DA. The role of chromosomal change in plant evolution. Oxford: Oxford University Press; 2002. [Google Scholar]
- Lewis WH. Polyploidy in species populations. In: Lewis WH, editor. Polyploidy – biological relevance. New York: Plenum; 1980. pp. 103–144. [Google Scholar]
- Li BH, Xu XM, Ridout MS. Modelling the establishment and spread of autotetraploid plants in a spatially heterogeneous environment. Journal of Evolutionary Biology. 2004;17:562–573. doi: 10.1111/j.1420-9101.2004.00700.x. [DOI] [PubMed] [Google Scholar]
- Lillie RD. Simplification of the manufacture of Schiff reagent for use in histochemical procedures. Stain Technology. 1951;25:163–165. doi: 10.3109/10520295109113200. [DOI] [PubMed] [Google Scholar]
- Lövkvist B, Hultgard UM. Chromosome numbers in south Swedish vascular plants. Opera Botanica. 1999;137:1–42. [Google Scholar]
- Ložek B. Neživá příroda ve vztahu k flóře a vegetaci. In: Hejný S, Slavík B, editors. Květena České socialistické republiky 1. Praha: Academia; 1988. pp. 31–35. [Google Scholar]
- Lumaret R, Guillerm JL, Delay J, Ait Lhaj Loutfi A, Izco J, Jay M. Polyploidy and habitat differentiation in Dactylis glomerata L. from Galicia (Spain) Oecologia. 1987;73:436–446. doi: 10.1007/BF00385262. [DOI] [PubMed] [Google Scholar]
- Mable BK. Breaking down taxonomic barriers in polyploid research. Trends in Plant Sciences. 2003;8:582–590. doi: 10.1016/j.tplants.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Mable BK. Polyploidy and self-compatibility: is there an association? New Phytologist. 2004;162:803–811. doi: 10.1111/j.1469-8137.2004.01055.x. [DOI] [PubMed] [Google Scholar]
- Májovský J, Murín A. Karyotaxonomický prehlad flóry Slovenska. Bratislava: Veda; 1987. [Google Scholar]
- Mandáková T, Münzbergová Z. Distribution and ecology of cytotypes of the Aster amellus aggregates in the Czech Republic. Annals of Botany. 2006;98:845–856. doi: 10.1093/aob/mcl165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson J. Stomatal size in fossil plants: evidence for polyploidy in the majority of angiosperms. Science. 1994;264:421–424. doi: 10.1126/science.264.5157.421. [DOI] [PubMed] [Google Scholar]
- McArthur ED, Sanderson SC. Cytogeography and chromosome evolution of subgenus Tridentatae of Artemisia (Asteraceae) American Journal of Botany. 1999;86:1754–1775. [PubMed] [Google Scholar]
- Mehlich A. New extractant for soil test evaluation of phosphorus, potassium, magnesium, calcium, sodium, manganese and zinc. Communications in Soil Science and Plant Analysis. 1978;9:477–492. [Google Scholar]
- Meirmans PG, Vlot EC, den Nijs JCM, Menken SBJ. Spatial ecological and genetic structure of mixed population of sexual diploid and apomictic triploid dandelions. Journal of Evolutionary Biology. 2003;16:343–352. doi: 10.1046/j.1420-9101.2003.00515.x. [DOI] [PubMed] [Google Scholar]
- Měsíček J, Jarolímová V. List of chromosome numbers of the Czech vascular plants. Praha: Academia; 1992. [Google Scholar]
- Meusel H, Jäger E, Weinert E. Vergleichende Chorologie der zentraleuropäischen Flora. Jena: Gustav Fischer Verlag; 1965. [Google Scholar]
- Moore RJ. Index to plant chromosome numbers 1967–1971. Regnum Vegetabile. 1973;90:1–539. [Google Scholar]
- Mráz P, Šingliarová B, Urfus T, Krahulec F. Cytogeography of Pilosella officinarum (Compositae): altitudinal and longitudinal differences in ploidy level distribution in the Czech Republic and Slovakia and general pattern in Europe. Annals of Botany. 2008;101:59–71. doi: 10.1093/aob/mcm282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murín A, Svobodová Z, Májovský J, Feráková V. Chromosome numbers of some species of the Slovak Flora. Thaiszia – Journal of Botany. 1999;9:31–40. [Google Scholar]
- Müntzing A. The evolutionary significance of autopolyploidy. Hereditas. 1936;21:263–378. [Google Scholar]
- Ohryzek J. Srovnávací biologie cytotypů česneku planého (Allium oleraceum). 2007 MSc thesis, Department of Botany, Faculty of Science, Palacký University, Olomouc, Czech Republic. [Google Scholar]
- Omelchuk-Myakushko TY. Flora Evropejskoj tschasti SSSR 4. Leningrad: Nauka; 1979. Sem. 167. Alliaceae J. G. Agardh – Lukovyje; pp. 261–276. [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annual Review of Genetics. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Pastor J. Karyology of Allium species from the Iberian peninsula. Phyton. 1982;22:171–200. (Horn) [Google Scholar]
- Pastor J, Valdés B. Revisión del género Allium (Liliaceae) en peninsula Ibérica e Islas Baleares. Sevilla: Universidad de Sevilla Press; 1983. [Google Scholar]
- Perný M, Kolarčík V, Majeský L, Mártonfi P. Cytogeography of the Phleum pratense group (Poaceae) in the Carpathians and Pannonia. Botanical Journal of the Linnean Society. 2008;157:475–485. [Google Scholar]
- Petit C, Thompson JD. Species diversity and ecological range in relation to ploidy level in the flora of the Pyrenees. Evolutionary Ecology. 1999;13:45–66. [Google Scholar]
- Petit C, Bretagnolle F, Felber F. Evolutionary consequences of diploid–polyploid hybrid zones in wild species. Trends in Ecology and Evolution. 1999;14:306–311. doi: 10.1016/s0169-5347(99)01608-0. [DOI] [PubMed] [Google Scholar]
- Petřík P, Wild J. Environmental correlates of the patterns of plant distribution at the meso-scale: a case study from Northern Bohemia (Czech Republic) Preslia. 2006;78:211–234. [Google Scholar]
- Pogan E, Janjun A, Malecka J, Wcislo H. Further studies in chromosome number of Polish Angiosperms. Part XIX. Acta Biologica Cracoviensia Series Botanica. 1986;28:79–81. [Google Scholar]
- Pogan E, Jankun A, Sawicka Z. Further studies in chromosome numbers of Polish Angiosperms. Part 22. Acta Biologica Cracoviensia Series Botanica. 1990;31:1–17. [Google Scholar]
- Quitt E. Klimatické oblasti Československa. Praha: Academia; 1971. [Google Scholar]
- Ramsey J, Schemske D. Pathways, mechamisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology and Systematics. 1998;29:467–501. [Google Scholar]
- Rivero-Guerra AO. Cytogenetics, geographical distribution, and pollen fertility of diploid and tetraploid cytotypes of Santolina pectinata Lag. (Asteraceae: Anthemideae) Botanical Journal of the Linnean Society. 2008;156:657–667. [Google Scholar]
- Rodriguez D. A model for the establishment of polyploidy in plants: viable but infertile hybrids, iteroparity, and demographic stochasticity. Journal of Theoretical Biology. 1996;180:189–196. [Google Scholar]
- Ronsheim ML. Dispersal distances and predation rates of sexual and asexual propagules of Allium vineale. American Midland Naturalist. 1994;131:55–64. [Google Scholar]
- Rothera S, Davy A. Polyploidy and habitat differentiation in Deschampsia cespitosa. New Phytologist. 1986;102:449–467. [Google Scholar]
- Sádlo J. Diverzita vegetace České republiky, její příčiny a historický vývoj. In: Chytrý M, editor. Vegetace České republiky 1. Travinná a keříčková vegetace. Praha: Academia; 2007. pp. 53–64. [Google Scholar]
- Schönswetter P, Lachmayer M, Lettner C, et al. Sympatric diploid and hexaploid cytotypes of Senecio carniolicus (Asteraceae) in the Eastern Alps are separated along an altitudinal gradient. Journal of Plant Research. 2007;120:721–725. doi: 10.1007/s10265-007-0108-x. [DOI] [PubMed] [Google Scholar]
- Segraves K, Thompson JN. Plant polyploidy and pollination: floral traits and insect visits to diploids and tetraploid Heuchera grossularifolia. Evolution. 1999;53:1114–1127. doi: 10.1111/j.1558-5646.1999.tb04526.x. [DOI] [PubMed] [Google Scholar]
- Segraves K, Thompson JN, Soltis PS, Soltis DE. Multiple origins of polyploidy and the geographic structure of Heuchera grossularia. Molecular Ecology. 1999;8:253–262. [Google Scholar]
- Seregin AP. Floristic materials and a key for the genus Allium L. (Alliaceae) species in European Russia. Bjulleten Moskovskogo Obschestva Ispytalej Prirody, Otdel Boilogicheskij. 2005;110:45–51. (in Russian) [Google Scholar]
- Soltis DE, Soltis PS. Molecular data and the dynamics nature of polyploidy. Critical Reviews in Plant Sciences. 1993;12:348–352. [Google Scholar]
- Soltis DE, Soltis PS. The dynamic nature of polyploid genomes. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:8089–8091. doi: 10.1073/pnas.92.18.8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS. Polyploidy: recurrent formation and genome evolution. Trends in Ecology and Evolution. 1999;14:348–352. doi: 10.1016/s0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. The role of genetic and genomic attributes in the success of polyploids. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7051–7057. doi: 10.1073/pnas.97.13.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Tate JA. Advances in the study of polyploidy since Plant speciation. New Phytologist. 2003;161:173–191. [Google Scholar]
- Speta F. Über Oberösterreiches wildwachsende Laucharten (Allium L., Alliaceae) Linzer Biologische Beiträge. 1984;16:45–81. [Google Scholar]
- Staňková H. Populační genetika polyploidního komplexu česneku planého (Allium oleraceum) na území České republiky. 2005 MSc thesis, Department of Botany, Faculty of Science, Palacký University, Olomouc, Czech Republic. [Google Scholar]
- Stearn WT. Allium L. In: Tutin TG, Heywood VH, Burges NA, et al., editors. Flora Europaea 5. Cambridge: Cambridge University Press; 1980. pp. 49–69. [Google Scholar]
- Stebbins GL. Variation and evolution in plants. New York: Columbia University Press; 1950. [Google Scholar]
- Stebbins GL. Polyploidy, hybridization and the invasion of new habitats. Annals of the Missouri Botanical Garden. 1985;72:824–832. [Google Scholar]
- Stuessy TF, Weiss-Schneeweiss H, Keil DJ. Diploid and polyploid cytotype distribution in Melampodium cinereum and M. leucanthum (Asteraceae, Heliantheae) American Journal of Botany. 2004;91:889–898. doi: 10.3732/ajb.91.6.889. [DOI] [PubMed] [Google Scholar]
- Stutz HC, Sanderson SC. Evolutionary studies of Atriplex: chromosome races of A. confertifolia (shadscale) American Journal of Botany. 1983;70:1536–1547. [Google Scholar]
- Suda J. Sympatric occurrences of various cytotypes of Vaccinium sect. Oxycoccus (Ericaceae) Nordic Journal of Botany. 2002;22:593–601. [Google Scholar]
- Suda J, Malcová R, Abazid D, et al. Cytotype distribution in Empetrum (Ericaceae) at various spatial scales in the Czech Republic. Folia Geobotanica. 2004;39:161–171. [Google Scholar]
- Suda J, Krahulcová A, Trávníček P, Krahulec F. Ploidy level vs. DNA ploidy level: an appeal for consistent terminology. Taxon. 2006;55:447–450. [Google Scholar]
- Suda J, Weiss-Schneeweiss H, Tribsch A, Schneeweiss GM, Trávníček P, Schönswetter P. Complex distribution patterns of di-, tetra-, and hexaploid cytotypes in the European high mountain plant Senecio carniolicus (Asteraceae) American Journal of Botany. 2007a;94:1391–1401. doi: 10.3732/ajb.94.8.1391. [DOI] [PubMed] [Google Scholar]
- Suda J, Kron P, Husband BC, Trávníček P. Flow cytometry and ploidy: applications in plant systematics, ecology and evolutionary biology. In: Doležel J, Greilhuber J, Suda J, editors. Flow cytometry with plant cells. Weinheim: Wiley-Vch Verlag; 2007b. pp. 103–130. [Google Scholar]
- Šopova M. Cytological study in the genus Allium from Macedonia. Godishen Zbornik na Biologickiot Fakultet Universitet Skopje, Prirodno-Matematichki. 1972;24:83–102. [Google Scholar]
- Španiel S, Marhold K, Hodálová I, Lihová J. Diploid and tetraploid cytotypes of Centaurea stoebe (Asteraceae) in Central Europe: morphological differentiation and cytotype distribution patterns. Folia Geobotanica. 2008;43:131–158. [Google Scholar]
- Štěpánková J. Non-adaptive hypothesis of allopatric cytotype distribution in Myosotis lamottiana (Boraginaceae) Folia Geobotanica. 2001;36:147–161. [Google Scholar]
- Thompson J, Lumaret R. The evolutionary dynamics of polyploid plants: origins, establishment and persistence. Trends in Ecology and Evolution. 1992;7:302–307. doi: 10.1016/0169-5347(92)90228-4. [DOI] [PubMed] [Google Scholar]
- Tschermak-Woess E. Über chromosomale Plastizität bei Wildformen von Allium carinatum und anderen Allium–Arten aus den Ostalpen. Chromosoma. 1947;3:66–87. [Google Scholar]
- Váchová M, Feráková V. Löve Á, editor. Taxon. 1978;27:375–392. [Report] IOPB chromosome number reports LXI. [Google Scholar]
- Vakhtina LI. Chromosome numbers in some species ot the genus Allium (Alliaceae) in the Flora of USSR. Botanicheskii Zhurnal. 1984;70:700–701. [Google Scholar]
- Vakhtina LI, Kudrjassova TL. Kariosistematitcheskoje issledovanie nekotorych vidov sekcii Codonoprasum roda Allium (Alliaceae) Botanicheskii Zhurnal. 1985;70:76–88. [Google Scholar]
- Vosa CG. Heterochromatic banding patterns in Allium. Chromosoma. 1976;57:119–133. [Google Scholar]
- Weiss H, Dobeš C, Schneeweiss GM, Greimler J. Occurrence of tetraploid and hexaploid cytotypes between and within populations in Dianthus sect. Plumaria (Caryophyllaceae) New Phytologist. 2002;156:85–94. [Google Scholar]
- Wendel J. Genome evolution in polyploids. Plant Molecular Biology. 2000;42:225–249. [PubMed] [Google Scholar]
- Wendelbo P. Alliaceae. In: Rechinger KH, editor. Flora Iranica No. 76. Graz: Akademische Druck- und Verlagsanstalt; 1971. 100. [Google Scholar]
- Wetschnig W. Chromosomenzahlen Kärntner Gefäßpflanzen (Teil 3): Karyologie und Verbreitun der Allium-Arten (Alliaceae) in Kärnten. Carinthia. 1992;182:497–533. [Google Scholar]
- Willmans O. On the significance of demographic processes in phytosociology. In: White J, editor. The population structure of vegetation. Dordrecht: Dr. W. Jung Publishers; 1985. pp. 15–31. [Google Scholar]
- Wittmann H. Beiträge zur Karyologie de Gattung Allium und zur Verbreitung der Arten im Bundesland Salzburg (Österreich) Linzer Biologische Beiträge. 1984;16:83–104. [Google Scholar]
- Xie-Kui C, Ao CQ, Zhang Q, Chen LT, Liu JQ. Diploid and tetraploid distribution of Allium przewalskianum Regel. (Liliaceae) in the Qinghai-Tibetan Plateau and adjacent regions. Caryologia. 2008;61:190–198. [Google Scholar]
- Yamauchi A, Hosokawa A, Nagata H, Shimoda M. Triploid bridge and role of parthenogenesis in the evolution of autopolyploidy. The American Naturalist. 2004;164:101–112. doi: 10.1086/421356. [DOI] [PubMed] [Google Scholar]
- Zahariady C. Le sous-genre Codonoprasum (Genre Allium L., Fam. Alliaceae Agard, 1858) en Grèce et en Roumanie. IIe partie. Biologia Gallo-Hellenica. 1975;6:27–64. [Google Scholar]
- Zar JH. Biostatistical analysis. 4th edn. Upper Saddle River, NJ: Prentice Hall; 1996. [Google Scholar]
- Zbíral J. Analýza půd I. Jednotné pracovní postupy. Brno: Státní kontrolní a zkušební ústav zemědělský; 1995. [Google Scholar]
- Zbíral J. Analýza půd III. Jednotné pracovní postupy. Brno: Ústřední kontrolní a zkušební ústav zemědělský; 1997. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.