Abstract
Background and Aims
Iron (Fe) is necessary for plant growth and development. Although it is well known that Fe deficiency causes chlorosis in plants, it remains unclear how the Fe homeostasis is regulated in mesophyll cells. The aim of this work was to identify a gene related to Fe homeostasis in leaves.
Methods
A spontaneous mutant irm1, which revealed typical Fe-deficiency chlorosis, was found from Arabidopsis thaliana. Using map-based cloning, the gene responsible for the altered phenotype of irm1 was cloned. The expression of genes was analysed using northern blot hybridization and multiplex RT-PCR analysis. Further, GUS staining with transgenic promoter-GUS lines and transient expression of the fusion protein with GFP were used for detecting the expression pattern of the gene in different tissues and at different developmental stages, and for the subcelluar localization of the gene product.
Key Results
A point mutation from G to A at nucleotide 2317 of ClpC1 on chromosome V of Arabidopsis is responsible for the irm1 phenotype. The leaf chlorosis of the mutant irm1 and clpc1 (a T-DNA-inserted null mutant of ClpC1) could be converted to green by watering the soil with Fe solution. The expression intensity of ferric reductase FRO8 in irm1 and clpc1 was disordered (significantly higher than that of wild type).
Conclusions
The glycine residue at amino acid 773 of ClpC1 is essential for its functions. In addition to its known functions reported previously, ClpC1 is involved in leaf Fe homeostasis, presumably via chloroplast translocation of some nuclear-encoded proteins which function in Fe transport.
Keywords: Arabidopsis, ClpC1, iron homeostasis, chloroplasts, Clp protease
INTRODUCTION
Iron (Fe), an essential mineral nutrient for plant growth and development, functions as a component of many important enzymes and proteins involved in fundamentally biochemical processes, such as respiration and photosynthesis (Briat et al., 1995). Deficiency of Fe leads to chlorosis in young leaves and retarded growth, consequently resulting in reduced crop productivity and food quality. Although Fe is one of the six most abundant elements in the earth's crust, Fe deficiency is a worldwide problem for crop production on calcareous soils because it exists primarily in an oxidized ferric form [Fe(III)], which is not available to plants due to its low solubility. Therefore, plants have to activate specific mechanisms to mobilize and take up Fe from the soil. Dicotyledonous plants such as Arabidopsis use a reduction mechanism known as the strategy I mechanism (Römheld and Marschner, 1986) to acquire Fe from soils under stress of Fe limitation effectively. The reduction of ferric to ferrous Fe on root surface and subsequent transfer of ferrous ion into root cells are mainly accomplished by plasmalemma ferric chelate reductase FRO2 (FERRIC REDUCTASE/OXIDASE 2) (Robinson et al., 1999) and metal transporter IRT1 (IRON-REGULATED TRANSPORTER 1) (Eide et al., 1996; Henriques et al., 2002; Varotto et al., 2002; Vert et al., 2002). The expression of the two major Fe uptake genes in Arabidopsis is regulated by transcription factors FIT, bHLH38 and bHLH39 (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005, 2008). FIT interacts with bHLH38 or bHLH39, forming heterodimer FIT-bHLH38 or FIT-bHLH39, which directly functions in the activation of IRT1 and FRO2 transcription (Yuan et al., 2008).
In the cell, Fe is compartmentalized into different organelles, such as chloroplasts, mitochondria and vacuoles, for utilization or storage. In rapidly growing leaves, about 80 % of the Fe is localized in the chloroplasts, where Fe is involved in electron transport, photosynthesis, haem biosynthesis and Fe–S clusters (Kim and Guerinot, 2007). Although the necessity for Fe in the chloroplast has been clearly established, the processes involved in Fe homeostasis in this organelle are still relatively unknown. Recently, it was shown that AtPIC1 is involved in chloroplast Fe transport and that AtFRO7 is responsible for Fe reduction in chloroplasts (Duy et al., 2007; Jeong et al., 2008). The mitochondrion is also an organelle with a high demand for Fe (Briat et al., 2007) but the Fe transporters and reductase(s) involved have not been identified. Generally, it is unclear how Fe is regulated across the envelopes of chloroplasts and mitochondria.
ClpC [the subfamily C of Clp (caseinolytic protease) proteins] is a component of ATP-dependent proteases. As a molecular chaperone it plays an important role in protein quality control (Adam and Clarke, 2002; Hengge and Bukau, 2003; Constan et al., 2004). In higher plants, there are two chloroplast-localized ClpC proteins (ClpC1 and ClpC2). These two proteins possess similar functions because overexpression of ClpC2 was able to complement the clpc1 mutant to wild type (Kovacheva et al., 2007). Knocking out ClpC2 by T-DNA insertion, the mutant did not show distinct phenotypic changes (Park and Rodermel, 2004; Kovacheva et al., 2007), whereas loss of function of ClpC1 in the clpc1 mutant showed as clearly stunted growth with pale green leaves and a substantial reduction in accumulation in PSI and PSII (Constan et al., 2004; Sjogren et al., 2004). These suggest that ClpC1 is more important than ClpC2 in their biological functions.
Here, a new ClpC1 mutant was characterized and the glycine at the position of amino acid 773 of ClpC1 was identified as essential because substitution of the glycine to arginine via a point mutation exhibited the same phenotypes as the loss-of-function mutant generated by T-DNA insertion. It was also found that the leaves in the two mutants could be changed from pale green to a normal shade of green by watering the soil with Fe, indicating that ClpC1 is involved in Fe homeostasis in leaves.
MATERIALS AND METHODS
Plant growth conditions
Seeds of Arabidopsis thaliana (ecotype Columbia), irm1 mutant, transgenic and T-DNA insertion lines clpc1 (SALK_014058, obtained from the Nottingham Arabidopsis Stock Center) were surface-sterilized with 3 % commercial bleach for 10 min and then washed three times with distilled water. After vernalization at 4 °C in the dark for 2 d, the seeds were sown on plates with MS basal salt medium (Murashige and Skoog, 1962) supplemented with 3 % sucrose and 0·6 % phytogel at pH 5·8 and germinated at 23 °C with a 16-h light period for 7 d. The seedlings were then used for further analysis.
For seed harvest and hybridization, Arabidopsis plants were grown in potting soil mixture and kept in growth chambers at 22 °C with illumination at 120 mmol m−2 s−1 for a 16-h daily light period. The relative humidity was approx. 70 % (±5 %). For nutrition experiments, seedlings were grown in potting soil watered with 100 µm FeEDTA, 100 µm MnSO4 or 30 µm ZnSO4 every 3 d.
Map-based cloning of IRM1
The irm1 (Columbia background) mutant was crossed with Landsberg erecta to create mapping populations. The plants with mutation phenotypes were selected and used for genetic mapping of IRM1 with simple sequence length polymorphism (SSLP) markers, which were designed based on information from the Monsanto Arabidopsis Polymorphism Database (Jander et al., 2002).
Vector constructions and Arabidopsis transformation
The PClpC1::GUS construction was generated by fusing the ClpC1 promoter (1·699 kb), which was amplified with primers 5′-ccatcatcttggGTCATCCTCTGTTTCTCAAG-3′ and 5′-cccgggGACTTCCTAAATAAATT-3′ from genomic DNA, in front of the β-glucuronidase (GUS) coding sequence in pCAMBIA1381 vector. To generate complementation lines of the irm1 mutant, a 7·3-kb fragment including the native promoter and the ClpC1 gene was cut out from BAC (bacterial artificial chromosome) clone K3K7 with restriction endonucleases SalI and BglII and cloned into pCAMBIA1391 vector. Arabidopsis transformation via Agrobacterium tumefaciens strain (GV3101) was carried out by the floral dip method (Clough and Bent, 1998).
Subcellular localization of ClpC1
To investigate the subcellular location of ClpC1, the coding sequence of ClpC1 was amplified from A. thaliana cDNA by PCR using the 5′-aagcttATGGCTATGGCCACAAGGGT-3′ and 5′-GGATCCAATGTAATTGTGACCAAG-3′ primers. The PCR product was cloned into the pGEM T-easy vector (Promega, USA). After verification by sequencing, the sequence of ClpC1 was excised by HindIII and BamHI and cloned into the pJIT163–hGFP vector to generate the ClpC1–GFP fusion protein (GFP at the C-terminus of ClpC1) expression plasmid pJIT163–ClpC1–hGFP. For transient expression in onion epidermis cells, 5 µg of the pJIT163–ClpC1–hGFP plasmid was used to bombard the onion epidermis cells according to a described procedure (Weigel and Glazebrook, 2002). For transient expression in cowpea protoplasts, 10–15 mg of the purified pJIT163–ClpC1–hGFP plasmid was used to transform cowpea mesophyll protoplasts by PEG transformation according to a described procedure (Shah et al., 2002). The GFP fluorescence in onion epidermis cells and in cowpea protoplasts were detected using a confocal laser scanning microscope (Olympus, FV500, Japan). The GFP fluorescence was observed at an excitation wavelength of 488 nm and an emission wavelength of 506–538 nm, and the autofluorescence of chloroplasts was observed at an excitation wavelength of 488 nm and an emission wavelength of 664–696 nm.
GUS activity assay
Five independent transgenic lines were selected for the GUS staining analysis. Seedlings were grown on MS plates without antibiotics until they reached three-to-four-true-leaf stage. One true leaf from a single plant was excised and examined by histochemical GUS staining. Seedlings with positive GUS signals were transferred to soil and grown further for GUS assays on the developing seeds. The progenies of these plants were plated on MS medium and sampled during germination and early seedling stages. For the metal-deficiency experiments, 4-d-old seedlings were transferred onto MS medium lacking Fe, zinc (Zn) or manganese (Mn). After growing for 4 d in the growth chamber with a 16-h light period, plants were harvested. For the histochemical GUS assay, plant tissues were stained by submersion in staining solution (Jefferson et al., 1987).
Northern blot analysis
Total RNA was extracted using the Trizol reagent from roots and shoots of plants cultivated in MS medium with and without Fe supply following the instructions of the manufacturer (Invitrogen, USA). For northern blot analysis, 10 µg of total RNA was denatured and electrophoresed on a 1·2 % 3-(N-morpholino)-propane-sulfonic acid/formaldehyde/agarose gel, and then blotted onto a nylon Hybond-N+ membrane according to the manufacturer's instructions (Amersham, Buckinghamshire, UK). The probes, which were amplified by PCR using primers (5′-GTTCCCAATGATGGTCGTAGC-3′ and 5′-TTGATGCTTGACTCCGAGGTT-3′ for AtFRO2, 5′-TCTCTCCAGCAACTTCAACT-3′ and 5′-AATGACTCGGTATCGCAAGA-3′ for AtIRT1, and 5′-CCGGAGATAGATAGAGAGAG-3′ and 5′-CGATTCCAGTACCCTCACCA-3′ for ClpC1) from cDNA, were labelled with [32P]dCTP. The hybridization, probe labelling and membrane washing were performed as described by Church and Gilbert (1984). Subsequently, the membranes were exposed to Fujifilm (BAS-SR2025, Tokyo, Japan) in an X-ray imaging plate under −70 °C for 1–3 d.
Multiplex RT-PCR analysis
For expression analysis of PIC1, FRO6, FRO7 and FRO8, total RNA was extracted from the leaf of 2-week-old seedlings grown on soil. For expression analysis of ClpC1, ClpC2, NRAMP3 and NRAMP4, RNA was extracted from 3-d-old seedlings grown on an MS plate. The expression pattern of these genes was detected using the GenomeLab GeXP analysis system multiplex RT-PCR assay (Beckman, Fullerton, CA, USA) according to the protocol described by Chen et al. (2007). The housekeeping gene ACTIN2 (At1g49240) was used as the control. The gene expression data obtained from multiplex RT-PCR were normalized via dividing the peak area value of each gene by the peak area of ACTIN2. The primers used in the multiplex RT-PCR reactions are listed in Table 1.
Table 1.
Primers used in the Multiple RT-PCR experiment in this paper
| Gene name | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| FRO6 | 5′-AGGTGACACTATAGAATACACATTTGTCTCCTCCAAGA-3′ | 5′-GTACGACTCACTATAGGGAATAGCCTAAACCTCGGGA-3′ |
| FRO7 | 5′-AGGTGACACTATAGAATATCGGTGATCAAATCCTCACA-3′ | 5′-GTACGACTCACTATAGGGACAGCTGCAGAAACAACTCCA-3′ |
| FRO8 | 5′-AGGTGACACTATAGAATAATGGCTTGCCACATTAGTCC-3′ | 5′-GTACGACTCACTATAGGGAATAGTTGCAGCGAAACCACC-3′ |
| PIC1 | 5′-AGGTGACACTATAGAATAACTGCTAGTGGGATTGCTGC-3′ | 5′-GTACGACTCACTATAGGGACCGAGTAGTGCTGATCCCAT-3′ |
| ClpC1 | 5′-AGGTGACACTATAGAATAGTGCGACAGGCAATGAATG-3′ | 5′-GTACGACTCACTATAGGGACCCTCACCAATAAGACCA-3′ |
| ClpC2 | 5′-AGGTGACACTATAGAATAGCTCTTCTTACACCTCCTTT-3′ | 5′-GTACGACTCACTATAGGGAATCATCTTCACACGCCCT-3′ |
| Nramp3 | 5′-AGGTGACACTATAGAATATTGCGTTTTTAGATCCAGGG-3′ | 5′-GTACGACTCACTATAGGGAAACCAAAAGACCCATTGCTG-3′ |
| Nramp4 | 5′-AGGTGACACTATAGAATAAGCAAATCATGGGCAGTTTC-3′ | 5′-GTACGACTCACTATAGGGAGAAGCCACTAGTTGCCAAGG-3′ |
| ACTIN2 | 5′-AGGTGACACTATAGAATACTGGATTCGCTGGAGATGAT-3′ | 5′-GTACGACTCACTATAGGGACTTCAGGAGCAATACGGAGC-3′ |
Elemental analysis
Wild-type and irm1 were grown on soil under a 16-h light period. Plant tissues were pooled from three plants and assayed as a group. Each experimental set was composed of five groups. Leaves were collected when plants started to bolt, and dried overnight in an oven at 65 °C after washing with ultra-pure water. Seeds were harvested when they were fully dry. The concentrations of various metals were determined using inductively coupled plasma spectroscopy (Perkin Elmer, USA) according to methods published previously (Yuan et al., 2008). The means and standard deviations were calculated from three biological replications.
Electron microscopy
For observation of chloroplast structure, new true leaves of the seedlings grown on soil were harvested. Leaves were cut into approx. 1-cm-long segments and then washed in 0·5 mm CaSO4. The segments were fixed overnight in 0·1 mm potassium phosphate buffer, pH 7·4, containing 0·2 % (w/v) glutaraldehyde and 1·5 % (w/v) paraformaldehyde. After being rinsed three times in 0·1 mm potassium phosphate buffer, pH 7·4, the tissue was dehydrated through a graded ethanol series of 20 % and 40 %, and post-fixed in 0·25 % (w/v) osmium tetroxide for 2 h in 40 % ethanol at 4 °C. Leaf segments were washed again in 40 % ethanol three times (each 10 min) and treated in a solution with 0·3 % (w/v) uranyl acetate in 40 % ethanol for 2 h at 4 °C. The material was washed again twice in 40 % ethanol (10 min each) and once in 50 % ethanol for 10 min. The samples were then dehydrated in an ethanol series of 75 % and 90 %, twice at 100 %, for 30 min each, infiltrated with LR White resin, and polymerized at 50 °C for 24 h in vacuum. Ultra-thin sections were cut with a microtome (Ultracut E; Reichert, Vienna) and stained with uranyl acetate and lead citrate. Semi-thin sections were stained with toluidine blue. Sections used for electron microscopy were examined in an electron microscope (EM 902; Zeiss, Jena, Germany).
Chlorophyll isolation and quantification
Whole plants of wild type, irm1 and clpc1 grown under normal and Fe-supplemented (100 µm FeEDTA) conditions were harvested 4 weeks after germination and weighed. They were then ground with sand in 80 % acetone. The ground tissue was spun at approx. 2000 g for 5 min to remove the sand and other debris from the extracted chlorophyll. The supernatant was then measured spectrophotometrically at 645 nm and 663 nm, respectively. The amount of chlorophyll (mg g−1) in each sample was determined using the equation given in Arnon (1949).
RESULTS
Isolation and characterization of irm1 mutant
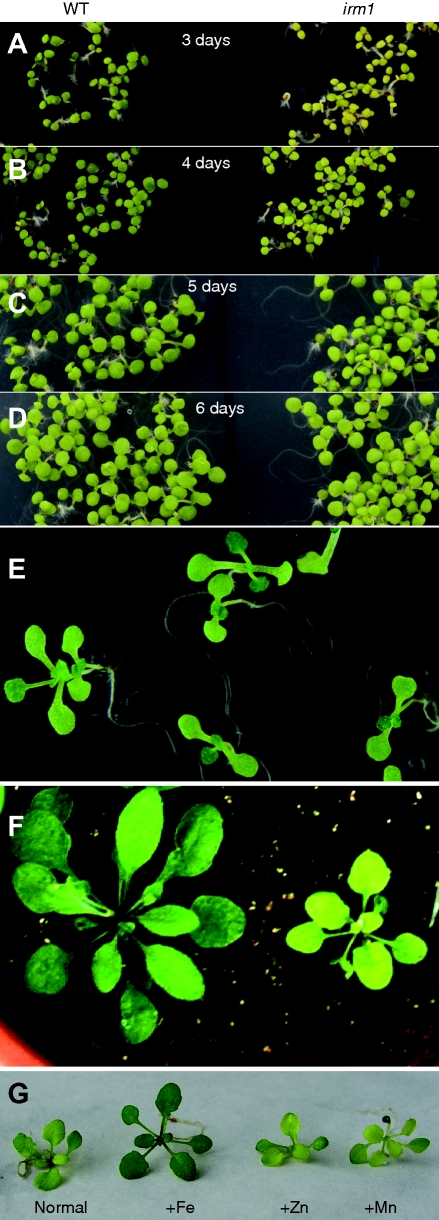
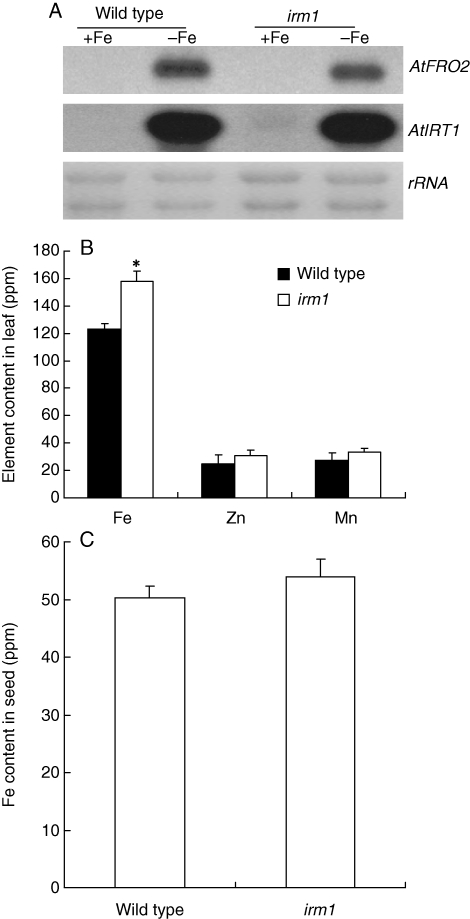
A plant with chlorotic leaves was found from the segregation population of a heterozygote T-DNA insertion line of AtFRO4 in A. thaliana ecotype Columbia (Col-0) grown in a growth chamber. PCR analysis showed that no T-DNA fragment was inserted in AtFRO4 of this plant (data not shown). To purify the genetic background, the mutant was backcrossed with Col-0 five times. Then, it was used for further analysis. The mutant displayed yellow cotyledons at an earlier stage of germination (3 d) on MS medium than wild type (Fig. 1A). With extension to germination, the cotyledons of the mutant turned increasingly green (Fig. 1B–D). By the 10th day, the mutant plant revealed green cotyledons and true leaves like the wild type on MS medium (Fig. 1E). After transplantation to soil, the mutant showed pale green leaves and retarded growth (Fig. 1F). To test whether the pale green of the mutant phenotype on soil is due to deficiency of some mineral nutrients, additional Fe, Zn and Mn were supplied to the soil by watering. Interestingly, the pale leaves of the mutant turned to green 3 d after watering with Fe, but not by watering with Zn or Mn (Fig. 1G). Therefore, this mutant was called iron-rescued mutant 1 (irm1). Subsequently, irm1 was crossed with wild-type Col-0. The F1 plants displayed normal growth like the wild type and segregation of wild type and mutant phenotype was observed in F2 at a ratio of 3 : 1 (data not shown), demonstrating that the altered phenotype in irm1 is caused by a recessive mutation in a single nuclear-encoded gene. To determine whether the mutation of IRM1 affects the expression of Fe-uptake genes, the expression of FRO2 and IRT1 was examined by northern-blot analysis. As shown in Fig. 2A, the expression pattern and intensity of FRO2 and IRT1 in irm1 was the same as the wild type, indicating that the mutation of IRM1 did not affect the expression of the two marker genes for Fe uptake. To demonstrate further whether IRM1 is involved in Fe homeostasis, the Fe content of mutant seeds and leaves was determined. The Fe content in leaves of irm1 was about 20 % higher than that of wild type (Fig. 2B) whereas no difference was observed in seeds between the mutant and wild type (Fig. 2C). The content of Zn and Mn in leaves was also measured. It was higher in leaves of irm1 than that of wild type, but the difference was not significant (Fig. 2B).
Fig. 1.
Phenotypic characterization of irm1 mutant: (A–D) phenotype comparison between irm1 and wild type at an early stage of germination (3, 4, 5 and 6 d) on an MS agar plate; (E) irm1 seedlings grown on an MS agar plate for 10 d; (F) wild type and irm1 grown on soil for 2 weeks; (G) phenotype of irm1 on soil (normal) and when watered with 100 µm FeEDTA (+Fe), 30 µm ZnSO4 (+Zn) or 100 µm MnSO4 (+Mn). The picture was taken on the 3rd day after treatment.
Fig. 2.
Fe-deficiency responses and Fe content in wild type and irm1: (A) northern blot analysis of Fe uptake genes FRO2 and IRT1 in roots of seedlings exposed to Fe deficiency (−) and sufficiency (+); (B) contents of Fe, Zn and Mn in leaves of seedlings growing on soil for 2 weeks; (C) Fe content in seeds. Data are shown as means ± standard error of the mean (n = 4).
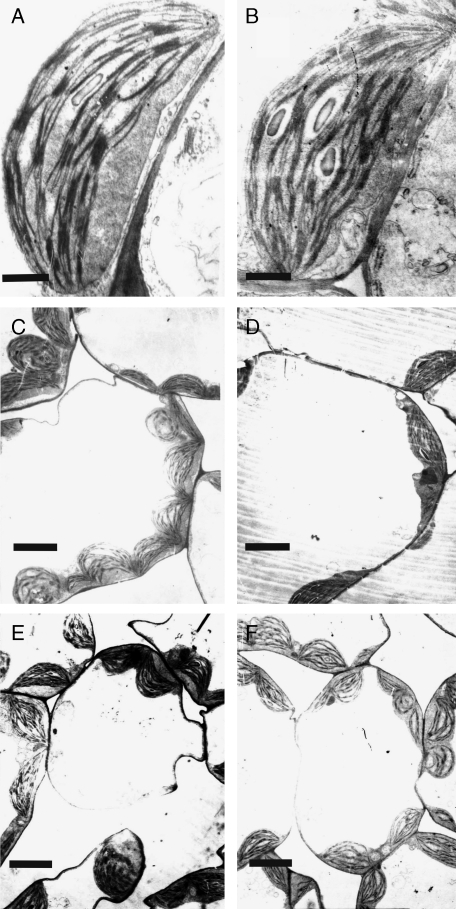
Considering the pale green phenotype of irm1 leaves of soil-grown plants, their chlorophyll content was determined. Consistent with the phenotype, the chlorophyll content of irm1 (132·3 ± 8·03 µg Chl g−1 f. wt) was approx. 43 % lower than that of wild type (232·3 ± 5·83 µg Chl g−1 f. wt). Further, the chloroplast ultrastructure in young leaves of irm1 plants grown on soil was investigated using transmission electron microscopy. Chloroplasts of irm1 revealed a shape similar to those seen in wild type, such as characteristic lens-like shape and well-developed thylakoid membrane system with numerous grana stacks (Fig. 3A, B). However, the leaves of the irm1 mutant revealed fewer chloroplasts per cell (4·2 ± 0·3) than those of the wild type (9 ± 0·8; Fig. 3C, D). When watered with Fe, the chloroplast number in leaf cells of irm1 was restored to the level of the wild type (Fig. 3E, F).
Fig. 3.
Transmission electron micrographs of chloroplast structure (A, B) and mesophyll cells (C–F) of irm1 and wild type: (A and B) chloroplast structure of young leaves of wild type (A) and irm1 (B); (C and D) an overview of a mesophyll cell of young leaves of wild type (C) and irm1 (D); (E, F) transmission electron micrographs of a mesophyll cell of wild type (E) and irm1 (F) grown on soil watered with Fe solution. Scale bar = 1 µm.
A point mutation in ClpC1 is responsible for the irm1 phenotype
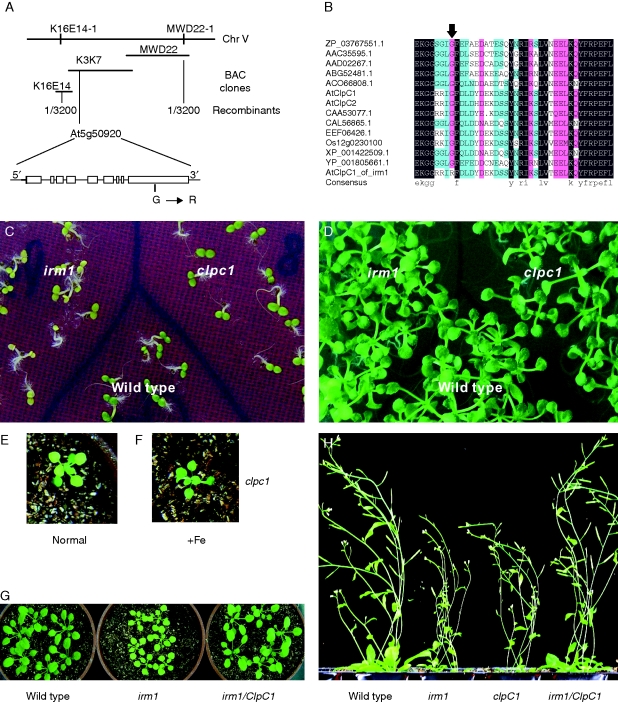
To identify the gene responsible for the irm1 phenotype, irm1 was crossed with Landsberg erecta and mapped using SSLP (single sequence length polymorphic) markers. Approximately 3200 chlorotic F2 mutant progenies were genotyped with SSLP markers, and irm1 was mapped to a 150-kb interval between the SSLP markers K16E14-1 and MWD22-1 at the bottom of chromosome V, which includes three BACs (K16E14, K3K7 and MWD22). To detect the candidate gene of irm1, the 42 predicted open reading frames in the interval were amplified and sequenced. Comparison with the known genomic sequence of Col-0 revealed that ClpC1 in irm1 showed a substitution of G to A at nucleotide 2317, resulting in conversation of glycine to arginine at amino acid 773 of the protein (Fig. 4A), whereas no sequence difference was observed among the other 41 open reading frames between irm1 and wild type, implying that ClpC1 is the candidate for irm1. The sequences of ClpC1 homologues from different phylogenetic kingdoms were obtained from the database and compared at amino acid level. It revealed that the glycine at position of amino acid 773 is highly conserved among the proteins of ClpC (Fig. 4B). The results indicate that the glycine at amino acid 773 may be essential for the function of ClpC1, strongly suggesting that the point mutation resulting in conversion of glycine to arginine at amino acid 773 in ClpC1 is the reason for the altered phenotypes in the mutant irm1.
Fig. 4.
(A) The schematic outline of genetic and physical maps of IRM1 and the structure of the candidate gene At5g50920 (ClpC1) localized on BAC clone K3K7 (the open boxes represent exons and the lines among the open boxes represent introns). The site of mutation G → R is shown. (B) Sequence alignment of ClpC proteins from Arabidopsis (AtClpC1 and AtClpC2), Nostoc azollae (ZP_03767551·1), Trichodesmium erythraeum (ABG52481·1), Guillardia theta (AAC35595·1), Rhodomonas salina (AAD02267·1), Micromonas sp. RCC299 (AC066808·1), Populus trichocarpa (EEF06426·1), Brassica napus (CAA53077·1), Ostreococcus tauri (CAL56865·1), Oryza sativa (Os12g0230100), Ostreococcus lucimarinus CCE9901 (XP_001422509·1) and Cyanothece sp. ATCC 51142 (YP_001805661·1). The amino acid indicated by arrow is the mutation site found in the irm1 mutant. (C and D) Phenotypic comparison of irm1, clpc1 (a T-DNA insertion mutant of ClpC1) and wild type grown on an MS agar plate for 4 d (C) and for 10 d (D). (E and F) Phenotypic recovery of leaf chlorosis of clpc1 in soil, 3 d after being watered with Fe solution (100 µm FeEDTA) (+Fe). (G and H) Phenotypic complementation by expression of ClpC1 in an irm1 mutant at early (G) and a late stage (H).
To confirm the hypothesis that ClpC1 is the gene of irm1, a 7·3-kb fragment including the native promoter and the coding sequence of ClpC1 was cut out from the BAC clone K3K7 and introduced into the irm1 genome by Agrobacterium tumefaciens-mediated transformation. The transgenic plants exhibited normal growth like the wild-type (Fig. 4G, H), indicating that the defect functions in irm1 was complemented by expression of ClpC1. The chlorophyll content of the transgenic plants also recovered to the same level as wild type (data not shown). These results demonstrate that a single base change (G to A) at nucleotide 2317 of ClpC1 in irm1 is responsible for the altered phenotypes (chlorosis and retarded growth).
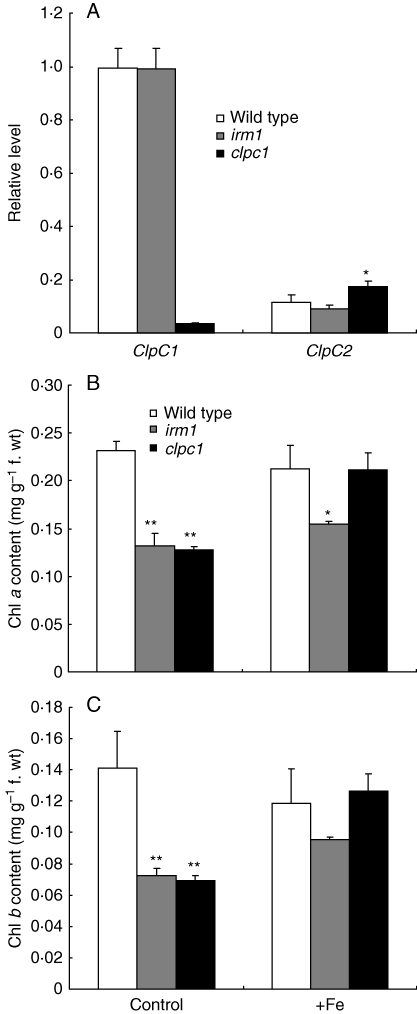
Further, T-DNA insertion ClpC1-mutant line clpc1 (hsp93-v-2, SALK_014058; Kovachva et al., 2005) was obtained from Nottingham Arabidopsis Stock Center and characterized. As shown in Fig. 4C–E, clpc1 displayed a similar phenotype to irm1 (yellow cotyledons at an early stage of germination, chlorotic leaves and retarded growth on soil), and its leaf chlorosis was also restored to normal by watering with Fe (Fig. 4E, F). However, irm1 showed much stronger mutation phenotypes than clpc1 (Fig. 4C, H). Considering the function redundancy of ClpC1 and ClpC2, it is speculated that the phenotypic difference between irm1 and clpc1 may be caused by the different expression levels of ClpC2 in the two mutant plants, so the ClpC2 expression levels in wild type, irm1 and clpc1 were analysed. As shown in Fig. 5A, the expression level of ClpC2 in plants of clpc1 is significantly higher than that of irm1 or wild type.
Fig. 5.
Analysis of gene expression and chlorophyll content in wild type, irm1 and clpc1: (A) ClpC1 and ClpC2 expression level in wild type and the irm1 and clpc1 mutants; (B) chlorophyll a content in wild type, irm1 and clpc1; (C) chlorophyll b content in wild type, irm1 and clpc1.
Further, the chlorophyll content was determined in wild type, irm1 and clpc1 plants under normal and Fe-supplemented (100 µm) conditions in the soil. The chlorophyll a and b content in irm1 and clpc1 was significantly lower than in wild type under normal growth conditions, but increased obviously when the soil was watered with Fe, especially in clpc1, where the chlorophyll a and b content even reached the same level as wild type (Fig. 5B, C). The ratio of chlorophyll a and b in wild type (1·82 ± 0·12), irm1 (1·82 ± 0·05) and clpc1 (1·85 ± 0·06) was not altered.
Expression profile of ClpC1 and the localization of its protein
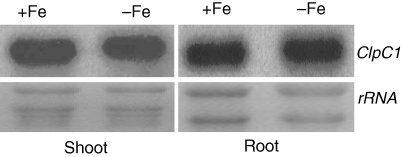
In previous studies of ClpC1, the focus was only on its role in chloroplast development (Constan et al., 2004; Sjogren et al., 2004; Kovacheva et al., 2005, 2007; Nakagawara et al., 2007). Given the Fe-related phenotype in irm1, it was of great interest to determine whether ClpC1 expression is related to Fe status. Therefore, RNA-blot analysis was performed with total RNA extracted from the roots and shoots of Arabidopsis seedlings exposed either to Fe-sufficient or -deficient conditions. ClpC1 mRNA was detected in shoots and roots, and it was more abundant in shoots than that in roots (Fig. 6). No difference in ClpC1 expression intensity was observed between the Fe-sufficient and -deficient conditions in either shoots or roots (Fig. 6).
Fig. 6.
Northern blot analysis of ClpC1 expression in roots and shoots under Fe-sufficient (+Fe) and -deficient conditions (−Fe).
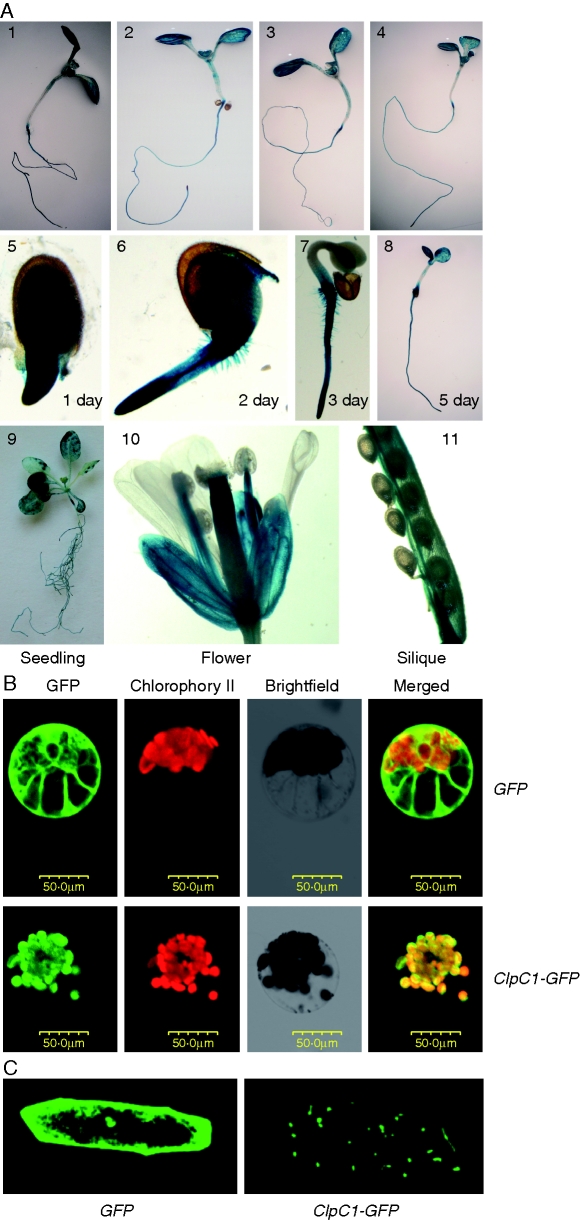
To localize ClpC1 expression in plant tissues, the putative promoter (1699 bp of genomic DNA sequence upstream of the transcription start site of ClpC1) was amplified and fused with the coding sequence of GUS. The ClpC1 promotor::GUS construct (PClpC1::GUS) was introduced into Arabidopsis and GUS activity of stable transformants was analysed. Consistent with the results of northern-blot analysis, GUS activity was detected in both root and shoot, and its expression abundance was not affected by the status of Fe, Zn or Mn (Fig. 7A1–4). GUS expression was also strongly detected in germinating seeds, cotyledons (Fig. 7A5–8), seedlings, filaments and siliques (Fig. 7A9–11).
Fig. 7.
Expression pattern analysis of ClpC1 and subcellular localization of the ClpC1 protein. (A) Histochemical analysis of ClpC1 expression pattern using ClpC1 promoter-GUS transgenic lines: (1–4) GUS staining of the seedlings treated with Fe (2), Zn (3) or Mn (4) deficiency for 1 week; (5–8) GUS staining in the seedlings at 1, 2, 3 and 5 d after germination; (9–11) GUS staining of seedlings at the five-leaf stage and flowers and siliques from plants grown on soil. (B) Subcellular localization of ClpC1 by transient expression of ClpC1–GFP in cowpea protoplasts and in onion epidermis cells. The gene GFP (control) and ClpC1–GFP (a fusion gene of ClpC1 and GFP) under control of the 35S promoter were separately introduced into cowpea protoplasts using PEG transformation and expressed. (C) GFP (control) and ClpC1–GFP were transiently expressed in onion epidermis cells introduced by bombardment. The GFP signal is localized in chloroplasts of cowpea protoplasts (B) and in plastids of onion epidermis cells (C) when transformed with 35S::ClpC1–GFP, whereas the GFP signal was spread through the whole cytoplasm in the cells that were transformed with 35S::GFP. Scale bar = 50 µm.
Previous biochemical work indicated that ClpC1 protein localized to the chloroplasts (see review, Adam et al., 2006). To confirm this, an expression plasmid containing a ClpC1–GFP fusion protein controlled by 35S promoter was created and transformed into cowpea protoplasts and onion epidermis cells for transient expression analysis. As shown in Fig. 7B, the ClpC1–GFP protein was clearly localized in chloroplasts of the transformed cowpea protoplasts (Fig. 7B) and in plastids of onion epidermis cells (Fig. 7C).
Expression of genes related to Fe homeostasis in the irm1 mutant
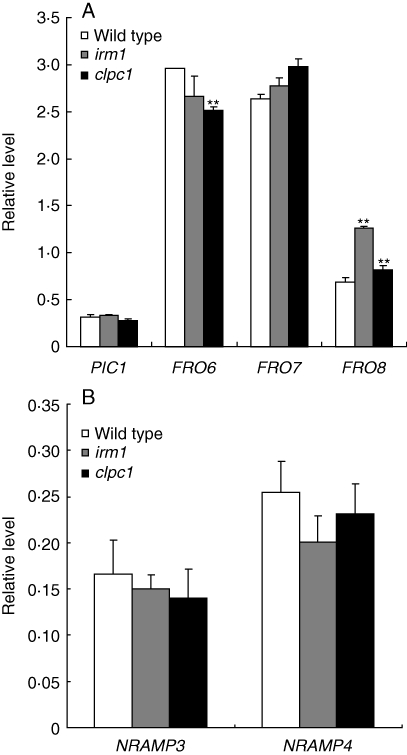
In A. thaliana, there exist eight ferric-chelate reductase genes. Of them, FRO6, FRO7 and FRO8 mainly expressed in leaves, are speculated to be involved in Fe reduction in the chloroplasts and mitochondria (Wu et al., 2005; Mukherjee et al., 2006; Jeong et al., 2008). PIC1 is localized to the inner envelope of the chloroplast and is critical for chloroplast development (Duy et al., 2007). So, the expression of the four genes in the seedling leaves of irm1 and wild type grown on soil were analysed by multiplex RT-PCR. As shown in Fig. 8A, the expression intensity of FRO8 in irm1 and clpc1 was significantly higher than that of wild type, whereas FRO6 displayed a lower expression in the two mutants than that of wild type, especially in clpc1. Same as FRO8, FRO7 also showed a higher expression in irm1 and clpc1 than that in wild type, but the difference was not significant. For PIC1, no expression difference was observed between the mutants and wild type (Fig. 8A).
Fig. 8.
Relative quantitative analysis of the expression intensities of PIC1, FRO6/7 and FRO8 in leaves of 2-week-old plants grown on soil (A), NRAMP3 and NRAMP4 in cotyledons of 3-d-old seedlings (B) by multiplex RT-PCR analysis.
NRAMP3 and NRAMP4 are two Fe transporter proteins localized on the vacuole membrane and involved in mobilization of Fe stored in the vacuole during germination (Thomine et al., 2003; Lanquar et al., 2005). Considering the yellow phenotype of irm1 at an early stage of germination, the expression of the two genes was checked in 3-d-old seedlings. The expression level of NRAMP3 and NRAMP4 in irm1 and clpc1 was clearly lower than that of wild type, although the difference was not significant (Fig. 8B).
DISCUSSION
The glycine residue at amino acid 773 of ClpC1 is essential for its functions
In this work, the spontaneous mutant irm1 found in the Columbia ecotype of A. thaliana, which showed yellow cotyledons at an early stage of germination, chlorotic phenotype and retarded growth on soil, was characterized. Map-based cloning confirmed that a single nucleotide change (G to A) at nucleotide 2317 of ClpC1, resulting in conversation of glycine to arginine at amino acid 773 of the protein, is the reason for the altered phenotypes of irm1. Sequence comparison revealed that the glycine residue at amino acid 773 of ClpC1 is highly conserved among ClpC proteins from different phylogenetic kingdoms (Fig. 4B). Additionally, irm1 exhibited similar phenotypes to clpc1, a null mutant of ClpC1 generated by T-DNA insertion. These results imply that the glycine at amino acid 773 is an essential amino acid residue for the function of ClpC1, and substitution of the glycine to arginine due to the point mutation from G to A in irm1 led to loss of function, although its expression abundance was not altered (Fig. 5A).
ClpC1 is involved in leaf Fe homeostasis of Arabidopsis
Previous studies demonstrated that ClpC1, as a stromal molecular chaperone, is required for importing protein to the chloroplast, and plays a vital role in chloroplast development and its functions. Knockout of ClpC1 by T-DNA insertion in Arabidopsis resulted in a lower protein translocation rate in isolated chloroplasts, reduced plant growth and chloroplast development (Constan et al., 2004; Sjögren et al., 2004; Kovacheva et al., 2007). Here, we strongly suggest that the ClpC1 protein is also involved in Fe homeostasis in Arabidopsis leaves for the following reasons: (a) the point mutation from G to A at nucleotide 2317 of ClpC1 is responsible for the altered phenotypes in the irm1 mutant because the expression of wild-type ClpC1 in irm1 could fully convert the mutant to wild type; (b) the mutant exhibited Fe-deficiency chlorosis, although the Fe content in its leaves was significantly higher than that of wild type; (c) watering with an Fe solution could turn the chlorotic leaves of the T-DNA knockout mutant clcp1 and the mutant irm1, growing on soil, from yellow to green; (d) the expression of the genes, such as FRO6, FRO7, FRO8, NRAMP3 and NRAMP4, related to Fe homeostasis in leaves and to Fe mobilization during germination was disordered. Considering the higher Fe content in irm1 leaves (Fig. 2B), and no changes in the expression pattern of the Fe uptake genes FRO2 and IRT1 in the irm1 mutant (Fig. 2A), we speculate that ClpC1 may be involved in Fe homeostasis in leaf cells other than absorption and translocation from roots to shoots. ClpC1 is localized to chloroplasts and plastids (Fig. 7B and C), accumulates mainly in the stroma (Shanklin et al., 1995), and is also associated with the chloroplast protein translocation machinery in the inner envelope (Nielsen et al., 1997). It functions by controlling protein quality and translocation of precursor proteins encoded by nuclear genes. Loss of ClpC1 results in lower protein translocation rates in chloroplasts (Constan et al., 2004; Kovacheva et al., 2005). As chlorosis of the mutant irm1 and clpc1 leaves was removed by watering the plants with Fe solution, it is reasonable to assume that ClpC1, as a molecular chaperone, is involved in translocating some nuclear-encoding proteins that function in Fe transport to the chloroplasts in leaf cells. In the mutants, the nuclear-encoding genes FRO6, FRO7 and FRO8, which are Fe3+-chelate reductase and mainly express in leaves (Wu et al., 2005; Mukherjee et al., 2006), revealed a disordered expression (lower for FRO6, higher for FRO7 and FRO8) in comparison with wild type (Fig. 8). Therefore, the disorder of Fe homeostasis in irm1 leaves might, directly or indirectly, be the result of translocation of these or some other unknown proteins, from the cytoplasm into the chloroplasts, being affected, and, consequently, reducing their ability to transport Fe into the chloroplasts, resulting in the Fe-deficiency phenotype even though there is a high Fe content in the leaves. However, to understand how ClpC1 functions in the regulation of Fe homeostasis in Arabidopsis leaves needs to be studied in more detail.
Mobilizing Fe from vacuoles and transferring it into the cytoplasm is an essential process for seed germination (Lanquar et al., 2005). NRAMP3 and NRAMP4, two metal transporters localized on the vacuole membrane, are involved in this process because knockout of the two genes leads to the arrest of germination under low Fe nutrition. In mutant irm1 and clpc1, the expression intensity of NRAMP3 and NRAMP4 was lower than that of wild type on the third day of germination (Fig. 8B). According to this, we speculate that the yellow cotyledons of irm1 and clpc1 at an early stage of germination might be due to reduced Fe mobilization from the vacuole via the reduced expression of NRAMP3 and NRAMP4 in seedlings. As Fe acquisition is shifted from seed to culture medium at the late stage of germination, the irm1 seedlings will restore their normal phenotype (Fig. 1A–E). As shown in Fig. 4C, irm1 displayed a stronger phenotype than clpc1. This can be explained by the reduced expression of NRAMP4 in clpc1 being much less than that in irm1 (Fig. 8B). It is possible to explain that the increased expression of ClpC2 in clpc1 (not in irm1) is the reason for its chlorotic cotyledon phenotype being alleviated (Figs 4C and 5A). ClpC2 is a homologue of ClpC1 and reveals functional redundancy with ClpC1 (Kovacheva et al., 2007). However, more experiments are needed to confirm it.
In summary, the spontaneous mutant irm1 is caused by a single nucleotide substitution of G to A at nucleotide 2317 of ClpC1, resulting in conversion of glycine to arginine at amino acid 773. This also demonstrated that the glycine at amino acid 773 is an essential amino acid residue for ClpC1 functions. Here, it has been discovered that ClpC1, in addition to its known functions reported previously, is involved in Fe homeostasis in leaf cells of Arabidopsis, based on the fact that chlorotic leaves of clpc1 and irm1 plants grown on soil could be turned green by watering them with an Fe solution. The observed results of this work should shed new light on understanding the functions of ClpC1.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (grant no. 30700487) and Chinese Academy of Sciences (grant no. KSCX2-YW-N-056).
LITERATURE CITED
- Adam Z, Clarke AK. Cutting edge of chloroplast proteolysis. Trends in Plant Science. 2002;7:451–456. doi: 10.1016/s1360-1385(02)02326-9. [DOI] [PubMed] [Google Scholar]
- Adam Z, Rudella A, van Wijk KJ. Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Current Opinion in Plant Biology. 2006;9:234–240. doi: 10.1016/j.pbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Aron DI. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in BETA VULGARIS. Plant Physiology. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat JF, Fobis-Loisy I, Grignon N, et al. Cellular and molecular aspects of iron metabolism in plants. Biology of the Cell. 1995;84:69–81. [Google Scholar]
- Briat JF, Curie C, Gaymard F. Iron utilization and metabolism in plants. Current Opinion in Plant Biology. 2007;10:276–282. doi: 10.1016/j.pbi.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Chen QR, Vansant G, Oades K, et al. Diagnosis of the small round blue cell tumors using multiplex polymerase chain reaction. Journal of Molecular Diagnostics. 2007;9:80–88. doi: 10.2353/jmoldx.2007.060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proceedings of the National Academy of Sciences of the USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML. The essential basic helix–loop–helix protein FIT1 is required for the iron deficiency response. The Plant Cell. 2004;16:3400–3412. doi: 10.1105/tpc.104.024315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constan D, Froehlich JE, Rangarajan S, Keegstra K. A stromal Hsp100 protein is required for normal chloroplast development and function in Arabidopsis. Plant Physiology. 2004;136:3605–3615. doi: 10.1104/pp.104.052928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duy D, Wanner G, Meda AR, von Wiren N, Soll J, Philippar K. PIC1, an ancient permease in Arabidopsis chloroplasts, mediates iron transport. The Plant Cell. 2007;19:986–1006. doi: 10.1105/tpc.106.047407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide DJ, Broderius M, Fett J, Guerinot ML. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proceedings of the National Academy of Sciences of the USA. 1996;93:5624–28. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R, Bukau B. Proteolysis in prokaryotes: protein quality control and regulatory principles. Molecular Microbiology. 2003;49:1451–1462. doi: 10.1046/j.1365-2958.2003.03693.x. [DOI] [PubMed] [Google Scholar]
- Henriques R, Jasik J, Klein M, et al. Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defects. Plant Molecular Biology. 2002;50:587–597. doi: 10.1023/a:1019942200164. [DOI] [PubMed] [Google Scholar]
- Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P. FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Letters. 2004;577:528–534. doi: 10.1016/j.febslet.2004.10.062. [DOI] [PubMed] [Google Scholar]
- Jander G, Norris S, Rounsley S, Bush D, Levin I, Last R. Arabidopsis map-based cloning in the post-genome era. Plant Physiology. 2002;129:440–450. doi: 10.1104/pp.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Cohu C, Kerkeb L, Pilon M, Connolly E, Guerinot Ml. Chloroplast Fe(III) chelate reductase activity is essential for seedling viability under iron limiting conditions. Proceedings of the National Academy of Sciences of the USA. 2008;105:10619–10624. doi: 10.1073/pnas.0708367105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Guerinot Ml. Mining iron: iron uptake and transport in plants. FEBS Letters. 2007;581:2273–2280. doi: 10.1016/j.febslet.2007.04.043. [DOI] [PubMed] [Google Scholar]
- Kovacheva S, Bedard J, Patel R, et al. In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. The Plant Journal. 2005;41:412–428. doi: 10.1111/j.1365-313X.2004.02307.x. [DOI] [PubMed] [Google Scholar]
- Kovacheva S, Bedard J, Wardle A, Patel R, Jarvis P. Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. The Plant Journal. 2007;50:364–379. doi: 10.1111/j.1365-313X.2007.03060.x. [DOI] [PubMed] [Google Scholar]
- Lanquar V, Lelievre F, Bolte S, et al. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO Journal. 2005;24:4041–4051. doi: 10.1038/sj.emboj.7600864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee I, Campbell NH, Ash JS, Connolly EL. Expression profiling of the Arabidopsis ferric chelate reductase (FRO) gene family reveals differential regulation by iron and copper. Planta. 2006;223:1178–1190. doi: 10.1007/s00425-005-0165-0. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Nakagawara E, Sakuraba Y, Yamasato A, Tanaka R, Tanaka A. Clp protease controls chlorophyll b synthesis by regulating the level of chlorophyllide a oxygenase. The Plant Journal. 2007;49:800–809. doi: 10.1111/j.1365-313X.2006.02996.x. [DOI] [PubMed] [Google Scholar]
- Nielsen E, Akita M, Davila-Aponte J, Keegstra K. Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO Journal. 1997;16:935–946. doi: 10.1093/emboj/16.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Rodermel SR. Mutations in ClpC2/Hsp100 suppress the requirement for FtsH in thylakoid membrane biogenesis. Proceedings of the National Academy of Sciences of the USA. 2004;101:12765–12770. doi: 10.1073/pnas.0402764101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML. A ferric-chelate reductase for iron uptake from soils. Nature. 1999;397:694–697. doi: 10.1038/17800. [DOI] [PubMed] [Google Scholar]
- Römheld V, Marschner H. Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiology. 1986;80:175–180. doi: 10.1104/pp.80.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K, Russinova E, Gadella TW, Willemse J, de Vries SC. The Arabidopsis kinase-associated protein phosphatase controls internalization of the somatic embryogenesis receptor kinase 1. Genes & Development. 2002;16:1707–1720. doi: 10.1101/gad.220402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J, DeWitt ND, Flanagan JM. The stroma of higher plant plastids contains ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: an archetypal two-component ATP-dependent protease. The Plant Cell. 1995;7:1713–1722. doi: 10.1105/tpc.7.10.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren LL, MacDonald TM, Sutinen S, Clarke AK. Inactivation of the ClpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiology. 2004;136:4114–4126. doi: 10.1104/pp.104.053835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomine S, Lelievre F, Debarbieux E. AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. The Plant Journal. 2003;34:685–695. doi: 10.1046/j.1365-313x.2003.01760.x. [DOI] [PubMed] [Google Scholar]
- Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F, Leister D. The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. The Plant Journal. 2002;31:589–599. doi: 10.1046/j.1365-313x.2002.01381.x. [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldechamp F, et al. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. The Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. Arabidopsis: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- Wu H, Lihua L, Du J, Yuan Y, Cheng X, Ling H-Q. Molecular and biochemical characterization of the Fe(III) chelate reductase gene family in Arabidopsis thaliana. Plant Cell Physiology. 2005;46:1505–1514. doi: 10.1093/pcp/pci163. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, et al. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Research. 2008;18:385–397. doi: 10.1038/cr.2008.26. [DOI] [PubMed] [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling H-Q. AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Research. 2005;15:613–621. doi: 10.1038/sj.cr.7290331. [DOI] [PubMed] [Google Scholar]