Summary
Despite the discovery of the tuberculosis (TB) bacillus over 100 years ago and the availability of effective drugs for over 50 years, there remain a number of formidable challenges for controlling Mycobacterium tuberculosis (MTb). Understanding the genetic and immunologic factors that influence human susceptibility could lead to novel insights for vaccine development as well as diagnostic advances to target treatment to those who are at risk for developing active disease. Although a series of studies over the past 50 years suggests that host genetics influences resistance to TB, a comprehensive understanding of which genes and variants are associated with susceptibility is only partially understood. In this article, we review recent advances in our understanding of human variation of the immune system and its effects on macrophage function and influence on MTb susceptibility. We emphasize recent discoveries in human genetic studies and correlate these findings with efforts to understand how these variants alter the molecular and cellular functions that regulate the macrophage response to MTb.
Keywords: innate immunity, tuberculosis, macrophage, human genetics
Introduction
Mycobacterium tuberculosis (MTb) latently infects approximately one-third of humanity and is currently only comparable to human immunodeficiency virus (HIV) as a leading infectious cause of mortality in the world. Despite the discovery of the tuberculosis (TB) bacillus over 100 years ago and the availability of effective drugs for over 50 years, there remain a number of formidable challenges for controlling MTb infection (1). These obstacles include lengthy treatment regimens of 6 to 9-month duration, drug resistance, the lack of a highly efficacious vaccine, and incomplete understanding of what controls infectivity and progression of disease. Although vaccination with Mycobacterium bovis bacille Calmette–Guérin (BCG) does offer some protection against MTb, its efficacy is suboptimal and not adequate for disease control (2–4). Development of a more effective vaccine is a high priority worldwide and depends on a thorough understanding of the host response to infection.
There are a number of critical questions about the host immune response to MTb that are not well understood. Although only 10% of individuals who are infected with MTb develop active disease, it is not known which immune responses are associated with susceptibility or resistance. In addition, it is not known why some individuals have disseminated TB that spreads to the meninges and central nervous system, while most people have localized disease in the lungs. Finally, we do not understand the wide variation in efficacy of the BCG vaccine that has been observed in many studies (2–4). The central topic of this review is to understand how common genetic variation in the innate immune response influences host susceptibility to MTb. The emphasis is twofold: first, to understand which variants have been identified through human genetic studies to be associated with TB susceptibility; and second, to understand how these polymorphisms alter the function of the encoded protein and the ensuing cellular immune response.
Of mice and men
Murine studies of TB have contributed significant advances to our understanding of the host immune response. Despite these insights, there remain important differences between mice and humans, and the relevance of murine infection studies needs to be interpreted with caution (5). Murine studies generally use high doses of MTb, often with strains that are attenuated by laboratory passage, and sometimes with artificial routes of inoculation. The mouse is usually inbred, and often only one strain is studied. Furthermore, experimental mice have a standardized age, are rarely vaccinated, and have a short life span. In addition, most mouse studies are performed on a C57Bl/6 background that models primary progressive TB rather than reactivation disease, which is the most common form of human TB. The mice develop chronic persistent TB, while 90% of humans contain their primary infection in a latent state without active symptoms. Furthermore, mouse granulomas do not caseate, a pathologic feature that is a hallmark of human granulomas. Rabbits form caseating granulomas, but studies in rabbits lack the powerful immunologic tools of the mouse (6). At the cellular level, nitric oxide killing mechanisms may differ between murine and human macrophages, although this point is controversial (7, 8). All of these factors influence the interpretation of murine results and their relevance for human disease. In this review, we emphasize findings from studies of human infection, including genetic studies of the role of the innate immune response in human MTb infection.
TB risk factors
In addition to genetic risk factors, there are other host factors as well as environmental influences associated with susceptibility to TB. A recent case–control study from West Africa investigated these potential risk factors in smear-positive individuals with TB that were matched with household or community controls (687 matched pairs). In a multivariate analysis, a number of risk factors were associated with TB susceptibility including male sex [odds ratio (OR) 1.43], HIV infection (OR 2.07), smoking (past, OR 1.53; current, OR 2.03), asthma (OR 0.28), single/widowed/divorced marital state (OR 1.48–1.67), family history of TB (OR 3.24), number of adults in the household (OR 1.37 for 6–10 adults; 2.80 for >10 adults), and house ownership (OR 1.42) (9). In a household contact study in Uganda (302 index cases and 1206 household contacts), risk factors associated with active TB in contacts were HIV infection (OR 4.03), age ≤5 years (OR 6.88), BCG vaccination (OR 0.51),≥18 h contact per day with index case (OR 2.39), house type (OR 2.80 for Muzigo style, a multifamily house that often has shared rooms and limited air ventilation), and cavitary lung disease in the index patient (OR 1.97) (10). Despite evidence of these host and environmental influences, many individuals have TB without any identifiable risk factors, suggesting that host genetic variation may influence susceptibility.
Human genetics and TB susceptibility
A series of studies over the past 50 years suggest that host genetic factors influence susceptibility to TB (11, 12). Although previous studies have uncovered some of the genes involved in human predisposition to mycobacterial infections, a comprehensive understanding of genetic susceptibility factors remains an elusive and important goal (11). There are four major lines of evidence to support a role for genetics in TB. First, twin studies indicate that TB rates among monozygotic twins are more than twice the rate of dizygotic twins (31.4 versus 14.9 TB cases per 100 twins for monozygotic versus dizygotic twins, P < 0.05, binary variable multiple regression analysis) (13). Second, several primary immunodeficiency disorders are associated with susceptibility to mycobacteria in a Mendelian fashion attributable to rare single gene mutations with high penetrance. These disorders include severe combined immunodeficiency, hyper-immunoglobulin (Ig) E syndrome, chronic granulomatous disease, anhidrotic ectodermal dysplasia with immunodeficiency, hyper-IgM syndrome, and Mendelian susceptibility to mycobacterial disease (MSMD) (11). This latter group of disorders is more selectively associated with mycobacterial infection and sometimes also with Salmonella but not with excessive susceptibility to other pathogens. In a series of elegant studies over the past 10 years, a number of single gene defects have been identified that center around the interleukin-12 (IL-12)/interferon-γ (IFN-γ) axis that culminates in macrophage activation by T cells and subsequent killing of organisms. The genes with identified mutations include IL-12Rβ1, IL-12p40, IFN-γR1, IFN-γR2, and signal transducer and activator of transcription-1 (STAT-1). Most of the infections associated with these Mendelian disorders have been from BCG or environmental bacteria. However, some of disorders are also associated with MTb susceptibility (IFN-γR2 and IL-12p40 among the MSMD). These disorders have been extensively reviewed elsewhere and are not the topic of the current review (11, 14).
The third line of evidence for host genetics and TB susceptibility comes from the study of complex inheritance patterns, where the assumption is that genetic influence is polygenic and attributable to alleles that are common in the population with low penetrance for any single allele. Several genome-wide studies of susceptibility to TB have been performed with family-based linkage studies. These studies have identified several loci that include 2q35 in a Canadian population (15), 8q12–13 in a study from Morocco (16), 17q11.2 in Brazil (17), and 15q and Xq in populations from The Gambia and South Africa (18). Fine mapping of the 15q locus in families from Africa (The Gambia, Guinea, South Africa) suggests that Ube3a or a closely linked gene may contain the causative locus (19). Efforts to identify the genes underlying each of these associations are ongoing.
The fourth line of evidence comes from candidate gene association studies. These studies evaluate whether common polymorphisms in candidate genes are associated with susceptibility to disease. The most common study design is a case–control format with comparison of polymorphism (single nucleotide, insertions, deletions, or microsatellite markers) frequencies between cases and controls. The strength of this study design is the capacity to enroll large cohort sizes. One disadvantage is the problem of population heterogeneity or admixture, where differences in ethnic composition of the cases and controls can lead to false associations that are not attributable to differences in disease susceptibility. Methods to control for population admixture include matching cases and controls for ethnicity and adjusting for ethnicity as a possible confounder in a multivariate logistic regression model. In addition, the population can be examined for subtle evidence of admixture by genotyping a set of ‘genomic controls’, which are unlinked single nucleotide polymorphisms (SNPs) that are chosen for their variation across ethnicities. This approach has recently been used for several case–control studies of TB (20, 21). An alternative study design that is not vulnerable to population admixture is the transmission disequilibrium test (TDT). This strategy is family based and looks for evidence of non-random transmission of the candidate allele from a heterozygous parent to an affected child. In addition to the problem of population admixture, candidate gene association studies are vulnerable to false-positive results from multiple comparisons. This is a particularly poignant problem in the current era of high throughput genotyping strategies. In light of this, replication and validation of findings with independent cohorts is an essential part of a careful study design. In addition to genetic evidence of association of a particular allele with susceptibility, the candidate gene approach can be linked to functional studies of the polymorphisms to determine whether there is biologic plausibility and mechanistic insight into disease pathogenesis.
Advances in genomic technology and immunology have accelerated the number of candidate gene association studies in infectious diseases. Table 1 summarizes the genes and variants that have been examined, whether there have been replication studies, and whether a functional defect is associated with the polymorphism. This review prioritizes discussion of the innate immune genes that have been most thoroughly examined with consistent and validated genetic findings and/or associated with well-defined functional defects. Studies of the human leukocyte antigen (HLA) locus are not discussed because they have been reviewed elsewhere (12, 22, 23). Together, these studies are providing insight into human susceptibility to TB and the underlying mechanisms of pathogenesis from genetically regulated variation of macrophage function and the innate immune response.
Table 1.
Candidate gene association studies and susceptibility to tuberculosis
| Gene |
Variants |
Validate* |
Function of varianty |
Comments |
|
|---|---|---|---|---|---|
| TB |
Other diseases |
||||
| Binding, receptors, and signaling | |||||
| MBL | Alleles B, C, and D and promoter | Variable | Variable | Y | Heterogeneous results from various studies |
| Surfactant protein | Multiple in Sp-A and Sp-D | N | N | N | |
| DC-SIGN | −871G, −336A | N | Y, dengue | P, expression | |
| P2XA7 | −762 | N | N | N | |
| A1513C (E469A) | Y | N | Y | Alters ATP-induced macrophage functions, including MTb killing | |
| TLR2 | G2258A (R753Q) | N | Y, lyme | Y, signaling | |
| Intron 2 GT microsatellite | N | N | P, expression | ||
| C597T (N199N) | N | Y, leprosy | N | ||
| TIRAP/MAL | C539T (S180L) | Y | Y, malaria, bacteremia, pneumococcal empyema or baceremia | Y, signaling | |
| C558T (A186A) | N | N | P, signaling | Associated with TB meningitis | |
| Cytokines/chemokine | |||||
| IFN-γ | þ874A, −1616T, þ3234T | 874, likely. Others: N | N | Y, 874 and IFN-γ expression | 874 variant affects function and has the most promising association with Tb, but not replicated in all studies |
| IFN-γR1 | Microsatellite | N | N | N | |
| T-56C | Y | N | N | ||
| IL-1β/IL-1RA | Multiple | N | Y, H pylori, HBV | P, for some alleles | |
| IL-12-Rβ1 | Q214R, G378R, and M365T | Variable | N | P, signaling | |
| IL-12B | Multiple | N | N | P | |
| IL-10 | A-1082G, A-592C | Variable | Y, leprosy | P, expression | |
| CCL2 (MCP1) | A-2518G | Y | N | Y, expression | Variant may inhibit IL-12 production |
| TNF-α | G-308A, G-238A | Variable | Y, malaria, leprosy | P | |
| TNFR1 | Multiple | Y | N | P, signaling | |
| IL-8 | T-251A | N | Y, RSV | N | |
| Other | |||||
| HLA | Multiple studies demonstrate an association. Reviewed elsewhere | ||||
| VDR | Fok1, BsmI, ApaI, TaqI | Variable | Y, bone mineral density | P | Heterogeneous results from various studies |
| Slc11a1 (NRAMP1) | 3′UTR, D543N, INT4, and 5′(GT)n | Y | Y, Neisseria, leishmania | P, transcription | Best documented association with 15 studies. Functional data are not clear |
| Ube3a | Multiple | Y | N | N | |
| SP110 | Two variants | Y | N | N | Identified from mouse sst1 locus |
See text for references and more detail.
N, no; Y, yes; P, possible effect.
Validation refers to whether the genetic findings were replicated in another MTb cohort or with a separate study design (family-based or case–control).
Validation is also noted if it occurred with another disease (other diseases).
Function refers to whether the variant is associated with altered function of the gene, either through regulation of expression levels or through a non-synonymous coding region polymorphism that alters the function of the gene.
Initial detection of MTb
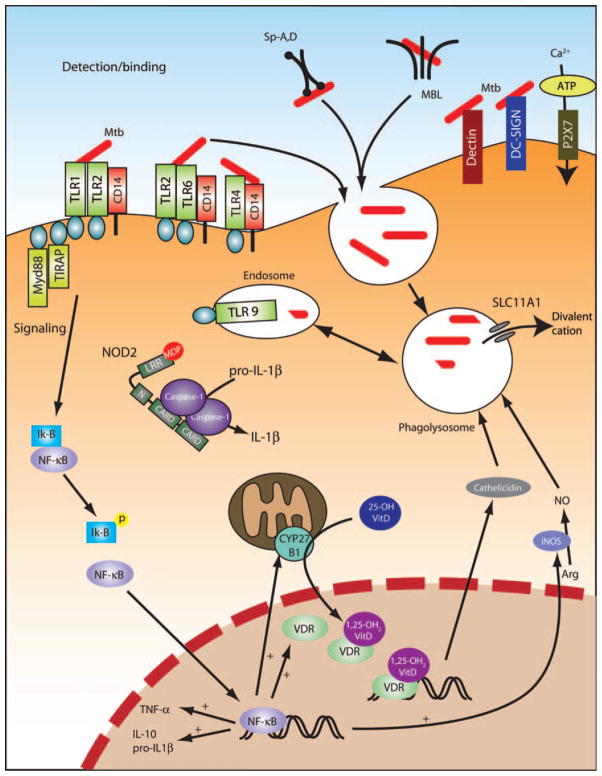
A number of receptors are critical for MTb detection by phagocytes, including the complement receptors, the mannose receptor (MR), dendritic cell-specific intercellular adhesion molecule-3 (ICAM-3)-grabbing non-integrin (DC-SIGN) (CD209), surfactant protein A (Sp-A) receptor, the class A scavenger receptor, mannose-binding lectin (MBL), and possibly also dectin-1 (24–26). In human macrophages, the primary receptors for MTb are the MR and complement receptor 3 (CR3) (27, 28)(Fig. 1). In contrast, human dendritic cells primarily use DC-SIGN with no significant role for CR3 or MR (29). Dectin-1 is a C-type lectin that recognizes β-glucans and has previously been shown to cooperate with Toll-like receptor 2 (TLR2) to recognize fungi (30–32). A recent study suggests that dectin-1 cooperates with TLR2 to recognize MTb and other mycobacteria (33). The significance of these findings is unclear at present because mycobacteria are not known to contain β-glucans. Among these receptors, studies of polymorphisms and TB susceptibility in MBL, surfactant protein, and DC-SIGN have been undertaken and are discussed in more detail.
Fig. 1. Macrophage response to MTb.
Depicted are the molecules that have been identified as important mediators of the macrophage response to MTb. iNOS, inducible nitric oxide synthase; LRR, leucine-rich region. MTb is recognized by several receptors and ingested by the alveolar macrophage. For example, MTb expresses several ligands that activate TLRs, in turn leading to signaling downstream of the MyD88 adapter protein. See text for more detail.
Mannose-binding lectin
MBL is an innate immune molecule that recognizes microbial surface carbohydrates and regulates opsonization, phagocytosis, and activation of the complement pathway (34). MBL is a 32-kDa protein with a collagen-like domain as well as a carbohydrate recognition domain (CRD) that binds to high mannose and N-acetylglucosamine oligosaccharides present on a range of pathogens, including MTb (35, 36). MBL activates the complement pathway in an antibody-independent manner in conjunction with the MBL-associated serine protease and leads to phagocytosis through the complement receptors or collectin receptor (37).
Common variant alleles of MBL exist and have been characterized with numerous functional and human genetic studies. Three polymorphisms are correlated with low levels of functional MBL. The wildtype allele is referred to as allele A, and the variant alleles are in codons 54 (allele B), 57 (allele C), and 52 (allele D). These variants disrupt the collagen domain and lower the levels of functional protein by 5 to 10-fold in heterozygotes and to nearly undetectable levels in homozygotes. There are also promoter region variants that influence MBL levels, including a variant G-221C, that is termed allele Y or X. In general, MBL deficiency leads to increased susceptibility to various infections. A small number of studies have examined whether MBL deficiency is associated with susceptibility to TB. The results have ranged from showing that MBL deficiency is associated with an increased risk [Indian cohort, frequency in controls (n = 109) 0.018 versus cases (n = 202) 0.109, OR 6.5, P = 0.008] (38), a decreased risk in South Africa [frequency in controls (n = 79) 0.278 versus cases (n = 155) 0.109, P = 0.001] (39) and Tanzania (36), mixed results in Denmark (depending on level of deficiency) (40), and a mild protective effect of the codon 57 mutation, but not other alleles, in The Gambia [allele frequency in controls (n = 794) 0.33 versus cases (n = 844) 0.28, P = 0.037]. The heterogeneity of these results could be attributable to several factors, including population admixture, incomplete genotyping (most of the studies did not genotype the promoter alleles), different clinical phenotypes, and variable allele penetrance in different ethnic backgrounds. Overall, the current data do not support a consistent or major effect from MBL deficiency on susceptibility to TB.
Dendritic cell-specific ICAM-3-grabbing non-integrin
DC-SIGN is a C-type lectin that recognizes a number of microbes, including HIV and MTb. MTb ligands for DC-SIGN include lipoarabinomannan (LAM), lipomannan, arabinomannan, and the 19-kDa antigen (29, 41, 42). Initial studies indicated that DC-SIGN was primarily a receptor on dendritic cells rather than macrophages. However, a more recent study showed that DC-SIGN expression is induced on alveolar macrophages and was found on macrophages from bronchoalveolar lavage fluid from TB patients but not from healthy adults, asthmatic patients, or sarcoid patients (43). These results present a model whereby infection of alveolar macrophages from naive hosts occurs through the complement and MRs. Then, further spread of infection may occur through DC-SIGN-expressing alveolar macrophages.
Genetic studies of human DC-SIGN in South Africa have identified two promoter variants, −871G and −336A, that are associated with TB susceptibility [SNP A-336G, AG/AA genotype frequency in cases (n = 351) 0.293 versus controls (n = 360) 0.381, P < 0.001; SNP A-871G, AG/GG genotype frequency in cases (n = 351) 0.168 versus controls (n = 360) 0.272, P = 0.01] (20). The study design used genomic controls to adjust for population admixture. The −336 SNP has also been found to be associated with Dengue fever and alters transcriptional activity at an Sp1-binding site, where the SNP resides (44). Several other polymorphisms were not associated with TB susceptibility, including variants that regulate the length of the neck region that separates the CRD from the transmembrane domain (45). In a study from Colombia, there was no association of the −336 SNP with TB susceptibility (46). Together, these results suggest a possible role for DC-SIGN variation and susceptibility to human TB, although the magnitude of the effect is small and has not been validated in a second TB population.
Surfactant proteins
Pulmonary surfactant is a complex structure with lipids and proteins that reduces surface tension of alveoli and promotes lung expansion (25). There are four surfactant proteins that are C-type lectins and members of the collectin family. Similar to MBL, collectins form multimeric structures with a collagenous N-terminal domain and a carbohydrate C-terminal domain. These proteins bind to pathogens, mediate uptake into phagocytes, and modulate effector mechanisms such as oxidant production, lung inflammation, and bacterial killing (47). Of the four surfactant proteins (Sp-A, Sp-B, Sp-C, Sp-D), two have been shown to modulate host interactions with MTb. Sp-A increases phagocytosis of MTb, probably through a direct action on the macrophage as well as through opsonic activity (48). SP-D binds to MTb and appears to reduce phagocytosis, possibly through a LAM–mannose interaction (49). In a study from Mexico, several variants of Sp-A and Sp-D were associated with susceptibility to TB (50). These results have not been verified in an independent cohort, and the functional significance of the variants has not been reported.
TLR pathway
TLRs and in vitro recognition of MTb
TLRs constitute a family of transmembrane proteins that are known as pattern recognition receptors for their detection of microbial ligands and subsequent initiation of inflammatory signaling pathways (51–54). The bacterial molecules that are recognized by TLRs include lipopeptides by TLR2 (as a heterodimer with TLR1 or TLR6), lipopolysaccharide by TLR4, flagellin by TLR5, and bacterial CpG DNA by TLR9. Several in vitro and in vivo studies suggest that TLRs are critical regulators of the immune response to TB. In vitro studies indicate that TB is recognized by several TLRs, including TLR2/1/6, TLR9, and possibly also TLR4 (55, 56). Initial studies documented a central role for TLR2 in TB recognition by macrophages (57–59). The mycobacterial cell wall contains a number of proinflammatory TLR2 ligands, including lipoproteins, mycolylarabinogalactan–peptidoglycan complex (the cell wall core structure), lipids, and LAM (57–61). LAM contains a mannose-rich core carbohydrate with highly branched arabinofuranosyl side chains, which are linked to acyl groups. LAM from MTb has mannose-capped ends and is non-stimulatory (Man-LAM). In contrast, rapidly growing mycobacterial species lack mannose end capping (ara-LAM) and are strong inducers of cytokine production. ara-LAM but not Man-LAM has been shown to be a TLR2 ligand (58, 59).
TLR2 works as a heterodimer with TLR1 or TLR6 in a complex with CD36 to recognize lipopeptides with different structures. TLR1 exhibits preferential recognition of triacylated lipopeptides, while TLR6 is activated by diacylated structures. The stimulatory activity is also affected by the terminal peptide composition, chirality of the diacylglycerol carbon, and the fatty acid composition (62–67). The 19-kDa lipoprotein of MTb is recognized by TLR1 (68). In addition, Mycobacterium leprae is recognized by TLR1 with an additional contribution by TLR6 (69). One study suggests that an extract of MTb (called soluble TB factor) is recognized by TLR6 (70). Together, these results suggest that both TLR1 and TLR6 cooperate with TLR2 to recognize mycobacteria.
Recent data indicate that TLR9 cooperates with TLR2 to recognize MTb in macrophages as well as splenic dendritic cells. When murine Tlr9−/− splenic dendritic cells were stimulated with live MTb, there was a partial reduction in IL-12p40. However, in Tlr2/9−/− cells, there was further inhibition of cytokine to background levels, suggesting that the majority of TLR-mediated mycobacterial signaling is through these two receptors (along with the heterodimerization with TLR1 or 6). Finally, there is one report of TLR4-mediated signaling in response to stimulation with a heat-labile cell-associated factor from viable mycobacteria (58). The identity of this ligand remains unknown and is predicted to be a non-canonical TLR4 ligand because mycobacteria do not contain endotoxin. There is also a substantial degree of macrophage activation by MTb that is myeloid differentiation factor 88 (MyD88) and TLR independent (71, 72).
TLRs and in vivo recognition of MTb
Although TLRs and the innate immune system are essential for defending the host against microbes, the degree of redundancy and specificity manifested in vivo among different TLR family members is only partially understood. Despite the overwhelming evidence showing a critical role for TLR mediation of MTb recognition in vitro, the in vivo significance of individual TLRs has been more difficult to show consistently. Three different studies have shown that MyD88 regulates a protective immune response to MTb in mice. In all three studies, Myd88−/− mice had increased numbers of bacteria in the lung in comparison with wildtype controls (100 to 1000-fold) (73–75). There were also important differences in these studies. One study, which used the Kurono strain, found no major differences in survival or lung histopathology (75). In contrast, the other two studies, which both used the H37Rv strain, showed increased mortality and lung pathology in the Myd88−/− strains (73, 74). The explanation for these differences is not currently known, although one possibility is the strain of MTb that was used. A critical role for individual TLRs during murine MTb or BCG infection has been more difficult to show consistently. Although some studies have shown a critical role for TLR2, others have not (76–79). In one study, increased susceptibility was only observed in high-dose infections (78). The preponderance of evidence suggests no significant in vivo role for TLR4 in mediating protective immunity to murine MTb (77, 78, 80), although there is one study that did find a difference (81). Tlr9−/− mice have increased mortality and increased lung bacterial burdens compared with wildtype mice, effects that were magnified in Tlr2/9−/− mice (82). Interestingly, TLR9 but not TLR2 was found to control production of IFN-γ from CD4+ T cells in infected mice. Together, these in vivo studies suggest an important though not absolute role for the TLR pathway in mediating host protection to murine MTb infection. Furthermore, the effects appear to be strongest when multiple TLRs are impaired.
Human genetics, TLRs, and MTb
Human genetic studies of TLR pathway variants indicate that there are several common variants that may influence disease susceptibility. A polymorphism in TLR2 in the Toll-IL-1 receptor (TIR) domain (G2258A), which changes an arginine to glutamine (R753Q), was originally described in 2000 and later found to influence signaling to diacylated and triacylated lipopeptides as measured by nuclear factor kappa B (NF-κB) luciferase assays (83, 84). Furthermore, in human whole-blood studies, there was a slight decrease in cytokine production to stimulation with Borrelia burgdorferi lysates in heterozygous individuals (753RQ) as compared with homozygous individuals (753RR). Heterozygosity was associated with protection from Lyme disease with a modest OR and marginal statistical significance (84). In a small study from Turkey (129 TB cases, 116 controls), the homozygous (753QQ) and heterozygous (753RQ) genotypes were associated with an increased risk of TB (753QQ: OR 6.4, P = 0.022; 753QR: OR 1.60, P = 0.019) (85). Although the statistical significance was marginal and this association has not yet been replicated, the findings are suggestive of a real and possible causal association, in light of the functional defect associated with this SNP. A second polymorphism in the TLR2 TIR domain (C2029T, R677W) was described originally in the context of an association with susceptibility to lepromatous leprosy (86). This SNP was later found to be an artifact that resulted from a variant in a duplicated region of TLR2 that is present several kilobases upstream of the true gene (87). In a study from Korea (176 TB cases original cohort, 82 TB cases validation cohort, 191 controls), a microsatellite GT dinucleotide repeat polymorphism in intron II was found to be associated with TB. Shorter GT repeats were associated with increased susceptibility to TB (OR 1.6–2.0, P = 0.009–0.02) as well as slightly altered TLR2 promoter activity measured in a luciferase assay (88, 89). We recently found a synonymous polymorphism (T597C, N199N) that was associated with susceptibility to TB meningitis in Vietnam [183 pulmonary TB, 175 TB meningitis, 392 controls; 597C allele frequency of 0.252 in controls versus 0.330 in cases (OR 1.47, P = 0.007); 597CC genotype frequency 0.048 in controls versus 0.14 in TBM cases (OR 3.26, P = 0.0002)] (T.T. Nguyen, T.R. Hawn, S.J. Dunstan, A. Aderem, unpublished observations). We also recently found that this SNP is associated with reversal reaction in leprosy (P.Y. Bochud, T.R. Hawn, G. Kaplan, A. Aderem, unpublished observations). There are no known functional defects associated with this polymorphism.
Recent studies of the adapter protein, TIR domain containing adapter protein/Myd88-adapter like (TIRAP/MAL) suggest an association with susceptibility to TB. TIRAP has two isoforms (221 and 235 amino acids), and both have a C-terminal TIR domain that mediates signals from TLR2 and TLR4. A polymorphism was recently described that changes a serine to a leucine at amino acid 180 (S180L, C539T) and impairs TLR2-mediated NF-κB signaling in reconstitution experiments (90). In addition, the 180L variant was less able to bind TLR2 in comparison with the 180S variant. The heterozygous state is associated with protection from several diseases, including malaria, invasive pneumococcal disease, bacteremia, and one study of TB from Guinea-Bissau. In the latter study (675 cases, 605 controls), the heterozygous frequency was 0.6% in cases versus 2.0% in controls (OR 0.23, P = 0.041). This association was confirmed in a family-based study with a TDT. Although the protective effect of this SNP for TB was marginal and the polymorphism was of low frequency in this population, this SNP appears to be important with its functional defect and number of replicated studies.
We recently examined TIRAP variants in a study from Vietnam (91) that included studies of overall susceptibility to TB as well as susceptibility to TB meningitis. Although we did not find an association of S180L with susceptibility to TB, the frequency of this SNP was too low for a proper statistical evaluation (2.3% in cases versus 1.7% in controls, P = 0.61). We did find that a synonymous SNP (C558T, A186A) was associated with increased susceptibility to TB with a 558T allele frequency of 0.035 in controls versus 0.074 in cases (OR 2.25, P < 0.001). Subgroup analysis showed that SNP 558T was more strongly associated with susceptibility to meningeal TB (OR 3.02, P < 0.001) than pulmonary TB (OR of 1.55, P = 0.22). In comparison with the 558CC genotype, the 558TT genotype was associated with decreased whole-blood IL-6 production, which suggested that TIRAP influences disease susceptibility by modulating the inflammatory response. We do not currently understand how this SNP alters cytokine protection, but possibilities include that it is in linkage disequilibrium with a coding region SNP or one that alters expression levels. These results suggest that the TLR pathway influences susceptibility to meningeal and pulmonary TB by different immune mechanisms.
Cytoplasmic sensors: NOD2 and NLRP12
The human nucleotide oligomerization domain (NOD)-like receptor (NLR) or NACHT [domain present in NAIP (neuronal apoptosis inhibitor protein), CIITA (class II transactivator), HET-E (plant het product involved in vegetative incompatibility) and TP-1 (telomerase-associated protein 1)-LRR (leucine-rich repeat)] family contains over 20 proteins. NLRs are cytoplasmic proteins that contain a leucine-rich region at the C-terminus and a central nucleotide-binding (NOD) domain (92–94). The N-terminus contains a protein interaction domain and varies within different family members. N-terminal motifs include caspase activation and recruitment domain (CARD), pyrin, or baculovirus inhibitor of apoptosis repeat domains. Two of these proteins have been shown to have effects when signaling with mycobacterial products: NOD2 and NLR family, pyrin domain containing 12 (NLRP12).
NOD2
NOD2 [also known as CARD15 (caspase recruitment domain family, member 15) or NLR family, CARD domain-containing 2)] contains a leucine-rich region, a central NACHT nucleotide-binding domain, and two CARD domains. Frameshift mutations in the N-terminal leucine-rich region have been associated with Crohn’s disease in adults (95). Controversy exists as to whether Crohn’s disease has an environmental or infectious trigger. Crohn’s disease itself resembles a chronic intestinal granulomatous infection seen in animals called Johne’s disease, which appears to be caused by Mycobacteria avium spp. paratuberculosis (96). Rare missense mutations in the NACHT domain of the CARD12 protein have also been linked to Blau’s syndrome, which is an uncommon autosomal dominant granulomatous arthritis with uveitis seen in children (97). NOD2 detects intracellular muramyl dipeptide, a component of the peptidoglycan molecule that is found on mycobacteria and Gram-positive bacteria (98). After stimulation, NOD2 mediates production of α-4 defensins or cryptdins, which have bactericidal activity against mycobacteria and may play a role in host immune defense to MTb infection (99). Despite support that NOD2 plays a non-redundant role in the recognition of MTb (100), there is no evidence that variation in NOD2 plays a role in susceptibility to human mycobacterial disease. Two studies, one in The Gambia and the other in South Africa, have failed to link known Crohn’s disease mutations with MTb susceptibility, but the frequency of the mutations in both studies were rather low in the study populations (no homozygous patients were noted) (101, 102). Furthermore, no differences were seen in the Gambian cohort when they examined a number of promoter region SNPs. Although studies to date suggest that there is no correlation with human SNPs and susceptibility to TB, further analysis needs to be carried out before definitive conclusions can be made.
NLRP12
NLRP12 (also known as NALP12, Monarch-1, or PYPAF7) is an NLR that mediates inflammatory responses to MTb in vitro. NLRP12 regulates expression of major histocompatibility complex class I genes and is thought to inhibit proinflammatory cytokine production to a number of stimuli, including MTb, by interacting with IL-1 receptor-associated kinase-2 (103, 104). Although it has been shown that NLRP12 can impair the immune response in cells stimulated with heat-killed MTb, there are no studies of the role of genetic variation in NLRP12 and susceptibility to MTb.
Other receptors: P2X7
The purinergic P2X7 receptor is highly expressed on macrophages and mediates adenosine triphosphate (ATP)-induced killing of mycobacteria. Stimulation of P2X7 causes opening of a divalent cation channel, influx of calcium, induction of caspases, and eventual apoptosis. Phospholipase D is activated, which promotes phagolysosomal fusion and killing of mycobacteria. Several polymorphisms exist in P2X7, including those in the promoter region, an ATP-binding domain, a trafficking domain, and an ankyrin-like repeat domain (A1513C, E496A). Studies of several of the coding region polymorphisms have shown functional defects with decreased ATP-induced uptake of ethidium as well as ATP-induced apoptosis (105, 106). Two case–control studies have found associations of P2X7 SNPs with TB susceptibility. A promoter region polymorphism (−762) with unknown functional significance was associated with pulmonary TB in The Gambia (allele frequency 0.254 in cases versus 0.329 in controls, OR 0.70, P = 0.003) (107). A recent study of the A1513C SNP showed an association with extrapulmonary TB but not pulmonary TB in Sydney, Australia (OR approximately 3.1–4.0 for extrapulmonary TB, P < 0.01) (108). One strength of this study is replication of the findings in two separate cohorts. Possible weaknesses include the small sample size and the level of population heterogeneity with subjects from a number of different countries. The A1513C polymorphism is also associated with impaired killing of MTb and BCG in ATP-stimulated macrophages (108, 109). This is the only SNP that is currently known to be associated with impaired effector function that alters the ability of the macrophage to kill MTb.
Cytokines, chemokines, and effector molecules
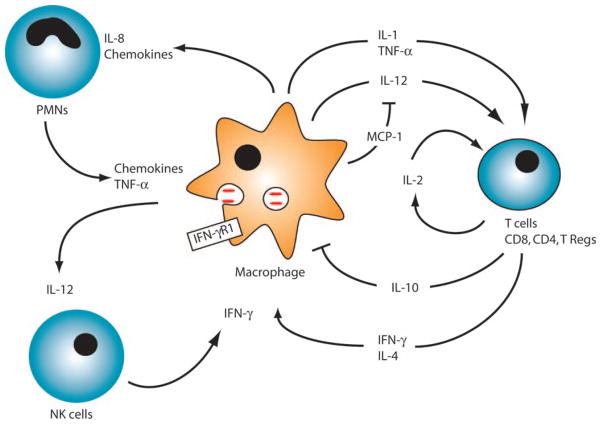
Innate immune recognition of MTb by phagocytic cells leads to cellular activation and rapid production of pro- and anti-inflammatory cytokines (Fig. 2). These cytokines and chemokines recruit inflammatory cells [T cells, neutrophils, and natural killer (NK) cells] to areas of infection, activate transmigrated cells, and coordinate the inflammatory and adaptive immune response to MTb. Successful outcome of mycobacterial infections depends upon cytokine networks established and maintained by macrophages. We review cytokines and chemokines important for the host to control MTb and then discuss genetic variations seen in these molecules that lead to enhanced susceptibility to active disease.
Fig. 2. Cellular immune response to MTb.
After infection, activated macrophages secrete cytokines and chemokines that activate macrophages, T cells, neutrophils, or NK cells. T cells and NK cells produce IFN-γ that acts with other cytokines to activate macrophages to produce NO, which contributes to clearance of MTb. See text for more detail.
Tumor necrosis factor-α
Tumor necrosis factor-α (TNF-α) is a prototypic proinflammatory cytokine that is produced by monocytes and macrophages when exposed to microbial products derived from MTb. TNF-α is produced as a trimeric surface molecule and is then cleaved by TNF-α-converting enzyme to release the trimeric molecule from the cell. In synergy with IFN-γ, TNF-α activates macrophages to produce nitric oxide synthase 2 (NOS2), allowing the macrophage to kill intracellularly replicating MTb (110, 111). Mice deficient in TNF-α and TNF-α receptor (TNFR) show increased susceptibility to MTb and impaired granuloma formation following infection with MTb (112, 113). Expression of TNF-α, which is unable to be cleaved from the cells, restores partial protection to mycobacterial infections and allows for normal granuloma formation (114). In humans, there is evidence that TNF-α plays an important role in host defense to MTb because patients receiving TNF-α inhibitors are more susceptible to TB (115). In humans, genetic control of TNF-α expression ma2y be associated with the development of TB, as suggested by work from Uganda, which showed that in MTb, smear-positive patients and their household contacts, TNF-α production from MTb-stimulated whole-blood assays had significant heritability (116). Variation in both the TNF-α gene and TNFR were associated in linkage and association studies with in vitro TNF-α production as well as the development of active TB disease (117). Further support that TNF-α plays a critical role in controlling human disease comes from increased rates of reactivation disease seen in patients receiving TNF-α inhibitors (115). There is also evidence that polymorphisms in the TNF-α reporter gene at the −308 position are associated with increased susceptibility to lepromatous leprosy (118), as well as other diseases such as malaria and septic shock (119, 120). However, the evidence for association of these polymorphisms with TB has been less consistent. The main polymorphisms that have been identified are found in the promoter region at −238 and −308. In a Colombian study, both the −238G (OR 1.8, P = 0.02) and −308G (OR 2.2, P < 0.0001) polymorphisms were associated with pulmonary TB. Interestingly, these polymorphisms were also associated with protection against autoimmune diseases such as systemic lupus erythematosus, rheumatoid arthritis, and Sjögren’s syndrome (121). An association with increased TB susceptibility was also seen in an Iranian cohort for −238GG homozygous but not for −238AG heterozygous carriers (122). Lack of association with pulmonary TB with either the −238 or −308 polymorphisms has also been documented in Turkish, Chinese, and Thai populations (123–125). In Chinese miners, an association for TB acquisition was identified in the −308GG persons only if a concurrent solute carrier family 11 member a1 (Slc11a1) [natural-resistance-associated macrophage protein (NRAMP)] mutation was noticed (OR 2.38, 95% CI 1.14–4.98). Functional studies of the SNP −308 have shown mixed results regarding its association with altered TNF-α levels (126, 127). Although the majority of evidence suggests that the TNFα promoter polymorphisms are not consistently associated with TB susceptibility, many of the studies have been insufficient in size to show minor differences. Further studies are needed before these common polymorphisms can be discredited.
Interleukin-1β
IL-1β is a proinflammatory cytokine that is produced by a myriad of infections and proinflammatory conditions (128). IL-1β is produced in the cell in an inactive precursor form (pro-IL-1β) and then is cleaved by Caspase-1 to an active form, which is secreted from the cell through an unknown mechanism. Conversion of pro-IL-1β to IL-1β can occur with numerous bacterial stimuli, including MTb, and can also occur when bacterial products are sensed within the cytoplasm by NLR proteins (i.e. NOD2), which contain a CARD domain important for Caspase-1 activation and inflammasome function. Once secreted, IL-1β acts primarily through IL-1R type I receptor, which further activates the expression of other proinflammatory cytokines. In some cases, however, IL-1β function is blocked by either binding to a membrane-bound decoy receptor (IL-1R type II) or by a soluble receptor antagonist (IL-1RA) that blocks the proinflammatory effects of IL-1β. Evidence that IL-1β plays an important role in the pathogenesis of MTb comes from knockout animal studies. Animals that have both IL-1β and IL-1α deleted form larger granulomas after infection and are unable to clear the mycobacteria as efficiently as wildtype mice (129). Furthermore, animals that lack the IL-1R type I have impaired survival and are unable to contain growth of the organism in vivo (130). This response may be because of impaired cell-mediated immunity, as measured by IFN-γ production. There is also evidence that susceptibility to clinical disease in an African cohort with MTb is associated with IL-1 gene cluster, specifically variation in the IL-1RA gene (131). In this study, other polymorphisms in the IL-1β and IL-1α gene did not show significant difference in TB susceptibility. Polymorphisms in non-coding regions of IL-1β have been linked to human variation in cytokine production and a −511 C allele in the promoter region of the gene is associated with protection from acquiring pulmonary TB (OR 0.63, 95% CI 0.44–0.90, P = 0.029) (132). Furthermore, studies carried out on emigrated Indians in London showed a relationship between polymorphisms in both IL-1β and IL-1RA and the functional ratio of expression and the acquisition of tuberculous pleurisy (P = 0.028), although multiple polymorphisms in either IL-1β or IL-1RA gene when analyzed individually were not associated with differences in TB resistance (123). Whether variation in genes of the IL-1 cluster confer risk to TB clinical disease continues to be preliminary, and further studies are needed before substantial conclusions can be made.
Interleukin-10
IL-10 is a cytokine produced by alternatively activated macrophages, dendritic cells, B cells, and regulatory T-cell subsets, and IL-10 is known to suppress proinflammatory cytokine response by the innate immune and adaptive immune systems. IL-10 is also thought to play an important regulatory role in many bacterial and viral infections (124). Studies carried out on mice have identified that IL-10 may not be important for susceptibility to initial infection with MTb but may play a role in reactivation of latent disease (125, 133). The contribution of IL-10 to the pathogenesis of mycobacterial disease in humans has focused on identification of polymorphisms in the promoter region, which are associated with increased production of IL-10 by monocytic cells in blood and enhanced susceptibility to MTb in certain populations (134). Evidence suggests that these polymorphisms are associated with disease severity in leprosy (135, 136). There is discrepant data regarding the effect of these polymorphisms on human TB susceptibility. Some have identified that those with the A-1082G have significant predisposition to TB in Sicilian and Cambodian patients and significant predisposition of the A-592C SNP in a Korean population, but the first two associations have been questioned because the control population were not in Hardy–Weinberg equilibrium (137–139). Others have shown no association with variation from these promoter SNPs in Spanish and African populations (131, 140). In the Ugandan study mentioned previously, there was also evidence of an association of a set of tagged haplotype IL-10 SNPs (that included the A-592C SNP) with acquisition of active disease from MTb (117). Some studies suggest that A-592C is associated with altered levels of IL-10 (141, 142) and impaired Sp-1 enhancer binding (139). The A-1082G variation has also been associated with altered levels of IL-10 (134). Whether IL-10 promoter polymorphisms play a role in susceptibility to TB continues to be controversial.
IL-12/IL-12Rβ1
IL-12 is a heterodimeric, covalently linked cytokine comprised of two subunits (p40 and p35). The p40 subunit is present in both IL-12 and IL-23, while the p35 subunit is specific for IL-12. IL-12 is mainly secreted by hematopoietic phagocytic cells (monocytes, macrophages, and neutrophils) and dendritic cells, and promotes T-cell differentiation into T-helper 1 (Th1) cells and production of IFN-γ by signaling through IL-12Rβ1 and IL-12Rβ2 (143). IL-12 plays an important role in stimulating IFN-γ production and establishing a potent Th1 response to intracellular pathogens such as MTb and Salmonella. Early studies showed that mice, when given exogenous IL-12, had increased resistance to MTb (144). Furthermore, mice deficient in IL-12p40 and IL-12p35 show enhanced susceptibility to MTb infection (145). The role of IL-12 in mycobacterial disease has been firmly established by the presence of patients with uncommon polymorphisms or mutations that predispose to severe disseminated mycobacterial infection in a Mendelian fashion (11). In addition to these Mendelian phenotypes, there is conflicting evidence that common variations in the IL-12Rβ1 gene confer susceptibility to MTb. In a Japanese cohort, there has been shown an increase in susceptibility [OR 2.45 (1.20–4.99), P = 0.013] to MTb when three missense non-synonymous polymorphisms (M365T, G378R, and Q214R) are present (146). These identical missense polymorphisms were present in a Moroccan cohort at high frequency, but no susceptibility was associated with these SNPs (147). The authors did identify additional non-coding region SNPs that were associated with increased susceptibility to TB acquisition. Lack of association with clinical TB was seen in a Korean population with these Q214R, G378R, and M365T polymorphisms (148). Studies suggesting an association of IL-12Rβ1 polymorphisms with impaired signaling are mixed. Some have shown impaired signaling through a STAT-4 pathway (146), while others have shown no effect on IFN-γ production (147). Because of the inconsistencies in the above data, the role of common polymorphisms of IL-12Rβ1 in TB susceptibility remains incompletely understood.
Similar discrepancies have been seen among various populations when attempting to identify clinically relevant variation in IL-12B, the gene encoding IL-12p40. In a Hong Kong patient population of 1030 people, four non-coding region polymorphisms were identified, and two haplotypes showed significant differences between those affected with disease versus healthy controls (149). The 3′ non-coding region SNP identified in this cohort has been associated with insulin-dependant diabetes as well as decreased production of IL-12 mRNA transcript and protein levels but not with TB in an African American cohort from Houston (150–153). All of this evidence points to an important role for IL-12 in the pathogenesis of TB. However, functional correlates are still being analyzed, and the overall consistency of the genetic data is still being resolved.
IFN-γ/IFN-γR
IFN-γ, the prototypic cytokine of the Th1 cell response, is a cytokine essential for the effective control of MTb in the host. IFN-γ is made of CD4+ T cells, CD8+ T cells, and NK cells, and it stimulates a mycobactericidal response in macrophages characterized by the induction of NOS (154, 155). Mice that fail to produce IFN-γ have disseminated mycobacterial infection whether challenged by aerosolized route or intravenously (156, 157). Furthermore, increased resistance to MTb by mice given IL-12 intravenously was abrogated when repeated in IFN-γ gene-disrupted mice (158). In humans, a series of uncommon genetic mutations in IFN-γR1 and R2 lead to MSMD in humans and have been extensively reviewed elsewhere (11, 159). There have been inconsistent findings using IFN-γR1 microsatellite markers and association studies with TB susceptibility. Only one small study reported a significant change in frequency in occurrence of a microsatellite marker, but this change was at low frequency and marginally significant (P = 0.02) (160). This finding was not replicated in a Gambian population (161). In a separate study from The Gambia, a T-56C polymorphism was marginally associated with susceptibility to TB (CC genotype: OR 0.75, P = 0.02) (162). A recent study in Uganda replicated this association of SNP T-56C with active TB as well as in vitro TNF-α production (see above) (117). Studies looking at IFN-γR2 have not shown any associations (162).
Studies of IFN-γ suggest that common polymorphisms in the IFN-γ gene play a role in TB susceptibility. There have been three polymorphisms in the IFN-γ gene that have been studied in various populations (A-1616G, T+874A, C+3234T) (160, 162–165). Variation in the promoter region of the IFN-γ gene disrupts an NF-κB binding site (T+874A) and is associated with an increased frequency of MTb in a study comparing 314 South Africans with pulmonary and meningeal TB versus 235 healthy controls (T allele, OR 1.64, 95% CI 1.16–2.30, P = 0.0055) (164). Susceptibility was confirmed in a Sicilian study, where there was a significantly lower frequency of +874TT in cases versus controls (163). Furthermore, a Spanish study also confirmed that the +874AA allele was associated with TB susceptibility and significantly decreased levels of IFN-γ in peripheral blood monocytic cells stimulated with TB antigens (140). These findings, however, were not repeated in another West African population from Guinea-Bissau, The Gambia, and Republic of Guinea for the T+874A variation, but significance was seen in two other promoter polymorphisms located in the IFN-γ gene A-1616G (OR 1.49 95% CI 1.11–2.00; P = 0.008) and C+3234T (OR 1.40, 95% CI 1.09–1.80, P = 0.009) (162). The T874A polymorphism was also not associated with susceptibility in a small south Indian study (165). Overall, the evidence that IFN-γ promoter variation is significantly associated with susceptibility to develop active TB disease is strong, while evidence that IFN-γR1 common variation is less convincing.
Chemokines: CCL2 and IL-8
CCL2, also called monocyte chemoattractant protein 1, is a β chemokine that is induced by monocytes infected with MTb (166). CCL2 causes chemotaxis of memory T lymphocytes, NK cells, and macrophages to sites of inflammation. In animal models, there is evidence that CCL2 may modulate disease severity. Mice that overexpress CCL2 showed high levels of CCL2 in all tissues, and these animals were susceptible to intracellular pathogens (167). Mice with targeted gene deletion of CCL2 failed to show increased susceptibility to MTb (168). CCL2 may act in mice to cause decreased IL-12p40 production and skew the T-cell response away from a Th1 response (169). In humans, variation in the promoter region of CCL2 causes significant changes in chemokine expression to IL-1β (170). Furthermore, genetic analysis in Brazilian patients has indicated that susceptibility to intracellular pathogens (Leishmania, MTb) are linked to 17q11–12 chromosome, which contains the CCL2 protein, along with many other chemokines and NOS2A (17). These authors, however, failed to show significant association of CCL2 promoter polymorphisms and increased TB susceptibility. In contrast, another study with both Mexican and Korean cohorts found increased susceptibility to TB in those with the A-2518G CCL2 promoter polymorphism [Mexican: OR 5.4, 95% CI 3.4–8.6, P = 0.00001; Korean: OR 2.63, 95% CI 1.9–3.7, P = 0.0001] (21). Patients with the affected GG genotype have increased amounts of CCL2 in their plasma and lower amounts of IL-12, and they make less CCL2 when their monocytes are stimulated with MTb antigens. These findings agree with the mouse studies and are consistent with the thought that elevated levels of CCL2 may favor Th2 cytokine response and decreased IL-12 production.
Another chemokine produced by phagocytic cells and tissue cells after simulation with MTb or its products is IL-8 (171–173). Increased levels of IL-8 are seen in the bronchoalveolar lavage fluid of humans with TB infection, and proportionately higher levels of IL-8 may be associated with increased mortality (174, 175). A polymorphism in the promoter region of IL-8, T-251A, is seen in high frequency in both Caucasian and African American populations. This polymorphism not only is associated with enhanced severity to bronchiolitis in infants with respiratory syncytial virus (176) but is also significantly more prevalent in two separate Caucasian and African American populations from Houston with MTb infection (177). Both A allele homozygotes and heterozygotes showed a higher frequency of MTb in these two populations [Caucasian: TA allele, OR 2.01 95% CI 1.06–3.80, P = 0.07, AA allele, OR 3.41 95% CI 1.52–7.64, P < 0.01; African American TA allele, OR 3.39, 95% CI 1.40–8.25, P < 0.01, AA allele, OR 3.46, 95% CI 1.48–8.08, P < 0.01] (177). In an African study, however, the T-251A was not associated with significant risk for pulmonary TB (178). A mechanism associated with this SNP is currently unknown but may involve increased IL-8 production. Further studies need to be carried out to identify the role IL-8 plays in MTb infection.
Other genes
Slc11a1
Slc11a1, better known by its former name NRAMP1, was originally identified in mice, where it regulates resistance to intracellular pathogens such as Salmonella typhimurium, BCG, and Leishmania donovani (179, 180). The murine susceptibility locus is associated with a single polymorphism in a non-conserved region, which encodes for a glycine to aspartic acid substitution at position 169. Slc11a1 has been extensively studied over the years and has complex functional phenotypes (181). Although the precise function of Slc11a1 continues to be actively pursued, it is thought to encode a protein which functions in transportation of divalent cations across phagosomal membranes and sequester essential cofactors (Mn2+, Fe2+, Cu2+, orMg2+) from bacteria and enhancing phagosomal killing (182). Other possible functions include regulation of chemokine and cytokine production, nitric oxide release, oxidative burst, and anti-microbial activity. A series of mouse studies have shown an influence of Slc11a1 on susceptibility to MTb infection (179, 180).
Many Slc11a1 polymorphisms have been associated with a number of diseases including TB, autoimmune disease, meningococcal meningitis, and leishmaniasis (181). Because of the known association in mouse models of TB susceptibility, many studies have attempted to associate genetic variation in the human Slc11a1 homolog with TB susceptibility in humans. Four polymorphisms [3′UTR, D543N, INT4, and 5′(GT)n] were examined in many of these studies, and the data have been summarized in recent reviews as well as a meta-analysis of the 15 known studies (22, 23, 137, 183–195). In the meta-analysis, subgroup analysis of the four separate polymorphisms indicates that three of the polymorphisms are associated with susceptibility to TB in Asian populations: 3′UTR (OR 1.46, 95% CI 1.1–1.9, P = 0.009), D543N (OR 1.65, 95% CI 1.29–2.12, P = 0.001), and 5′(GT)n (OR 1.86, 95% CI 1.33–2.62, P < 0.01). No significance was seen when European studies were grouped. African populations showed significance in all studies, except the D543N (OR 1.69, 95% CI 1.14–2.50, P = 0.01), INT4 (OR 1.50, 95% CI 1.17–1.91, P = 0.001), and 5′(GT)n (OR 1.31, 95% CI 1.05–1.64, P = 0.02). When all of the studies were pooled, significant associations were seen with the 3′UTR (OR 1.33, 95% CI 1.08–1.63; P = 0.008), D543N(OR 1.67, 95%CI 1.36–2.05, P < 0.001), and 5′ (GT)n (OR 1.32, 95% CI 1.03–1.68; P = 0.026). Only one of these variations, the promoter 5′(GT)n polymorphic z-DNA repeat, has been associated with changes in levels of mRNA transcription and possibly altered protein levels with various alleles (196). In aggregate, these studies indicate that Slc11a1 variants are consistently associated with TB susceptibility in Asian and African populations, but the role of Scl11a1 variation to TB susceptibility in European populations is questionable.
Vitamin D receptor
The possible role of vitamin D in TB pathogenesis has a long history, including hypotheses regarding the benefits of cod liver in the 1800s, effects of hypocalcemia, and the benefits of sunlight exposure in sanatoriums (197, 198). A large number of studies indicate that vitamin D influences the immune response, including effects on lymphocytes and macrophages (199). Furthermore, several studies have shown that vitamin D suppresses the growth of MTb in macrophages (200–202). Recent work has further elucidated the mechanism of this anti-tuberculous property of vitamin D and linked it to TLR signaling (203). Activation of human macrophages with the 19 kDa MTb lipopeptide (TLR2/TLR1 ligand) induced expression of the vitamin D receptor (VDR) as well as Cyp27B1, a vitamin D-1-hydroxylase that converts inactive provitamin D [25(OH)D3] into the active 1,25(OH)2D3. Treatment of macrophages with 1,25(OH)2D3 induced expression of the anti-microbial peptide cathelicidin, which was sufficient for killing of MTb.
These mechanistic studies broaden our current understanding of genetic studies of vitamin D and TB susceptibility. Previous work has shown an association of vitamin D deficiency with susceptibility to TB [OR for undetectable 25(OH)D3 was 9.9 (P = 0.009) and for relative 25(OH)D3 deficiency was 2.9 (P = 0.008)] (196). There have also been a number of studies of VDR polymorphisms and susceptibility to TB. There are several VDR polymorphisms with nomenclature derived from their restriction enzyme cleavage sites. The FokI (alleles F and f) polymorphism creates an alternative initiation codon in the f allele, which results in a protein that is 3 amino acids longer and is associated with less efficient transcription and decreased bone mineral density (204, 205). The TaqI polymorphism (alleles T and t) is associated with altered bone mineral density and may influence transcription levels (206). There are two additional SNPs, BsmI and ApaI, which are non-functional and in linkage disequilibrium with FokI and TaqI (207). A number of studies have been performed on VDR polymorphisms and susceptibility to MTb. Overall, there is substantial heterogeneity of the results that makes it difficult to make definitive conclusions (137, 208–212). The heterogeneity of these results could be attributable to several factors including population admixture, different case and control definitions, small sample sizes for some of the studies, different ethnic backgrounds, and incomplete genotyping of the VDR locus, as the various studies examined different numbers of the known VDR SNPs. This latter possibility was further examined in a case–control and family study in West Africa, which examined FokI–BsmI–ApaI–TaqI haplotypes. The family study (using a TDT) found stronger associations of haplotypes with TB susceptibility in comparison with individual polymorphisms. Together, these results suggest a possible role for VDR polymorphisms and TB susceptibility, although further examination is still warranted, preferably with complete analyses with large sample sizes and family-based designs with haplotype analysis.
SP110
The sst1 locus controls resistance to MTb in mice and was found in fine mapping studies to be attributable to the gene Ipr1 (intracellular pathogen resistance 1) (213). Two polymorphisms in SP110, the nearest human homologue to Ipr, were found to be associated with susceptibility to TB in a family-based study in The Gambia and were also replicated in cohorts from Guinea-Bissau and the Republic of Guinea. The function of SP110 is not well understood, but it may be involved with myeloid cell differentiation or transcriptional regulation (214).
Conclusions and future trends
Recent advances in immunology and genetics are rapidly expanding our understanding of the role of common variation on susceptibility to TB. A number of candidate genes have been identified in case–control studies for their possible role in TB susceptibility. The most promising candidates that have shown consistent effects in their association with TB susceptibility in multiple studies include HLA and Slc11a1. Other genes with strongly suggestive associations include IFN-γ, TIRAP/MAL, and CCL2. In the genetics field, technologic advances are redefining the scope of studies that are feasible. Current efforts are using large-scale genome-wide association studies with hundreds of thousands of polymorphisms and case–control study designs. With rigorous study designs that address problems of population admixture, multiple comparisons, and adequate sample sizes, the ensuing years will provide an extraordinary wealth of information. In parallel, advances in our understanding of innate immunity and macrophage function are equally rapid and leading to detailed molecular and cellular pathogenesis models of MTb infection. Furthermore, a number of variants in candidate genes that are associated with disease susceptibility have been shown to alter the molecular and cellular function of the gene, either through effects on expression level or direct alteration of protein function through non-synonymous variants in the coding region. For several genes, there is evidence that allelic variants are associated with altered expression at the mRNA or protein level (DC-SIGN, IFN-γ, IL-1β, IL-12Rβ1, IL-10, CCL2, and TNF-α). However, for most of these genes, the effects are modest, inconsistent, or not replicated in multiple studies. Further work is needed to clarify the functional role of the effect of these polymorphisms on expression levels or signaling pathways. The evidence is compelling that several genes contain variants that directly alter the functional properties of the encoded protein. These examples include MBL (alleles B, C, and D), P2XA7_A1513C (E469A), TLR2_G2258A (R753Q), and MAL/TIRAP_C539T (S180L). Overall, there is a small and growing list of genetic variants that appear to regulate function. The combined analysis of the association of genetic variants with susceptibility to TB in vivo and altered functional effects in vitro suggests that common variation in macrophage genes does influence host susceptibility to TB. Future discoveries in genetics and immunology should further clarify these mechanisms and substantially illuminate our understanding of genetic regulation of host susceptibility to TB.
Acknowledgments
We would like to thank Sarah Dunstan, Thuong Thuong Nguyen, Annelies Verbon, Jeremy Farrar, Willem Hanekom, Gilla Kaplan, Alan Aderem, and Lue Ping Zhao for their outstanding collaborative support.
References
- 1.Small PM, Fujiwara PI. Management of tuberculosis in the United States. N Engl J Med. 2001;345:189–200. doi: 10.1056/NEJM200107193450307. [DOI] [PubMed] [Google Scholar]
- 2.Colditz GA, et al. The efficacy of bacillus Calmette-Guerin vaccination of newborns and infants in the prevention of tuberculosis: meta-analyses of the published literature. Pediatrics. 1995;96:29–35. [PubMed] [Google Scholar]
- 3.Colditz GA, et al. Efficacy of BCG vaccine in the prevention of tuberculosis. Meta-analysis of the published literature. JAMA. 1994;271:698–702. [PubMed] [Google Scholar]
- 4.Fine PE. BCG: the challenge continues. Scand J Infect Dis. 2001;33:243–245. doi: 10.1080/003655401300077144. [DOI] [PubMed] [Google Scholar]
- 5.Casanova JL, Abel L. The human model: a genetic dissection of immunity to infection in natural conditions. Nat Rev Immunol. 2004;4:55–66. doi: 10.1038/nri1264. [DOI] [PubMed] [Google Scholar]
- 6.Dorman SE, et al. Susceptibility to tuberculosis: clues from studies with inbred and outbred New Zealand White rabbits. Infect Immun. 2004;72:1700–1705. doi: 10.1128/IAI.72.3.1700-1705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneemann M, Schoedon G, Hofer S, Blau N, Guerrero L, Schaffner A. Nitric oxide synthase is not a constituent of the antimicrobial armature of human mononuclear phagocytes. J Infect Dis. 1993;167:1358–1363. doi: 10.1093/infdis/167.6.1358. [DOI] [PubMed] [Google Scholar]
- 8.MacMicking J, Xie Q, Nathan C. Nitric oxide and macrophage function. Ann Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 9.Lienhardt C, et al. Investigation of the risk factors for tuberculosis: a case-control study in three countries in West Africa. Int J Epidemiol. 2005;34:914–923. doi: 10.1093/ije/dyi100. [DOI] [PubMed] [Google Scholar]
- 10.Guwatudde D, et al. Tuberculosis in household contacts of infectious cases in Kampala, Uganda. Am J Epidemiol. 2003;158:887–898. doi: 10.1093/aje/kwg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casanova JL, Abel L. Genetic dissection of immunity to mycobacteria: the human model. Annu Rev Immunol. 2002;20:581–620. doi: 10.1146/annurev.immunol.20.081501.125851. [DOI] [PubMed] [Google Scholar]
- 12.Cooke GS, Hill AV. Genetics of susceptibility to human infectious disease. Nat Rev Genet. 2001;2:967–977. doi: 10.1038/35103577. [DOI] [PubMed] [Google Scholar]
- 13.Comstock GW. Tuberculosis in twins: a re-analysis of the Prophit survey. Am Rev Respir Dis. 1978;117:621–624. doi: 10.1164/arrd.1978.117.4.621. [DOI] [PubMed] [Google Scholar]
- 14.Dorman SE, et al. Clinical features of dominant and recessive interferon gamma receptor 1 deficiencies. Lancet. 2004;364:2113–2121. doi: 10.1016/S0140-6736(04)17552-1. [DOI] [PubMed] [Google Scholar]
- 15.Greenwood CM, et al. Linkage of tuberculosis to chromosome 2q35 loci, including NRAMP1, in a large aboriginal Canadian family. Am J Hum Genet. 2000;67:405–416. doi: 10.1086/303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baghdadi JE, et al. An autosomal dominant major gene confers predisposition to pulmonary tuberculosis in adults. J Exp Med. 2006;203:1679–1684. doi: 10.1084/jem.20060269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jamieson SE, et al. Evidence for a cluster of genes on chromosome 17q11-q21 controlling susceptibility to tuberculosis and leprosy in Brazilians. Genes Immun. 2004;5:46–57. doi: 10.1038/sj.gene.6364029. [DOI] [PubMed] [Google Scholar]
- 18.Bellamy R, et al. Genetic susceptibility to tuberculosis in Africans: a genome-wide scan. Proc Natl Acad Sci USA. 2000;97:8005–8009. doi: 10.1073/pnas.140201897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cervino AC, et al. Fine mapping of a putative tuberculosis-susceptibility locus on chromosome 15q11-13 in African families. Hum Mol Genet. 2002;11:1599–1603. doi: 10.1093/hmg/11.14.1599. [DOI] [PubMed] [Google Scholar]
- 20.Barreiro LB, et al. Promoter variation in the DC-SIGN-encoding gene CD209 is associated with tuberculosis. PLoS Med. 2006;3:e20. doi: 10.1371/journal.pmed.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flores-Villanueva PO, et al. A functional promoter polymorphism in monocyte chemoattractant protein-1 is associated with increased susceptibility to pulmonary tuberculosis. J Exp Med. 2005;202:1649–1658. doi: 10.1084/jem.20050126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernando SL, Britton WJ. Genetic susceptibility to mycobacterial disease in humans. Immunol Cell Biol. 2006;84:125–137. doi: 10.1111/j.1440-1711.2006.01420.x. [DOI] [PubMed] [Google Scholar]
- 23.Remus N, Alcais A, Abel L. Human genetics of common mycobacterial infections. Immunol Res. 2003;28:109–129. doi: 10.1385/IR:28:2:109. [DOI] [PubMed] [Google Scholar]
- 24.Ernst JD. Macrophage receptors for Mycobacterium tuberculosis. Infect Immun. 1998;66:1277–1281. doi: 10.1128/iai.66.4.1277-1281.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferguson JS, Schlesinger LS. Pulmonary surfactant in innate immunity and the pathogenesis of tuberculosis. Tuber Lung Dis. 2000;80:173–184. doi: 10.1054/tuld.2000.0242. [DOI] [PubMed] [Google Scholar]
- 26.Koppel EA, van Gisbergen KP, Geijtenbeek TB, van Kooyk Y. Distinct functions of DC-SIGN and its homologues L-SIGN (DC-SIGNR) and mSIGNR1 in pathogen recognition and immune regulation. Cell Microbiol. 2005;7:157–165. doi: 10.1111/j.1462-5822.2004.00480.x. [DOI] [PubMed] [Google Scholar]
- 27.Kang PB, et al. The human macrophage mannose receptor directs Mycobacterium tuberculosis lipoarabinomannan-mediated phagosome biogenesis. J Exp Med. 2005;202:987–999. doi: 10.1084/jem.20051239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlesinger LS. Macrophage phagocytosis of virulent but not attenuated strains of Mycobacterium tuberculosis is mediated by mannose receptors in addition to complement receptors. J Immunol. 1993;150:2920–2930. [PubMed] [Google Scholar]
- 29.Tailleux L, et al. DC-SIGN is the major Mycobacterium tuberculosis receptor on human dendritic cells. J Exp Med. 2003;197:121–127. doi: 10.1084/jem.20021468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown GD. Dectin-1: a signalling non-TLR pattern-recognition receptor. Nat Rev Immunol. 2006;6:33–43. doi: 10.1038/nri1745. [DOI] [PubMed] [Google Scholar]
- 31.Brown GD, Gordon S. Immune recognition. A new receptor for beta-glucans. Nature. 2001;413:36–37. doi: 10.1038/35092620. [DOI] [PubMed] [Google Scholar]
- 32.Gantner BN, Simmons RM, Canavera SJ, Akira S, Underhill DM. Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J Exp Med. 2003;197:1107–1117. doi: 10.1084/jem.20021787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yadav M, Schorey JS. The beta-glucan receptor dectin-1 functions together with TLR2 to mediate macrophage activation by mycobacteria. Blood. 2006;108:3168–3175. doi: 10.1182/blood-2006-05-024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eisen DP, Minchinton RM. Impact of mannose-binding lectin on susceptibility to infectious diseases. Clin Infect Dis. 2003;37:1496–1505. doi: 10.1086/379324. [DOI] [PubMed] [Google Scholar]
- 35.Garred P, Harboe M, Oettinger T, Koch C, Svejgaard A. Dual role of mannan-binding protein in infections: another case of heterosis? Eur J Immunogenet. 1994;21:125–131. doi: 10.1111/j.1744-313x.1994.tb00183.x. [DOI] [PubMed] [Google Scholar]
- 36.Garred P, et al. Mannan-binding lectin in the sub-Saharan HIV and tuberculosis epidemics. Scand J Immunol. 1997;46:204–208. doi: 10.1046/j.1365-3083.1997.d01-111.x. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi K, Ezekowitz RA. The role of the mannose-binding lectin in innate immunity. Clin Infect Dis. 2005;41 (Suppl):S440–S444. doi: 10.1086/431987. [DOI] [PubMed] [Google Scholar]
- 38.Selvaraj P, Narayanan PR, Reetha AM. Association of functional mutant homozygotes of the mannose binding protein gene with susceptibility to pulmonary tuberculosis in India. Tuber Lung Dis. 1999;79:221–227. doi: 10.1054/tuld.1999.0204. [DOI] [PubMed] [Google Scholar]
- 39.Hoal-Van Helden EG, et al. Mannose-binding protein B allele confers protection against tuberculous meningitis. Pediatr Res. 1999;45:459–464. doi: 10.1203/00006450-199904010-00002. [DOI] [PubMed] [Google Scholar]
- 40.Soborg C, Madsen HO, Andersen AB, Lillebaek T, Kok-Jensen A, Garred P. Mannose-binding lectin polymorphisms in clinical tuberculosis. J Infect Dis. 2003;188:777–782. doi: 10.1086/377183. [DOI] [PubMed] [Google Scholar]
- 41.Geijtenbeek TB, et al. Mycobacteria target DC-SIGN to suppress dendritic cell function. J Exp Med. 2003;197:7–17. doi: 10.1084/jem.20021229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maeda N, et al. The cell surface receptor DC-SIGN discriminates between Mycobacterium species through selective recognition of the mannose caps on lipoarabinomannan. J Biol Chem. 2003;278:5513–5516. doi: 10.1074/jbc.C200586200. [DOI] [PubMed] [Google Scholar]
- 43.Tailleux L, et al. DC-SIGN induction in alveolar macrophages defines privileged target host cells for mycobacteria in patients with tuberculosis. PLoS Med. 2005;2:e381. doi: 10.1371/journal.pmed.0020381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakuntabhai A, et al. A variant in the CD209 promoter is associated with severity of dengue disease. Nat Genet. 2005;37:507–513. doi: 10.1038/ng1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barreiro LB, et al. Length variation of DC-SIGN and L-SIGN neck-region has no impact on tuberculosis susceptibility. Hum Immunol. 2007;68:106–112. doi: 10.1016/j.humimm.2006.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez LM, Anaya JM, Sierra-Filardi E, Cadena J, Corbi A, Martin J. Analysis of DC-SIGN (CD209) functional variants in patients with tuberculosis. Hum Immunol. 2006;67:808–811. doi: 10.1016/j.humimm.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 47.LeVine AM, et al. Distinct effects of surfactant protein A or D deficiency during bacterial infection on the lung. J Immunol. 2000;165:3934–3940. doi: 10.4049/jimmunol.165.7.3934. [DOI] [PubMed] [Google Scholar]
- 48.Gaynor CD, McCormack FX, Voelker DR, McGowan SE, Schlesinger LS. Pulmonary surfactant protein A mediates enhanced phagocytosis of Mycobacterium tuberculosis by a direct interaction with human macrophages. J Immunol. 1995;155:5343–5351. [PubMed] [Google Scholar]
- 49.Ferguson JS, Voelker DR, McCormack FX, Schlesinger LS. Surfactant protein D binds to Mycobacterium tuberculosis bacilli and lipoarabinomannan via carbohydrate-lectin interactions resulting in reduced phagocytosis of the bacteria by macrophages. J Immunol. 1999;163:312–321. [PubMed] [Google Scholar]
- 50.Floros J, et al. Surfactant protein genetic marker alleles identify a subgroup of tuberculosis in a Mexican population. J Infect Dis. 2000;182:1473–1478. doi: 10.1086/315866. [DOI] [PubMed] [Google Scholar]
- 51.Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 52.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 53.Beutler B, et al. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu Rev Immunol. 2006;24:353–389. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- 54.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 55.Heldwein KA, Fenton MJ. The role of Toll-like receptors in immunity against mycobacterial infection. Microbes Infect. 2002;4:937–944. doi: 10.1016/s1286-4579(02)01611-8. [DOI] [PubMed] [Google Scholar]
- 56.Quesniaux V, et al. Toll-like receptor pathways in the immune responses to mycobacteria. Microbes Infect. 2004;6:946–959. doi: 10.1016/j.micinf.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 57.Brightbill HD, et al. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science. 1999;285:732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 58.Means TK, Wang S, Lien E, Yoshimura A, Golenbock DT, Fenton MJ. Human toll-like receptors mediate cellular activation by Mycobacterium tuberculosis. J Immunol. 1999;163:3920–3927. [PubMed] [Google Scholar]
- 59.Underhill DM, Ozinsky A, Smith KD, Aderem A. Toll-like receptor-2 mediates mycobacteria-induced pro-inflammatory signaling in macrophages. Proc Natl Acad Sci USA. 1999;96:14459–14463. doi: 10.1073/pnas.96.25.14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Means TK, Lien E, Yoshimura A, Wang S, Golenbock DT, Fenton MJ. The CD14 ligands lipoarabinomannan and lipopolysaccharide differ in their requirement for Toll-like receptors. J Immunol. 1999;163:6748–6755. [PubMed] [Google Scholar]
- 61.Thoma-Uszynski S, et al. Induction of direct antimicrobial activity through mammalian toll-like receptors. Science. 2001;291:1544–1547. doi: 10.1126/science.291.5508.1544. [DOI] [PubMed] [Google Scholar]
- 62.Buwitt-Beckmann U, et al. Toll-like receptor 6-independent signaling by diacylated lipopeptides. Eur J Immunol. 2005;35:282–289. doi: 10.1002/eji.200424955. [DOI] [PubMed] [Google Scholar]
- 63.Morr M, Takeuchi O, Akira S, Simon MM, Muhlradt PF. Differential recognition of structural details of bacterial lipopeptides by toll-like receptors. Eur J Immunol. 2002;32:3337–3347. doi: 10.1002/1521-4141(200212)32:12<3337::AID-IMMU3337>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 64.Muller SD, et al. Triacyl-lipopentapeptide adjuvants: TLR2-dependent activation of macrophages and modulation of receptor-mediated cell activation by altering acyl-moieties. Int J Immunopharmacol. 2004;4:1287–1300. doi: 10.1016/j.intimp.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 65.Okusawa T, et al. Relationship between structures and biological activities of mycoplasmal diacylated lipopeptides and their recognition by toll-like receptors 2 and 6. Infect Immun. 2004;72:1657–1665. doi: 10.1128/IAI.72.3.1657-1665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Omueti KO, Beyer JM, Johnson CM, Lyle EA, Tapping RI. Domain exchange between human toll-like receptors 1 and 6 reveals a region required for lipopeptide discrimination. J Biol Chem. 2005;280:36616–36625. doi: 10.1074/jbc.M504320200. [DOI] [PubMed] [Google Scholar]
- 67.Takeuchi O, et al. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int Immunol. 2001;13:933–940. doi: 10.1093/intimm/13.7.933. [DOI] [PubMed] [Google Scholar]
- 68.Takeuchi O, et al. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol. 2002;169:10–14. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- 69.Krutzik SR, et al. Activation and regulation of Toll-like receptors 2 and 1 in human leprosy. Nat Med. 2003;9:525–532. doi: 10.1038/nm864. [DOI] [PubMed] [Google Scholar]
- 70.Bulut Y, Faure E, Thomas L, Equils O, Arditi M. Cooperation of Toll-like receptor 2 and 6 for cellular activation by soluble tuberculosis factor and Borrelia burgdorferi outer surface protein A lipoprotein: role of Toll-interacting protein and IL-1 receptor signaling molecules in Toll-like receptor 2 signaling. J Immunol. 2001;167:987–994. doi: 10.4049/jimmunol.167.2.987. [DOI] [PubMed] [Google Scholar]
- 71.Fortune SM, et al. Mycobacterium tuberculosis inhibits macrophage responses to IFN-gamma through myeloid differentiation factor 88-dependent and -independent mechanisms. J Immunol. 2004;172:6272–6280. doi: 10.4049/jimmunol.172.10.6272. [DOI] [PubMed] [Google Scholar]
- 72.Schnappinger D, Schoolnik GK, Ehrt S. Expression profiling of host pathogen interactions: how Mycobacterium tuberculosis and the macrophage adapt to one another. Microbes Infect. 2006;8:1132–1140. doi: 10.1016/j.micinf.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 73.Fremond CM, Yeremeev V, Nicolle DM, Jacobs M, Quesniaux VF, Ryffel B. Fatal Mycobacterium tuberculosis infection despite adaptive immune response in the absence of MyD88. J Clin Invest. 2004;114:1790–1799. doi: 10.1172/JCI21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scanga CA, Bafica A, Feng CG, Cheever AW, Hieny S, Sher A. MyD88-deficient mice display a profound loss in resistance to Mycobacterium tuberculosis associated with partially impaired Th1 cytokine and nitric oxide synthase 2 expression. Infect Immun. 2004;72:2400–2404. doi: 10.1128/IAI.72.4.2400-2404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sugawara I, Yamada H, Mizuno S, Takeda K, Akira S. Mycobacterial infection in MyD88-deficient mice. Microbiol Immunol. 2003;47:841–847. doi: 10.1111/j.1348-0421.2003.tb03450.x. [DOI] [PubMed] [Google Scholar]
- 76.Drennan MB, et al. Toll-like receptor 2-deficient mice succumb to Mycobacterium tuberculosis infection. Am J Pathol. 2004;164:49–57. doi: 10.1016/S0002-9440(10)63095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heldwein KA, et al. TLR2 and TLR4 serve distinct roles in the host immune response against Mycobacterium bovis BCG. J Leukoc Biol. 2003;74:277–286. doi: 10.1189/jlb.0103026. [DOI] [PubMed] [Google Scholar]
- 78.Reiling N, et al. Cutting edge: Toll-like receptor (TLR)2- and TLR4-mediated pathogen recognition in resistance to air-borne infection with Mycobacterium tuberculosis. J Immunol. 2002;169:3480–3484. doi: 10.4049/jimmunol.169.7.3480. [DOI] [PubMed] [Google Scholar]
- 79.Sugawara I, Yamada H, Li C, Mizuno S, Takeuchi O, Akira S. Mycobacterial infection in TLR2 and TLR6 knockout mice. Microbiol Immunol. 2003;47:327–336. doi: 10.1111/j.1348-0421.2003.tb03404.x. [DOI] [PubMed] [Google Scholar]
- 80.Shim TS, Turner OC, Orme IM. Toll-like receptor 4 plays no role in susceptibility of mice to Mycobacterium tuberculosis infection. Tuberculosis (Edinb) 2003;83:367–371. doi: 10.1016/s1472-9792(03)00071-4. [DOI] [PubMed] [Google Scholar]
- 81.Abel B, et al. Toll-like receptor 4 expression is required to control chronic Mycobacterium tuberculosis infection in mice. J Immunol. 2002;169:3155–3162. doi: 10.4049/jimmunol.169.6.3155. [DOI] [PubMed] [Google Scholar]
- 82.Bafica A, Scanga CA, Feng CG, Leifer C, Cheever A, Sher A. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J Exp Med. 2005;202:1715–1724. doi: 10.1084/jem.20051782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lorenz E, Mira JP, Cornish KL, Arbour NC, Schwartz DA. A novel polymorphism in the toll-like receptor 2 gene and its potential association with staphylococcal infection. Infect Immun. 2000;68:6398–6401. doi: 10.1128/iai.68.11.6398-6401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schroder NW, et al. Heterozygous Arg753Gln polymorphism of human TLR-2 impairs immune activation by Borrelia burgdorferi and protects from late stage Lyme disease. J Immunol. 2005;175:2534–2540. doi: 10.4049/jimmunol.175.4.2534. [DOI] [PubMed] [Google Scholar]
- 85.Ogus AC, et al. The Arg753GLn polymorphism of the human toll-like receptor 2 gene in tuberculosis disease. Eur Respir J. 2004;23:219–223. doi: 10.1183/09031936.03.00061703. [DOI] [PubMed] [Google Scholar]
- 86.Kang TJ, Chae GT. Detection of Toll-like receptor 2 (TLR2) mutation in the lepromatous leprosy patients. FEMS Immunol Med Microbiol. 2001;31:53–58. doi: 10.1111/j.1574-695X.2001.tb01586.x. [DOI] [PubMed] [Google Scholar]
- 87.Malhotra D, Relhan V, Reddy BS, Bamezai R. TLR2 Arg677Trp polymorphism in leprosy: revisited. Hum Genet. 2005;116:413–415. doi: 10.1007/s00439-004-1249-9. [DOI] [PubMed] [Google Scholar]
- 88.Yim JJ, Ding L, Schaffer AA, Park GY, Shim YS, Holland SM. A microsatellite polymorphism in intron 2 of human Toll-like receptor 2 gene: functional implications and racial differences. FEMS Immunol Med Microbiol. 2004;40:163–169. doi: 10.1016/S0928-8244(03)00342-0. [DOI] [PubMed] [Google Scholar]
- 89.Yim JJ, et al. The association between microsatellite polymorphisms in intron II of the human Toll-like receptor 2 gene and tuberculosis among Koreans. Genes Immun. 2006;7:150–155. doi: 10.1038/sj.gene.6364274. [DOI] [PubMed] [Google Scholar]
- 90.Khor CC, et al. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nat Genet. 2007;39:523–528. doi: 10.1038/ng1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hawn TR, et al. A polymorphism in toll-interleukin 1 receptor domain containing adaptor protein is associated with susceptibility to meningeal tuberculosis. J Infect Dis. 2006;194:1127–1134. doi: 10.1086/507907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol. 2006;7:1250–1257. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- 93.Martinon F, Tschopp J. NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 2005;26:447–454. doi: 10.1016/j.it.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 94.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 95.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 96.Greenstein RJ. Is Crohn’s disease caused by a mycobacterium? Comparisons with leprosy, tuberculosis, and Johne’s disease. Lancet Infect Dis. 2003;3:507–514. doi: 10.1016/s1473-3099(03)00724-2. [DOI] [PubMed] [Google Scholar]
- 97.Miceli-Richard C, et al. CARD15 mutations in Blau syndrome. Nat Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 98.Girardin SE, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 99.Kobayashi KS, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 100.Ferwerda G, et al. NOD2 and toll-like receptors are nonredundant recognition systems of Mycobacterium tuberculosis. PLoS Pathog. 2005;1:279–285. doi: 10.1371/journal.ppat.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Moller M, Nebel A, Kwiatkowski R, van Helden PD, Hoal EG, Schreiber S. Host susceptibility to tuberculosis: CARD15 polymorphisms in a South African population. Mol Cell Probes. 2007;21:148–151. doi: 10.1016/j.mcp.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 102.Stockton JC, Howson JM, Awomoyi AA, McAdam KP, Blackwell JM, Newport MJ. Polymorphism in NOD2, Crohn’s disease, and susceptibility to pulmonary tuberculosis. FEMS Immunol Med Microbiol. 2004;41:157–160. doi: 10.1016/j.femsim.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 103.Williams KL, et al. The CATERPILLER protein monarch-1 is an antagonist of toll-like receptor-, tumor necrosis factor alpha-, and Mycobacterium tuberculosis-induced pro-inflammatory signals. J Biol Chem. 2005;280:39914–39924. doi: 10.1074/jbc.M502820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Williams KL, Taxman DJ, Linhoff MW, Reed W, Ting JP. Cutting edge: Monarch-1: a pyrin/nucleotide-binding domain/leucine-rich repeat protein that controls classical and nonclassical MHC class I genes. J Immunol. 2003;170:5354–5358. doi: 10.4049/jimmunol.170.11.5354. [DOI] [PubMed] [Google Scholar]
- 105.Fernando SL, Saunders BM, Sluyter R, Skarratt KK, Wiley JS, Britton WJ. Gene dosage determines the negative effects of polymorphic alleles of the P2X7 receptor on adenosine triphosphate-mediated killing of mycobacteria by human macrophages. J Infect Dis. 2005;192:149–155. doi: 10.1086/430622. [DOI] [PubMed] [Google Scholar]
- 106.Shemon AN, et al. A Thr357 to Ser polymorphism in homozygous and compound heterozygous subjects causes absent or reduced P2X7 function and impairs ATP-induced mycobacterial killing by macrophages. J Biol Chem. 2006;281:2079–2086. doi: 10.1074/jbc.M507816200. [DOI] [PubMed] [Google Scholar]
- 107.Li CM, et al. Association of a polymorphism in the P2X7 gene with tuberculosis in a Gambian population. J Infect Dis. 2002;186:1458–1462. doi: 10.1086/344351. [DOI] [PubMed] [Google Scholar]
- 108.Fernando SL, et al. A polymorphism in the P2X7 gene increases susceptibility to extrapulmonary tuberculosis. Am J Respir Crit Care Med. 2007;175:360–366. doi: 10.1164/rccm.200607-970OC. [DOI] [PubMed] [Google Scholar]
- 109.Saunders BM, Fernando SL, Sluyter R, Britton WJ, Wiley JS. A loss-of-function polymorphism in the human P2X7 receptor abolishes ATP-mediated killing of mycobacteria. J Immunol. 2003;171:5442–5446. doi: 10.4049/jimmunol.171.10.5442. [DOI] [PubMed] [Google Scholar]
- 110.Ding AH, Nathan CF, Stuehr DJ. Release of reactive nitrogen intermediates and reactive oxygen intermediates from mouse peritoneal macrophages. Comparison of activating cytokines and evidence for independent production. J Immunol. 1988;141:2407–2412. [PubMed] [Google Scholar]
- 111.Flesch IE, Hess JH, Oswald IP, Kaufmann SH. Growth inhibition of Mycobacterium bovis by IFN-gamma stimulated macrophages: regulation by endogenous tumor necrosis factor-alpha and by IL-10. Int Immunol. 1994;6:693–700. doi: 10.1093/intimm/6.5.693. [DOI] [PubMed] [Google Scholar]
- 112.Bean AG, et al. Structural deficiencies in granuloma formation in TNF gene-targeted mice underlie the heightened susceptibility to aerosol Mycobacterium tuberculosis infection, which is not compensated for by lymphotoxin. J Immunol. 1999;162:3504–3511. [PubMed] [Google Scholar]
- 113.Flynn JL, et al. Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity. 1995;2:561–572. doi: 10.1016/1074-7613(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 114.Roach DR, Briscoe H, Saunders B, France MP, Riminton S, Britton WJ. Secreted lymphotoxin-alpha is essential for the control of an intracellular bacterial infection. J Exp Med. 2001;193:239–246. doi: 10.1084/jem.193.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Keane J, et al. Tuberculosis associated with infliximab, a tumor necrosis factor alpha-neutralizing agent. N Engl J Med. 2001;345:1098–1104. doi: 10.1056/NEJMoa011110. [DOI] [PubMed] [Google Scholar]
- 116.Stein CM, et al. Heritability analysis of cytokines as intermediate phenotypes of tuberculosis. J Infect Dis. 2003;187:1679–1685. doi: 10.1086/375249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stein CM, et al. Linkage and association analysis of candidate genes for TB and TNF alpha cytokine expression: evidence for association with IFNGR1, IL-10, and TNF receptor 1 genes. Hum Genet. 2007 doi: 10.1007/s00439-007-0357-8. (in press) [DOI] [PubMed] [Google Scholar]
- 118.Roy S, et al. Tumor necrosis factor promoter polymorphism and susceptibility to lepromatous leprosy. J Infect Dis. 1997;176:530–532. doi: 10.1086/517282. [DOI] [PubMed] [Google Scholar]
- 119.McGuire W, Hill AV, Allsopp CE, Greenwood BM, Kwiatkowski D. Variation in the TNF-alpha promoter region associated with susceptibility to cerebral malaria. Nature. 1994;371:508–510. doi: 10.1038/371508a0. [DOI] [PubMed] [Google Scholar]
- 120.Mira JP, et al. Association of TNF2, a TNF-alpha promoter polymorphism, with septic shock susceptibility and mortality: a multicenter study. JAMA. 1999;282:561–568. doi: 10.1001/jama.282.6.561. [DOI] [PubMed] [Google Scholar]
- 121.Correa PA, Gomez LM, Cadena J, Anaya JM. Autoimmunity and tuberculosis. Opposite association with TNF polymorphism. J Rheumatol. 2005;32:219–224. [PubMed] [Google Scholar]
- 122.Amirzargar AA, et al. Cytokine single nucleotide polymorphisms in Iranian patients with pulmonary tuberculosis. Eur Cytokine Netw. 2006;17:84–89. [PubMed] [Google Scholar]
- 123.Wilkinson RJ, et al. Influence of polymorphism in the genes for the interleukin (IL)-1 receptor antagonist and IL-1beta on tuberculosis. J Exp Med. 1999;189:1863–1874. doi: 10.1084/jem.189.12.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mege JL, Meghari S, Honstettre A, Capo C, Raoult D. The two faces of interleukin 10 in human infectious diseases. Lancet Infect Dis. 2006;6:557–569. doi: 10.1016/S1473-3099(06)70577-1. [DOI] [PubMed] [Google Scholar]
- 125.Mohan VP, et al. Effects of tumor necrosis factor alpha on host immune response in chronic persistent tuberculosis: possible role for limiting pathology. Infect Immun. 2001;69:1847–1855. doi: 10.1128/IAI.69.3.1847-1855.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Brinkman BM, Zuijdeest D, Kaijzel EL, Breedveld FC, Verweij CL. Relevance of the tumor necrosis factor alpha (TNF alpha)-308 promoter polymorphism in TNF alpha gene regulation. J Inflamm. 1995;46:32–41. [PubMed] [Google Scholar]
- 127.Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci USA. 1997;94:3195–3199. doi: 10.1073/pnas.94.7.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dinarello CA. Biologic basis for interleukin-1 in disease. Blood. 1996;87:2095–2147. [PubMed] [Google Scholar]
- 129.Yamada H, Mizumo S, Horai R, Iwakura Y, Sugawara I. Protective role of interleukin-1 in mycobacterial infection in IL-1 alpha/beta double-knockout mice. Lab Invest. 2000;80:759–767. doi: 10.1038/labinvest.3780079. [DOI] [PubMed] [Google Scholar]
- 130.Juffermans NP, et al. Interleukin-1 signaling is essential for host defense during murine pulmonary tuberculosis. J Infect Dis. 2000;182:902–908. doi: 10.1086/315771. [DOI] [PubMed] [Google Scholar]
- 131.Bellamy R, Ruwende C, Corrah T, McAdam KP, Whittle HC, Hill AV. Assessment of the interleukin 1 gene cluster and other candidate gene polymorphisms in host susceptibility to tuberculosis. Tuber Lung Dis. 1998;79:83–89. doi: 10.1054/tuld.1998.0009. [DOI] [PubMed] [Google Scholar]
- 132.Awomoyi AA, et al. Polymorphism in IL1B: IL1B-511 association with tuberculosis and decreased lipopolysaccharide-induced IL-1beta in IFN-gamma primed ex-vivo whole blood assay. J Endotoxin Res. 2005;11:281–286. doi: 10.1179/096805105X58706. [DOI] [PubMed] [Google Scholar]
- 133.North RJ. Mice incapable of making IL-4 or IL-10 display normal resistance to infection with Mycobacterium tuberculosis. Clin Exp Immunol. 1998;113:55–58. doi: 10.1046/j.1365-2249.1998.00636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24:1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 135.Moraes MO, et al. Interleukin-10 promoter single-nucleotide polymorphisms as markers for disease susceptibility and disease severity in leprosy. Genes Immun. 2004;5:592–595. doi: 10.1038/sj.gene.6364122. [DOI] [PubMed] [Google Scholar]
- 136.Santos AR, et al. Role of tumor necrosis factor-alpha and interleukin-10 promoter gene polymorphisms in leprosy. J Infect Dis. 2002;186:1687–1691. doi: 10.1086/345366. [DOI] [PubMed] [Google Scholar]
- 137.Delgado JC, Baena A, Thim S, Goldfeld AE. Ethnic-specific genetic associations with pulmonary tuberculosis. J Infect Dis. 2002;186:1463–1468. doi: 10.1086/344891. [DOI] [PubMed] [Google Scholar]
- 138.Scola L, et al. IL-10 and TNF-alpha polymorphisms in a sample of Sicilian patients affected by tuberculosis: implication for ageing and life span expectancy. Mech Ageing Dev. 2003;124:569–572. doi: 10.1016/s0047-6374(03)00038-1. [DOI] [PubMed] [Google Scholar]
- 139.Shin HD, Park BL, Kim YH, Cheong HS, Lee IH, Park SK. Common interleukin 10 polymorphism associated with decreased risk of tuberculosis. Exp Mol Med. 2005;37:128–132. doi: 10.1038/emm.2005.17. [DOI] [PubMed] [Google Scholar]
- 140.Lopez-Maderuelo D, et al. Interferon-gamma and interleukin-10 gene polymorphisms in pulmonary tuberculosis. Am J Respir Crit Care Med. 2003;167:970–975. doi: 10.1164/rccm.200205-438BC. [DOI] [PubMed] [Google Scholar]
- 141.Lowe PR, Galley HF, Abdel-Fattah A, Webster NR. Influence of interleukin-10 polymorphisms on interleukin-10 expression and survival in critically ill patients. Crit Care Med. 2003;31:34–38. doi: 10.1097/00003246-200301000-00005. [DOI] [PubMed] [Google Scholar]
- 142.Rosenwasser LJ, Borish L. Genetics of atopy and asthma: the rationale behind promoter-based candidate gene studies (IL-4 and IL-10) Am J Respir Crit Care Med. 1997;156:S152–S155. doi: 10.1164/ajrccm.156.4.12tac-14. [DOI] [PubMed] [Google Scholar]
- 143.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 144.Cooper AM, Magram J, Ferrante J, Orme IM. Interleukin 12 (IL-12) is crucial to the development of protective immunity in mice intravenously infected with Mycobacterium tuberculosis. J Exp Med. 1997;186:39–45. doi: 10.1084/jem.186.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Khader SA, et al. IL-23 compensates for the absence of IL-12p70 and is essential for the IL-17 response during tuberculosis but is dispensable for protection and antigen-specific IFN-gamma responses if IL-12p70 is available. J Immunol. 2005;175:788–795. doi: 10.4049/jimmunol.175.2.788. [DOI] [PubMed] [Google Scholar]
- 146.Akahoshi M, et al. Influence of interleukin-12 receptor beta1 polymorphisms on tuberculosis. Hum Genet. 2003;112:237–243. doi: 10.1007/s00439-002-0873-5. [DOI] [PubMed] [Google Scholar]
- 147.Remus N, et al. Association of IL12RB1 polymorphisms with pulmonary tuberculosis in adults in Morocco. J Infect Dis. 2004;190:580–587. doi: 10.1086/422534. [DOI] [PubMed] [Google Scholar]
- 148.Lee HW, et al. Lack of an association between interleukin-12 receptor beta1 polymorphisms and tuberculosis in Koreans. Respiration. 2005;72:365–368. doi: 10.1159/000086249. [DOI] [PubMed] [Google Scholar]
- 149.Tso HW, Lau YL, Tam CM, Wong HS, Chiang AK. Associations between IL12B polymorphisms and tuberculosis in the Hong Kong Chinese population. J Infect Dis. 2004;190:913–919. doi: 10.1086/422693. [DOI] [PubMed] [Google Scholar]
- 150.Ma X, et al. No evidence for association between the polymorphism in the 3′ untranslated region of interleukin-12B and human susceptibility to tuberculosis. J Infect Dis. 2003;188:1116–1118. doi: 10.1086/378674. [DOI] [PubMed] [Google Scholar]
- 151.Morahan G, et al. Association of IL12B promoter polymorphism with severity of atopic and non-atopic asthma in children. Lancet. 2002;360:455–459. doi: 10.1016/S0140-6736(02)09676-9. [DOI] [PubMed] [Google Scholar]
- 152.Morahan G, et al. Linkage disequilibrium of a type 1 diabetes susceptibility locus with a regulatory IL12B allele. Nat Genet. 2001;27:218–221. doi: 10.1038/84872. [DOI] [PubMed] [Google Scholar]
- 153.Seegers D, Zwiers A, Strober W, Pena AS, Bouma G. A TaqI polymorphism in the 3′UTR of the IL-12 p40 gene correlates with increased IL-12 secretion. Genes Immun. 2002;3:419–423. doi: 10.1038/sj.gene.6363919. [DOI] [PubMed] [Google Scholar]
- 154.Feng CG, et al. NK cell-derived IFN-gamma differentially regulates innate resistance and neutrophil response in T cell-deficient hosts infected with Mycobacterium tuberculosis. J Immunol. 2006;177:7086–7093. doi: 10.4049/jimmunol.177.10.7086. [DOI] [PubMed] [Google Scholar]
- 155.Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 156.Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, Orme IM. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J Exp Med. 1993;178:2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Flynn JL, Chan J, Triebold KJ, Dalton Dk, Stewart TA, Bloom BR. An essential role for interferon γ in resistance to Mycobacterium tuberculosis infection. J Exp Med. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Flynn JL, Goldstein MM, Triebold KJ, Sypek J, Wolf S, Bloom BR. IL-12 increases resistance of BALB/c mice to Mycobacterium tuberculosis infection. J Immunol. 1995;155:2515–2524. [PubMed] [Google Scholar]
- 159.Ottenhoff TH, Verreck FA, Lichtenauer-Kaligis EG, Hoeve MA, Sanal O, van Dissel JT. Genetics, cytokines and human infectious disease: lessons from weakly pathogenic mycobacteria and salmonellae. Nat Genet. 2002;32:97–105. doi: 10.1038/ng0902-97. [DOI] [PubMed] [Google Scholar]
- 160.Fraser DA, et al. Interferon-gamma receptor-1 gene polymorphism in tuberculosis patients from Croatia. Scand J Immunol. 2003;57:480–484. doi: 10.1046/j.1365-3083.2003.01253.x. [DOI] [PubMed] [Google Scholar]
- 161.Newport MJ, Awomoyi AA, Blackwell JM. Polymorphism in the interferon-gamma receptor-1 gene and susceptibility to pulmonary tuberculosis in The Gambia. Scand J Immunol. 2003;58:383–385. doi: 10.1046/j.1365-3083.2003.01328.x. [DOI] [PubMed] [Google Scholar]
- 162.Cooke GS, et al. Polymorphism within the interferon-gamma/receptor complex is associated with pulmonary tuberculosis. Am J Respir Crit Care Med. 2006;174:339–343. doi: 10.1164/rccm.200601-088OC. [DOI] [PubMed] [Google Scholar]
- 163.Lio D, et al. Genotype frequencies of the +874T–>A single nucleotide polymorphism in the first intron of the interferon-gamma gene in a sample of Sicilian patients affected by tuberculosis. Eur J Immunogenet. 2002;29:371–374. doi: 10.1046/j.1365-2370.2002.00327.x. [DOI] [PubMed] [Google Scholar]
- 164.Rossouw M, Nel HJ, Cooke GS, van Helden PD, Hoal EG. Association between tuberculosis and a polymorphic NFkappaB binding site in the interferon gamma gene. Lancet. 2003;361:1871–1872. doi: 10.1016/S0140-6736(03)13491-5. [DOI] [PubMed] [Google Scholar]
- 165.Vidyarani M, Selvaraj P, Prabhu Anand S, Jawahar MS, Adhilakshmi AR, Narayanan PR. Interferon gamma (IFNgamma) & interleukin-4 (IL-4) gene variants & cytokine levels in pulmonary tuberculosis. Indian J Med Res. 2006;124:403–410. [PubMed] [Google Scholar]
- 166.Lin Y, Gong J, Zhang M, Xue W, Barnes PF. Production of monocyte chemoattractant protein 1 in tuberculosis patients. Infect Immun. 1998;66:2319–2322. doi: 10.1128/iai.66.5.2319-2322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rutledge BJ, et al. High level monocyte chemoattractant protein-1 expression in transgenic mice increases their susceptibility to intracellular pathogens. J Immunol. 1995;155:4838–4843. [PubMed] [Google Scholar]
- 168.Lu B, et al. Abnormalities in monocyte recruitment and cytokine expression in monocyte chemoattractant protein 1-deficient mice. J Exp Med. 1998;187:601–608. doi: 10.1084/jem.187.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chensue SW, Warmington KS, Ruth JH, Sanghi PS, Lincoln P, Kunkel SL. Role of monocyte chemoattractant protein-1 (MCP-1) in Th1 (mycobacterial) and Th2 (schistosomal) antigen-induced granuloma formation: relationship to local inflammation, Th cell expression, and IL-12 production. J Immunol. 1996;157:4602–4608. [PubMed] [Google Scholar]
- 170.Rovin BH, Lu L, Saxena R. A novel polymorphism in the MCP-1 gene regulatory region that influences MCP-1 expression. Biochem Biophys Res Commun. 1999;259:344–348. doi: 10.1006/bbrc.1999.0796. [DOI] [PubMed] [Google Scholar]
- 171.Riedel DD, Kaufmann SH. Chemokine secretion by human polymorphonuclear granulocytes after stimulation with Mycobacterium tuberculosis and lipoarabinomannan. Infect Immun. 1997;65:4620–4623. doi: 10.1128/iai.65.11.4620-4623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wickremasinghe MI, Thomas LH, Friedland JS. Pulmonary epithelial cells are a source of IL-8 in the response to Mycobacterium tuberculosis: essential role of IL-1 from infected monocytes in a NF-kappa B-dependent network. J Immunol. 1999;163:3936–3947. [PubMed] [Google Scholar]
- 173.Zhang Y, et al. Enhanced interleukin-8 release and gene expression in macrophages after exposure to Mycobacterium tuberculosis and its components. J Clin Invest. 1995;95:586–592. doi: 10.1172/JCI117702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Friedland JS, Hartley JC, Hartley CG, Shattock RJ, Griffin GE. Inhibition of ex vivo proinflammatory cytokine secretion in fatal Mycobacterium tuberculosis infection. Clin Exp Immunol. 1995;100:233–238. doi: 10.1111/j.1365-2249.1995.tb03659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sadek MI, Sada E, Toossi Z, Schwander SK, Rich EA. Chemokines induced by infection of mononuclear phagocytes with mycobacteria and present in lung alveoli during active pulmonary tuberculosis. Am J Respir Cell Mol Biol. 1998;19:513–521. doi: 10.1165/ajrcmb.19.3.2815. [DOI] [PubMed] [Google Scholar]
- 176.Hull J, Thomson A, Kwiatkowski D. Association of respiratory syncytial virus bronchiolitis with the interleukin 8 gene region in UK families. Thorax. 2000;55:1023–1027. doi: 10.1136/thorax.55.12.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ma X, et al. Association between interleukin-8 gene alleles and human susceptibility to tuberculosis disease. J Infect Dis. 2003;188:349–355. doi: 10.1086/376559. [DOI] [PubMed] [Google Scholar]
- 178.Cooke GS, et al. Interleukin-8 polymorphism is not associated with pulmonary tuberculosis in the gambia. J Infect Dis. 2004;189:1545–1546. doi: 10.1086/382489. author reply 1546. [DOI] [PubMed] [Google Scholar]
- 179.Govoni G, Vidal S, Gauthier S, Skamene E, Malo D, Gros P. The Bcg/Ity/Lsh locus: genetic transfer of resistance to infections in C57BL/6J mice transgenic for the Nramp1 Gly169 allele. Infect Immun. 1996;64:2923–2929. doi: 10.1128/iai.64.8.2923-2929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Vidal SM, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate for BCG. Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 181.Blackwell JM, et al. SLC11A1 (formerly NRAMP1) and disease resistance. Cell Microbiol. 2001;3:773–784. doi: 10.1046/j.1462-5822.2001.00150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jabado N, Jankowski A, Dougaparsad S, Picard V, Grinstein S, Gros P. Natural resistance to intracellular infections: natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J Exp Med. 2000;192:1237–1248. doi: 10.1084/jem.192.9.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Li HT, Zhang TT, Zhou YQ, Huang QH, Huang J. SLC11A1 (formerly NRAMP1) gene polymorphisms and tuberculosis susceptibility: a meta-analysis. Int J Tuberc Lung Dis. 2006;10:3–12. [PubMed] [Google Scholar]
- 184.Awomoyi AA, Marchant A, Howson JM, McAdam KP, Blackwell JM, Newport MJ. Interleukin-10, polymorphism in SLC11A1 (formerly NRAMP1), and susceptibility to tuberculosis. J Infect Dis. 2002;186:1808–1814. doi: 10.1086/345920. [DOI] [PubMed] [Google Scholar]
- 185.Bellamy R, Ruwende C, Corrah T, McAdam K, Whittle H, Hill A. Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africans. N Engl J Med. 1998;338:640–644. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- 186.Duan HF, et al. A study on the association of 3′UTR polymorphisms of NRAMP1 gene with susceptibility to tuberculosis in Hans. Zhonghua Jie He He Hu Xi Za Zhi. 2003;26:286–289. [PubMed] [Google Scholar]
- 187.Fitness J, et al. Large-scale candidate gene study of tuberculosis susceptibility in the Karonga district of northern Malawi. Am J Trop Med Hyg. 2004;71:341–349. [PubMed] [Google Scholar]
- 188.Gao PS, et al. Genetic variants of NRAMP1 and active tuberculosis in Japanese populations. International Tuberculosis Genetics Team Clin Genet. 2000;58:74–76. doi: 10.1034/j.1399-0004.2000.580113.x. [DOI] [PubMed] [Google Scholar]
- 189.Kim JH, et al. NRAMP1 genetic polymorphisms as a risk factor of tuberculous pleurisy. Int J Tuberc Lung Dis. 2003;7:370–375. [PubMed] [Google Scholar]
- 190.Liaw YS, et al. Variations in the NRAMP1 gene and susceptibility of tuberculosis in Taiwanese. Int J Tuberc Lung Dis. 2002;6:454–460. [PubMed] [Google Scholar]
- 191.Liu W, et al. VDR and NRAMP1 gene polymorphisms in susceptibility to pulmonary tuberculosis among the Chinese Han population: a case-control study. Int J Tuberc Lung Dis. 2004;8:428–434. [PubMed] [Google Scholar]
- 192.Ma X, et al. 5′ dinucleotide repeat polymorphism of NRAMP1 and susceptibility to tuberculosis among Caucasian patients in Houston, Texas. Int J Tuberc Lung Dis. 2002;6:818–823. [PubMed] [Google Scholar]
- 193.Puzyrev VP, Freidin MB, Rudko AA, Strelis AK, Kolokolova OV. Polymorphisms of the candidate genes for genetic susceptibility to tuberculosis in the Slavic population of Siberia: a pilot study. Mol Biol (Mosk) 2002;36:788–791. [PubMed] [Google Scholar]
- 194.Ryu S, Park YK, Bai GH, Kim SJ, Park SN, Kang S. 3′UTR polymorphisms in the NRAMP1 gene are associated with susceptibility to tuberculosis in Koreans. Int J Tuberc Lung Dis. 2000;4:577–580. [PubMed] [Google Scholar]
- 195.Soborg C, Andersen AB, Madsen HO, Kok-Jensen A, Skinhoj P, Garred P. Natural resistance-associated macrophage protein 1 polymorphisms are associated with microscopy-positive tuberculosis. J Infect Dis. 2002;186:517–521. doi: 10.1086/341775. [DOI] [PubMed] [Google Scholar]
- 196.Searle S, Blackwell JM. Evidence for a functional repeat polymorphism in the promoter of the human NRAMP1 gene that correlates with autoimmune versus infectious disease susceptibility. J Med Genet. 1999;36:295–299. [PMC free article] [PubMed] [Google Scholar]
- 197.Davies PD. A possible link between vitamin D deficiency and impaired host defence to Mycobacterium tuberculosis. Tubercle. 1985;66:301–306. doi: 10.1016/0041-3879(85)90068-6. [DOI] [PubMed] [Google Scholar]
- 198.Davies PD, Brown RC, Woodhead JS. Serum concentrations of vitamin D metabolites in untreated tuberculosis. Thorax. 1985;40:187–190. doi: 10.1136/thx.40.3.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Deluca HF, Cantorna MT. Vitamin D: its role and uses in immunology. FASEB J. 2001;15:2579–2585. doi: 10.1096/fj.01-0433rev. [DOI] [PubMed] [Google Scholar]
- 200.Rockett KA, Brookes R, Udalova I, Vidal V, Hill AV, Kwiatkowski D. 1,25-Dihydroxyvitamin D3 induces nitric oxide synthase and suppresses growth of Mycobacterium tuberculosis in a human macrophage-like cell line. Infect Immun. 1998;66:5314–5321. doi: 10.1128/iai.66.11.5314-5321.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Rook GA, et al. Vitamin D3, gamma interferon, and control of proliferation of Mycobacterium tuberculosis by human monocytes. Immunology. 1986;57:159–163. [PMC free article] [PubMed] [Google Scholar]
- 202.Crowle AJ, Ross EJ, May MH. Inhibition by 1,25(OH)2-vitamin D3 of the multiplication of virulent tubercle bacilli in cultured human macrophages. Infect Immun. 1987;55:2945–2950. doi: 10.1128/iai.55.12.2945-2950.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Liu PT, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. 2006;311:1770–1773. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- 204.Arai H, et al. A vitamin D receptor gene polymorphism in the translation initiation codon: effect on protein activity and relation to bone mineral density in Japanese women. J Bone Miner Res. 1997;12:915–921. doi: 10.1359/jbmr.1997.12.6.915. [DOI] [PubMed] [Google Scholar]
- 205.Gross C, Eccleshall TR, Malloy PJ, Villa ML, Marcus R, Feldman D. The presence of a polymorphism at the translation initiation site of the vitamin D receptor gene is associated with low bone mineral density in postmenopausal Mexican-American women. J Bone Miner Res. 1996;11:1850–1855. doi: 10.1002/jbmr.5650111204. [DOI] [PubMed] [Google Scholar]
- 206.Morrison NA, et al. Prediction of bone density from vitamin D receptor alleles. Nature. 1994;367:284–287. doi: 10.1038/367284a0. [DOI] [PubMed] [Google Scholar]
- 207.Bornman L, et al. Vitamin D receptor polymorphisms and susceptibility to tuberculosis in West Africa: a case-control and family study. J Infect Dis. 2004;190:1631–1641. doi: 10.1086/424462. [DOI] [PubMed] [Google Scholar]
- 208.Bellamy R, et al. Tuberculosis and chronic hepatitis B virus infection in Africans and variation in the vitamin D receptor gene. J Infect Dis. 1999;179:721–724. doi: 10.1086/314614. [DOI] [PubMed] [Google Scholar]
- 209.Liu W, et al. A case-control study on the vitamin D receptor gene polymorphisms and susceptibility to pulmonary tuberculosis. Zhonghua Liu Xing Bing Xue Za Zhi. 2003;24:389–392. [PubMed] [Google Scholar]
- 210.Selvaraj P, Kurian SM, Uma H, Reetha AM, Narayanan PR. Influence of non-MHC genes on lymphocyte response to Mycobacterium tuberculosis antigens & tuberculin reactive status in pulmonary tuberculosis. Indian J Med Res. 2000;112:86–92. [PubMed] [Google Scholar]
- 211.Selvaraj P, Narayanan PR, Reetha AM. Association of vitamin D receptor genotypes with the susceptibility to pulmonary tuberculosis in female patients & resistance in female contacts. Indian J Med Res. 2000;111:172–179. [PubMed] [Google Scholar]
- 212.Wilkinson R, et al. Influence of vitamin D receptor and vitamin D receptor polymorphisms on tuberculosis among Gujarati Asians in west London: a case-control study. Lancet. 2000;355:618–621. doi: 10.1016/S0140-6736(99)02301-6. [DOI] [PubMed] [Google Scholar]
- 213.Pan H, et al. Ipr1 gene mediates innate immunity to tuberculosis. Nature. 2005;434:767–772. doi: 10.1038/nature03419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Bloch DB, et al. Sp110 localizes to the PML-Sp100 nuclear body and may function as a nuclear hormone receptor transcriptional coactivator. Mol Cell Biol. 2000;20:6138–6146. doi: 10.1128/mcb.20.16.6138-6146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]