Abstract
Objectives
Pancreatitis occurs as extraintestinal complication of IBD but the cause is poorly understood. MUC1 is overexpressed in an abnormal, hypoglycosylated form on the colonic epithelium in human IBD where it contributes to inflammation. MUC1 is also expressed on pancreatic ductal epithelia. We tested the possibility that in IBD, MUC1 expression on pancreatic ducts is also abnormal leading to inflammation and pancreatitis.
Methods
We used MUC1+/IL-10-/- mice that develop IBD. We imaged abnormal MUC1 expression in these mice by adoptively transferring T cells from T cell receptor transgenic mice specific for abnormal MUC1. Cells were labeled with a novel perfluorocarbon tracer reagent and quantified and visualized in vivo using high-throughput 19F NMR spectroscopy and MRI.
Results
MUC1-specific T cells migrated to the colon in mice with IBD, and also to the pancreas. Immunohistochemistry confirmed increased expression on the pancreatic ducts of the abnormal MUC1 seen in the colon and the presence of cellular infiltrate.
Conclusions
Migration of MUC1-specific T cells to the colon and the pancreas in diseased mice suggests that pancreatitis is an extraintestinal site of IBD, characterized by pro-inflammatory abnormal expression of MUC1. Therapies directed against abnormal MUC1 have the potential of targeting the disease in both sites.
Keywords: Inflammatory bowel disease MUC1, Pancreatitis, MRI, cell tracking
Introduction
In the US, over 1 million individuals have inflammatory bowel disease (IBD) with 30,000 new cases being diagnosed every year.1 Crohn's disease and ulcerative colitis, the two main entities of IBD, are systemic inflammatory diseases of unknown etiology and primarily involving the gastrointestinal tract.2 Multiple secondary problems, termed extraintestinal manifestations (EIM), develop in 20% to 40% of IBD patients and can involve almost every organ system.3,4 In particular IBD patients have an elevated risk of developing pancreatitis as well as pancreatic insufficiency 5,6. However, pancreatitis and pancreatic insufficiency have not been categorized as true EIM, because of the potential that they could also be side effects of drugs used in IBD therapy, such as 5-aminosalicyclic acid (5-ASA), sulfasalazine, azathioprine or its metabolite 6-mercaptopurine.7
MUC1 mucin is a large transmembrane glycoprotein that is expressed on the apical surface of ductal epithelial cells in many organs. The peptide backbone of MUC1 is dominated by a variable number of tandem repeats (VNTR) region composed on an average of 80-200 repeats that are 20 amino acids long.8 On healthy epithelia, the MUC1 VNTR region is heavily glycosylated with long branched O-linked carbohydrates. In contrast, MUC1 is highly overexpressed and its tandem repeats profoundly hypoglycosylated on the majority of human adenocarcinomas and their precursor lesions.8-11 We and others have previously shown that this “tumor form” of MUC1 is also present on inflamed colonic epithelium in human IBD.12,13 In addition, we confirmed in a mouse model that this form of MUC1 has an enhancing effect on IBD development, degree of inflammation, and progression to colon cancer.12 This is due to the known ability of abnormally expressed and hypoglycosylated MUC1 to attract cells of the innate immune system and promote acute inflammation that, if unresolved, over time develops into chronic inflammation.14-17 Furthermore, overexpression of abnormal MUC1 has been reported to have oncogenic effects.18-20
Our overall goal is to design MUC1 specific immunotherapy that would target cells expressing abnormal MUC1, removing them from the colon early in disease, and thus preventing the pro-IBD and pro-tumor effects of this molecule. The aim of the present study was to investigate whether or not T cells specific for the abnormal form of MUC1 would migrate preferentially to the IBD affected colon, and in addition, to other unknown sites of inflammation. We also wanted to determine whether their migration could be quantified and imaged.
We used MUC1 peptide-specific T cells (VFT) from a newly developed T cell receptor (TCR)-transgenic mouse (Ryan et al. submitted for publication), in which both CD4+ and CD8+ T cells are specific for the abnormal form of MUC1. The VFT T cells were labeled with a novel perfluorocarbon reagent (perfluoropolyether, PFPE) ex vivo 21-23 and adoptively transferred into mice with and without MUC1+ IBD. The biodistribution of labeled VFT T cells was accurately quantified using 19F NMR spectroscopy of intact organs and also visualized using in vivo 19F MRI. As expected, the injected cells migrated in a MUC1 specific manner to the colon in mice with MUC1+ IBD. The MUC1-specific cells did not migrate to the colon in healthy control mice. Interestingly, a large number of MUC1-specific T cells migrated also to the pancreas in mice with MUC1+ IBD. This suggested abnormal MUC1 expression at an extraintestinal site and we confirmed this finding using immunochemistry. Our findings therefore indicate that in IBD, changes in MUC1 expression seen in the colno occur also in the pancreas and cellular infiltration in response to those changes, rather than therapy, may be the cause of IBD associated pancreatitis.
Materials and Methods
Labeling T cells
T cells were isolated from spleen, thymus and lymph nodes of VFT mice using a pan T cell isolation kit (Miltenyi Biotec, CA) following the manufacturer's instructions. Cells were resuspended at 1-2×106 cells/mL in 10 mL of 20% FBS media and labeled by simple co-incubation with the PFPE reagent, as previously described.22 After three hour incubation at 37 °C, the cells were spun down at 300 g and washed twice with serum free media. Cells were resuspended in Hanks' Balanced Salt Solution (HBSS) prior to adoptive transfer. A portion of the cell suspension was used for cell number estimates by Cell Titer Glo (Promega, Madison, WI).
19F NMR calibration of labeled cells
In order to calibrate the mean 19F content present in cells after T cell labeling (above), we performed quantitative 19F NMR measurements in lysed cell pellets. A known number of labeled cells (∼106) were spun down, resuspended in 0.2 mL of Igepal solution in 1× HBSS (1% v/v) and incubated at room temperature for 20 minutes to lyse the cells. The labeled cell lysate (0.2 mL) was mixed with 100 μl of a calibrated 19F reference solution, trifluoroacetic acid (TFA) at 0.2 % v/v in PBS, and placed in a 5 mm NMR borosilicate tube. The 19F NMR measurements were performed using a Bruker AVANCE 500 MHz spectrometer (Bruker BioSpin, Inc., Billerica, MA). The average fluorine content per cell was calculated from the ratio of the integrated areas of the TFA and PFPE 19F spectra, normalized to the total cell number in the lysate.
Adoptive transfer of T cells
PFPE labeled T cells (15-20×106) suspended in HBSS were injected i.p into recipient mice under aseptic conditions. Mice were sacrificed at 72 hours post-transfer to harvest spleen, liver, pancreas, lung, thymus, colon, small intestine, inguinal lymph nodes, mesenteric lymph nodes and kidney. The organs were fixed in 4% paraformaldehyde and used for NMR spectroscopy.
NMR spectroscopy of excised organs
The 19F NMR measurements were performed on intact, excised organs to determine apparent T cell content using similar methods as above. The organs were weighed using a microbalance before sample preparation. A sealed capillary tube containing 5 μl of 2% TFA was place inside the NMR tube alongside the organ as a 19F reference. The density of apparent T cells tracking to a particular organ was calculated using the tissue weight and by dividing the number of 19F per organ by the amount of 19F per cell. The total number of apparent T cells per organ was then calculated.
MRI
Mice were anesthetized with an IP ketamine/xylazine cocktail, intubated and connected to a mechanical ventilator (150 strokes/minute, 300 μl/stroke) delivering an O2 / N2O mixture (70% / 30%), and mouse body temperature was maintained at 37 °C. The MRI data were acquired using an 11.7 Tesla Bruker micro-imaging system. Mice were imaged in a volume coil that can be tuned to either 19F or 1H in situ. Image acquisitions were gated to breathing to minimize motional artifacts. In a given imaging session, images were acquired for both the 19F and 1H with the same field of view and slice orientation. A conventional 1H image was used to provide an anatomical background for the 19F image. The 1H image images were obtained using a spin-echo sequence (parameters: TR/TE = 1200/15 ms, 512×256 matrix, 2 averages) and 19F images were obtained using a fast spin-echo sequence (parameters: TR/TE = 1500/11.5 ms, RARE factor = 8, 64×32 matrix, 512 averages). The co-registered 1H and 19F were fused, and the 19F was rendered in pseudo-color.
Histology and Immunohistochemistry
Tissues were fixed in 5% buffered formalin, embedded in paraffin, and stained with hematoxylin and eosin (H&E). Staining for MUC1 was performed using anti-MUC1-specific antibodies VU-3C6 (recognize all forms of MUC1 by binding to the epitope PDTRPAP in the VNTR region in a glycosylation-independent manner) and VU-4H5 (recognizes the epitope APDTRPAP in the VNTR region of hypoglycosylated tumor form of MUC1). Both antibodies were purified by us from hybridomas VU-3C6 and VU-4H5 kindly provided by Dr J. Hilgers (Vrije Universiteit, Amsterdam, The Netherlands). Briefly, 5 μm thick paraffin sections on glass slides were deparaffinized by baking overnight at 59 °C and nonspecific binding blocked with 2% normal goat serum and 5% BSA for 30 minutes. The sections were incubated with primary antibodies diluted with 1% BSA for 1 hour in a humidified chamber at room temperature. Following three washes, sections were incubated for 30 minutes in goat anti-mouse secondary antibody. Optimal concentrations of primary and secondary antibodies were determined empirically. Amplification of the signal was done by the avidin-biotin-peroxidase complex (ABC) method with a commercial kit (Vectastain ABC kit, Vector laboratories, Berlingame, CA). Color was developed using a 3,3′diaminobenzidine (DAB) kit (BD Pharmingen, San Jose, CA).
Immunofluorescence Staining
Frozen sections dried on glass slides were hydrated with 1× PBS and cell membranes permeablized for 15 minutes with 0.1% Triton X. Sections were washed with 0.5% BSA in 1× PBS, blocked for 45 minutes with 2% BSA in 1× PBS, and incubated for 1 hour with primary antibody, FITC conjugated anti-human CD227 (BD Pharmingen, San Jose, CA). This antibody recognizes the core peptide of MUC1 in a glycosylation independent manner. Sections were incubated for 1 hour with secondary antibody, cy5 Phalloidin, washed with 0.5% BSA in 1× PBS, and incubated with Hoechst nuclear stain for 30 seconds.
Results
Migration of adoptively transferred MUC1-specific T cells can be quantitated and imaged in vivo revealing expected and unexpected disease sites associated with IBD
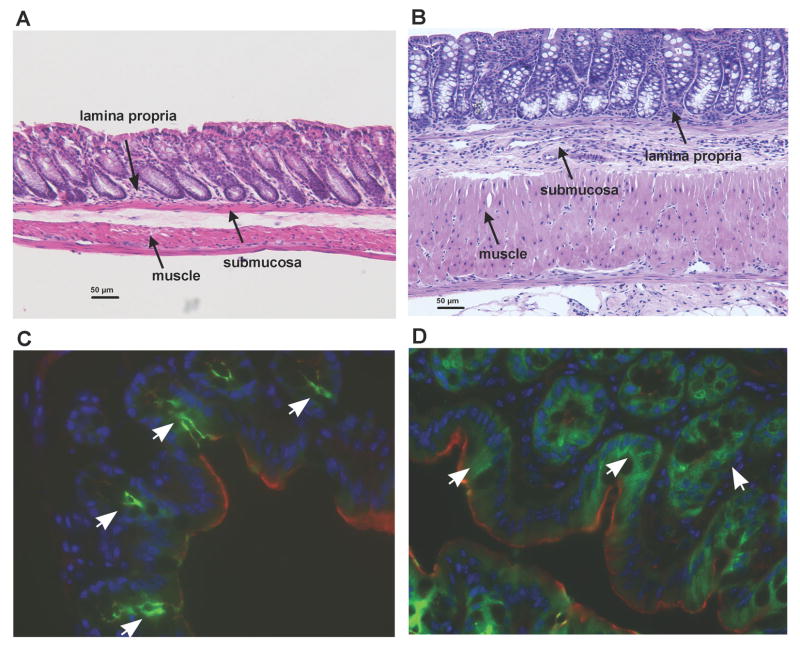
T cells were isolated from VFT mice having a TCR specific for an MHC Class II-restricted MUC1 epitope, a 12 amino acid peptide GVTSAPDTRPAP, derived from the tandem repeat region of the unglycosylated form of MUC1. This region was previously shown to be abnormally expressed on the tumor form of MUC1 and also MUC1 in IBD affected colonic epithelium. T cells were PFPE-labeled and injected into several different groups of mice. IL10-/- mice, raised in a specific pathogen-free environment, develop spontaneous chronic colitis with similarities to human IBD.24 MUC1 Tg mice express human MUC1 under its endogenous promoter and thus in the same spatial and tissue distribution seen in humans.25 These mice do not develop spontaneous IBD. When these two strains are crossed, the progeny that is MUC1+/IL10-/- develops IBD characterized by high level of hypoglycosylated MUC1 expression at sites of severe inflammation.12 Figure 1 shows healthy colon of MUC1 Tg mice (A and C) and IBD affected colons in MUC1+/IL10-/- mice (B and D). In comparison to the normal colon of the MUC1 Tg mouse (Fig 1A) the colon from MUC1+/IL10 -/- (Fig. 1B) mouse reveals pronounced thickening of the submucosa and muscle layers, and inflammatory infiltrate in the lamina propria and submucosa. Consistent with healthy colonic epithelium (Fig. 1C), MUC1 (green) is expressed apically at low levels, while in the diseased colon (Fig. 1D) MUC1 is expressed at high levels and can be visualized over the entire plasma membrane and in the cytoplasm of colonic epithelia lesions (Fig. 1D).
Figure 1. Inflammation and MUC1 expression in MUC1 Tg mice compared to MUC1+/IL10-/- mice.
Hematoxylin and eosin (H&E) stained mouse colon sections in (A) MUC1 Tg mice and (B) MUC1+/IL10-/- mice. Scale bar, 50 μm. Immunofluorecence stained mouse colons in (C) MUC1 Tg mice and (D) MUC1+/IL10-/- mice. Colon sections are stained with antibodies against MUC1 (green), nuclear stain (blue), and Phalloidin (red) for visualization of colonic tissue architecture.
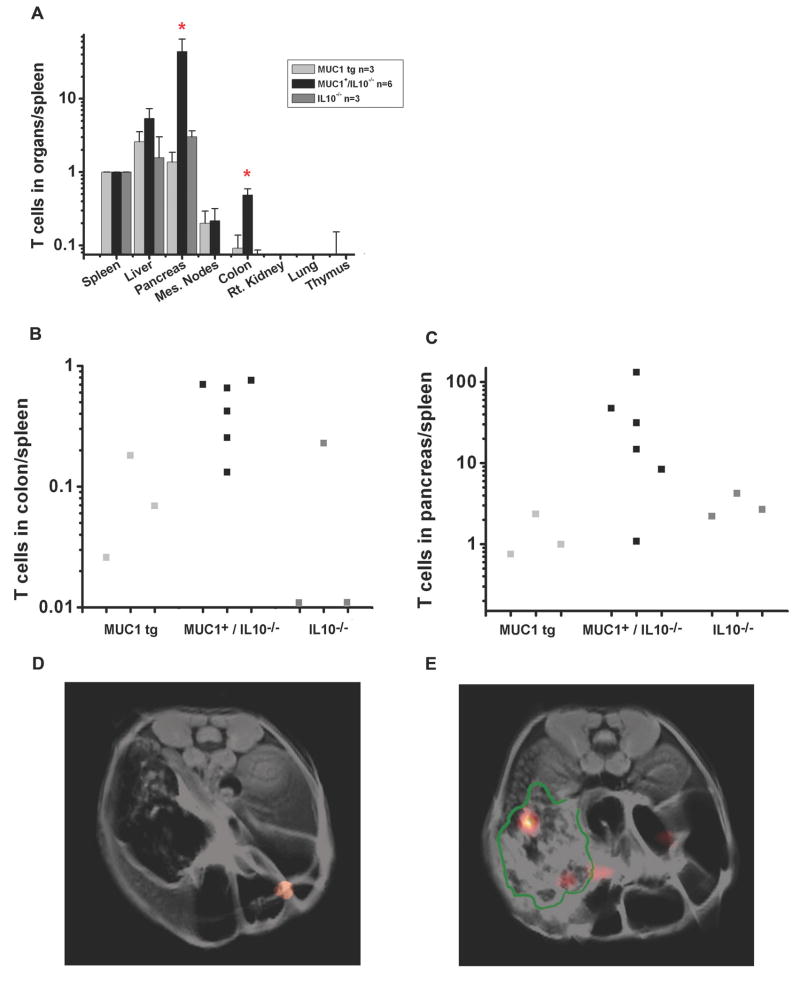
Three days after T cell transfer, the recipient mice were sacrificed and organs excised for analysis by 19F NMR spectroscopy. Prior knowledge of the amount of 19F per cell enabled us to calculate the number of injected T cells that had migrated to each organ. For each animal, the organ cell numbers were normalized to the number of cells measured in the spleen to account for individual variance in the transferred cell number that ended up in circulation. Figure 2A shows a log-scale histogram of labeled VFT T cells migrating into the spleen, liver, pancreas, mesenteric lymph nodes, colon, right kidney, lung, and thymus. As expected, we found a statistically higher number (p < 0.05, Wilcoxon rank-sum test) of labeled VFT T cells in the colon of MUC1+/IL10-/- mice with IBD compared to healthy MUC1 Tg and diseased IL10-/- mice but MUC1 negative (Fig. 2A).
Figure 2. MUC1-specific TCR-transgenic T cells (VFT cells) preferentially migrate to the colon and pancreas in MUC1+/IL10-/- mice.
(A) 19F NMR spectroscopy of the excised organs 72 hours after adoptive transfer of labeled VFT T cells into spleen, liver, pancreas, mesenteric lymph nodes, colon, kidney, lung, and thymus of MUC1 Tg mice, MUC1+/IL10-/- mice, and IL10-/- mice. Cell trafficking is normalized to the spleen (p<0.05, Wilcoxon rank sum test). (B) Cell trafficking to the colon is normalized to the spleen in individual mice. (C) Cell trafficking to the pancreas is normalized to the spleen in individual mice. (D and E) 1H/19F MRI composite image through the abdomen of MUC1+/IL10-/- mice 72 hours after adoptive transfer. 1H image is rendered in grey scale and 19F image is rendered in pseudo-color. The border of the pancreas is delineated in green.
While migration to the colon was not unexpected for T cells with T cell receptors for the antigen present at that site, the interesting result was that a large number of labeled VFT T cells migrated to the pancreas of MUC1+/IL10-/- mice with IBD (p < 0.05, Wilcoxon rank-sum test). This was not seen in either healthy MUC1 Tg mice or diseased IL10-/- mice (Fig. 2A). Migration of transferred cells to the liver and mesenteric lymph nodes was the same in all groups. Figures 2B and 2C show the quantification of labeled T cells in the colon and the pancreas relative to the spleen from individual mice. Representative axial MRI scans through the abdomen of the MUC1+/IL10-/- mice show 19F ‘hot-spots’, indicative of significant T cell accumulation in the colon (Fig. 2D) and the pancreas (Fig. 2E) of the MUC1+/IL10-/- mice. The boundaries of pancreas are delineated in green.
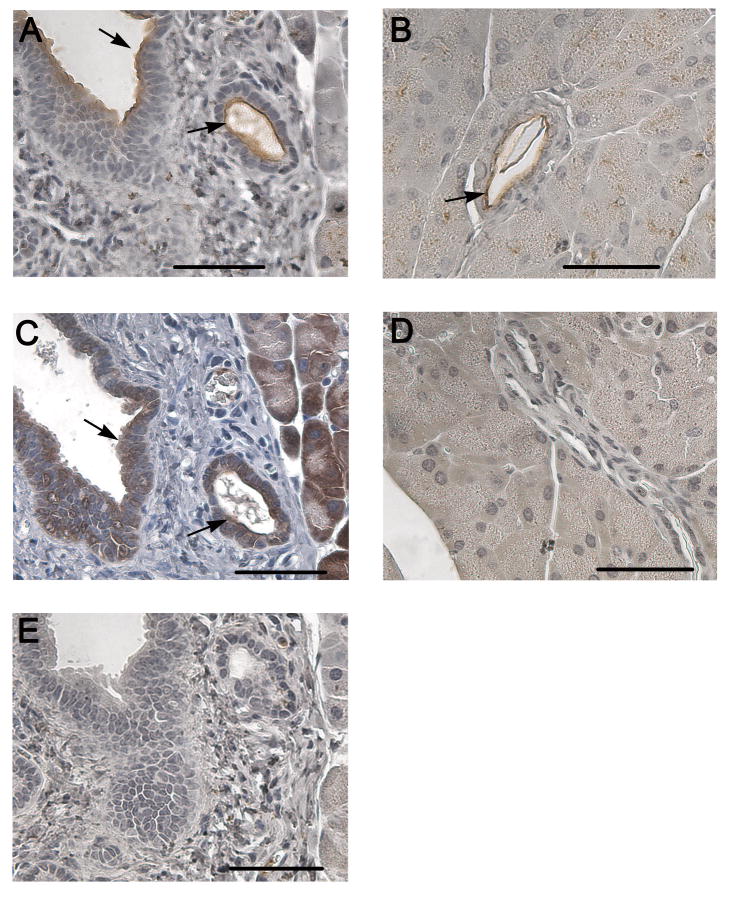
The pancreata from MUC1+/IL10-/- mice showed a pale white appearance at the time of sacrifice compared to the healthy MUC1 Tg mice and diseased IL10-/- mice. Subsequent histology sections stained with H&E showed increased infiltration of inflammatory cells in the pancreatic duct and acini (data not shown). Using immunohistochemistry we compared expression levels of normal and hypoglycosylated MUC1 in the pancreas of MUC1+/IL10-/- (Fig. 3A and C) and MUC1 Tg (Fig. 3B and D) mice. Both MUC1+/IL10-/- (Fig. 3A) and MUC1 Tg (Fig. 3B) mice exhibited the typical apical staining pattern of normal MUC1 expression in the pancreatic ducts, seen as dark brown staining around the edge of the duct facing the lumen (indicated by arrows).9 However, the aberrant hypoglycosylated form of MUC1 expressed in the pancreas of MUC1+/IL10-/- mice (Fig. 3C) is seen as diffuse brown staining around the nuclei of cuboidal epithelial cells lining the duct (indicated by arrow). The diffuse staining pattern is consistent with the loss of apical MUC1 expression in the ductal epithelial cells and is very different than in healthy MUC Tg mice (Fig. 3D). The isotype control is negative for MUC1 staining (Fig. 3E).
Figure 3. MUC1 expression in the pancreatic ducts of MUC1 Tg mice compared to MUC1+/IL10-/- mice.
Immunostaining (hematoxylin counterstained) of mouse pancreas in MUC1+/IL10-/- mice (A) and MUC1 Tg mice (B) with anti-MUC1 antibody (3C6). Immunostaining (hematoxylin counterstained) of mouse pancreas in MUC1+/IL10-/- mice (C) and MUC1 Tg mice (D) with anti-MUC1 antibody (4H5). Isotype control is negative for MUC1 staining (E). Scale bar, 50 μm.
Discussion
IBD patients often present with EIM that can involve different organ system, thus adding to the morbidity and mortality in a significant percentage of these patients. The etiology of EIM is not well understood, however, systemic inflammatory mediators and shared autoantigens are among proposed mechanisms.26 Recent evidence from clinical studies and results from a mouse model of chemical-induced colitis suggest that the association between pancreatitis and IBD is more than coincidental and difficult to ascribe to drug-induced pancreatic damage.27,28 Our findings indicate that inflammation in response to, or driven by, the expression of the abnormal form of MUC1 on pancreatic ductal epithelia may be a critical factor implicating pancreas as a true EIM of IBD.
The use of 19F fluorocarbon labels for tracking T cells and other cell types is an emerging technique that can accelerate the laborious task of accurate inflammation quantification in tissue specimens. This approach uses 19F NMR of intact tissues, thereby minimizing the time-intensive tissue preparation and analysis associated with traditional microscopy or FACs analysis. Additionally, 19F NMR is a bulk measurement, thus eliminating the potential for histological sampling bias and error, resulting in smaller, higher quality data sets. NMR analysis of excised tissues is non-destructive, and the same tissues may undergo conventional histological or biochemical analysis following NMR. As we demonstrate, in vivo 19F MRI detection is also possible in tissue regions containing concentrated deposits of labeled cells. Generally, the 19F MRI sensitivity enables one to detect 104 to 105 labeled cells per image voxel.21,23 Using 19F NMR in tissue samples, the minimum cell number detection limit is approximately an order of magnitude smaller.
In the future, in vivo imaging strategies will likely play an important role in preclinical studies and perhaps in clinical translation of live cell therapy against IBD and other diseases. A common need for cellular therapeutics is a non-invasive way to image the behavior and movement of cells post-injection. Non-invasive cell tracking using PFPE labeling and 19F MRI is potentially a powerful tool for providing critical feedback regarding the optimal delivery routes and therapeutic doses for individual patients.
Due to its apical topology in glandular and ductal epithelia, MUC1 is largely inaccessible to the immune system under healthy conditions. In contrast, MUC1 is quantitatively and qualitatively altered on the majority of human adenocarcinomas and their precursor lesions.8,10,11 This includes a large increase in the amount of MUC1 expressed on cells as well as a significant decrease in glycosylation revealing immunogenic peptide and glycopeptide neoepitopes.29,30 MUC1 expression can be modulated by inflammatory cytokines such as interferon γ (IFN-γ) and tumor necrosis factor (TNF-α).31,32 In addition, we have previously shown that the presence of MUC1 has a profound effect on the time of IBD occurrence, degree of inflammation, and progression to colon cancer in a mouse model of IBD.12 We propose that regardless of the etiology of IBD, which is multifactorial, there ensues an inflammatory cascade that includes increased TNF-α production and subsequent increase in MUC1 expression that may serve initially to protect the gut epithelium. However, the repetitive cycles of inflammation, damage, and regeneration result in the loss of cellular polarity and increased expression of the hypoglycosylated form of MUC1. This aberrant form of MUC1 has been shown to have chemotactic properties for innate inflammatory cells and oncogenic properties in the diseased colonocytes. This process apparently occurs not only in the colon but also in other sites such as the pancreas, where normal MUC1 expression converts to the overexpression of the abnormal form. It is possible that the site of the original inflammation, such as the colon in IBD, may produce cytokines and other soluble mediators that can act systemically and affect MUC1 expression at distal sites. Some of the candidates would be soluble TNF and IL-6. It remains to be seen if therapies directed against TNF, might be effective in part through lowering expression of the pro-inflammatory MUC1.
Acknowledgments
Financial Support: This study was funded in part by NIH grants R01-EB003453, P01-HD047675, P41-EB001977 (E.T.A.), Cancer Research and Prevention Foundation (P.L.B.), and RO1 56103 (O.J.F.).
Contributor Information
Deepak K. Kadayakkara, Department of Biological Sciences, Carnegie Mellon University, Pittsburgh, PA.
Pamela L. Beatty, Department of Immunology, University of Pittsburgh School of Medicine, Pittsburgh, PA.
Michael S. Turner, Department of Immunology, University of Pittsburgh School of Medicine, Pittsburgh, PA.
Jelena M. Janjic, Department of Biological Sciences, Carnegie Mellon University, Pittsburgh, PA.
Eric T. Ahrens, Department of Biological Sciences, Carnegie Mellon University, Pittsburgh, PA.
Olivera J. Finn, Department of Immunology, University of Pittsburgh School of Medicine, Pittsburgh, PA.
References
- 1.McFarland LV. State-of-the-art of irritable bowel syndrome and inflammatory bowel disease research in 2008. World J Gastroenterol. 2008;14(17):2625–9. doi: 10.3748/wjg.14.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448(7152):427–34. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 3.Rothfuss KS, Stange EF, Herrlinger KR. Extraintestinal manifestations and complications in inflammatory bowel diseases. World J Gastroenterol. 2006;12(30):4819–31. doi: 10.3748/wjg.v12.i30.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su CG, Judge TA, Lichtenstein GR. Extraintestinal manifestations of inflammatory bowel disease. Gastroenterol Clin North Am. 2002;31(1):307–27. doi: 10.1016/s0889-8553(01)00019-x. [DOI] [PubMed] [Google Scholar]
- 5.Gurian LE, Keeffe EB. Pancreatic insufficiency associated with ulcerative colitis and pericholangitis. Gastroenterology. 1982;82(3):581–5. [PubMed] [Google Scholar]
- 6.Herrlinger KR, Stange EF. The pancreas and inflammatory bowel diseases. Int J Pancreatol. 2000;27(3):171–9. doi: 10.1385/IJGC:27:3:171. [DOI] [PubMed] [Google Scholar]
- 7.Bermejo F, Lopez-Sanroman A, Taxonera C, et al. Acute pancreatitis in inflammatory bowel disease, with special reference to azathioprine-induced pancreatitis. Aliment Pharmacol Ther. 2008;28(5):623–8. doi: 10.1111/j.1365-2036.2008.03746.x. [DOI] [PubMed] [Google Scholar]
- 8.Vlad AM, Kettel JC, Alajez NM, Carlos CA, Finn OJ. MUC1 immunobiology: from discovery to clinical applications. Adv Immunol. 2004;82:249–93. doi: 10.1016/S0065-2776(04)82006-6. [DOI] [PubMed] [Google Scholar]
- 9.Adsay NV, Merati K, Andea A, et al. The dichotomy in the preinvasive neoplasia to invasive carcinoma sequence in the pancreas: differential expression of MUC1 and MUC2 supports the existence of two separate pathways of carcinogenesis. Mod Pathol. 2002;15(10):1087–95. doi: 10.1097/01.MP.0000028647.98725.8B. [DOI] [PubMed] [Google Scholar]
- 10.Ho SB, Ewing SL, Montgomery CK, Kim YS. Altered mucin core peptide immunoreactivity in the colon polyp-carcinoma sequence. Oncol Res. 1996;8(2):53–61. [PubMed] [Google Scholar]
- 11.Reis CA, David L, Correa P, et al. Intestinal metaplasia of human stomach displays distinct patterns of mucin (MUC1, MUC2, MUC5AC, and MUC6) expression. Cancer Res. 1999;59(5):1003–7. [PubMed] [Google Scholar]
- 12.Beatty PL, Plevy SE, Sepulveda AR, Finn OJ. Cutting edge: transgenic expression of human MUC1 in IL-10-/- mice accelerates inflammatory bowel disease and progression to colon cancer. J Immunol. 2007;179(2):735–9. doi: 10.4049/jimmunol.179.2.735. [DOI] [PubMed] [Google Scholar]
- 13.Longman RJ, Poulsom R, Corfield AP, Warren BF, Wright NA, Thomas MG. Alterations in the composition of the supramucosal defense barrier in relation to disease severity of ulcerative colitis. J Histochem Cytochem. 2006;54(12):1335–48. doi: 10.1369/jhc.5A6904.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aarnoudse CA, Garcia Vallejo JJ, Saeland E, van Kooyk Y. Recognition of tumor glycans by antigen-presenting cells. Curr Opin Immunol. 2006;18(1):105–11. doi: 10.1016/j.coi.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Carlos CA, Dong HF, Howard OM, Oppenheim JJ, Hanisch FG, Finn OJ. Human tumor antigen MUC1 is chemotactic for immature dendritic cells and elicits maturation but does not promote Th1 type immunity. J Immunol. 2005;175(3):1628–35. doi: 10.4049/jimmunol.175.3.1628. [DOI] [PubMed] [Google Scholar]
- 16.Hiltbold EM, Vlad AM, Ciborowski P, Watkins SC, Finn OJ. The mechanism of unresponsiveness to circulating tumor antigen MUC1 is a block in intracellular sorting and processing by dendritic cells. J Immunol. 2000;165(7):3730–41. doi: 10.4049/jimmunol.165.7.3730. [DOI] [PubMed] [Google Scholar]
- 17.Monti P, Leone BE, Zerbi A, et al. Tumor-derived MUC1 mucins interact with differentiating monocytes and induce IL-10highIL-12low regulatory dendritic cell. J Immunol. 2004;172(12):7341–9. doi: 10.4049/jimmunol.172.12.7341. [DOI] [PubMed] [Google Scholar]
- 18.Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein blocks glycogen synthase kinase 3beta-mediated phosphorylation and degradation of beta-catenin. Cancer Res. 2005;65(22):10413–22. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 19.Raina D, Kharbanda S, Kufe D. The MUC1 oncoprotein activates the anti-apoptotic phosphoinositide 3-kinase/Akt and Bcl-xL pathways in rat 3Y1 fibroblasts. J Biol Chem. 2004;279(20):20607–12. doi: 10.1074/jbc.M310538200. [DOI] [PubMed] [Google Scholar]
- 20.Schroeder JA, Adriance MC, Thompson MC, Camenisch TD, Gendler SJ. MUC1 alters beta-catenin-dependent tumor formation and promotes cellular invasion. Oncogene. 2003;22(9):1324–32. doi: 10.1038/sj.onc.1206291. [DOI] [PubMed] [Google Scholar]
- 21.Ahrens ET, Flores R, Xu H, Morel PA. In vivo imaging platform for tracking immunotherapeutic cells. Nat Biotechnol. 2005;23(8):983–7. doi: 10.1038/nbt1121. [DOI] [PubMed] [Google Scholar]
- 22.Janjic JM, Srinivas M, Kadayakkara DK, Ahrens ET. Self-delivering nanoemulsions for dual fluorine-19 MRI and fluorescence detection. J Am Chem Soc. 2008;130(9):2832–41. doi: 10.1021/ja077388j. [DOI] [PubMed] [Google Scholar]
- 23.Srinivas M, Morel PA, Ernst LA, Laidlaw DH, Ahrens ET. Fluorine-19 MRI for visualization and quantification of cell migration in a diabetes model. Magn Reson Med. 2007;58(4):725–34. doi: 10.1002/mrm.21352. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75(2):263–74. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 25.Rowse GJ, Tempero RM, VanLith ML, Hollingsworth MA, Gendler SJ. Tolerance and immunity to MUC1 in a human MUC1 transgenic murine model. Cancer Res. 1998;58(2):315–21. [PubMed] [Google Scholar]
- 26.Das KM. Relationship of extraintestinal involvements in inflammatory bowel disease: new insights into autoimmune pathogenesis. Dig Dis Sci. 1999;44(1):1–13. doi: 10.1023/a:1026629528233. [DOI] [PubMed] [Google Scholar]
- 27.Barthet M, Dubucquoy L, Garcia S, et al. Pancreatic changes in TNBS-induced colitis in mice. Gastroenterol Clin Biol. 2003;27(10):895–900. [PubMed] [Google Scholar]
- 28.Barthet M, Lesavre N, Desplats S, et al. Frequency and characteristics of pancreatitis in patients with inflammatory bowel disease. Pancreatology. 2006;6(5):464–71. doi: 10.1159/000094564. [DOI] [PubMed] [Google Scholar]
- 29.Vlad AM, Finn OJ. Glycoprotein tumor antigens for immunotherapy of breast cancer. Breast Dis. 2004;20:73–9. doi: 10.3233/bd-2004-20109. [DOI] [PubMed] [Google Scholar]
- 30.Vlad AM, Muller S, Cudic M, et al. Complex carbohydrates are not removed during processing of glycoproteins by dendritic cells: processing of tumor antigen MUC1 glycopeptides for presentation to major histocompatibility complex class II-restricted T cells. J Exp Med. 2002;196(11):1435–46. doi: 10.1084/jem.20020493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagow EL, Carson DD. Synergistic stimulation of MUC1 expression in normal breast epithelia and breast cancer cells by interferon-gamma and tumor necrosis factor-alpha. J Cell Biochem. 2002;86(4):759–72. doi: 10.1002/jcb.10261. [DOI] [PubMed] [Google Scholar]
- 32.Shirasaki H, Kanaizumi E, Watanabe K, et al. Tumor necrosis factor increases MUC1 mRNA in cultured human nasal epithelial cells. Acta Otolaryngol. 2003;123(4):524–31. doi: 10.1080/00016480310001268. [DOI] [PubMed] [Google Scholar]