Abstract
The metazoan nuclear pore complex (NPC) disassembles during mitosis, and many of its constituents distribute onto spindles and kinetochores, including the Nup107-160 sub-complex1,2. We have found that Nup107-160 interacts with the γ-tubulin ring complex (γ-TuRC), an essential and conserved microtubule (MT) nucleator3,4, and recruits γ-TuRC to unattached kinetochores. Unattached kinetochores nucleate MTs in a manner that is regulated by the Ran GTPase5; such MTs contribute to the formation of kinetochore fibers (k-fibers)6, MT bundles connecting kinetochores to spindle poles. Our data indicate that Nup107-160 and γ-TuRC act cooperatively to promote spindle assembly through MTs nucleation at kinetochores: HeLa cells lacking Nup107-160 or γ-TuRC were profoundly deficient in kinetochore-associated MT nucleation. Moreover, co-precipitated Nup107-160/γ-TuRC complexes nucleate MT formation in assays using purified tubulin. While Ran did not regulate MTs nucleation by γ-TuRC alone, Nup107-160/γ-TuRC complexes required Ran-GTP for MT nucleation. Our observations collectively show that Nup107-160 promotes spindle assembly through Ran-GTP-regulated nucleation of MT by γ-TuRC at kinetochores, and reveal a novel relationship between nucleoporins and the MT cytoskeleton.
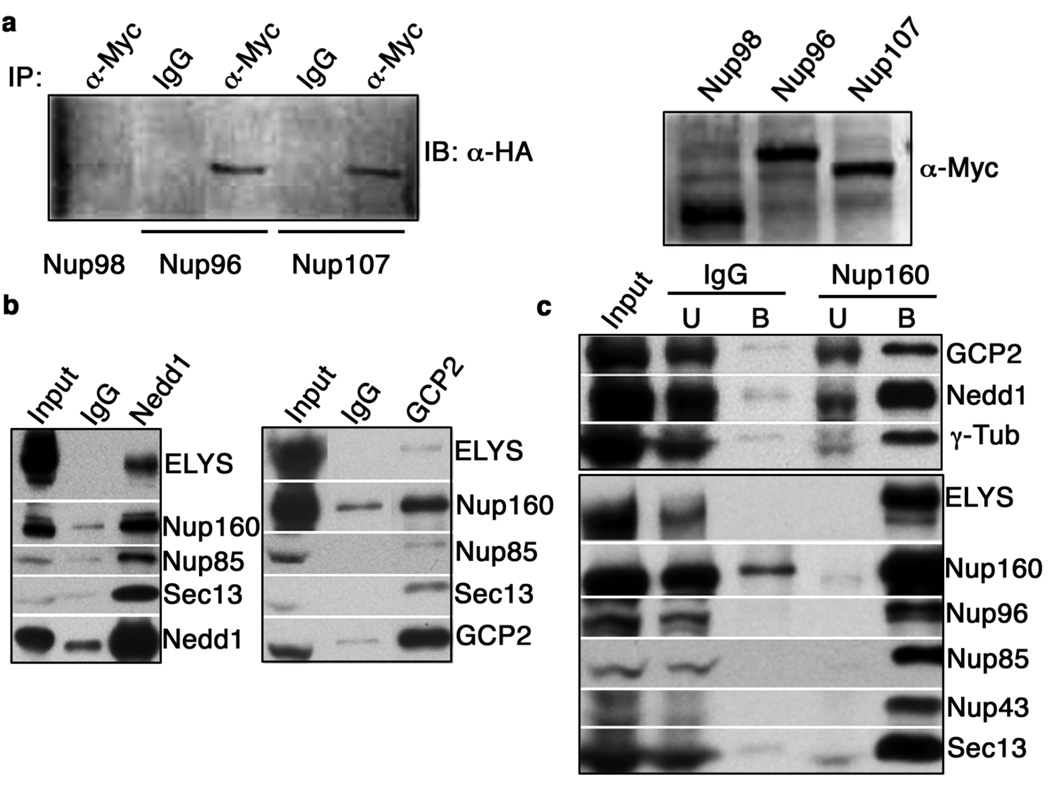
Nup107-160 is a stable complex that consists of nine nuclear pore complex (NPC) proteins (nucleoporins)7: Nup160, Nup133, Nup107, Nup96, Nup85, Nup43, Nup37, Sec13, and Seh1, as well as a sub-stochiometric component, ELYS. Nup107-160 remains kinetochore-bound throughout mitosis and shows enhanced accumulation on unattached kinetochores1. Nup107-160 has been implicated in spindle assembly in yeast8, mammalian tissue culture cells9 and Xenopus egg extracts (XEEs)1. To understand the nature of spindle defects in the absence of kinetochore-bound Nup107-160, we sought to identify proteins that interact with Nup107-160. We performed immunoprecipitation from HeLa cell extracts with anti-Nup96 antibodies, and analyzed the co-precipitated proteins by mass spectrometry. Among the precipitated proteins, we found GCP2 and GCP3, core members of γ-TuRC4. To confirm the association between these complexes in mammalian cells, we co-expressed HA-tagged GCP3 (HA-GCP3) and myc-tagged versions of Nup107-160 members (myc-Nup96, myc-Nup107) or other nucleoporins (myc-Nup98) in 293T cells. We performed immunoprecipitations from lysates of these cells using anti-myc antibodies, and Western blotted the precipitated proteins with antibodies against the HA tag. HA-GCP3 clearly co-precipitated with myc-Nup96 and myc-Nup107, but not with myc-Nup98 (Fig. 1a), indicating its specific association to the Nup107-160 complex.
Figure 1. Nup107-160 binds γ-TuRC.
(a) 293T cells were co-transfected with plasmids encoding HA-GCP3 and myc-Nup98, myc-Nup96 or myc-Nup107. Immunoprecipitations were performed with anti-myc antibodies or non-specific IgG, and precipitates were subjected to Western blotting with anti-HA antibodies (left panel). A Western blot of input extracts with anti-myc antibodies is shown in the right panel.
(b) Anti-xGCP2 and -xNedd1 antibodies were used for immunoprecipitation from XEEs. In each case, untreated XEE is shown on the left, the precipitated fraction is shown on the right, and the center lane shows proteins eluted from control IgG-coated beads. Uncropped images of the blots are shown in Supplementary Information, Fig. S3.
(c) XEEs were depleted of xNup107-160 using anti-xNup160 antibodies. Co-precipitating proteins (B) and the unbound fraction (U) were analyzed by Western blotting against xNup107-160 or against γ-TuRC components, as indicated. For comparison, similar fractions associated with IgG-coated beads are shown, as well as untreated extracts (Input). Uncropped images of the blots are shown in Supplementary Information, Fig. S3.
To examine whether this interaction is conserved in other vertebrate species, we cloned cDNAs of Xenopus laevis Nup107-160 components and generated polyclonal antibodies against them (Supplementary Information, Fig. S1). Throughout this report, Xenopus nucleoporins will be distinguished as xNups. We similarly raised antibodies against the Xenopus laevis homologues of GCP2, as well as Nedd1, another γ-TuRC component3,4. In immunoprecipitation experiments, anti-xGCP2 and -xNedd1 antibodies co-precipitated xNup107-160 components from XEEs (Fig. 1b; Supplementary Information, Fig. S3). Conversely, anti-xNup160 antibodies co-precipitated xGCP2, xNedd1 and γ-tubulin (Fig. 1c; Supplementary Information, Fig. S3). γ-TuRC components were co-precipitated with all other antibodies against xNup107-160 complex members, although the extent of co-precipitation varied amongst antibodies (data not shown). We did not observe co-precipitation with multiple other nucleoporins that are not components of Nup107-160 (Fig. 1a and data not shown). These findings confirmed that Nup107-160 and γ-TuRC interact in amphibians as well as in mammals.
Depletion of Nup107-160 or γ-TuRC blocks spindle assembly around sperm chromosomes in M-phase (CSF) XEEs1,10. Chromatin-coated beads within CSF XEEs can also form MTs and spindles through mechanisms that depend on Ran11. We found that the assembly of these structures was similarly dependent upon both xNup107-160 and γ-TuRC (data not shown). We hypothesized that γ-TuRC acts with Nup107-160 to nucleate MTs during early stages of k-fiber assembly. We made three predictions based upon this idea: First, γ-TuRC should localize to kinetochores with Nup107-160. Second, kinetochore-associated MT nucleation should require both Nup107-160 and γ-TuRC. Third, complexes containing xNup107-160 and γ-TuRC should have the capacity to nucleate MTs.
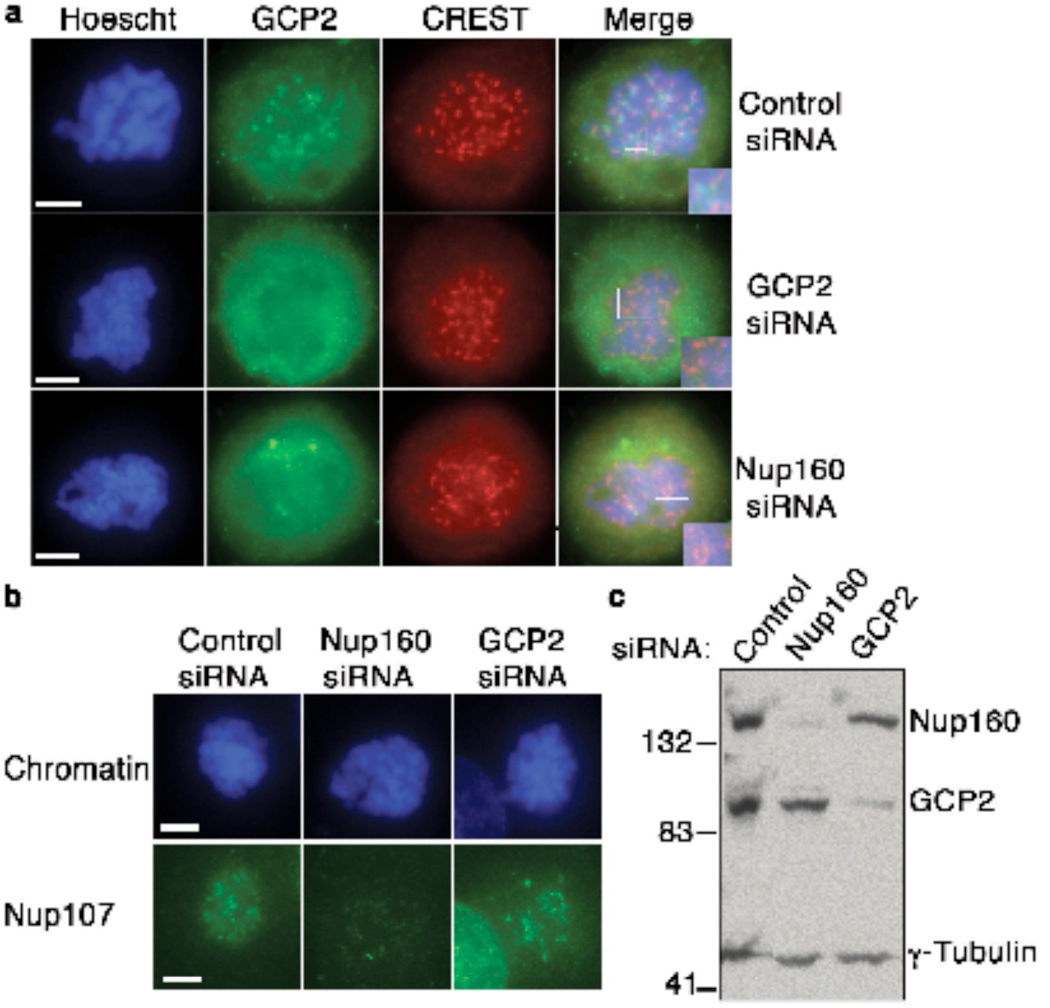
To test the first prediction, we examined γ-TuRC localization in nocodazole-treated HeLa cells by immunostaining with human auto-antibodies against centromeric antigens (CREST sera)12 and antibodies against GCP2 (Fig. 2a). GCP2 localized in foci that were juxtaposed to CREST antigens at mitotic kinetochores, in a pattern that was similar to that pattern of Nup107-160 localization (Supplementary Information, Fig. S2a). We likewise stained kinetochores of mitotic chromosomes assembled in XEEs treated with nocodazole, using antibodies against a kinetochore marker (Bub1) and against GCP2, Nedd1, γ-Tubulin or components of Nup107-160 (Supplementary Information, Fig. S2b). We observed co-localization of Bub1 and γ-TuRC, indicating that γ-TuRC is recruited to unattached kinetochores in a manner similar to Nup107-160.
Figure 2. γ-TuRC is recruited to kinetochores by Nup107-160.
(a) HeLa cells were depleted of GCP2 (second row) or Nup160 (third row) by siRNA. The cells were treated with nocodazole for 16 hrs and processed for indirect immunofluorescence with antibodies against GCP2 (second column, green) and kinetochore marker CREST serum (third column, red) and counterstained with Hoechst 33342 dye to visualize chromosomal DNA (left, blue). Inset shows enlarged, merged image of kinetochores. Top row shows HeLa cells transfected with a control siRNA, treated in an identical manner. Scale bar = 5 µm.
(b) HeLa cells were treated as in (a), stained with antibodies against Nup107 (second row, green) and counterstained with Hoechst 33342 dye to visualize chromosomal DNA (top row, blue).
(c) Whole cell extracts from control HeLa cells (left), cells depleted of Nup160 (center) or cells depleted of GCP2 (right) were analyzed by Western blotting with the indicated antibodies to monitor the overall level of depletion through siRNAs.
We further wished to determine whether γ-TuRC and Nup107-160 are recruited to kinetochores in a coordinated fashion. To address this question, we transfected HeLa cells with control oligonucleotides or siRNAs designed to inhibit synthesis of GCP2 or Nup160, and stained with antibodies against components of the γ-TuRC and Nup107-160 complexes. In nocodazole-arrested cells depleted of GCP2, we observed a clear loss of GCP2 from kinetochores (Fig. 2a). Notably, we did not observe changes in the levels Nup107 at kinetochores of GCP2-depleted cells (Fig. 2b), indicating that the Nup107-160 complex is recruited in a manner that does not require γ-TuRC. By contrast, when we depleted Nup160 we observed a loss of both Nup107 and γ-TuRC from kinetochores (Fig. 2a,b), clearly indicating that the Nup107-160 complex is essential for accumulation of γ-TuRC on kinetochores. Similar patterns were observed with oligonucleotides directed against other members of the Nup107-160 complex (Nup133, Nup107, Seh1), but not with control oligonucleotides (data not shown).
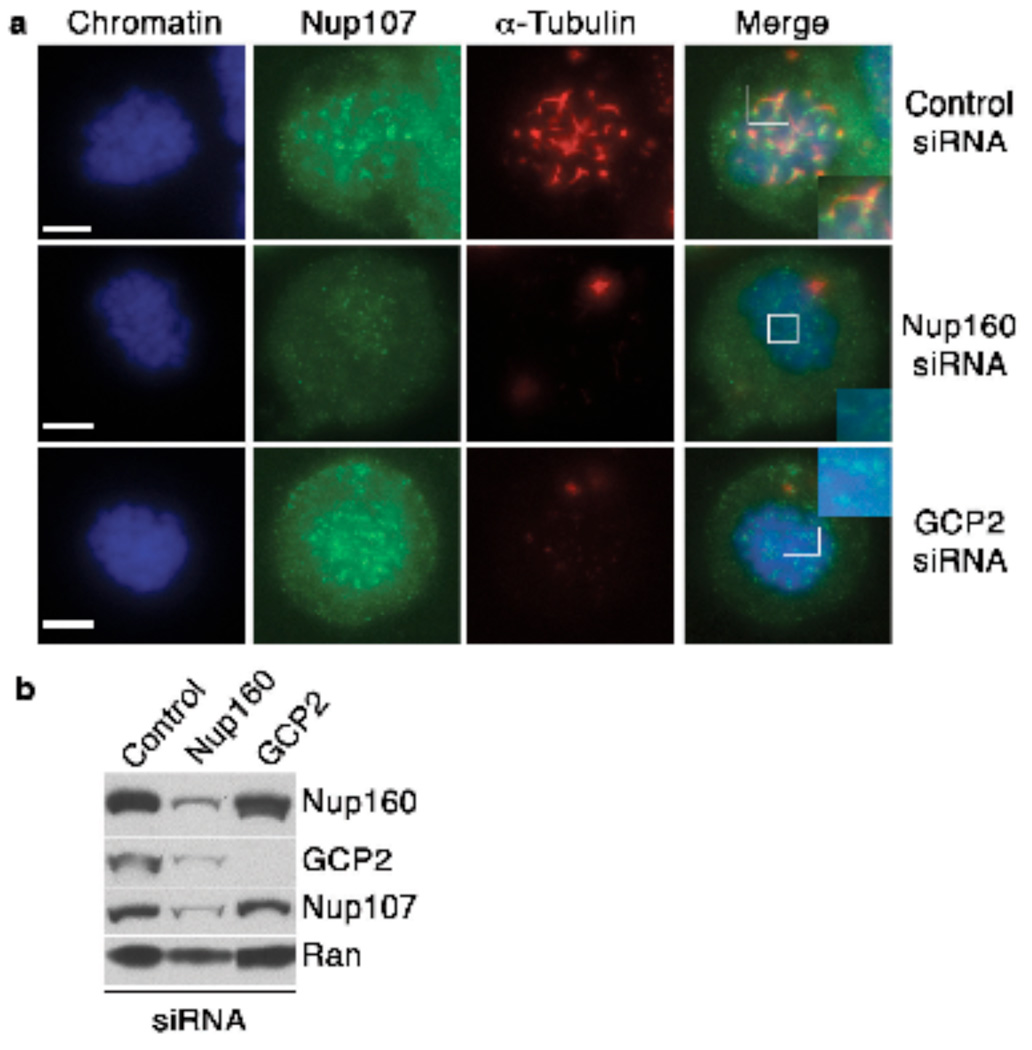
To test whether kinetochore-associated MT nucleation requires Nup107-160 or γ-TuRC, we performed a nocodazole wash-out assay13 using HeLa cells transfected with control oligonucleotides or siRNAs against Nup160 or GCP2. (Fig. 3a,b; Supplementary Information, Fig. S3). We synchronized cells in mitosis with nocodazole, which was removed by washing with fresh, cold media, maintaining MTs in a de-polymerized state. The cells were warmed to 37°C, followed by fixation and staining with anti-α-tubulin antibodies (Fig. 3a). Control cells showed MTs associated to kinetochores (kMTs) as early as 60 seconds after temperature shift, which grew to give rise to MT fibers within 5 minutes. However, cells depleted of Nup160 or GCP2 showed no kMT seeds adjacent to chromatin or kinetochores at early times, and a striking deficiency of kMTs at later times. Similar results were obtained after depletion of other Nup107-160 components (data not shown). These finding validate our second prediction, that both Nup107-160 and γ-TuRC are indispensable for MT nucleation at kinetochores.
Figure 3. Nup107-160 is critical for MT nucleation at kinetochores in HeLa cells.
(a) HeLa cells transfected with a control siRNA (top panels), a siRNA against Nup160 (middle panels) or an siRNA directed against GCP2 (bottom panels). The cells were analyzed in a MT re-growth assay at 2 minutes after being returned to 37°C, with staining for α-tubulin (red) and Nup107 (green). Mitotic cells were identified by chromosome morphology through counterstaining with Hoechst 33342 dye (blue). Scale bar= 5 µm
(b) Whole cell extracts were made from control HeLa cells (left), or cells depleted of Nup107-160 (center) or of GCP2 (right). The extracts were analyzed by Western blotting with the indicated antibodies to monitor the overall level of depletion through siRNAs. ). Uncropped images of the blots are shown in Supplementary Information, Fig. S3.
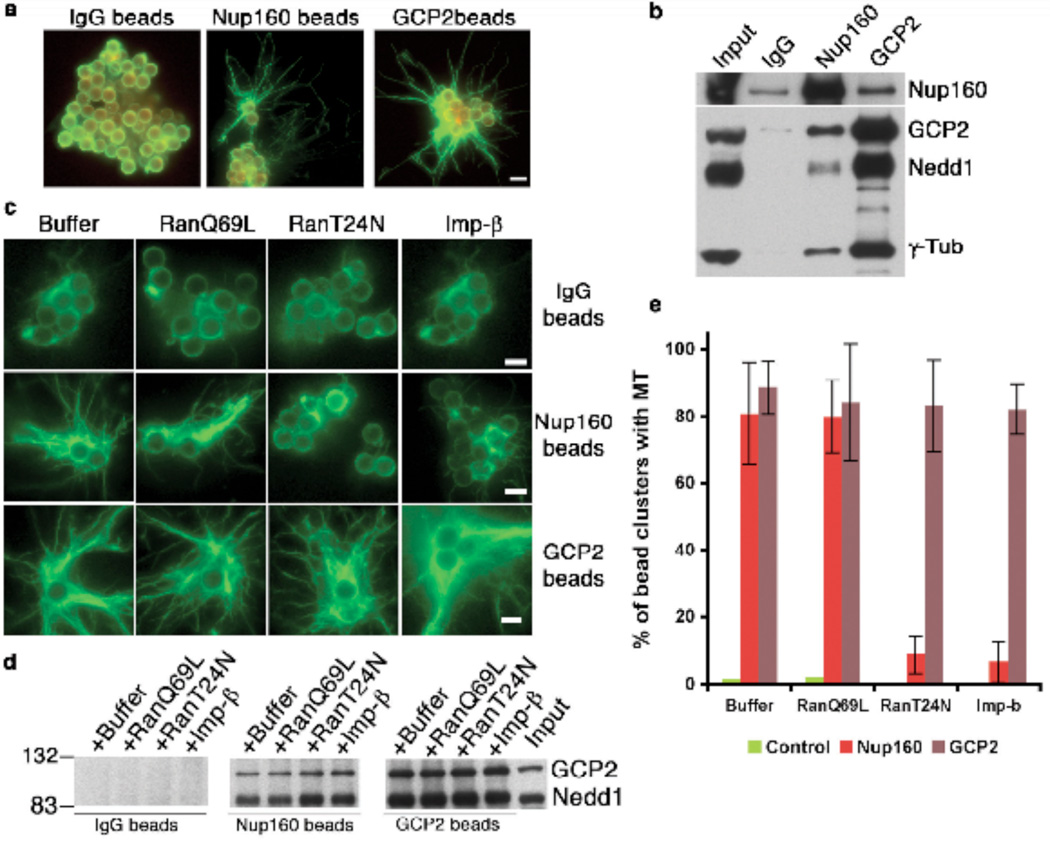
Our findings showed that γ-TuRC localizes to kinetochores in a Nup107-160-dependent manner and that it is required for MT nucleation at kinetochores. We wished to test directly whether γ-TuRC might be a MT-nucleating agent at that site. To address this question, we tested whether supercomplexes containing xNup107-160 and γ-TuRC retain the capacity to nucleate MTs. We incubated magnetic beads coated with anti-xNup160, anti-xGCP2 or control IgG in CSF XEEs, followed by isolation and washing. The beads were incubated in vitro with purified tubulin14. The reactions were fixed after 10 minutes, followed by immunostaining for α-tubulin. As expected, the anti-xGCP2 antibody coated beads formed robust MTs, giving an aster-like appearance, while there were no MTs associated with IgG-coated beads (Fig. 4a). Importantly, anti-xNup107-160 antibody-coated beads also nucleated MTs, proportional to the amount of γ-TuRC that was co-precipitated (Fig. 4b; Supplementary Information, Fig. S3). These findings clearly validate the third prediction of our hypothesis, that γ-TuRC can nucleate MTs in association with Nup107-160.
Figure 4. Ran-GTP regulates MT nucleation by xNup107-160 / γ-TuRC complexes.
(a) Magnetic beads coated with control IgG, anti-xNup160 or anti-xGCP2 antibodies were incubated in CSF XEEs for 90 minutes. The beads were re-isolated, washed and incubated with tubulin and GTP for 10 minutes. After fixation, the beads were pelleted onto coverslips and stained with antibodies against α-tubulin. Scale bar = 2.8 µm
(b) Proteins associated with beads prepared as in (a) were eluted with low pH. Eluted fractions and untreated XEE (input) were analyzed by Western blotting for xNup160 and γ-TuRC components. Uncropped images of the blots are shown in Supplementary Information, Fig. S3.
(c) Magnetic beads coated with control IgG, anti-xGCP2 or anti-xNup160 antibodies were incubated for 90 minutes in Control, Ran-T24N, Ran-Q69L or Importin-b-treated CSF XEEs. The beads were re-isolated, washed and incubated with tubulin and GTP for 10 minutes. After fixation, the beads were pelleted onto coverslips and stained with antibodies against α-tubulin. Scale bar = 2.8 µm
(d) Proteins bound to beads as in (c) were eluted with low pH and analyzed along with untreated XEE (input) by western blotting for GCP2 and Nedd1 (γ-TuRC components). Uncropped images of the blots are shown in Supplementary Information, Fig. S3.
(e) To quantitate the extent of MT nucleation, magnetic beads coated with control IgG, anti-xGCP2 or anti-xNup160 antibodies were in incubated in untreated, Ran-T24N-, Ran-Q69L or Importin-β-treated CSF XEEs, as in (c). In three independent experiments, we scored clusters containing three to six beads for associated MTs. The graph shows the mean percentage of clusters showing MTs in each case, +/− SD.
Ran is a critical regulator of trafficking through interphase NPCs15 and it has also been directly implicated in the assembly of MTs at kinetochores5. Ran-GTP depletion blocks MT nucleation at kinetochores, while preventing association of Ran’s GTPase activating protein (RanGAP1) to kinetochores enhances MT nucleation5. Thus, we asked whether Ran might control MT assembly through xNup107-160-bound γ-TuRC. To test this idea, we precipitated complexes on magnetic beads coated with anti-xNup160, anti-xGCP2 or IgG after manipulation of Ran-GTP levels within XEEs. Ran-GTP levels were increased through the addition of a constitutively GTP-bound Ran mutant (Ran-Q69L), decreased through addition of a mutant that blocks Ran’s guanine nucleotide exchange factor (Ran-T24N) or decreased by the addition of the Ran-GTP binding protein Importin-β15,16.
Manipulation of Ran-GTP levels did not alter the recruitment of γ-TuRC to anti-xNup160-coated beads suggesting that the binding of γ-TuRC to xNup107-160 was not sensitive to Ran-GTP (Fig. 4d; Supplementary Information, Fig. S3). When assayed for their capacity to form MTs, IgG-coated beads did not show nucleation of MTs, as expected (Fig. 4c). Anti-xGCP2 beads showed MT nucleation that was equivalent in all samples, suggesting that γ-TuRC is not directly sensitive to Ran-GTP levels. Importantly, anti-xNup160-coated beads from both Ran-T24N and Importin-β-treated XEEs showed a dramatic suppression of MT assembly, indicating that Ran regulates the capacity of the γ-TuRC/xNup107-160 supercomplex to nucleate MTs (Fig. 4c,e).
Nup107-160 plays a central role in the assembly of NPC-containing nuclear envelopes (NE)17,18 and in kinetochore-associated MT nucleation (Fig. 3). Notably, Ran-GTP controls its activity in both of these contexts: The Nup107-160 complex becomes bound to Importin-β during mitosis, negatively regulating the association of nucleoporins and thus NPC assembly19. As cells return to interphase, NPC assembly is facilitated by the local generation of Ran-GTP near chromatin, which releases the inhibition by Importin-β. Our findings indicate that Nup107-160 mediates kinetochore-associated MT nucleation through recruitment of γ-TuRC to kinetochores (Fig. 2). The MT nucleation activity of xNup107-160-bound γ-TuRC was high in the presence of Ran-GTP, but low when Ran-GTP was depleted (Fig. 4). Since Ran-GTP does not control the activity of γ-TuRC in the absence of Nup107-160, one possibility that would account for these findings is that Nup107-160 binding inhibits γ-TuRC activity, but that this inhibition is released by Ran-GTP.
It is possible that Ran-GTP may release one or more Importin-β-family proteins (karyopherins), whose binding to the Nup107-160/γ-TuRC supercomplex inhibits its activity in MT nucleation. Importin-β itself does not show a pattern of accumulation that would be consistent with it acting as the effector in this model (data not shown), but other karyopherins would be attractive effector candidates. Alternatively, Ran-GTP may promote the release of a cargo from soluble karyopherin-cargo complexes, and this cargo may activate MT nucleation by the Nup107-160/γ-TuRC supercomplex (Fig. 5).
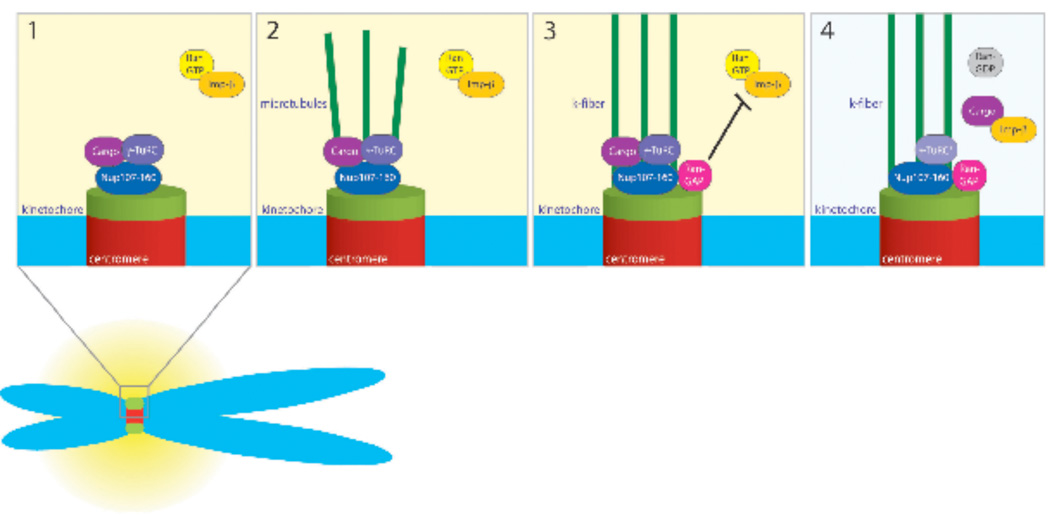
Figure 5. Model for Nup107-160 and γ-TuRC regulation at kinetochores.
Step 1: Nup107-160 (dark blue) and active γ-TuRC (purple) bind to unattached kinetochores. We postulate that a transport cargo (plum) may be released in the high Ran-GTP environment that maintains γ-TuRC in an active state.
Step 2: Nup107-160 and γ-TuRC nucleate MTs (green lines) at kinetochores, promoting the assembly of k-fibers.
Step 3: RanGAP1 (magenta) is recruited along k-fibers. Its recruitment lowers Ran-GTP levels near kinetochores. The lower Ran-GTP levels in turn allow Importin-b or another import receptor to bind the cargo protein.
Step 4: After the cargo molecule is sequestered, γ-TuRC becomes inactive (pale lilac), but k-fibers remain stable.
Notably, Ran’s GTPase activating protein (RanGAP1) is targeted to mammalian kinetochores in a manner that is closely coupled to the formation of initial MT attachments20, and this targeting is critical for suppressing nucleation at later stages of spindle assembly5. It is possible that Ran-dependent MT nucleation at kinetochores is important for the early assembly or capture of MTs, but less desirable as k-fibers mature. It could therefore be inhibited through RanGAP1 recruitment and suppression of Ran-GTP levels in the vicinity of kinetochores.
Utilization of γ-TuRC at kinetochores poses conundrum because kinetochore-anchored γ-TuRC should nucleate kMTs of polarity opposite to mature k-fibers. We envision three possible resolutions of this issue: First, MTs may be nucleated near kinetochores without anchorage, and become subsequently attached and stabilized by interactions of their plus-ends with kinetochores21. Second, γ-TuRC might be anchored during nucleation, but would be subsequently released, allowing plus-end attachments to form. Either of these ideas is consistent with γ-TuRC localization throughout Drosophila spindles, and with the essential role of non-centrosomal γ-TuRC in k-fiber assembly22. Finally, kinetochore-nucleated MTs may be anchored with an inverse polarity as an assembly intermediate, perhaps to facilitate centrosomal MT capture through anti-parallel MT-MT interactions. Failure to disassemble such an intermediate could contribute to k-fibers defects when RanGAP1 association to kinetochores is blocked5,23.
In summary, we have demonstrated that Nup107-160 and γ-TuRC associate with each other and promote Ran-dependent MT nucleation on mitotic kinetochores. Notably, this finding may reflect a broader relationship between the MT cytoskeleton and the NE that is shared among vertebrates24, insects25, plants26 and fungi27. Since Nup107-160 and γ-TuRC are both found throughout spindles during prometaphase1,28, this mechanism may also be operative in other parts of the spindle. The utilization of γ-TuRC unifies MT nucleation at kinetochores with mechanisms previously documented at other sites, particularly at centrosomes, and establishes an intimate linkage between the dynamics of the NE and MT cytoskeleton.
METHODS
Antibodies and reagents
Xenopus laevis cDNA corresponding to the following fragment of Nup107-160 and γ-TuRC components were sub-cloned into bacterial expression vector pET28a+ for bacterial expression: Nup160 (aa 1100-1400), Nup133 (aa 865-1138), Nup107 (aa 216-516), Nup96 (aa 679-924), Nup85 (aa 352-653), Sec13 (aa 1-320), Seh1 (aa 195-360), ELYS (aa 2106-2408), GCP2 (aa 1-260) and Nedd1 (aa 351-651). Proteins were expressed in E. coli Bl21 (DE3) cells. Except for xNedd1 and xSec13, all expressed fragments were insoluble. Insoluble proteins were purified from inclusion body and gel bands were used for immunizing rabbits. Antibodies against human Nup160 (aa 994-1316) and human GCP2 (aa 286-486) were also raised similarly. Rabbit polyclonal antibodies were generated by Pacific Immunology. To affinity purify antibodies, the recombinant fragments were solubilized in urea and cross-linked to NHS-Sepharose beads. In Supplementary Information, Fig. S1, these affinity purified antibodies recognized correct size protein on western blots. CREST sera were a gift from I. Ouspensky (National Institutes of Health, Bethesda, MD). Mouse monoclonal anti-α-tubulin and rabbit polyclonal anti-γ-tubulin were purchased from Sigma-Aldrich. Fluorescent secondary antibodies were from Molecular Probes. All other reagents were obtained from Sigma-Aldrich unless otherwise stated.
Xenopus egg extract preparation and use
CSF-arrested XEEs were prepared as described29. For immunodepletion, 500 µg of control IgG or indicated antibodies were immobilized on Protein A Sepharose beads and cross-linked with 5 mg ml−1 of dimethyl pimelidate (DMP) in 0.2 M triethanolamine (pH=8.3). Immunodepletions were performed as described1.
Immunofluorescence of XEE samples
In Supplementary Information, Fig. S2b, CSF XEEs containing sperm chromatin were driven into interphase by addition of 0.6 mM CaCl2. After 60 minutes at room temperatures, CSF arrest was reinstated through addition of 0.5 volumes of fresh CSF extract, with simultaneous addition of 0.04 mg ml−1 nocodazole. The samples were incubated at room temperature for 40 minutes. Reactions were diluted 10-fold in dilution buffer 1 (0.8X CSF-XB, 250 mM sucrose) and fixed with an equal volume of fixation buffer (4% formaldehyde in dilution buffer 1) for 30 minutes. The chromatin was spun onto coverslips through 40% glycerol cushion (in CSF-XB and 0.1% Triton X-100) and post-fixed in chilled methanol for 5 minutes. The coverslips were re-hydrated in 0.1% Triton X-100 containing PBS and processed for immunofluorescence as described elsewhere29.
MT nucleation by immunopurified complexes
25 µg anti-xNup160, anti-xGCP2 antibodies or non-specific rabbit IgG controls were bound to 100 µl magnetic beads, crosslinked, and incubated in CSF XEEs for 90 minutes at 25°C with rotation. The beads were separated from the extract, washed three times with cold CSF-XB and once with BRB80. The beads were re-suspended in 200 µl cold BRB80 buffer.
15 µl beads containing proteins precipitated from XEEs by anti-xNup160, anti-xGCP2 antibodies or non-specific rabbit IgG controls were incubated at 37°C for 10 minutes in reactions containing 30 µM unlabeled tubulin and 5 mM GTP. The samples were fixed with 1% glutaradehyde for 5 minutes, diluted 5-fold with 60% glycerol in BRB80 29 and spun through 25% glycerol cushion in BRB80 at 6,000 rpm for 6 minute at room temperature. The beads were once washed with BRB80, resuspended and spread onto coverslips and processed for immunofluorescence with anti-α-tubulin antibodies14. In Fig. 4c, beads were incubated in CSF XEEs that contained buffer or 0.2 mg ml−1 of RanQ69L, RanT24N or Importin-β, as indicated. Magnetic beads measure 2.8 µm.
Cell culture, siRNA, immunoprecipitation and immunofluorescence
293T and HeLa cells were maintained in DMEM medium with 10% fetal calf serum (DMEM complete medium) and 5% CO2 at 37°C. Lipofectamine 2000 was used for trasfection experiments. For siRNA, cells were grown on coverslips and tranfected using Lipofectamine siRNA Max or Oligofectamine reagents (Invitrogen) with oligonucleotides directed against Nup160 (AAC GAG GAC AGT TCT GCA TTT), GCP2 (set of 3 siRNA oligo’s for GCP2 from Invitrogen) or control oligonucleotides (AAC TGT CAG TCA GTC GTA GTA). All oligonucleotides were obtained from Qiagen, except GCP2 oligonucleotides were from Invitrogen.
In Fig. 2a and Supplementary Information, Fig. S2a, HeLa cells were incubated in 0.1 µg ml−1 of nocodazole in DMEM complete medium for 16hrs at 37°C. For immunofluorescence, cells were permeabilized with PHEM buffer for 5 minutes, followed with fixation for 15 minutes with 4% formaldehyde in PBS.
In Fig. 3, 0.1 µg ml−1 of nocodazole in DMEM complete medium was added 56 hours after transfection, and incubation was continued for 16 hours at 37°C. The nocodazole-containing medium was removed, and the cells were washed twice with cold complete DMEM medium, with further incubation in DMEM medium for 45 minutes on ice. The medium was replaced with fresh 37°C complete DMEM medium and incubation was continued at 37°C for 2 minutes. The cells were processed for immunofluorescence. Cells were permeabilized with PHEM buffer for 5 minutes, followed with fixation for 5 minutes with 4% formaldehyde in BRB80 and for 10 minutes with 4% formaldehyde in PBS. Images were taken on Axioscope microscope with OPEN LAB software.
Supplementary Material
ACKNOWLEDGEMENTS
RKM, AA and MD were supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development Intramural funds (Z01 HD008816 and Z01 HD008740). BMAF and PC were supported by National Institutes of Health R01 GM07159 and Texas HEB (Higher Education Board) 010019-0022-2006.
Footnotes
AUTHOR CONTRIBUTIONS: All authors contributed to experimental design. RKM, AA and PC performed experiments and analyzed data. MD wrote the manuscript.
REFERENCES
- 1.Orjalo AV, et al. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell. 2006;17:3806–3818. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loiodice I, et al. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luders J, Stearns T. Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol. 2007;8:161–167. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- 4.Wiese C, Zheng Y. Microtubule nucleation: gamma-tubulin and beyond. J Cell Sci. 2006;119:4143–4153. doi: 10.1242/jcs.03226. [DOI] [PubMed] [Google Scholar]
- 5.Torosantucci L, De Luca M, Guarguaglini G, Lavia P, Degrassi F. Localized RanGTP Accumulation Promotes Microtubule Nucleation at Kinetochores in Somatic Mammalian Cells. Mol Biol Cell. 2008 doi: 10.1091/mbc.E07-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Connell CB, Khodjakov AL. Cooperative mechanisms of mitotic spindle formation. J Cell Sci. 2007;120:1717–1722. doi: 10.1242/jcs.03442. [DOI] [PubMed] [Google Scholar]
- 7.Belgareh N, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aitchison JD, Blobel G, Rout MP. Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J Cell Biol. 1995;131:1659–1675. doi: 10.1083/jcb.131.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuccolo M, et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. Embo J. 2007;26:1853–1864. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran [see comments] Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- 11.Heald R, et al. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts [see comments] Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 12.Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 13.Tulu US, Fagerstrom C, Ferenz NP, Wadsworth P. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr Biol. 2006;16:536–541. doi: 10.1016/j.cub.2006.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- 15.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 16.Klebe C, Bischoff FR, Ponstingl H, Wittinghofer A. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry. 1995;34:639–647. doi: 10.1021/bi00002a031. [DOI] [PubMed] [Google Scholar]
- 17.Harel A, et al. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- 18.Walther TC, et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- 19.Kutay U, Hetzer MW. Reorganization of the nuclear envelope during open mitosis. Curr Opin Cell Biol. 2008;20:669–677. doi: 10.1016/j.ceb.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol. 2004;14:611–617. doi: 10.1016/j.cub.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 21.Maiato H, Rieder CL, Khodjakov A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol. 2004;167:831–840. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goshima G, et al. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnaoutov A, et al. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat Cell Biol. 2005;7:626–632. doi: 10.1038/ncb1263. [DOI] [PubMed] [Google Scholar]
- 24.Resendes KK, Rasala BA, Forbes DJ. Centrin 2 localizes to the vertebrate nuclear pore and plays a role in mRNA and protein export. Mol Cell Biol. 2008;28:1755–1769. doi: 10.1128/MCB.01697-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rebollo E, Llamazares S, Reina J, Gonzalez C. Contribution of noncentrosomal microtubules to spindle assembly in Drosophila spermatocytes. PLoS Biol. 2004;2:E8. doi: 10.1371/journal.pbio.0020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seltzer V, et al. Arabidopsis GCP2 and GCP3 are part of a soluble gamma-tubulin complex and have nuclear envelope targeting domains. Plant J. 2007;52:322–331. doi: 10.1111/j.1365-313X.2007.03240.x. [DOI] [PubMed] [Google Scholar]
- 27.Knop M, Pereira G, Schiebel E. Microtubule organization by the budding yeast spindle pole body. Biol Cell. 1999;91:291–304. [PubMed] [Google Scholar]
- 28.Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desai A, Murray A, Mitchison TJ, Walczak CE. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 1999;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.