Abstract
The expression of recombinant proteins in E. coli often leads to inactive aggregated proteins known as the inclusion bodies. To date, the best available tool has been the use of fusion tags, including the carbohydrate-binding protein; e.g., the maltose-binding protein (MBP) that enhances the solubility of recombinant proteins. However, none of these fusion tags work universally with every partner protein. We hypothesized that galectins, which are also carbohydrate-binding proteins, may help as fusion partners in folding the mammalian proteins in E. coli. Here we show for the first time that a small soluble lectin, human galectin-1, one member of a large galectin family, can function as a fusion partner to produce soluble folded recombinant human glycosyltransferase, β1,4-galactosyl-transferase-7 (β4Gal-T7), in E. coli. The enzyme β4Gal-T7 transfers galactose to xylose during the synthesis of the tetrasaccharide linker sequence attached to a Ser residue of proteoglycans. Without a fusion partner, β4Gal-T7 is expressed in E. coli as inclusion bodies. We have designed a new vector construct, pLgals1, from pET-23a that includes the sequence for human galectin-1, followed by the Tev protease cleavage site, a 6xHis-coding sequence, and a multi-cloning site where a cloned gene is inserted. After lactose affinity column purification of galectin-1-β4Gal-T7 fusion protein, the unique protease cleavage site allows the protein β4Gal-T7 to be cleaved from galectin-1 that binds and elutes from UDP-agarose column. The eluted protein is enzymatically active, and shows CD spectra comparable to the folded β4Gal-T1. The engineered galectin-1 vector could prove to be a valuable tool for expressing other proteins in E. coli.
Keywords: Protein expression in E. coli; Fusion tags; MBP; Galectins; Protein folding; Glycosyltransferases; beta-1,4-galactosyltransferase-T7
Introduction
Although a number of expression hosts are available for the production of recombinant proteins, E. coli remains the most favored host. However, the expression of the recombinant protein in E. coli often leads to the production of protein aggregates named inclusion bodies. Since E. coli is easy to use as a host, it is essential to find ways to overcome the formation of inclusion bodies during protein expression [1-4]. The best available tools to date have been the fusion tags that enhance the solubility of the expressed protein [5] such as DsbC, thioredoxin, T7pk, SUMO, GST, NusA, and a carbohydrate-binding protein, e.g., maltose-binding protein (MBP) [6-9]. None of these tags work universally with every protein partner. Even the most used fusion partner MBP, that is an excellent solubility enhancer, does not guarantee to give an active form of protein. At times, for purification purposes, MBP-fusion protein does not bind to the amylose-affinity resin. Other times, after release from the affinity resin, protein precipitates out, possibly because the protein is misfolded or prone to aggregation in its native state [7, 10]. Here we show that another carbohydrate-binding protein, human galectin-1, can be an efficient fusion partner.
Galectins are a family of animal lectins defined by shared consensus amino acid sequences and affinity for β-galactose-containing oligosaccharides [11-14]. The galectin family members 1-15, are composed of one or two carbohydrate-recognition domains (CRDs). Among these galectins, galectin-1 has one CRD and binds preferentially to glycoconjugates with N-acetyllactosamine (Galβ1-3/4 GlcNAc) and to polylactosamine chains and α1-3 sialylated glycans with very high affinity [15]. It is differentially expressed by various normal and pathological tissues and is present both inside and outside the cells. It binds to a number of extracellular matrix (ECM) components in a carbohydrate-dependent manner, such as: thrombospondin, fibronectin and vitronectin [12)], whereas in the intracellular form it associates with proteins, such as Gemin4 and Ras involving carbohydrate-independent interactions [16, 17].
The β1,4 galactosyltransferase-7 (β4Gal-T7), that belongs to β4Gal-T family, T1-T7, [18, 19], transfers galactose from UDP-α-Gal to xylose bound to a serine residue in the core protein of proteoglycans, forming the disaccharide sequence, Galβ1-4Xylβ1-, in the common tetrasaccharide linker sequence GlcAβ1-3Galβ1-3Galβ1-4Xylβ1-R. Protein expression of β4Gal-T family members, including human β4Gal-T7, produce in E. coli insoluble inclusion bodies. Although a method of in vitro folding of inclusion bodies of one member of the family, β4Gal-T1, has been successfully developed in our laboratory [20], the same method does not fold in vitro the human β4Gal-T7 inclusion bodies. The human β4Gal-T7 has been expressed in E. coli as MBP fusion protein [21]. The fusion protein, however, shows poor solubility and aggregates after cleavage with protease to release β4Gal-T7 from MBP. This encouraged us to test another carbohydrate-binding protein such as galectin-1 as a fusion partner to increase folding, stability, and solubility of recombinant human β4Gal-T7, which, without any fusion partner, accumulates as inclusion bodies. We show here human galectin-1 is an efficient folding partner.
Materials and Methods
The DNA clones containing human β4Gal-T7 and human galectin-1 sequences were obtained from Open Biosystem; pET23a vector and BL21(DE3)pLysS cells from Novagen; XL2 blue ultracompetent cells from Stratagene; Taq DNA polymerase, PCR nucleotide mix, and rapid DNA ligation kit from Roche Pharmaceuticals; DNA miniprep spin columns, PCR purification and low-melting agarose extraction kits from Qiagen; Restriction enzymes from New England Biolabs, Inc; Ampicillin, UDP-Gal and xylose from Sigma-Aldrich; UDP-[6-3H] galactose from American Radiolabeled Chemicals; AG 1-X8 chloride resin 200-400 mesh from Bio-Rad; UDP-agarose gel from Calbiochem. DNA primers were synthesized by Integrated Technologies, Inc. Tev (Tobacco etch virus) protease was from Invitrogen.
Construction of galectin-1 vector with supplemental Tev cleavage site and the N-terminal His-tag (pLgals1-Tev) of the passenger proteins
The pLgals1-Tev vector was constructed using plasmid pET-23a. The human galectin-1 DNA sequence (Lgals1) was PCR amplified using follow primers:
Left primer: 5′ CGCGGATCCGCTTGTGGTCTGGTCGCCAGCAACCTGAATCTC 3′
Right primer: 5′CGCCCCAAGCTTGGGTCAGTCAAAGGCCACACATTTGATCTTGAAGTC3′
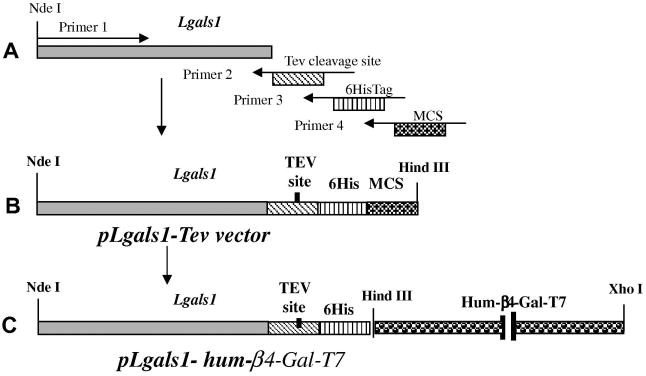
The PCR-amplified fragment of Lgals1 was digested by BamHI and HindIII restriction enzymes and inserted into the pET-23a vector, generating a resulting vector pLgals1 (Figure 1(A)). The vector was used to amplify fragment of the Lgals1 (galectin-1) sequence with following primers that have coding sequence for the Tev protease cleavage site, 6xHisTag, and multi-cloning site (MCS) for the restriction sites BamHI, EcoRI, SacI, SalI, HincII, HindIII, NotI, EagI, and XhoI (Figure 1(B)):
Figure 1.
(A). Schematics of PCR amplification of gals1 DNA sequence with Tev cleavage site, 6HisTag, and multicloning site (MCS). (B). Schema of pLgals1-Tev vector construct. (C). Schema of pLgals1-hum-βGal-T7 construct, the pLgals1-Tev sequence fused to human β4Gal-T7 sequence.
Left primer: 5′GATATACATATGGCTAGCATGACTGGTGGACAGCAAATGGGTCGCGGTTCCGCTTGT 3′
Right Primer 2 that inserts Tev cleave site: 5′ACCCTGGAAGTACAGGTTCTCGTCAAAGGCCACACATTT 3′
Right Primer 3 that inserts 6HisTag: 5′GTGGTGGTGGTGGTGGTGACCCTGGAAGTACAGGTTCTCGTCAAAGGC 3′
Right Primer 4 that inserts multiple cloning site (MCS): 5′ TGCGGCCGCAAGCTTGTCGACGAGCTCGAATTCGGATCCGTGGTGGTGGTGGTGGTGACCCTGGAAGTA 3′
The PCR fragment was digested by NdeI and HindIII restriction enzymes and inserted into pET23a. A schematic representation of expression pLgals1-Tev vector is given in Figure1 (A-B). This pLgals1-Tev vector clone was used for construction of human β4Gal-T7 fused to galectin-1 (Figure 1(C)).
Cloning and expression of a soluble form of human β4Gal-T7 with human galectin-1 as fusion partner
For the construction of plasmid containing β4Gal-T7 DNA sequence fused with galectin-1 DNA sequence in the pLgals1-Tev vector, the β4Gal-T7 sequence was PCR amplified using following primers:
Left primer: 5′ GTCCCAGCCAAGCTTAGCTGCTCTGGGGACGTGGCCCGGGCAGTC 3′
Right primer: 5′GTCCAAGCCCTCGAGTCAGCTGAATGTGCACCAGGGTGTGGCGGT 3′
The PCR fragment was digested with HindIII and XhoI enzymes and sub-cloned into the pLgals1-Tev vector. The sequence of galectin1-hum-β4Gal-T7 DNA was confirmed by sequence analysis.
Expression and purification of galectin-1-hum-β4Gal-T7 fusion protein
To express the soluble galectin-1–hum-β4Gal-T7 fusion protein, E. coli BL21(DE3)pLysS cells were transformed with the plasmid containing galectin-1–hum-β4Gal-T7 sequence and grown at 37°C in LB supplemented with ampicillin (100 μg/ml) to OD600 nm = 0.6–0.8. Isopropyl β-D thiogalactopyranoside (IPTG) was then added to a final concentration of 1 mM and the cells were further incubated overnight at 23 °C. Cells were harvested by centrifugation at 3000g × 10 min and resuspended in 100 mM PBS buffer, pH 7.4, containing 0.5 M NaCl, and cells were disrupted by sonication (6 × 30 s). The solution was centrifuged at 15000g × 20 min at 4°C. The resulting supernatant was loaded on to a 2 ml bed volume of alpha-lactose column, which was first equilibrated with 100 mM PBS (pH 7.4) buffer containing 0.5 M NaCl (equilibration/wash buffer). Next, the column was washed with equilibration/wash buffer 5 times the bed volume and the bound protein eluted with 100 mM lactose.
Purification of human β4Gal-T7 on UDP-agarose column after Tev protease cleavage of the galectin-1-hum-β4GalT7 fusion protein
Four mg of galectin-1-hum-β4Gal-T7 fusion protein was cleaved with 100 units of Tev protease in 2.5 ml 50mM Tris/HCl, pH 8.0, 0.5 mM EDTA and 1mM DTT at 4°C for 48 hours. After adjusting Mn2+ concentration to 25 mM, the protein was loaded on a UDP-agarose column, and human β4Gal-T7 was purified and analysed as described for β4Gal-T1 [20].
Cloning of the human β4Gal-T7 using MBP as a fusion partner
For expression of human β4Gal-T7 fused with MBP, the DNA sequence coding the residues 52 to 327 of human β4Gal-T7 was cloned in to pET23a vector between the BamHI and EcoRI restriction sites using the following primers:
Left Primer: 5′ GTCCCAGCCGGATCCAGCTGCTCTGGGGACGTGGCCCGGGCAGTC 3′
Right Primer: 5′GTCCAAGCCGAATTCTCAGCTGAATGTGCACCAGGGTGTGGCGGT 3′
The DNA fragment released from the pET23a-β4Gal-T7 vector by digestion with the restriction enzymes BamHI and HindIII, was sub-cloned into the pMAL-c2x vector between BamH I and HindIII restriction sites. The sequence of the fusion construct was confirmed by DNA sequence analysis.
Expression and purification of MBP-hum-β4Gal-T7 fusion protein
To express the soluble MBP–hum-β4Gal-T7 fusion protein, E. coli BL21 cells were transformed with the pMAL-hum-β4Gal-T7 plasmid and grown at 37°C in LB supplemented with ampicillin (100 μg/ml) to OD600 nm = 0.6–0.8. Isopropyl β-D thiogalactopyranoside (IPTG) was then added to a final concentration of 1 mM and the cells were further incubated overnight at 23°C. Cells were harvested by centrifugation at 3000 g for 10 min and resuspended in 10 mM Tris/HCl buffer, pH 8.0, containing 150 mM NaCl. The resuspended cells were disrupted by sonication (6 × 30 s). The solution was centrifuged at 15000 g × 20 min at 4°C. The resulting supernatant was purified on a 4 ml amylose column as described [7].
Purification of human β4Gal-T7 on UDP-agarose column after Factor Xa proteolytic cleavage of the MBP-hum-β4Gal-T7 fusion protein
Four mg of MBP-hum-β4GalT-7 fusion protein was cleaved with 10 μg of Factor Xa protease in a total volume of 2.5 ml containing 20 mM Tris/HCl, pH 8.0, 100 mM NaCl, 2 mM CaCl2 at 4°C for 48 hours. The solution was adjusted to 25 mM MnCl2, loaded on a UDP-agarose column, and purified as described above.
CD spectroscopy of human β4Gal-T7 and bovine β4Gal-T1
The near UV (250-320) and far UV (200-250) CD spectra of human β4Gal-T7 and bovine β4Gal-T1 proteins at a concentration of 1 mg/ml in 100 mM PBS pH 7.4 were determined with AVIV Mod.202 CD Spectropolarimeter (Aviv Instruments).
Enzyme activity assay and kinetic analysis of human β4Gal-T7
For specific activity measurements, reactions were carried out at 37°C for 30 min in a 100-μL volume containing 10 mM MnCl2, 25 mM Tris/HCl (pH 7.0), 500 μM UDP-Gal, 0.5 μCi of 3H-labeled sugar nucleotide, and 5 mM xylose with 1 μg of enzyme. The reactions were terminated and analyzed as described [20]. The kinetic analysis was carried out as described previously for β4Gal-T1 [22].
Results
1.1 Expression and purification of the fusion protein galectin-1-hum-β4Gal-T7
Since without a fusion partner human β4Gal-T7 is produced as inclusion bodies in E. coli which did not fold in vitro by the method developed in our laboratory [20], we inserted the human β4Gal-T7 DNA sequence into the pLgals1-Tev vector resulting into the expression vector pLgals1-hum-β4Gal-T7 (See methods and Figure 1(C)). This construct expressed in E. coli BL21(DE3)pLysS cells the soluble fusion protein galectin-1-hum-β4Gal-T7 as a 53 kDa protein (Figure S1). After sonication of the cells, the supernatant with a soluble protein was loaded on the alpha lactose column. Bound fusion protein was eluted with a yield of 15 mg from one liter bacterial culture (Table 1). The fusion protein galectin-1-hum-β4Gal-T7, in contrast to MBP–hum-β4Gal-T7, did not aggregate and was stable for a long time.
Table 1.
Comparison of the yields of soluble and folded β4Gal-T7 protein per 1 liter of bacterial culture using MBP and galectin-1 as fusion partners. The yield of the fusion proteins from 1 liter of bacterial culture is 40 mg for MBP-hum-β4Gal-T7 and 15 mg for galectin1-hum-β4Gal-T7. MBP-hum-β4Gal-T7, in contrast to galectin-1-hum-β4Gal-T7, precipitates out of solution before and after protease cleavage and cannot be stored for longer periods of time.
| Alpha-lactose | UDP-agarose column | |
|---|---|---|
| hum-Galectin-1-hum-β4Gal-T7 | 15 mg | 7.5 mg |
| Alpha-amylose | UDP-agarose column | |
| MBP-hum-β4Gal-T7 | 40 mg | 5 mg |
1.2 Expression and purification of MBP–hum-β4Gal-T7 fusion protein
The pmal-2x-hum-β4Gal-T7 construct was expressed in E. coli BL21 cells as MBP-hum-β4Gal-T7 fusion protein. Analysis of the total protein on the SDS-PAGE gels shows a predominant band of 71 kDa (Figure S1). After expression of the soluble fusion protein, supernatant from sonicated cells was loaded on the alpha amylose column. Bound fusion protein was eluted with a yield of 40 mg from one liter of bacterial culture (Table 1). Fusion protein aggregated and was difficult to store for long time.
1.3 Solubility of human β4Gal-T7 after release from the fusion partners: galectin-1 or maltose binding protein
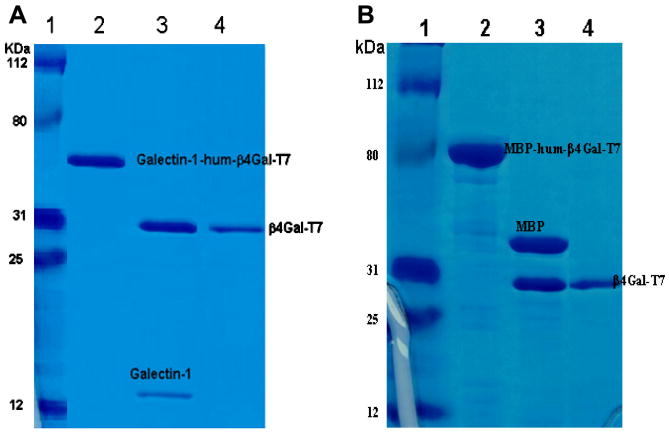
Fifteen mg of lactose column purified galectin-1-hum-β4Gal-T7, produced from a liter of bacterial culture, after cleavage with Tev protease to release the β4Gal-T7, was very stable solution and did not aggregate. After purification on UDP-agarose column about 7.5 mg of purified and soluble β4Gal-T7 was obtained (Table 1, Figure 2). On the other hand, after factor Xa proteolytic cleavage of 40 mg of MBP-hum-β4Gal-T7 fusion protein, obtained from a liter of bacterial culture, the protein solution aggregates and clumps. Only 5 mg of β4Gal-T7 was recovered after UDP-agarose column purification, which was also prone to aggregation (Table 1). β4Gal-T7 obtained after purification on UDP-agarose column from both fusion proteins was detected as a single band of about 31 kDa on SDS-PAGE gel electrophoresis (Figure 2).
Figure 2.
SDS–PAGE analyses of fusion proteins, Galectin-1-hum-β4GalT-7 (A) and MBP-hum-β4GalT-7 (B). Markers (A1 and B1). Galectin-1-hum-β4Gal-T7 after purification on alpha lactose column (A2), and MBP-hum-β4Gal-T7 after purification on alpha amylose column (B2). Galectin-1-hum-β4Gal-T7 after cleavage with Tev (A3), and MBP-hum-β4Gal-T7 after cleavage with Factor Xa (B3). β4Gal-T7 after purification on UDP-agarose column from galectin-1-fusion protein (A4) and from MBP-fusion protein (B4).
1.4 The near and far UV CD spectra, enzyme activity and kinetic analysis of human β4Gal-T7
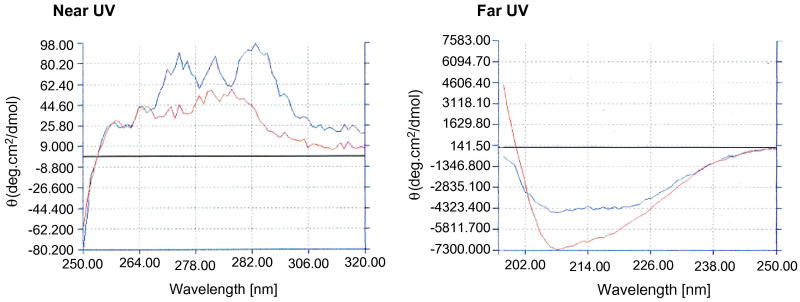
The β4Gal-T7 purified on the UDP-agarose column from the Tev cleaved galectin-1-hum-β4Gal-T7, showed near and far UV CD spectra comparable to the folded bovine β4Gal-T1 (Figure 3) for which structure is known [23]. The human β4Gal-T7, produced using galectin-1 as a fusion tag, has a specific activity of 600 nM/min per mg of protein, and protein shows no loss of activity at least for an hour at 37°C (Figure 4 and insert). The data of the kinetic analysis with UDP-Gal as donor substrate and xylose as an acceptor substrate are presented in Table 2.
Figure 3.
Comparison of near and far UV CD spectra of hum-β4Gal-T7 with hum-β4Gal-T1.
Figure 4.
Activity at different concentrations of β4Gal-T7 at 5 mM Xyl, 500 μM UDP-Gal. The insert shows time course of Gal transfer with 1 μg of β4Gal-T7 at 5 mM Xyl, 500 μM UDP-Gal.
Table 2.
Kinetics parameters of human β4Gal-T7 for donor UDP-Gal and the acceptor substrate xylose.
| Enzyme | KA (mM) UDP-Gal |
KB mM) xylose |
Kia (mM) | Kib (mM) | kcat (s-1) |
|---|---|---|---|---|---|
| Hum-β4Gal-T7 | 0.519±0.02 | 16.0±1.50 | 0.034±0.002 | 5.6±0.050 | 0.62 |
Discussion
Insolubility of recombinant proteins in E. coli is a major impediment to their production for structural and functional studies. There is not a single general method that can be used to express proteins in soluble and folded form in high yields. Using a fusion partner such as GST or MBP, or SUMO, etc., with a passenger protein helps in producing soluble and folded fusion protein. However, they do not work universally [5-9]. The best studied and most thoroughly validated solubility-enhancing protein is the maltose-binding protein (MBP). Structural studies of MBP have shown that it is an efficient enhancer of solubility only in the open conformation of the carbohydrate recognition domain (CRD) [10]. It is most likely that the CRD of the maltose binding protein is involved in folding of the passenger protein.
The carbohydrate recognition domain represents part of the lectin molecule, which is responsible for the specific carbohydrate–protein interactions, which generally involve hydrogen bonding, interactions of the hydrophobic-face of the carbohydrate with the face of an aromatic side chain, the carbohydrate–π and CH–π stacking interactions, and cation–π interactions [24-26]. Since the hydrophobic effect, hydrogen bonds and salt bridges, all play roles in folding a protein, we hypothesized that carbohydrate recognition domains may provide such a platform during folding of proteins. Thus, based on this rationale, we attempted to test if galectin-1, a carbohydrate-binding protein, can assist as a fusion partner in the folding of the passenger protein, β4Gal-T7. We observed it acts as a better solubilizing and folding agent than MBP.
Galectins are a family of highly conserved glycan-binding proteins with affinity for β-galactoside-containing oligosaccharides. Galectins can be found inside and outside cells and have distinct functions in each location [16, 17]. In addition to carbohydrates, galectins also recognize protein ligands. For example, galectin-1 and galectin-3 interact directly with protein Gemin4, one of the components of snRNPs participating in pre-mRNA splicing and Ras protein, which communicates signals from outside the cell to the nucleus [3, 14, 17]. It was shown that the galactose-dependent binding of galectins, including galectin-1, can be interfered with by pentapeptide ligands [27]. It is also interesting to note that galectin-1 interacts preferentially with the unfolded vitronectin multimers than with the inactive folded monomers [12].
Here for the first time we show that galectin-1 as a fusion partner can act as a chaperone for the expression of soluble and folded passenger protein human β4Gal-T7 in E. coli. Galectin-1 is about 14 kD protein, which is expressed as soluble protein in E. coli and is very easily purified by the alpha-lactose column. Human β4Gal-T7 is an important enzyme involved in the synthesis of proteoglycans, a major component of the extracellular matrix. Mutation of β4Gal-T7 is responsible for human diseases like mascular cornealdystrophy and a progeroid variant of the Ehlers-Danlos syndrome [28, 29]. Expression of the recombinant human β4Gal-T7 alone, without any fusion partner, results in the production of insoluble aggregates, the inclusion bodies. As a fusion protein with galectin-1 it is expressed as soluble and active protein in good yields (15 mg of fusion protein/liter bacterial culture). Recently, human β4Gal-T7 has also been expressed as MBP-fused protein by Daligault F. et al. [21]. Comparison of the efficiency of production of soluble and folded human β4Gal-T7 using either MBP or galectin-1 as fusion partners has shown that with the latter the final yield of β4Gal-T7 is higher per liter of culture than with the former, 7.5 mg compared to 5 mg. Although the initial yield of MBP-hum-β4Gal-T7 fusion protein is high, the protein aggregates, particularly while releasing β4Gal-T7 from the fusion protein using Factor Xa. Most of it also aggregates during further purification steps.
The CD spectra of human β4Gal-T7 prepared from the galectin-1 fusion protein, after cleavage with Tev protease and further purification on the UDP-agarose column, was comparable to the folded bovine β4Gal-T1 [23], confirming that the protein is folded. Furthermore, the kinetics parameters for the donor substrate UDP-Gal (Km = 0.519 mM) and for the acceptor xylose (Km = 16 mM) are comparable with the data obtained by Daligault et al. [21] with the MBP-human β4Gal-T7 fusion protein, using 4-nitrophenyl-β-D-xylopyranoside as an acceptor substrate (Km for the acceptor, 1.27 mM and Km for UDP-Gal, 0.23 mM).
The crystal structures of the wild-type galectin-1 and mutant Cys2Ser- and Arg111His-galectin-1 show the same overall fold except for a set of delineated alternations in distinct loop regions and turns of mutant protein [30]. It was clearly shown that these single-site mutations have an effect on the local conformational changes in the loop, which works as a lid and traps sugar in the CRD. Furthermore, it has been shown that mutations of all six cysteines to serines, including Cys2, increase the stability of human galectin-1 expressed in E. coli [31]. We have mutated those six cysteins to serines and used the mutant galectin-1 as a fusion partner. We observed that mutant galectin-1 by itself was expressed as soluble protein in E. coli, but as a fusion protein with β4Gal-T7, it gave a very low yield of the soluble fusion protein (data not shown). A similar observation has been observed with MBP. It has been reported that mutations of the conserved residues in the CRD domain of MBP produced insoluble mutated MPB-fusion protein in E. coli [10]. However, the mutated MBP itself is expressed as soluble folded protein. These observations are consistent with the hypothesis that the carbohydrate-binding domain plays an important role in assisting the folding of the carrier protein by involving hydrogen bonding and hydrophobic interactions in the CRD pocket during folding process. This is further supported by the observation that the peptides with a core sequence of Tyr-Xxx-Tyr can bind to CRD of galectin-1 [32], suggesting that the CRD domain has a potential to form CH-π stacking interactions, which play a role in the folding of proteins.
Conclusion
Our results indicate that proteins with CRD domains, such as galectins, may function as useful fusion partners for folding proteins such as glycosyltransferases that are difficult to be expressed as soluble folded proteins in E. coli. Our results with human β4Gal-T7 and preliminary results with other glycosyltransferases show that galectin-1 can be more effective in enhancing the solubility and folding of a passenger protein than MBP.
Supplementary Material
Acknowledgments
We thank Dr. Natalia Mercer for the critical reading of the manuscript. This project has been funded in whole or in part with Federal funds from the NCI, NIH under contract HHSN261200800001E. The content of this publication does not necessarily reflect the view or policies of the Department of HHS, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government. This research was supported [in part] by the Intramural Research Program of the NIH, NCI, CCR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sahdev S, Khattar SK, Saini KS. Production of active eukaryotic proteins through bacterial expression systems: a review of the existing biotechnology strategies. Mol Cell Biochem. 2008;307:249–264. doi: 10.1007/s11010-007-9603-6. [DOI] [PubMed] [Google Scholar]
- 2.Martínez-Alonso M, González-Montalbán N, García-Fruitós E, Villaverde A. Learning about protein solubility from bacterial inclusion bodies. Microb Cell Fact. 2009;8:4. doi: 10.1186/1475-2859-8-4. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol. 2004;22:1399–408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- 4.Sørensen HP, Mortensen KK. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J Biotechnol. 2005;115:113–28. doi: 10.1016/j.jbiotec.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Esposito D, Chatterjee DK. Enhancement of soluble protein expression through the use of fusion tags. Curr Opin Biotechnol. 2006;17:353–8. doi: 10.1016/j.copbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Bach H, Mazor Y, Shaky S, Shoham-Lev A, Berdichevsky Y, Gutnick DL, Benhar I. Escherichia coli maltose-binding protein as a molecular chaperone for recombinant intracellular cytoplasmic single-chain antibodies. J Mol Biol. 2001;312:79–93. doi: 10.1006/jmbi.2001.4914. [DOI] [PubMed] [Google Scholar]
- 7.Nallamsetty S, Waugh DS. A generic protocol for the expression and purification of recombinant proteins in Escherichia coli using a combinatorial His6-maltose binding protein fusion tag. Nat Protoc. 2007;2:383–91. doi: 10.1038/nprot.2007.50. [DOI] [PubMed] [Google Scholar]
- 8.Butt TR, Edavettal SC, Hall JP, Mattern MR. SUMO fusion technology for difficult-to-express proteins. Protein Expr Purif. 2005;43:1–9. doi: 10.1016/j.pep.2005.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Marco V, Stier G, Blandin S, de Marco A. The solubility and stability of recombinant proteins are increased by their fusion to NusA. Biochem Biophys Res Commun. 2004;322:766–71. doi: 10.1016/j.bbrc.2004.07.189. [DOI] [PubMed] [Google Scholar]
- 10.Nallamsetty S, Waugh DS. Mutations that alter the equilibrium between open and closed conformations of Escherichia coli maltose-binding protein impede its ability to enhance the solubility of passenger proteins. Biochem Biophys Res Commun. 2007;364:639–44. doi: 10.1016/j.bbrc.2007.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leffler H, Carlsson S, Hedlund M, Qian Y, Poirier F. Introduction to galectins. Glycoconj J. 2004;19:433–40. doi: 10.1023/B:GLYC.0000014072.34840.04. [DOI] [PubMed] [Google Scholar]
- 12.Camby I, Le Mercier M, Lefranc F, Kiss R. Galectin-1: a small protein with major functions. Glycobiology. 2006:137R–157R. doi: 10.1093/glycob/cwl025. [DOI] [PubMed] [Google Scholar]
- 13.Rabinovich GA. Galectin-1 as a potential cancer target. Br J Cancer. 2005;92:1188–92. doi: 10.1038/sj.bjc.6602493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasta GR. Roles of galectins in infection. Nat Rev Microbiol. 2009;7:424–38. doi: 10.1038/nrmicro2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stowell SR, Arthur CM, Mehta P, Slanina KA, Blixt O, Leffler H, Smith DF, Cummings RD. Galectin-1, -2, and -3 exhibit differential recognition of sialylated glycans and blood group antigens. J Biol Chem. 2008;283:109–23. doi: 10.1074/jbc.M709545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu FT, Rabinovich GA. Galectins as modulators of tumour progression. Nat Rev Cancer. 2005;5:29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- 17.Liu FT, Patterson RJ, Wang JL. Intracellular functions of galectins. Biochim Biophys Acta. 2002;1572:263–73. doi: 10.1016/s0304-4165(02)00313-6. [DOI] [PubMed] [Google Scholar]
- 18.Almeida R, Levery SB, Mandel U, Kresse H, Schwientek T, Bennett EP, Clausen H. Cloning and expression of a proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I. A seventh member of the human beta4-galactosyltransferase gene family. J Biol Chem. 1999;274:26165–71. doi: 10.1074/jbc.274.37.26165. [DOI] [PubMed] [Google Scholar]
- 19.Hennet T. The galactosyltransferase family. Cell Mol Life Sci. 2002;59:1081–1095. doi: 10.1007/s00018-002-8489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boeggeman E, Ramakrishnan B, Qasba PK. The N-terminal stem regions of bovine and human β1,4-galactosyltransferase I increases the in vitro folding efficiency of their catalytic domain from inclusion bodies. Prot Exp & Puri J. 2003;30:219–229. doi: 10.1016/s1046-5928(03)00093-7. [DOI] [PubMed] [Google Scholar]
- 21.Daligault F, Rahuel-Clermont S, Gulberti S, Cung MT, Branlant G, Netter P, Magdalou J, Lattard V. Thermodynamic insights into the structural basis governing the donor substrate recognition by human beta1,4-galactosyltransferase 7. Biochem J. 2009;418:605–14. doi: 10.1042/BJ20081093. [DOI] [PubMed] [Google Scholar]
- 22.Boeggeman E, Qasba PK. Studies on the metal binding sites in the catalytic domain of beta1,4-galactosyltransferase. Glycobiology. 2002;12:395–407. doi: 10.1093/glycob/cwf045. [DOI] [PubMed] [Google Scholar]
- 23.Ramakrishnan B, Boeggeman E, Ramasamy V, Qasba PK. Structure and catalytic cycle of beta-1,4-galactosyltransferase. Curr Opin Struct Biol. 2004;14:593–600. doi: 10.1016/j.sbi.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Laughrey ZR, Kiehna SE, Riemen AJ, Waters ML. Carbohydrate-π Interactions: What Are They Worth? J Am Chem Soc. 2008;130:14625–14633. doi: 10.1021/ja803960x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallivan JP, Dougherty DA. Cation pi interaction. Chem Rev. 1997;97:1303–1324. doi: 10.1021/cr9603744. [DOI] [PubMed] [Google Scholar]
- 27.Arnusch CJ, André S, Valentini P, Lensch M, Russwurm R, Siebert HC, Fischer MJ, Gabius HJ, Pieters RJ. Interference of the galactose-dependent binding of lectins by novel pentapeptide ligands. Bioorg Med Chem Lett. 2004;22:1437–40. doi: 10.1016/j.bmcl.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 28.Perrimon N, Bernfield M. Specificities of heparan sulphate proteoglycans in developmental processes. Nature. 2000;404:725–8. doi: 10.1038/35008000. [DOI] [PubMed] [Google Scholar]
- 29.Faiyaz-Ul-Haque M, Zaidi SH, Al-Ali M, Al-Mureikhi MS, Kennedy S, Al-Thani G, Tsui LC, Teebi AS. A novel missense mutation in the galactosyltransferase-I (B4GALT7) gene in a family exhibiting facioskeletal anomalies and Ehlers-Danlos syndrome resembling the progeroid type1. Am J Med Genet. 2004;28A:39–45. doi: 10.1002/ajmg.a.30005. [DOI] [PubMed] [Google Scholar]
- 30.López-Lucendo MF, Solís D, André S, Hirabayashi J, Kasai K, Kaltner H, Gabius HJ, Romero A. Growth-regulatory human galectin-1: crystallographic characterization of the structural changes induced by single-site mutations and their impact on the thermodynamics of ligand binding. J Mol Biol. 2004;343:957–70. doi: 10.1016/j.jmb.2004.08.078. [DOI] [PubMed] [Google Scholar]
- 31.Nishi N, Abe A, Iwaki J, Yoshida H, Itoh A, Shoji H, Kamitori S, Hirabayashi J, Nakamura T. Functional and structural bases of a cysteine-less mutant as a long-lasting substitute for galectin-1. Glycobiology. 2008;18:1065–73. doi: 10.1093/glycob/cwn089. [DOI] [PubMed] [Google Scholar]
- 32.Wéber E, Hetényi A, Váczi B, Szolnoki E, Fajka-Boja R, Tubak V, Monostori E, Martinek TA. Galectin-1–asialofetuin interaction is inhibited by peptides containing the Tyr-Xxx-Tyr motif acting on the glycoprotein. Chembiochem. 2009;11:228–34. doi: 10.1002/cbic.200900502. 2010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.