Summary
Aromatic amino acid ammonia-lyases catalyze the deamination of L-His, L-Phe, and L-Tyr, yielding ammonia plus aryl acids bearing an α,β-unsaturated propenoic acid. We report crystallographic analyses of unliganded Rhodobacter sphaeroides tyrosine ammonia-lyase (RsTAL) and RsTAL bound to p-coumarate and caffeate. His 89 of RsTAL forms a hydrogen bond with the p-hydroxyl moieties of coumarate and caffeate. His 89 is conserved in TALs but replaced in phenylalanine ammonia-lyases (PALs) and histidine ammonia-lyases (HALs). Substitution of His 89 by Phe, a characteristic residue of PALs, yields a mutant with a switch in kinetic preference from L-Tyr to L-Phe. Structures of the H89F mutant in complex with the PAL product, cinnamate, or the PAL-specific inhibitor, 2-aminoindan-2-phosphonate (AIP), support the role of position 89 as a specificity determinant in the family of aromatic amino acid ammonia-lyases and aminomutases responsible for β-amino acid biosynthesis.
Introduction
Tyrosine ammonia-lyase (TAL) catalyzes the nonoxidative elimination of ammonia from L-Tyr, yielding trans-p-coumaric acid (trans-p-hydroxycinnamic acid). TAL is a member of a family of ammonia-lyases that deaminate the aromatic amino acids, L-His, L-Phe, and L-Tyr (Figure 1) (reviewed in [1]). In plants and fungi, a dedicated TAL has not been identified, but instead phenylalanine ammonia-lyase (PAL) occurs widely. PAL produces transcinnamic acid, leading to a variety of specialized phenolic plant and fungal metabolites. PAL from dicotyledonous plants efficiently deaminates L-Phe, while PAL from some monocots including maize efficiently deaminates both L-Phe and L-Tyr [2]. Similarly, PAL from the yeast Rhodosporidium toruloides turns over both L-Phe and L-Tyr [3]. Thus, PAL-derived TAL activity in monocots and fungi provides an alternative route to the phenylpropanoid precursor p-coumaric acid, in lieu of hydroxylation of cinnamic acid by the membrane-bound cytochrome-P450 monooxygenase, cinnamate-4-hydroxylase.
Figure 1.
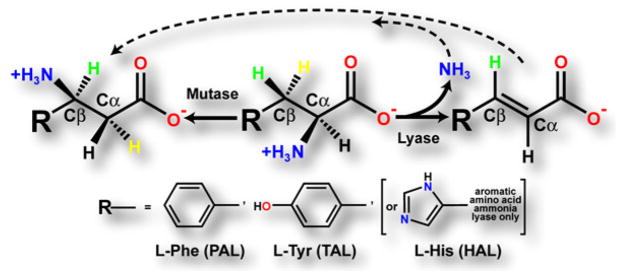
Reactions Catalyzed by the Aromatic Amino Acid Ammonia-Lyases and the Related Aminomutases
Tyrosine ammonia-lyase (TAL), phenylalanine ammonia-lyase (PAL), and histidine ammonia-lyase (HAL) catalyze the nonoxidative deamination of their respective amino acid substrates, yielding the corresponding α-β unsaturated aryl-acid product plus ammonia. The aminomutases catalyze the α-β migration of the amino group of the α-amino acid substrate. Labeling experiments [1, 15] have shown that the yellow β-proton (pro-S in L-Phe and L-Tyr, pro-R in L-His) of the substrate is stereospecifically abstracted. The dashed arrows emphasize the intermediacy of the lyase reaction in the overall reaction catalyzed by the aminomutases, which invoke Michael addition of ammonia to Cβ of the aryl-acid intermediate [1, 14–16].
In bacteria, phenylpropanoids are rare, and accordingly PAL and TAL are poorly represented (at least based upon gene annotation). To date, PALs have been identified in Streptomyces maritimus [4], Photorhabdus luminescens [5], Sorangium cellulosum [6], and Streptomyces verticillatus [7]. In these bacteria, the PAL product cinnamic acid serves as an intermediate in the biosynthesis of antibiotic or antifungal compounds (enterocin, 3,5-dihydroxy-4-isopropyl-stilbene, soraphen A and cinnamamide). PALs have also been identified in Anabaena variabilis and Nostoc punctiforme, although the ultimate fate of cinnamic acid in these cyanobacteria is currently unknown (M.C.M., unpublished data). The only confirmed bacterial sources of TAL are several species of purple phototropic bacteria (Rhodobacter capsulatus, Rhodobacter sphaeroides, and Halorhodospira halophila), in which p-coumarate is a precursor of the chromophore of photoactive yellow protein [8, 9], and the actinomycete Saccharothrix espanaensis [10], where coumarate is used for saccharomicin antibiotic biosynthesis.
The aromatic amino acid ammonia lyases contain a 4-methylidene-imidazole-5-one (MIO) cofactor formed by the spontaneous (autocatalytic) cyclization and dehydration of an internal Ala-Ser-Gly tripeptide segment [11]. Two alternative mechanisms have been suggested for the role of the electrophilic MIO cofactor in catalyzing the elimination of the α-amino group and the stereo-specific abstraction of a β-proton from the L-amino acid substrate [12, 13]. Both mechanisms are intensely debated and must account for the activity of the recently characterized tyrosine- and phenylalanine aminomutases (TAM and PAM, respectively) [14–16]. These MIO-dependent enzymes are closely related to the aromatic amino acid ammonia-lyases forming intermediate aryl acids, but they ultimately catalyze a second reactive step in which the α-amino group removed from the substrate is transferred to the β-carbon, yielding a β-amino acid product (Figure 1).
Crystal structures are available for PALs from R. toruloides [13, 17] and Petroselinum crispum [18], as well as histidine ammonia-lyase (HAL) from the bacterium Pseudomonas putida [19]. Nevertheless, the enzymatic mechanism and determinants of substrate specificity of the aromatic amino acid ammonia-lyases remain to be fully defined. Earlier studies have been hindered in part by a lack of accurate information pertaining to the mode of substrate/product binding. We now report the crystal structure of TAL from the bacterium R. sphaeroides. The structure provides a unique view of an aromatic amino acid ammonia-lyase with a kinetic preference for L-Tyr. We also describe the structures of TAL complexed with the products of the TAL-catalyzed reactions using L-Tyr and L-DOPA substrates, namely p-coumaric and caffeic acids, respectively. These structures provide definitive views of the binding of products to aromatic amino acid ammonia-lyases, thus identifying the substrate selectivity determinants of this family of biosynthetic enzymes. Based upon these high-resolution structures, RsTAL was successfully engineered into a kinetically authentic PAL, and additional structures of mutant RsTAL were obtained with the product of the PAL-catalyzed reaction using L-Phe as a substrate, namely cinnamic acid and with the PAL-specific inhibitor, AIP.
Results and Discussion
X-Ray Crystal Structure of R. sphaeroides TAL
The monoclinic crystal form (space group P21) of R. sphaeroides TAL (RsTAL) contains two complete homotetramers of the polypeptide chain per asymmetric unit. The initial structure solution was obtained by molecular replacement, with a search model derived from a tetramer of PpHAL (PDB entry 1GKM). The rotation function identified two orientations for the tetramer, with peak heights of 13.4 and 12.5 σ; translation-function searches then positioned the two tetramers, with an R factor/correlation coefficient of 0.524/0.146 for the first tetramer and, subsequently, 0.514/0.182 for the second tetramer. The atomic model of RsTAL was refined to 1.50 Å resolution (Table 1). The two tetramers of RsTAL are nearly identical, as the root-mean-squared positional deviation (rmsd) between equivalent backbone atoms is only 0.12 Å. The eight distinct monomers also exhibit nearly identical backbone conformations; for pair-wise comparisons between individual monomers, the rmsd values range from 0.10 to 0.18 Å. Only residues 1 to 7 and the C-terminal residue 523 of each of the eight RsTAL monomers are poorly ordered in electron-density maps.
Table 1.
Summary of Data Collection and Refinement Statistics
| RsTAL Unliganded | RsTAL-Coumarate | RsTAL-Caffeate | H89F TAL-Cinnamate | H89F TAL-Coumarate | H89F TAL-AIP | |
|---|---|---|---|---|---|---|
| Space group | P21 | |||||
| Unit cell parameters | ||||||
| a (Å) | 87.9 | 87.4 | 87.6 | 87.6 | 87.6 | 87.6 |
| b (Å) | 155.5 | 154.8 | 155.0 | 155.0 | 155.0 | 154.8 |
| c (Å) | 164.3 | 164.1 | 164.2 | 164.0 | 164.1 | 164.5 |
| β (°) | 94.1 | 94.1 | 94.1 | 94.2 | 94.1 | 94.2 |
| Monomers per asymmetric unit | 8 | |||||
| Resolution rangea (Å) | 100–1.50 (1.58–1.50) | 100–1.58 (1.68–1.58) | 100–1.90 (2.00–1.90) | 100–1.90 (2.00–1.90) | 100–2.00 (2.10–2.00) | 100–1.75 (1.84–1.75) |
| Number of reflections measured | 3,870,665 | 1,901,422 | 1,195,768 | 1,133,848 | 891,554 | 1,396,599 |
| Merging R factora | 0.085 (0.618) | 0.068 (0.570) | 0.112 (0.546) | 0.135 (0.511) | 0.139 (0.503) | 0.089 (0.487) |
| Mean (I/σ I)a | 12.2 (2.3) | 15.6 (2.0) | 8.5 (2.4) | 7.5 (2.2) | 7.5 (2.5) | 9.3 (1.9) |
| Completenessa | 0.933 (0.859) | 0.873 (0.494) | 0.989 (0.975) | 0.992 (0.973) | 0.962 (0.959) | 0.964 (0.822) |
| Redundancya | 5.9 (3.6) | 3.7 (2.7) | 3.5 (3.4) | 3.3 (2.6) | 3.2 (2.7) | 3.3 (2.2) |
| Number of reflections used | 644,928 | 509,395 | 338,602 | 337,722 | 282,486 | 420,387 |
| R factora | 0.174 (0.254) | 0.192 (0.298) | 0.175 (0.253) | 0.172 (0.265) | 0.172 (0.247) | 0.189 (0.293) |
| Free R factora | 0.189 (0.276) | 0.208 (0.319) | 0.198 (0.283) | 0.196 (0.285) | 0.201 (0.281) | 0.210 (0.321) |
| Number of amino acid residues | 4,130 | 4,130 | 4,130 | 4,130 | 4,130 | 4,130 |
| Number of water molecules | 3,914 | 3,482 | 3,224 | 3,288 | 3,110 | 3,089 |
| Residues with most favorable conformation (%) | 93.3 | 93.4 | 93.4 | 93.0 | 93.1 | 92.9 |
Merging R factor = Σhkl Σi | Ii(hkl) − 〈I(hkl)〉 |/Σhkl Σi Ii(hkl).
Values in parentheses describe the highest resolution shell.
Structure of RsTAL
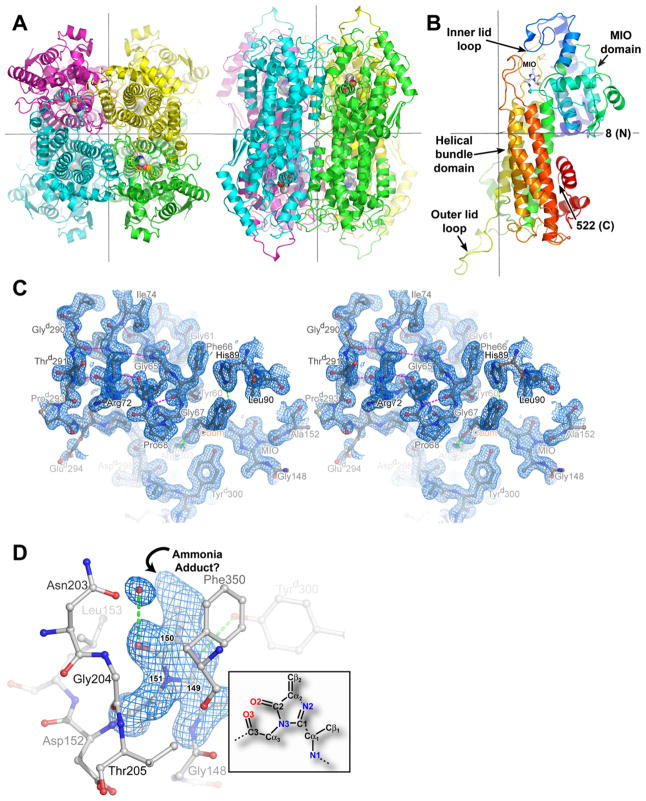
Both the tertiary and quaternary structures (Figures 2A and 2B) of RsTAL are similar to those described previously for related ammonia-lyases, PpHAL (PDB entry 1GKM), and PALs from parsley (P. crispum; 1W27) and yeast (R. toruloides; 1T6P, 1Y2M). These proteins form homotetrameric oligomers with 222-point symmetry. The homotetramer contains four active sites, with three distinct monomers participating in formation of each active-site cavity. Each monomer adopts a predominantly α-helical fold (Figure 2B), organized around an up-down bundle of five α helices. The flanking regions of these helices, together with the extended hairpin loop linking helices 4 and 5 of the helical bundle, are responsible for forming the monomer-monomer interfaces at the core of the tetramer. The N-terminal region of the polypeptide chain contributes to a domain that carries the MIO cofactor (Figure 2B), and at the opposite end of the bundle, the C-terminal segment forms a peripheral α-helical layer. The C-terminal domain participates in additional intersubunit contacts that stabilize the homotetramer, and it provides the outer-lid loop (Figures 2B and 2C, and discussion below), which caps the active-site cavity of an adjacent monomer. Like bacterial HAL, RsTAL lacks an additional domain that is inserted into the C-terminal domain of both yeast and parsley PAL. The function of this additional domain is uncertain; regulatory roles in shielding access to the active-site tunnel [18] or modulating the flexibility of an active-site lid loop [20] have been suggested as functional roles.
Figure 2.
Three-Dimensional Structure of RsTAL
(A) Ribbon representations of the RsTAL homotetramer, with the polypeptide chains of the individual monomers colored green (a), cyan (b), magenta (c), and yellow (d). The atoms of the four MIO cofactors are drawn as color-coded van der Waals spheres with red for oxygen, light gray for carbon, and blue for nitrogen. Orthogonal views from the top (left) and front (right) of the homotetramer are shown. The 222-point symmetry of the homotetramer is generated by three mutually orthogonal and intersecting 2-fold rotational axes, shown as gray lines. In each orientation, two of the axes are visible, with the third axis perpendicular to the page.
(B) Ribbon representation of the RsTAL monomer. The polypeptide chain is colored according to a gradient, with blue and red serving as extremes for the N- and C termini, respectively. The atoms of the MIO cofactor formed by the tripeptide segment Ala 149-Ser 150-Gly 151 are drawn as balls and sticks color-coded by atom type. The 2-fold axes that relate this monomer to the other monomers in the homotetramer are shown as gray lines.
(C) Electron density and interactions of the active-site lid loops of RsTAL complexed with coumarate shown as a stereo pair. The three-stranded β sheet is shown at the upper left. Three residues of the inner loop, Tyr 60, Phe 66, and Gly 67, encompass the bound coumarate product. Backbone hydrogen-bonding interactions of the lid loop are shown as magenta dashed lines; hydrogen-bonding interactions involving coumarate are represented as green dashed lines. The blue-colored contours envelop regions greater than 1.0σ in the final 2Fobs − Fcalc electron-density map calculated at 1.58 Å resolution.
(D) Methylidene-imidazolone (MIO) cofactor. MIO and protein residues are shown as balls and sticks colored by atom type. Hydrogen-bonding interactions are represented as green dashed lines. An oxyanion hole is formed by the backbone amides of Leu 153 and Gly 204. The 149-150-151 numbering indicates the amino acid origin of the MIO cofactor. The blue-colored contours envelop regions greater than 3σ in the MIO-omit Fobs − Fcalc electron-density map. The inset shows the atom nomenclature of the native MIO cofactor, with the atom names colored according to atom type and numbered according to the originating residue within the 149–151 tripeptide (Ala 149 is 1, Ser 150 is 2 and Gly 151 is 3).
From a comparison of the superposed polypeptide-chain backbones of the known ammonia-lyase structures, RsTAL differs from P. putida HAL by 1.9 Å rmsd for 464 equivalent residues (with 34% sequence identity), from parsley PAL by 2.0 Å rmsd for 445 equivalent residues (with 27% sequence identity), and from yeast PAL by 1.9 Å rmsd for 446 equivalent residues (with 30% sequence identity). The greater overall structural similarity of RsTAL with the bacterial HAL, in comparison to the eukaryotic PALs, is consistent with the higher overall sequence identity of RsTAL with the bacterial HALs and the absence of additional polypeptide segments (at the N terminus and within the shielding domain) that occur in the eukaryotic PALs. These characteristics have led to suggestions that TAL and eukaryotic PAL diverged from bacterial HAL in separate evolutionary lineages [18].
Methylidene-Imidazolone Cofactor of RsTAL
The MIO cofactor of RsTAL, formed by Ala 149, Ser 150, and Gly 151, resides in well-defined electron density (Figure 2D). The imidazolone ring stacks against the side chain of Phe 353. The MIO N2 atom, derived from Ser 150, accepts hydrogen bonds from the hydroxyl moiety of Tyrd300 and the side-chain amide of Gln 436. Residue numbering refers to the polypeptide chain of a single monomer designated “a”; residues designated with superscripted “b,” “c,” or “d” are contributed by one of the dyad-related monomers of the homotetramer. The MIO keto group oxygen, O2, sits in a pocket lined by the backbone amides of Leu 153 and Gly 204 (the oxyanion hole, see below) but does not make any direct contacts with protein residues and instead forms a hydrogen bond with a water molecule (Figure 2D).
MIO appears to carry an adduct attached to the electrophilic methylidene carbon Cβ2 (Figure 2D). The precise identity of this adduct is uncertain, but likely corresponds to the covalent attachment of an ammonium group. This posited adduct supports MIO derivatization by ammonia derived from ammonium acetate used for crystallization and present at 0.3 M. The presence of a nucleophilic adduct is supported by the planar sp2 configuration of the MIO N3 atom, associated with aromaticity of the imidazolone ring. Similarly, crystal structures of other ammonia-lyases bearing a covalent adduct indicate sp2 hybridization of N3 [13, 18, 19, 21]. In contrast, in structures of both PAL [17] and HAL [19] with unmodified MIO, the N3 atom assumes an sp3 hybridization state, indicative of a nonaromatic imidazolone ring.
Binding of Coumarate and Caffeate Reaction Products to RsTAL
The binding of coumarate observed in RsTAL crystals supports product inhibition of the ammonia-lyases [22]. However, attempts to complex L-Tyr or observe turnover of L-Tyr in RsTAL crystals were unsuccessful. The high concentration of ammonium ion in the crystallization medium and the attendant covalent modification of MIO likely prevent both L-Tyr binding and TAL activity in crystals. Notably, HAL and PAL samples bearing modified cofactors are catalytically inactive [18, 19]. In crystal soaking experiments of RsTAL with the PAL product cinnamic acid or the PAL-specific inhibitor, 2-aminoindan-2-phosphonic acid (AIP) [23], no significant binding was observed, in accord with the poor turnover of L-Phe by RsTAL and the weak inhibition of RsTAL activity by AIP (see discussion below and Table 2).
Table 2.
Kinetic Parameters for Wild-Type and H89F RsTAL and Comparison with Other TAL/PAL Enzymes
| Enzyme [Reference] | Substrate | Km (μM) | kcat (s−1) | kcat/Km (s−1 μM−1) | AIP Ki (μM) |
|---|---|---|---|---|---|
| WT RsTAL [this study] | L-Tyr | 74.2 | 4.32 | 0.058 | NM |
| L-Phe | 11,400 | 13.10 | 0.0011 | 16.3 | |
| H89F RsTAL [this study] | L-Tyr | NM | NM | NM | NM |
| L-Phe | 434 | 8.15 | 0.019 | 0.60 | |
| RcTAL [8] | L-Tyr | 15.6 | 27.7 | 1.77 | ND |
| L-Phe | 1277 | 15.1 | 0.0118 | ND | |
| SeTAL [10] | L-Tyr | 15.5 | 0.015 | 9.68 × 10−4 | ND |
| L-Phe | 2,860 | 0.0038 | 1.3 × 10−6 | ND | |
| ZmPAL [2] | L-Tyr | 19 | 0.9 | 0.0473 | ND |
| L-Phe | 270 | 10 | 0.037 | ND | |
| PcPAL [22] | L-Tyr | 2,500 | 0.3 | 1.2 × 10−4 | ND |
| L-Phe | 17.2 | 22 | 1.28 | 0.025 | |
| SmPAL [4] | L-Phe | 23 | 0.0048 | 2.1 × 10−3 | 1.81 |
| PlPAL [5] | L-Phe | 320 | 0.8 | 2.5 × 10−3 | ND |
Rc, Se, Zm, Pc, Sm, and Pl are R. capsulatus, S. espanaensis, Z. mays, P. crispum, S. maritimus, and P. luminescens, respectively. NM is not measurable, and ND is not determined.
The RsTAL-coumarate cocrystal structure provides eight independent views (per asymmetric unit) of the enzyme-product complex. Notably, all of the protein residues that form interactions with coumarate are invariant in the TALs functionally characterized to date (Figure 3A). The coumarate conformation and interactions with RsTAL are nearly identical in the eight copies of the complex in the asymmetric unit. In terms of the observed propenoate conformation, the alkene double bond is not conjugated with either the hydroxyphenyl ring (approximately 30° out of plane) or the carboxylate group (approximately 50° out of plane). Most notably, in all eight instances, the same (si) face of the alkene double bond is directed toward MIO. The hydroxyphenyl ring of coumarate is roughly orthogonal to the plane of MIO. The closest approach to the electrophilic methylidene carbon of MIO—3.6 Å—is by the coumarate carbon atom equivalent to the Cβ atom in the L-Tyr substrate (Figure 3B).
Figure 3.
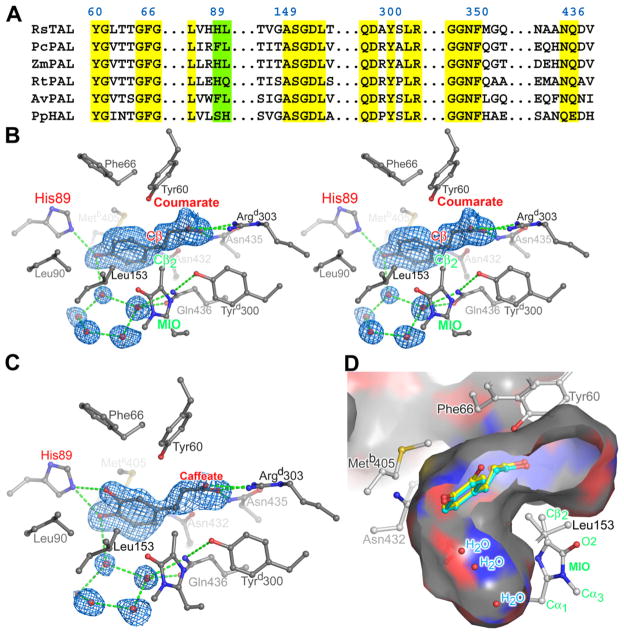
Active Site of RsTAL
(A) Partial amino acid (single letter codes) sequence alignment of RsTAL with representative members of the aromatic amino acid ammonia-lyase family discussed in the text. Only regions that form the active site of the enzymes are shown. Numbering is according to RsTAL. Yellow boxes highlight conserved catalytic and binding residues, while the green box highlights the specificity determining residues.
(B) Electron density and interactions of the coumarate product bound in the active site of wild-type RsTAL shown as a stereo pair. The coumarate, MIO cofactor, and protein side chains that line the active-site pocket are rendered as balls and sticks and colored according to atom type. Hydrogen-bonding interactions are shown as green dashed lines. The blue-colored contours envelop regions greater than 2.5σ in the initial Fobs − Fcalc electron-density map calculated at 1.58 Å resolution with phases derived from the unliganded model. The closest atom of coumarate to the MIO cofactor (labels colored green) is labeled Cβ and colored red.
(C) Electron density and interactions of the caffeate product bound in the active site of wild-type RsTAL. The phenyl ring of caffeate adopts primarily the conformation shown; a second, lower-occupancy conformation that differs only in the (inward) position of the meta-hydroxyl group is also observed. The blue-colored contours envelop regions greater than 2.5σ in the initial Fobs − Fcalc electron-density map calculated at 1.90 Å resolution with phases derived from the unliganded model.
(D) Product binding pocket in RsTAL. The depicted surface represents the area accessible to a probe sphere 1.4 Å in radius and the surface is color-coded according to the identity of the underlying protein atom (carbon is gray; nitrogen is blue; oxygen is red). The front portion of the RsTAL tetramer has been cut away to reveal the internal cavity in the vicinity of the MIO cofactor. The coumarate molecule (shown in cyan) was excluded in the calculation of the molecular surface. The position of a caffeate molecule bound to RsTAL is shown in yellow. MIO is labeled (green) as shown in the inset of Figure 2D.
The carboxylate group of the propenoate moiety forms hydrogen bonds with the side-chain amide of Asn 435 and a salt bridge with the δ-guanido group of Argd303 from the dyad related polypeptide chain. The aliphatic portion of the propenoate segment packs against two residues from the inner lid-loop, Tyr 60 and Gly 67. The nonpolar side chains of Leu 90, Leu 153, and Metb405, and the hydrophilic side chains of Asn 432 and Gln 436 surround the phenyl ring of the bound coumarate. Finally, the p-hydroxyl group of the hydroxyphenyl ring forms hydrogen bonds with the side chain of His 89 and a water molecule that resides in a hydrogen-bonding network involving four other water molecules (Figure 3B). Caffeic acid carries an additional meta-hydroxyl group on the phenyl ring, and the crystal structure of the complex with RsTAL demonstrates that this hydroxyl moiety sits in the space residing above the cofactor (Figure 3C). Caffeic acid is the product of the deamination of the α-amino-acid L-DOPA (3,4-dihydroxy-L-Phe), and, interestingly, RsTAL exhibits activity with L-DOPA (data not shown).
The L-shaped active-site cavity is only partially filled by coumarate and extends above the methylidene carbon of MIO and into space occupied by the network of water molecules described above (Figures 3B and 3D). The excess space in the vicinity of the coumarate binding site is available to phenyl rings with larger substituents, as shown for the caffeic acid complex (Figures 3C and 3D). These results collectively raise intriguing possibilities for the deployment of RsTAL for in vivo generation of bioactive phenylpropanoids and for the rational engineering of TALs to accept diverse amino-acid substrates.
Previous crystallographic analyses of PAL and HAL have yielded (with one exception) only unliganded enzyme structures, but these structures have served as a starting point for predictive modeling of substrate-, product-, and reaction-intermediate-bound states. These docking attempts assumed attack on the MIO methylidene-carbon by either the α-amino group or the electron-rich aromatic ring of the substrate; and thus, substrate docking targeted a conjectured relative positioning of substrate and cofactor, as well as the formation of favorable substrate-protein interactions. Although some of these modeled complexes (in particular those described [1]) agree roughly with the binding mode observed in the TAL-coumarate complex, in general, the ligand-enzyme interactions have not been accurately predicted. The mode of substrate/product binding, in particular the positioning of the aromatic ring, suggested most recently in Ritter and Schulz [18], deviates significantly from the product complexes reported here. This recent docking study was updated from a previous model [19] to maximize interaction between the substrate and PcPAL-Phe 400. Ironically, the previous model [19] more closely approximates the experimentally determined complexes reported here.
A weakly bound cinnamate molecule described in a crystal structure of R. toruloides PAL [13] was modeled in a reverse orientation, with its phenyl and carboxylate groups approximately transposed relative to those of coumarate bound to TAL. Finally, there is no apparent involvement of metal ions in the RsTAL-coumarate interaction, as proposed for HAL-substrate interactions [24].
The structures of RsTAL-coumarate and RsTAL-caffeate complexes provide conclusive identification of the active-site residues that form the binding pocket for the product. Together with the pattern of amino acid conservation of the substrate-recognition site (Figure 3A) and the results of site-directed mutagenesis [24, 25] of active-site residues, this structural information provides a rational basis for interpreting the roles that these residues play in determining substrate specificity and in catalyzing the enzyme reactions.
Tyr 60, Gly 67, Tyr 300, and Arg 303 (RsTAL numbering), which interact with the backbone atoms of the amino acid substrate, are highly conserved in HAL, PAL, and TALs. Indeed, the salt bridge between the Arg 303 δ-guanido moiety and the substrate’s α-carboxylate group served as an anchoring interaction in most of the earlier modeling attempts. Replacement of this conserved Arg with Ala in PAL [25] or Ile in HAL [24] caused large decreases in enzyme activity, whereas a Lys substitution in HAL minimally affected activity [24]. Phe substitution of the Tyr corresponding to RsTAL Tyr 300 in both HAL [24] and PAL [25] also resulted in significant losses of activity. Tyr 60, from the inner-lid loop (see below), likely resides near the Cβ atom of the amino acid substrates, and, consistent with a postulated role as a general base for β-proton abstraction, substitution of this Tyr by Phe severely debilitated enzyme activity for both HAL [24] and PAL [25]. The residues that interact with the aromatic ring of the substrate are more variable among the ammonia-lyases, but are more similar between the PAL and TAL families than between these two families and the HALs.
Notably, the His at the position corresponding to His 89 in RsTAL is found in other functionally characterized bacterial TALs, as well as maize and yeast PALs, the latter of which possess significant TAL activity [2, 3]. In contrast, in PALs, a Phe occurs almost invariably at the position equivalent to RsTAL His 89, and Phe would support favorable nonpolar interactions with the phenyl group of the L-Phe amino acid substrate. Furthermore, the pivotal two-residue His-Leu sequence (89 and 90 in RsTAL), which characterizes the TALs and is replaced by Phe-Leu in the PALs, is instead Ser-His in the HALs (Figure 3A). The HAL/PAL residue-substitution indicated previously [19] resides not at RsTAL His 89 but at the position adjacent to His 89 in RsTAL, namely Leu 90. RsTAL Leu 90 corresponds to His 89 in PpHAL and Leu 138 in PcPAL [18]. Notably, the L138H mutant (together with Q488E) was generated in PcPAL, but this double mutant did not result in a switch from PAL to HAL activity, leading the authors to state that substrate specificity preferences cannot be readily explained by amino acid differences in the respective ammonia-lyases [1, 25].
Structure-Based Switch of L-Tyr/L-Phe Substrate Preferences of RsTAL
These structures point to RsTAL His 89 as a key determinant of the substrate specificity of the aromatic amino acid ammonia-lyases. We reasoned that replacement of His 89, which forms a favorable hydrogen bond with the side-chain hydroxyl group of L-Tyr, with Phe would result in considerable turnover of L-Phe. From an assessment of the kinetic properties of the RsTAL mutants (Table 2), wild-type RsTAL displays a marked kinetic preference for L-Tyr, as both the Km (150-fold) and kcat/Km (53-fold) are substantially better for L-Tyr as compared to L-Phe. These values are similar to the steady-state kinetic constants for the homologous TAL from Rhodobacter capsulatus [8].
In contrast, the engineered RsTAL variant with the single amino acid substitution H89F lacks activity with L-Tyr and, as predicted, efficiently turns over L-Phe. With L-Phe, the H89F point mutant exhibits a 26-fold decrease in Km and a 17-fold increase in kcat/Km compared to wild-type RsTAL. Indeed, the catalytic efficiency of the PAL activity for the H89F mutant (kcat/Km 0.019 s−1 μM−1) is only slightly lower than that of TAL activity for wild-type RsTAL (0.058 s−1 μM−1), and it exceeds the catalytic efficiency of some native PALs (Table 2). In addition, the differing kinetic specificities of the wild-type and mutant RsTALs are further substantiated by the relative susceptibilities to inhibition by AIP, a PAL-specific inhibitor [23]. For wild-type RsTAL, activity with L-Tyr is unaffected by AIP, whereas activity with LPhe is inhibited (Ki=16.3 μM). For the H89F mutant with L-Phe, the Ki = 0.60 μM and is 27-fold lower (greater inhibitory activity) than for wild-type RsTAL.
We next determined crystal structures of the H89F mutant complexed with the reaction product cinnamate (Figure 4A), as well as with coumarate (Figure 4B) and AIP (Figures 4C and 4D). The phenyl ring of Phe 89 occupies the same space as the His 89 imidazole ring of wild-type RsTAL with little active site perturbation. Likewise, cinnamate aligns with coumarate bound to wildtype RsTAL. The phenyl rings of cinnamate and Phe 89 are roughly coplanar and within van der Waals distances. Unexpectedly, binding of coumarate to the H89F mutant was observed. The position and orientation of the bound coumarate differs from that observed in wild-type RsTAL, relieving expected steric clashes with protein side chains (Figure 4E). Moreover, electron-density maps indicate that the coumarate molecule is poorly ordered in the H89F RsTAL active site. With the H89F mutant, although the salt bridge between the coumarate carboxylate group and the δ-guanido moiety of Argd303 is preserved, the p-hydroxyl group is pushed away from the phenyl ring of Phe 89 and lacks a hydrogen-bonding partner. Moreover, the opposite face of the propenoate portion of coumarate is oriented toward the MIO cofactor (Figure 4B). This disrupted binding mode likely explains the inactivity of the H89F RsTAL mutant with L-Tyr as substrate.
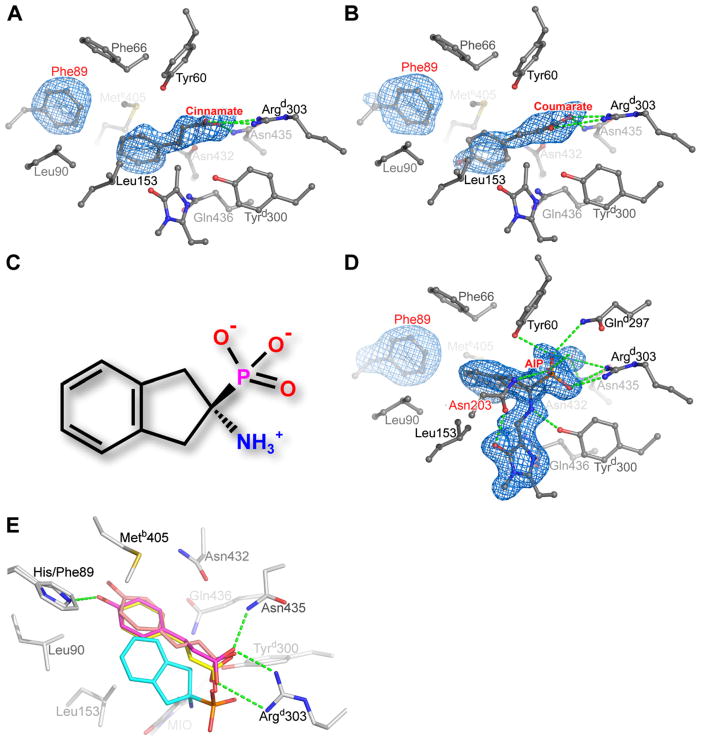
Figure 4.
Product Complexes of H89F RsTAL
(A) Electron density and interactions of the cinnamate product bound in the active site of H89F RsTAL. For the Fobs − Fcalc electron-density map (1.9 Å resolution and shown contoured at 2.5σ), cinnamate and the side chain of Phe 89 were excluded from all calculations.
(B) Electron density and interactions of coumarate bound to the H89F RsTAL active site. For the Fobs − Fcalc electron-density map (2.0 Å resolution and shown contoured at 2.5σ), coumarate and the side chain of Phe 89 were excluded from all calculations.
(C) Chemical structure of AIP.
(D) Electron density and interactions of the PAL inhibitor AIP bound covalently to the MIO cofactor of H89F RsTAL. For the Fobs − Fcalc electron-density map (1.75 Å resolution and shown contoured at 2.5σ), AIP, MIO, and the side chain of Phe 89 were excluded from all calculations. The residue labeled in red, Asn 203, is involved in a backbone conformational rearrangement that allows the Asn 203 side chain to engage the MIO cofactor upon AIP binding.
(E) Comparison of the binding modes of cinnamate (yellow), coumarate (orange-tan), and AIP (cyan) with the H89F RsTAL active site. Also shown is coumarate (magenta) bound to wild-type RsTAL, with the hydrogen-bonding interactions between the coumarate product and wild-type RsTAL represented as green dashed lines.
Most intriguing is the complex of the H89F mutant with AIP, in which the amino group of the inhibitor covalently attaches to the methylidene carbon of MIO (Figures 4C and 4D). Analogous to the cinnamate complex (Figure 4A), the salt bridge between the Argd303 δ-guanido moiety and the carboxylate mimic, namely the phosphonate group of AIP, and the edge-edge interaction between the phenyl rings of AIP and Phe 89 occur in a conserved fashion. However, because of the covalent attachment of the AIP moiety to the cofactor, the relative positions of AIP atoms with respect to TAL residues differ in comparison to cinnamate (Figures 4A and 4D). Furthermore, the formation of the covalent AIP adduct causes a significant rearrangement of a nearby segment of the polypeptide-chain-spanning residues 194–205 and points to possible mechanistic roles for some of these residues.
Active-Site Loops of RsTAL
In previously published HAL and PAL crystal structures, two loops situated near the entrance to the active sites exhibit high mobility, as evidenced by the variability in loop conformations, high crystallographic temperature factors, complete disorder, and the presence of proteolytically sensitive cleavage sites [13, 17–19]. Flexibility of these loops has been suggested to be a functional requirement for substrate binding and catalysis [20]. One active-site loop, termed the inner lid-loop, originates from the MIO domain of the same polypeptide chain that provides the MIO cofactor, and it contains a number of highly conserved residues. The second loop (the outer lid-loop) projects from the C-terminal domain of a dyad-related monomer in the homotetramer. The RsTAL structure is unique in that these lid loops are not only well ordered but form a compact arrangement within the active-site cavity (Figures 2C and 5A).
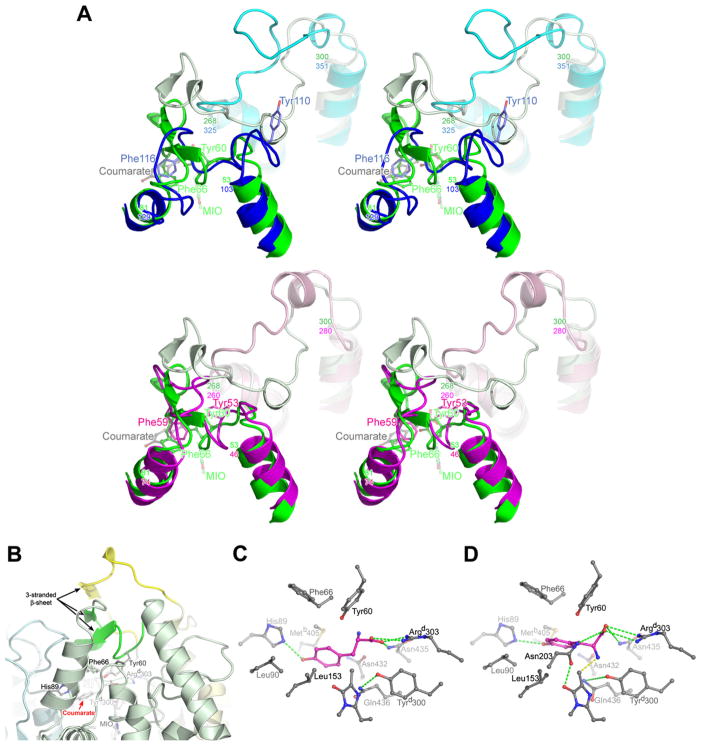
Figure 5.
Active-Site Lid Loops and a Model for L-Tyr Binding to RsTAL
(A) Stereo comparison of the lid-loop conformations of RsTAL (green), PcPAL (blue), and PpHAL (magenta). Two panels, top (RsTAL versus PcPAL) and bottom, (RsTAL versus PpHAL), are shown to simplify viewing. For each protein, the inner loops (RsTAL residues 53–81, PcPAL residues 103–129, and PpHAL residues 46–74) are shown in darker shades, and the outer loops (RsTAL residues 268–300, PcPAL residues 325–351, and PpHAL residues 260–280) are shown in lighter shades. The side chains of two invariant aromatic residues from the inner loop are shown (RsTAL Tyr 60 and Phe 66, PcPAL Tyr 110 and Phe 116, and PpHAL Tyr 53 and Phe 59).
(B) The RsTAL homotetramer in the vicinity of the active-site pocket of monomer a is shown in ribbon representation. The polypeptide chains of the individual monomers are colored as in Figure 1A, with the active-site lid loops shaded darker (green, inner loop of monomer a; yellow, outer loop of monomer d). The MIO cofactor, bound coumarate, and protein residues that interact with the coumarate are drawn as balls and sticks and colored by atom type.
(C) The L-Tyr substrate (magenta) was modeled with minimal modifications from the binding mode of the coumarate product shown in Figure 3B.
(D) The L-Tyr substrate was modeled based upon the binding mode of the AIP inhibitor shown in Figure 4D, which places the α-amino group within covalent bonding distance of the Cβ2 methylidene carbon of the MIO cofactor (yellow dashed line). Note that a hydrogen bond between the L-Tyr-OH and the His 89-NE2 is preserved despite the shifted position of the L-Tyr substrate.
Three Gly residues, 61, 65, and 67, facilitate formation of several interactions in this region, including a short three-stranded β sheet, which involves polypeptide segments from both the inner and outer lid-loops (Figure 5B). The environment of MIO in RsTAL is therefore sequestered from the bulk solvent, in contrast to the relatively open active-site access observed in other aromatic amino acid ammonia-lyase structures with disordered or more externally positioned lid loops. In the structure of the RsTAL-coumarate complex, the inner lid-loop comes in close contact with the coumarate molecule (Figures 2C and 3B).
In previous crystallographic studies of aromatic amino acid ammonia-lyases, attempts to obtain complexes with substrate or product were unsuccessful. Our observation of well-ordered coumarate binding in RsTAL crystals is likely due to the positioning of the lid loops. Substrate binding may also contribute to stabilizing this arrangement. Thus, the internalized position of the inner-lid loop within an enzyme-substrate complex would explain the observation that L-Tyr inhibits proteolytic cleavage at two sites along this loop in R. toruloides PAL [17]. Comparison of the crystal structures of unliganded and coumarate-complexed RsTAL indicates that coumarate binding is accommodated within the active site with no significant structural perturbation to the surrounding protein. This observation is somewhat surprising, in light of the close packing of residues around the coumarate product and the closed lid-loops which apparently occlude access to the active-site pocket. These results suggest a dynamic role of the active-site lid loops in substrate binding, sequestering of reaction intermediates, and/or catalysis.
Implications for the Reaction Mechanism of RsTAL
The structures of the RsTAL-coumarate, -caffeate and -cinnamate complexes allow for modeling of the binding of the substrate L-Tyr (Figures 5C and 5D). A sizable translational shift of the substrate would be required to bring the L-Tyr ring Cδ atom (C2 relative to C4 bearing the phenolic hydroxyl moiety) within bonding distance of the MIO methylidene, as suggested by the Friedel-Crafts-type mechanism [1]. Such a movement would disrupt the interactions formed by the α-carboxylate and para-hydroxylgroups toTAL(Figure 3B). On the other hand, although the substrate’s α-amino group in the modeled complex is oriented away from the cofactor, a modest change in conformation of the propenoate group would position the α-amino group appropriately for attack on the MIO methylidene carbon (Figures 5C and 5D).
If such an attack occurs, the negative charge formed on the MIO carbonyl oxygen (O2) would be stabilized by water-mediated protonation (enol tautomer) in the oxyanion hole (Figure 2D) and a newly formed interaction with the side-chain amide of Asn 203 (Figure 5D). Notably, Asn 203 is brought into proximity of the cofactor by a rearrangement of the segment of polypeptide-chain spanning residues 194–205 (as observed in the TAL-AIP covalent complex) (Figure 4D). Substitution of this highly conserved residue by Ala causes large decreases in kcat in both HAL and PAL [24, 25].
The covalent binding of the inhibitor AIP to H89F RsTAL observed crystallographically here does not necessarily support a mechanism beginning with amino-directed attack. Nevertheless, AIP does serve as a very useful geometrically constrained mimic of one L-Phe conformer (Figures 4E and 5D). While this structural study does not unconditionally resolve the mechanistic debate, the elucidation of these product and inhibitor complexes provides a useful structural perspective for considering additional mechanistic alternatives to the two currently described mechanisms.
Significance
The structures of RsTAL complexed with coumarate provide definitive experimental pictures of substrate or product binding to aromatic amino acid ammonia-lyases. The binding of coumarate in the RsTAL active site involves interactions of the propenoate moiety with protein residues that are highly conserved in aromatic amino acid ammonia-lyases including Tyr 60 and Arg 303. The residues that interact with the aromatic ring are more variable, as expected, given the differences in side-chain selectivities within the family of MIO-dependent enzymes. Notably, the RsTAL His89-imidazole group, which hydrogen bonds with the coumarate p-hydroxyl moiety, discriminates between L-Tyr and L-Phe or L-His as substrates. This discovery is the selectivity filter in PAL/TAL/HALs. Replacement of His 89 by Phe, a residue more characteristic of the PALs, yields a mutant RsTAL with a switch in substrate preference from L-Tyr to L-Phe. Structures of the H89F mutant RsTAL in complex with the reaction product, cinnamic acid, or the PAL inhibitor, 2-aminoindan-2-phosphonic acid, revealed binding modes in which the phenyl rings of Phe 89 and the ligands interact edge to edge. Based upon a comparison of available X-ray crystal structures, it appears that two loops capping the active site of the aromatic amino acid ammonia-lyases play a role in both substrate binding and catalysis. Our combined structural and functional studies provide a starting point for understanding the reaction mechanism of this enzyme family and also the aromatic amino acid aminomutases, which biosynthesize β-amino acids in nature. Engineering of alternative specificities in aromatic amino acid ammonia-lyases and the related aminomutases can now be undertaken using both site-directed approaches and focused combinatorial methods to expand the technological utility of these MIO-dependent enzymes.
Experimental Procedures
Synthetic Gene Expression and Purification, and Mutagenesis of RsTAL
A synthetic gene encoding the amino acid sequence of RsTAL was synthesized by GenScript (www.genscript.com). The gene sequence was optimized for codon preferences in Escherichia coli and bracketed by 5′-NcoI and 3′-BamH1 restriction sites. The gene was inserted between the NcoI and BamH1 sites of the expression vector pHIS8 under control of a T7 promoter to yield the target protein fused to a thrombin-cleavable N-terminal octahistidine tag [26].
Briefly, E. coli cultures in TB medium were grown at 37°C to an optical density (600 nm) of 1.5, induced with 1 mM isopropyl-β-D-thiogalactoside, and allowed to grow for an additional 6 hr at 20°C. Bacterial cells were harvested by centrifugation, resuspended in lysis buffer (50 mM TrisHCl [pH 8.0], 0.5 M NaCl, 20 mM imidazole, 1% [v/v] Tween 20, 10% [v/v] glycerol and 20 mM 2-mercaptoethanol), and lysed by sonication. RsTAL was isolated from the E. coli lysate by affinity chromatography with Ni2+-NTA agarose and eluted with lysis buffer supplemented with 0.25 M imidazole. Nearly homogeneous RsTAL was treated with thrombin for cleavage of the octahistidine tag, and then further purified by gel-exclusion chromatography using a Superdex 200 HR26/60 column (Pharmacia Biosystems). Site-directed mutants of the RsTAL gene were created in the plasmid pHIS8-RsTAL using the QuikChange protocol (Stratagene), and mutant proteins were expressed and purified as described for wild-type RsTAL. The DNA sequences of the mutant constructs were confirmed by sequencing of the entire RsTAL gene insert in both the forward and reverse directions.
Enzyme Assays
TAL activity was measured spectrophotometrically by monitoring formation of a conjugated aryl-acid product. The conversion of L-Tyr to p-coumarate was followed at 310 nm and that of L-Phe to cinnamate at 280 nm. The assay mixture (total volume 0.5 ml) contained 0.1MTrisHCl (pH 8.5) and, for each fixed amount of TAL, eight different initial concentrations of amino acid substrate. After preincubation of the enzyme at 37°C for 2 min, reactions were initiated by the addition of substrate, and formation of product was monitored for 5 min. For wild-type RsTAL, activity was assayed with 7.5 μg protein and initial L-Tyr concentrations between 0.01 and 2.4 mM, and PAL activity was assayed with 30 μg protein and initial L-Phe concentrations between 3.2 and 51.2 mM. For the RsTAL H89F mutant, PAL activity was assayed with 10 μg protein and initial L-Phe concentrations between 0.05 and 5.2 mM. Each series of assays were performed in triplicate, and a Hanes plot was used for the estimation of steady-state kinetic constants.
The inhibition of enzyme activity by AIP was measured with a similar assay system, except that the enzyme was preincubated for 7 min in the presence of fixed concentrations of AIP. Inhibition of PAL activity of wild-type RsTAL was measured with 15 μg protein, six AIP concentrations between 0 and 20 μM, and initial L-Phe concentrations between 9.6 and 25.6 mM. Inhibition of PAL activity of the H89F mutant was measured with 10 μg protein, five AIP concentrations between 0 and 5 μM, with initial L-Phe concentrations between 0.4 and 1.6 mM. Duplicate sets of enzyme assays were performed, and a Dixon plot was used for the estimation of the AIP inhibition constant, Ki.
Crystallization and X-Ray Structure Elucidation of RsTAL
Crystals of RsTAL (space group P21) were grown from a 1:1 mixture of protein solution (20 mg/ml in 12.5 mM Tris-HCl [pH 7.5], 50 mM NaCl) and a reservoir solution (0.1 M MOPSO-Na+ [pH 7.0], 7% [w/v] polyethylene glycol 8000, 0.3 M ammonium acetate, 2 mM dithiothreitol, 35 mM cyclohexylbutanoyl-N-hydroxyethylglucamide) using vapor diffusion at 4°C. Crystal growth occurred over a period of 1–3 weeks and was expedited through microseeding. The crystals exhibit a rhomboid morphology and grow to a maximum size of 0.4 × 0.1 × 0.1 mm. Crystals of RsTAL in complex with small-molecule ligands were produced by soaking crystals for 24–48 hr in reservoir solutions supplemented with 10–20 mM coumaric acid, caffeic acid, cinnamic acid, or AIP.
Crystals were transferred briefly to a cryo-protectant solution (consisting of reservoir solution supplemented with 15%–20% [v/v] polyethylene glycol 400) prior to immersion in liquid nitrogen. X-ray diffraction data were measured from frozen crystals at beamline 8.2.2 of the Advanced Light Source (Lawrence Berkeley National Laboratory) on an ADSC Quantum 315 CCD detector or at beamline 11.1 of the Stanford Synchrotron Radiation Laboratory on an ADSC Quantum 315 CCD detector. Diffraction intensities were indexed, integrated, and scaled with the programs XDS and XSCALE [27], or Mosflm [28] and Scala [29], as summarized in Table 1.
RsTAL crystals contain two complete homotetramers per asymmetric unit and diffract up to 1.50 Å resolution. Initial phases were determined by molecular replacement using the program Molrep [30]. A homology model for RsTAL was constructed with the program Modeller [31], based on the structure of P. putida HAL (PDB entry 1GKM). The program ARP/wARP [32] was used for automated rebuilding of the initial structure using an 8-fold, noncrystallographic symmetry (NCS) averaged map. Subsequent structural refinement used the program CNS [33] with NCS restraints applied until the final stages where they were removed. Xfit [34] was used for map inspection and manual rebuilding of the atomic model. Programs from the CCP4 suite [29] were employed for all other crystallographic calculations. For refinement of the coumarate, caffeate, and cinnamate molecules, no conformational restraints were applied to the freely rotatable dihedral angles of the propenoate moiety or to enforce similarity of NCS-related copies. Figures were drawn with the program Pymol (Delano Scientific, San Carlos, CA).
Acknowledgments
We are grateful to Kevin Watts and Claudia Schmidt-Dannert (University of Minnesota) for their communication of complementary results prior to publication, and to Michael Austin for useful discussions. We deeply appreciate the gift of AIP generously provided by Dr. Jerzy Zon (Wroclaw University, Poland). We thank the staff of the Advanced Light Source, Berkeley, California for assistance during data collection at beamline 8.2.2. This material is based upon work supported by the National Science Foundation under Grant No. 0236027 to JPN and by the National Institutes of Health grant AI47818 to BSM. J.P.N. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Accession Numbers
The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2O6Y (RsTAL), 2O7B (RsTAL-coumarate), 2O7D (RsTAL-caffeate), 2O78 (H89F RsTAL-cinnamate), 2O7F (H89F RsTAL-coumarate) and 2O7E (H89F RsTAL-AIP)].
References
- 1.Poppe L, Rétey J. Friedel-Crafts-type mechanism for the enzymatic elimination of ammonia from histidine and phenylalanine. Angew Chem Int Ed Engl. 2005;44:3668–3688. doi: 10.1002/anie.200461377. [DOI] [PubMed] [Google Scholar]
- 2.Rosler J, Krekel F, Amrhein N, Schmid J. Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol. 1997;113:175–179. doi: 10.1104/pp.113.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang H, Wood KV, Morgan JA. Metabolic engineering of the phenylpropanoid pathway in Saccharomyces cerevisiae. Appl Environ Microbiol. 2005;71:2962–2969. doi: 10.1128/AEM.71.6.2962-2969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiang L, Moore BS. Biochemical characterization of a prokaryotic phenylalanine ammonia lyase. J Bacteriol. 2005;187:4286–4289. doi: 10.1128/JB.187.12.4286-4289.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams JS, Thomas M, Clarke DJ. The gene stlA encodes a phenylalanine ammonia-lyase that is involved in the production of a stilbene antibiotic in Photorhabdus luminescens TT01. Microbiol. 2005;151:2543–2550. doi: 10.1099/mic.0.28136-0. [DOI] [PubMed] [Google Scholar]
- 6.Hill AM, Thompson BL, Harris JP, Segret R. Investigation of the early stages in soraphen A biosynthesis. Chem Bio Chem. 2003;4:1358–1359. doi: 10.1039/b303542p. [DOI] [PubMed] [Google Scholar]
- 7.Emes AV, Vining LC. Partial purification and properties of 1-phenylalanine ammonia lyase from Streptomyces verticillatus. Can J Biochem. 1970;48:613–622. doi: 10.1139/o70-099. [DOI] [PubMed] [Google Scholar]
- 8.Kyndt JA, Meyer TE, Cusanovich MA, Van Beeumen JJ. Characterization of a bacterial tyrosine ammonia lyase, a biosynthetic enzyme for the photoactive yellow protein. FEBS Lett. 2002;512:240–244. doi: 10.1016/s0014-5793(02)02272-x. [DOI] [PubMed] [Google Scholar]
- 9.Watts KT, Lee PC, Schmidt-Dannert C. Exploring recombinant flavonoid biosynthesis in metabolically engineered Escherichia coli. Chem Bio Chem. 2004;5:500–507. doi: 10.1002/cbic.200300783. [DOI] [PubMed] [Google Scholar]
- 10.Berner M, Krug D, Bihlmaier C, Vente A, Müller R, Bechthold A. Genes and enzymes involved in caffeic acid biosynthesis in the actinomycete Saccharothrix espanaensis. J Bacteriol. 2006;188:2666–2673. doi: 10.1128/JB.188.7.2666-2673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baedeker M, Schulz GE. Autocatalytic peptide cyclization during chain folding of histidine ammonia-lyase. Structure. 2002;10:61–67. doi: 10.1016/s0969-2126(01)00692-x. [DOI] [PubMed] [Google Scholar]
- 12.Rétey J. Discovery and role of methylidene imidazolone, a highly electrophilic prosthetic group. Biochim Biophys Acta. 2003;1647:179–184. doi: 10.1016/s1570-9639(03)00091-8. [DOI] [PubMed] [Google Scholar]
- 13.Calabrese JC, Jordan DB, Boodhoo A, Sariaslani S, Vannelli T. Crystal structure of phenylalanine ammonia lyase: multiple helix dipoles implicated in catalysis. Biochemistry. 2004;43:11403–11416. doi: 10.1021/bi049053+. [DOI] [PubMed] [Google Scholar]
- 14.Christenson SD, Liu W, Toney MD, Shen B. A novel 4-methylideneimidazole-5-one-containing tyrosine aminomutase in enediyne antitumor antibiotic C-1027 biosynthesis. J Am Chem Soc. 2003;125:6062–6063. doi: 10.1021/ja034609m. [DOI] [PubMed] [Google Scholar]
- 15.Walker KD, Klettke K, Akiyama T, Croteau R. Cloning, heterologous expression, and characterization of a phenylalanine aminomutase involved in taxol biosynthesis. J Biol Chem. 2004;279:53947–53954. doi: 10.1074/jbc.M411215200. [DOI] [PubMed] [Google Scholar]
- 16.Steele CL, Chen Y, Dougherty BA, Li W, Hofstead S, Lam KS, Xing Z, Chiang SJ. Purification, cloning, and functional expression of phenylalanine aminomutase: the first committed step in Taxol side-chain biosynthesis. Arch Biochem Biophys. 2005;438:1–10. doi: 10.1016/j.abb.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Gamez A, Sarkissian CN, Straub M, Patch MG, Han GW, Striepeke S, Fitzpatrick P, Scriver CR, Stevens RC. Structure-based chemical modification strategy for enzyme replacement treatment of phenylketonuria. Mol Genet Metab. 2005;86:134–140. doi: 10.1016/j.ymgme.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 18.Ritter H, Schulz GE. Structural basis for the entrance into the phenylpropanoid metabolism catalyzed by phenylalanine ammonia-lyase. Plant Cell. 2004;16:3426–3436. doi: 10.1105/tpc.104.025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwede TF, Rétey J, Schulz GE. Crystal structure of histidine ammonia-lyase revealing a novel polypeptide modification as the catalytic electrophile. Biochemistry. 1999;38:5355–5361. doi: 10.1021/bi982929q. [DOI] [PubMed] [Google Scholar]
- 20.Pilbák S, Tomin A, Rétey J, Poppe L. The essential tyrosine-containing loop conformation and the role of the C-terminal multi-helix region in eukaryotic phenylalanine ammonia-lyases. FEBS J. 2006;273:1004–1019. doi: 10.1111/j.1742-4658.2006.05127.x. [DOI] [PubMed] [Google Scholar]
- 21.Baedeker M, Schulz GE. Structures of two histidine ammonia-lyase modifications and implications for the catalytic mechanism. Eur J Biochem. 2002;269:1790–1797. doi: 10.1046/j.1432-1327.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- 22.Appert C, Logemann E, Hahlbrock K, Schmid J, Amrhein N. Structural and catalytic properties of the four phenylalanine ammonia-lyases from parsley (Petroselinum crispum Nym) Eur J Biochem. 1994;225:2177–2185. doi: 10.1111/j.1432-1033.1994.00491.x. [DOI] [PubMed] [Google Scholar]
- 23.Appert C, Zón J, Amrhein N. Kinetic analysis of the inhibition of phenylalanine ammonia-lyase by 2-aminoindan-2-phosphonic acid and other phenylalanine analogues. Phytochemistry. 2003;62:415–422. doi: 10.1016/s0031-9422(02)00561-7. [DOI] [PubMed] [Google Scholar]
- 24.Rother D, Poppe L, Viergutz S, Langer B, Rétey J. Characterization of the active site of histidine ammonia-lyase from Pseudomonas putida. Eur J Biochem. 2001;268:6011–6019. doi: 10.1046/j.0014-2956.2001.02298.x. [DOI] [PubMed] [Google Scholar]
- 25.Rother D, Poppe L, Morlock G, Viergutz S, Rétey J. An active site homology model of phenylalanine ammonia-lyase from Petroselinum crispum. Eur J Biochem. 2002;269:3065–3075. doi: 10.1046/j.1432-1033.2002.02984.x. [DOI] [PubMed] [Google Scholar]
- 26.Jez JM, Ferrer JL, Bowman ME, Dixon RA, Noel JP. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry. 2000;39:890–902. doi: 10.1021/bi991489f. [DOI] [PubMed] [Google Scholar]
- 27.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 28.Leslie AGW. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;(26) [Google Scholar]
- 29.CCP4 (Collaborative Computational Project Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallog D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 31.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 32.Lamzin VS, Wilson KS. Automated refinement of protein models. Acta Crystallogr D Biol Crystallogr. 1993;49:129–149. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 33.Brunger AT, Warren GL. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 34.McRee DE. XtalView/Xfit: a versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]