Abstract
We recently demonstrated that the mechanism of processing of an HLA-A*0201-restricted peptide epitope, Tyr369(D), derived from the membrane protein tyrosinase, involves retrotranslocation of glycosylated molecules from the endoplasmic reticulum to the cytosol; removal of an N-linked carbohydrate from Asn371 by peptide N-glycanase; proteolysis by the proteasome and other proteases; and retransport of the resulting peptides into the endoplasmic reticulum for association with HLA-A*0201. Carbohydrate removal results in deamidation of Asn371 to Asp. The Asn-containing homolog of this peptide, Tyr369(N), is not presented by tyrosinase-expressing cells, and this has been presumed to be due to quantitative glycosylation of Asn371. While examining cytosolic intermediates that accumulated in human melanoma cells treated with proteasome inhibitors, we were surprised to find both molecules that had been deglycosylated by peptide N-glycanase and a large number of molecules that had not been previously glycosylated. The failure of Tyr369(N) to be processed and presented from these latter molecules may be partially due to a process of deamidation independent of glycosylation. However, we also established that proteasomes degrade tyrosinase molecules that are still glycosylated, giving rise to a set of discrete intermediates that are not observed when unglycosylated molecules are degraded. We propose that Tyr369(N) fails to be presented because unglycosylated tyrosinase is degraded rapidly and relatively non-selectively. In contrast, glycosylation alters the selectivity of tyrosinase processing by the proteasome, enhancing the production or survival of Tyr369(D).
Keywords: antigen presentation/processing, antigen/peptide/epitope, human, tumor immunity
Introduction
Tyrosinase is a membrane-associated glycoprotein that is synthesized and folds in the endoplasmic reticulum (ER)5, and is subsequently transported via the trans-Golgi network to melanosomes in melanoma cells and melanocytes (1, 2). Interest in its degradation stems from the fact that tyrosinase derived peptides associate with class I molecules of the MHC, and act as antigens for the immunological recognition of melanomas (3-6). Previous work established that tyrosinase was degraded via a proteasome dependent pathway (7-9) that led to peptides presented by class I MHC molecules (8). The level of tyrosinase epitope presentation is determined in part by the level of unfolded/misfolded full-length tyrosinase molecules (10). We recently demonstrated that the processing of an HLA-A*0201-restricted peptide epitope derived from tyrosinase, Tyr369(D), involves retrotranslocation of glycosylated molecules from the endoplasmic reticulum to the cytosol; removal of an N-linked carbohydrate from Asn371 by peptide N-glycanase; proteolysis by the proteasome and other proteases; and retransport of the resulting peptides into the endoplasmic reticulum for association with HLA-A*0201 (11).
An unusual aspect of this system is the observation that one of the tyrosinase derived epitopes presented by HLA-A*0201 has undergone a deamidation. Thus, HLA-A*0201+ melanoma cells present the peptide YMDGTMSQV (Tyr369(D)), with an Asp residue at the 3rd position. However, the genetically encoded peptide YMNGTMSQV (Tyr369(N)), which contains Asn at this same position, is undetectable using either mass-spectrometry or specific T cells (4). The absence of Tyr369(N) on tyrosinase-expressing cells has been presumed to be due to quantitative glycosylation of Asn371. However, in all of previous studies where tyrosinase molecules were detected in the cytosol after inhibition of proteasome activity, their glycosylation status was not defined. We recently demonstrated that deamidation of Tyr369(D) is partially due to glycosylation of Asn371 in the ER and subsequent deglycosylation by peptide N-glycanase (PNGase) in the cytosol (11). However, our results also suggested the existence of a second pathway for deamidation of unglycosylated Asn371. Therefore, the absence of the Tyr369(N) expression on melanoma cells might reflect a lack of unglycosylated molecules that could give rise to this epitope, or a processing of unglycosylated tyrosinase molecules that prevents presentation of Tyr369(N).
The present work sought to provide greater insight into the forms of tyrosinase expressed in cells, their ability to be degraded by the proteasome, and their ability to function as epitope precursors. We also addressed whether deglycosylation is required for processing by the proteasome of glycosylated molecules and presentation of an MHC class I restricted epitope derived from tyrosinase. Our work supports a model in which N-glycosylation influences the processing of tyrosinase by the proteasome and determines the availability of peptides for presentation by MHC class I molecules.
Materials and Methods
Tyrosinase gene constructs and transfectants
Expression constructs containing the genes encoding Wild Type (WT) tyrosinase and R402Q tyrosinase in which R has been substituted to Q at position 402 in the p3Xflag –CMV-14 vector (Sigma-Aldrich) were a kind gift from R. Halaban (Yale University) (12). Mutations encoding T373V and N371D were introduced into p3XflagWT or both p3XflagWT and p3XflagR402Q tyrosinase, respectively, using the QuickChange XL site-directed mutagenesis kit (Stratagene). The melanoma cell line DM331 (tyrosinaseneg, HLA-A*0201+) transfected with WT human tyrosinase tagged with a flag epitope at the C-terminus has been previously described (10). DM331 was also transfected with plasmids encoding R402Q, R402Q N371D, or T373V tyrosinase using the Nucleofectamine kit V (Amaxa). These bulk transfectant lines were selected in 600 μg/ml G418 (Invitrogen) and used from 5 days to one month after transfection.
SDS-PAGE analyses, immunoblotting and glycosylation status
Melanoma cells were incubated in medium containing 50 μM z-vad-fmk (Promega) where indicated. The proteasome inhibitor epoxomicin (Calbiochem) was added 30min later where indicated. In some experiments, melanoma cells expressing WT tyrosinase were transfected with siRNA oligonucleotides targeting human PNGase (13) (Dharmacon), or control non-targeting siRNA (Dharmacon) using Oligofectamine (Invitrogen) following manufacturer’s protocol and used in proteasome blocking experiment 4 days following transfection. Cells (5 × 105) were harvested in 2 mM EDTA, 20 mM Hepes, 1 mM amino ethyl benzene sulfonyl fluoride, 10 μg/ml aprotinin, 10 μM pepstatin A, 10 μg/ml leupeptin, and 100 μM iodoacetamide 4h after addition of epoxomicin, and lysed by repetitive freeze-thawing. Lysates were separated by centrifugation at 125,000 × g for 1 h at 4°C into supernatant (cytosolic) and particulate (membrane) fractions. Half of each sample was treated in vitro with 1000U recombinant PNGase F or Endoglycosidase H (New England Biolabs) according to the manufacturer’s directions. Samples were separated on 8-20% SDS gels, transferred to Immobilon-P membranes (Millipore) and blocked in 5% nonfat dried milk in PBS with 0.05% Tween 20. Blots were probed overnight with mouse anti-flag antibody M2 (Sigma-Aldrich) washed, and probed with horseradish peroxidase-conjugated anti-mouse Fab’2 fragments. In some experiments, blots were probed with rabbit antibody specific for the carboxyl-terminal peptide of human tyrosinase (CPLLMEKEDYHSLLYQSHL) (Anaspec) and HRP conjugated anti-rabbit Fab’2 fragments. The immunoblots were developed according to the Amersham ECL protocol (Amersham Biosciences). After probing for tyrosinase, the membranes were stripped and blotted again with rabbit anti-calnexin (StressGen) or mouse anti-tubulin (Sigma-Aldrich) antibodies.
Epitope recognition by T cells
Presentation of Tyr369(D) and Tyr369(N) epitopes by melanoma cells was detected using specific T cell lines generated from mice expressing a chimeric HLA-A*0201/H-2Dd MHC class I molecule (8, 11). Melanoma cells expressing tyrosinase were incubated with epoxomicin and z-vad-fmk where indicated for 30 min, stripped of MHC class I associated epitopes with 300 mM glycine HCl /1% BSA, pH 2.5 (14), and incubated with epoxomicin and z-vad-fmk where indicated for an additional 6 h. Cycloheximide was added overnight before treatment with other inhibitors. Cells were fixed with glutaraldehyde 0.05% in PBS for 1 min. Recognition by T cells was evaluated by release of IFN-γ as measured by ELISA (10) (eBiosciences).
Results
A major proteasome substrate derived from tyrosinase is found in an unglycosylated form
HLA-A*0201+ melanoma cells present the Tyr369(D) peptide epitope, but not the Tyr369(N) peptide that corresponds to the genomic sequence of tyrosinase (4). We previously showed that the conversion of N to D can occur after glycosylation of Asn371 in the ER and subsequent deglycosylation by PNGase in the cytosol (11). Thus, the lack of presentation of Tyr369(N) has been taken to indicate that the entire pool of tyrosinase molecules is glycosylated at N371. To clarify the glycosylation status of proteasome substrates derived from tyrosinase, we utilized an amelanotic melanoma, DM331, transfected to express human tyrosinase tagged with a C-terminal flag epitope. This molecule was found as a broad doublet centered at ~78-kDa in the membrane fraction of melanoma cells (Fig. 1, lane 1), and as a 61-kDa species after in vitro deglycosylation with recombinant PNGase F (Fig. 1, lane 2). Inhibition of proteasome activity with epoxomicin led to the accumulation of a new, slightly smaller tyrosinase band of 58-kDa in both the membrane and in the cytosol (Fig. 1, lane 3). The size of this band did not change when samples were digested in vitro with PNGase F, demonstrating that it was not glycosylated.
Figure 1. A major proteasome substrate derived from tyrosinase is unglycosylated.
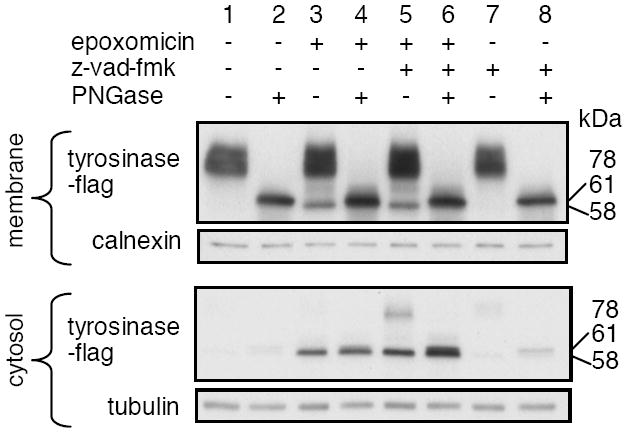
DM331 melanoma cells expressing flag-tagged WT tyrosinase were incubated in medium containing 50 μM z-vad-fmk, an inhibitor of PNGase, as indicated. Epoxomicin (2 μM) was added 30 min later as indicated. Cells were harvested 4 h later and cytosolic and membrane fractions prepared. Half of each sample was treated with PNGase in vitro, and samples were analyzed by SDS-PAGE and immunoblotting with anti-flag Ab.
To determine whether the 58-kDa band was produced from a glycosylated precursor, we used z-vad-fmk, which blocks the removal of N-glycans by PNGase in vivo (15). Incubation of cells with a combination of z-vad-fmk and epoxomicin did not lead to a reduction or loss of this band in either the membrane or cytosol, compared to cells treated with epoxomicin alone (Fig. 1, lanes 5 and 3). Treatment with z-vad-fmk plus epoxomicin also caused the accumulation of an additional 78-kDa tyrosinase band in the cytosol, which corresponds to the molecular weight of full-length glycosylated tyrosinase. After in vitro digestion with PNGase F, this band disappeared and there was a coordinate appearance of a 61-kDa band (Fig 1, lane 6). These results indicate that the 58-kDa band is an unglycosylated form of tyrosinase, while the 61-kDa band is produced by deglycosylation. Because the 61-kDa band was derived from mature glycosylated tyrosinase by in vitro deglycosylation with recombinant PNGase, and was detected using a C-terminal epitope tag, it is likely to represent deglycosylated full-length or nearly full-length tyrosinase, which has a molecular weight of 62, 610 ((16). The 58-kDa unglycosylated form appears to be slightly less than full-length, but may also migrate differently in SDS_PAGE due to the presence of Asn rather than Asp residues at the positions of the glycosylation sites. Regardless, these results show that unglycosylated molecules represent a major fraction of the proteasome substrates derived from tyrosinase.
Unglycosylated, deglycosylated and glycosylated forms of tyrosinase are proteasome substrates in both transfectants and melanoma cells expressing endogenous tyrosinase
Decreasing the translation rate using low dose of cycloheximide has been shown to increase the efficiency of tyrosinase glycosylation (17). To determine whether translation rate also affected the relative amounts of deglycosylated and unglycosylated tyrosinase-derived proteasome substrates, we examined their accumulation in the presence of low-dose cycloheximide and epoxomicin. The addition of cycloheximide decreased the amount of 78 and 58-kDa tyrosinase molecules in the cytosol, consistent with a decrease in translation rate (Fig 2A). However, there was a concomitant appearance of a band at 61-kDa, together with additional bands at 34, 47, and 52-kDa (Fig 2A, B). The 61, 52, 47 and 34-kDa bands did not shift after digestion with PNGase in vitro, showing they were not glycosylated (Fig 2B, lane 4). Conversely, when cells were treated with z-vad-fmk in addition to epoxomicin and cycloheximide, each of these bands was substantially diminished, and there were accompanying increases in bands at 78-kDa, 56-kDa, and a ladder of bands centered around a major species of 41-kDa (Fig 2B, lane 5). This pattern was reversed by treatment with PNGase, demonstrating that the cytosolic tyrosinase proteasome substrates of 34, 47, 52, and 61-kDa were each derived from corresponding glycosylated precursors.
Figure 2. Unglycosylated, deglycosylated and glycosylated forms of tyrosinase are proteasome substrates in a melanoma transfectant expressing WT flag tyrosinase.
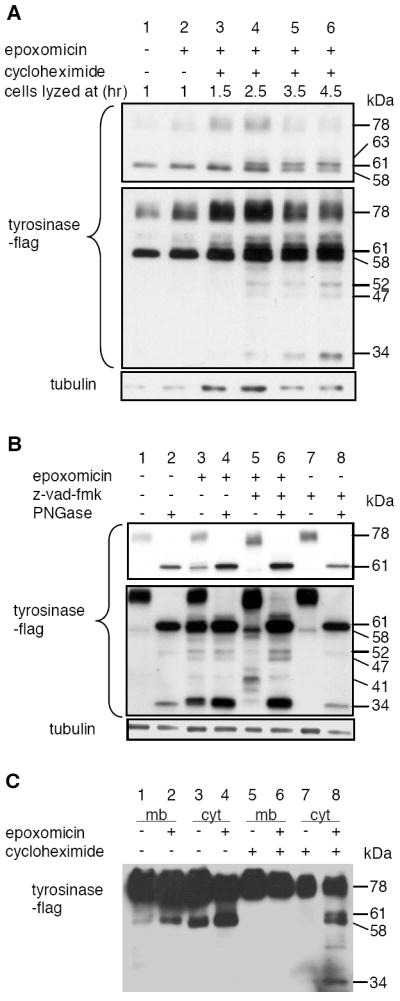
A, DM331 melanoma cells expressing flag-tagged WT tyrosinase were incubated in medium containing 2 μM epoxomicin and 1 μg/ml cycloheximide was added after 1h. Cells were harvested at the indicated times after addition of cycloheximide and cytosolic fractions were prepared. Samples were analyzed by SDS-PAGE and immunoblotting with anti-flag Ab. The amounts analyzed from cells treated with cycloheximide (lane 3-6) were 5 fold more than the amounts from untreated cells (lane 1, 2). B, DM331 melanoma cells expressing flag-tagged WT tyrosinase were incubated in medium containing 1 μg/ml cycloheximide in all samples and 50 μM z-vad-fmk as indicated. Epoxomicin (2 μM) was added 30 min later as indicated. Cytosolic fractions were prepared and analyzed as described in the legend to Fig 1. C, DM331 WT melanoma cells were incubated for 4h in medium in absence or presence of 2 μM epoxomicin and 1 μg/ml cycloheximide as indicated and described in the legend to Fig 1. Membrane (mb) and cytosolic (cyt) fractions were prepared and analyzed as described in the legend to Fig 1.
Based on their appearance in epoxomicin treated cells and their detection using the C-terminally localized flag epitope, the deglycosylated 52, 47, and 34-kDa bands represent tyrosinase forms that have been truncated at the N-terminus by the direct action of the proteasome. However, it is noteworthy that these discrete intermediates are only observed when glycosylation of tyrosinase is enhanced (Figure 2C). This suggests that glycosylation of these intermediates alters their processing by the proteasome, and suggests in turn that their glycosylated precursors are the real substrates for proteasomal processing. In keeping with this, treatment with z-vad-fmk in the absence of epoxomicin did not induce the accumulation of any glycosylated intermediates (Fig 2B, compare lane 5 and 7). These observations strongly suggest that these glycosylated intermediates are direct substrates for the proteasome.
Because the DM331 transfectant expressed a relatively high level of tyrosinase, we determined the representation of deglycosylated and unglycosylated tyrosinase proteasome substrates in DM93, a melanoma cell expressing endogenous tyrosinase (8). Endogenous tyrosinase was found as a 70-kDa doublet in the membrane fraction (Fig. 3A, lane 1) and as a 58-kDa band after deglycosylation with PNGase in vitro (Fig. 3A, lane 2). In the cytosol, tyrosinase was found as a 56-kDa species, whose appearance was blocked by treatment with cycloheximide (Fig. 3B, lane 2). This band did not shift upon treatment with z-vad-fmk (Fig. 3B, lane 5), indicating that it represented unglycosylated tyrosinase. Inhibition of proteasome activity with epoxomicin, in both the presence and absence of cycloheximide, led to the accumulation of 58-kDa species in both membrane (Fig. 3A, lane 3) and cytosol (Fig. 3B, lane 3). Epoxomicin also led to an increase in a 56-kDa species in the membrane (Fig. 3A, lane 3), which again did not shift upon treatment with z-vad-fmk (Fig 3A, lane 5). Epoxomicin treatment also led to the accumulation of a series of molecular weight bands ranging between 31 and 54-kDa (Fig. 3A, 3B, lane 3). Based on their increases in molecular weight in cells treated with epoxomicin and z-vad-fmk (Fig. 3A, lane 5 and Fig 3B, lane 4), and its reversal by PNGase treatment (Fig. 3A, lane 6), all originated from glycosylated precursors. While many of these precursors were evident in cells treated with z-vad-fmk and epoxomicin, they were not found in cells treated with z-vad-fmk alone (Fig. 3A, lanes 5 and 7). These results confirmed that both deglycosylated and unglycosylated tyrosinase are present in cells expressing the protein normally, and that both are substrates for the proteasome. However, they also establish that glycosylated precursors can be degraded by the proteasome, giving rise to a set of distinct smaller molecular weight intermediates. Thus, glycosylation of tyrosinase affects the pattern of proteasomal cleavage.
Figure 3. Unglycosylated, deglycosylated, and N-terminally truncated proteasome substrates are found in cells expressing endogenous tyrosinase.
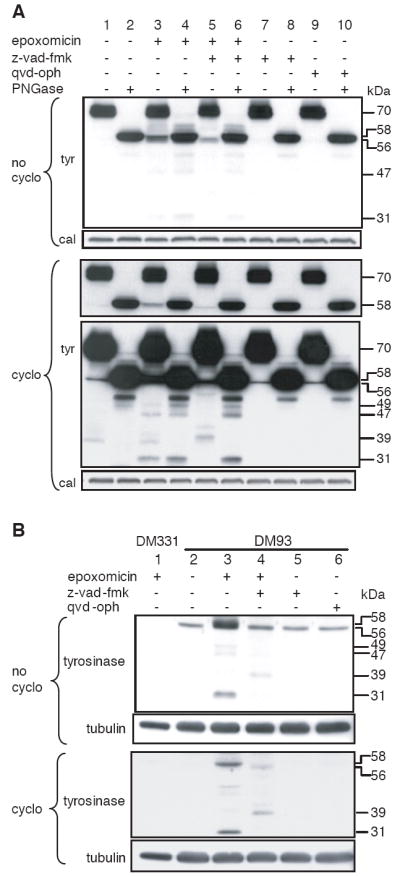
DM93 melanoma cells were incubated in medium containing cycloheximide and z-vad-fmk as indicated. Epoxomicin was added 30 min later as indicated. Cells were harvested 4h later and membrane (A) and cytosolic (B) fractions were prepared. Samples were analyzed by SDS-PAGE and immunoblotting with an Ab specific for the carboxyl-terminus of tyrosinase. Tyr, tyrosinase; cal, calnexin.
PNGase is not required for processing and presentation of Tyr369(D) in a transfectant expressing N371D mutant tyrosinase
We next wished to determine whether Tyr369(D) is derived from direct processing of a glycosylated tyrosinase precursor by the proteasome. Because, the epitope is created by deglycosylation leading to deamidation, we used a R402Q N371D tyrosinase mutant in which N371 was already substituted with D. As was observed with WT tyrosinase, all tyrosinase proteasome substrates that accumulated in presence of cycloheximide and epoxomicin were glycosylated (Fig 4A, lanes 3 and 4). However, the distribution of lower molecular weight species was centered at 38-kDa rather than 41-kDa, consistent with the loss of a glycosylation site and the presence of the N371D sequence in these fragments. DM331 cells transfected with either R402Q or R402Q N371D tyrosinase were incubated overnight in 1μg/ml cycloheximide, which prevented the production of unglycosylated tyrosinase (Fig. 4A, lane 3 and data not shown), and presentation of Tyr369(D) was assessed after epitope stripping in acid and reexpression in the presence of z-vad-fmk or epoxomicin. While presentation of Tyr369(D) from melanoma cells expressing normally glycosylated tyrosinase was substantially blocked by z-vad-fmk (Fig. 4B, left panel), its presentation from melanoma cells expressing N371D tyrosinase was not decreased at all (Fig. 4B, right panel). The observation that Tyr369(D) presentation is undiminished by PNGase blockade demonstrates that tyrosinase molecules can be degraded and processed by the proteasome for MHC class I presentation while they still bear N-glycans.
Figure 4. The Tyr369(D) epitope can be produced by the proteasome from glycosylated N371D tyrosinase without requirement of deglycosylation by PNGase.
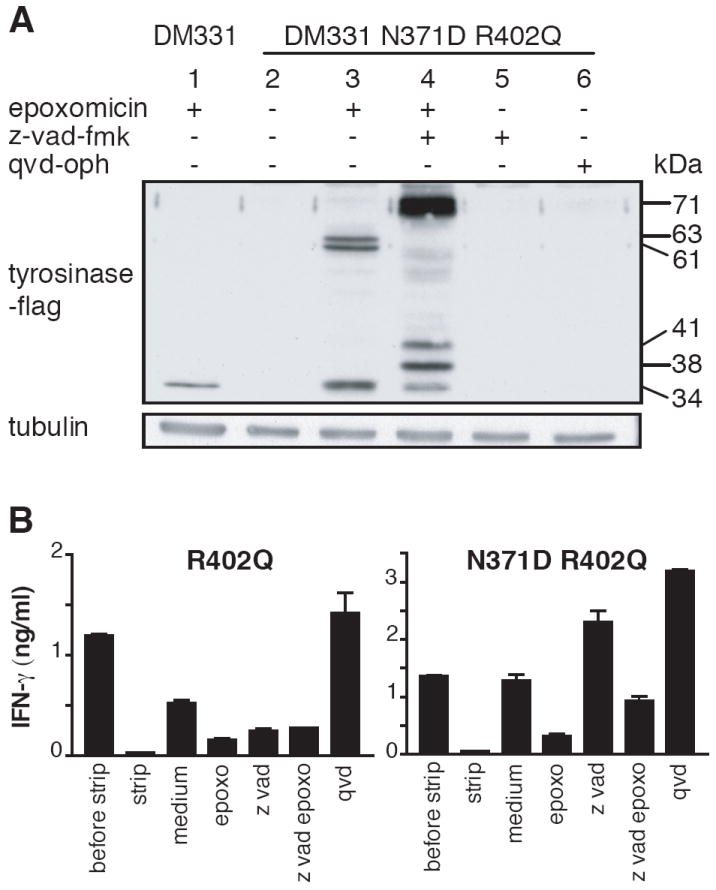
A, DM331 cells transfected to express N371D R402Q tyrosinase were incubated in medium containing 1 μg/ml cycloheximide in all samples and 50 μM z-vad-fmk, and 2 μM epoxomicin as indicated. Cytosolic fractions were analyzed by SDS-PAGE and immunoblotting with anti-flag Ab. B, Melanoma cells expressing R402Q (left panels) or N371D R402Q (right panels) tyrosinase were incubated in presence of 1 μg/ml cycloheximide overnight before and throughout the experiment with other inhibitors. Dependence of Tyr369(D) presentation on PNGase and proteasome activity was assessed after epitope stripping in acid and re-expression in the presence of the indicated inhibitors, as described in Materials and Methods.
Discussion
In the present work we investigated the glycosylation status of proteasome substrates derived from tyrosinase and their relevance for MHC class I epitope presentation. We suspected that glycosylation status might influence the efficiency of antigen processing and epitope presentation since melanoma cells have been shown to present the Tyr369(D) epitope, which is produced by deamidation, but not Tyr369(N), which corresponds to the genomic sequence (4). Production of Tyr369(D) requires glycosylation in the ER and subsequently deglycosylation by PNGase (11). The complete absence of the Tyr369(N) epitope suggested that N-glycans were quantitatively added at position 371 in the entire pool of tyrosinase molecules. However, while examining cytosolic intermediates that accumulated in cells treated with proteasome inhibitors, we were surprised to find a large number of molecules that had not been previously glycosylated. The failure of Tyr369(N) to be processed and presented from these unglycosylated molecules may be partially due to a process of deamidation independent of glycosylation (11). However, we also established that proteasomes degrade tyrosinase molecules that are still glycosylated, giving rise to a set of discrete intermediates that are not observed when unglycosylated molecules are degraded. This suggests that glycosylation of these intermediates alters their processing by the proteasome, and in turn that their glycosylated precursors are the real substrates for proteasomal processing. We propose that Tyr369(N) fails to be presented at least in part because unglycosylated tyrosinase is degraded rapidly and relatively non-selectively. In contrast, we hypothesize that glycosylation alters the selectivity of tyrosinase processing by the proteasome, enhancing the production or survival of Tyr369(D).
The observation that unglycosylated tyrosinase molecules were present as proteasome substrates in melanoma cells, often in larger quantities than deglycosylated tyrosinase, is surprising. Unglycosylated molecules could be differentiated from deglycosylated molecules because their status was not affected by blocking PNGase-mediated deglycosylation in vivo with z-vad-fmk. We have also obtained similar results using PNGase-specific targeting siRNA (data not shown). While initially observed in transfectants, these unglycosylated molecules were also found in melanoma cells expressing tyrosinase normally, demonstrating their biological relevance. The factors responsible for the production of these completely unglycosylated molecules remain unclear. One possibility is they have been translated in the cytosol rather than the ER. However, one would also expect these molecules to retain their signal sequences, and to show a higher molecular weight than deglycosylated tyrosinase (17). In addition, we detected unglycosylated tyrosinase molecules in the membrane fraction in melanoma cells as well as the cytosol. These observations suggest that unglycosylated tyrosinase molecules are normally translated in the ER, but fail to access the oligosaccharide transferase complex.
A second important observation in this work was that glycosylated tyrosinase and degradation intermediates derived from it did not accumulate when only PNGase activity was blocked, but required simultaneous inactivation of the proteasome. This indicates that full-length and N-truncated intermediates derived from tyrosinase could be degraded by the proteasome without prior deglycosylation by PNGase. This is surprising, since structural analyses of the proteasome have indicated that the opening of the catalytic chamber is too narrow to allow direct entry of a glycosylated N residue in the context of a polypeptide (18, 19). In understanding how glycosylated tyrosinase might be degraded, an important finding was that blockade of the proteasome resulted in the accumulation of several discrete tyrosinase degradation intermediates truncated at the N-terminus. By blocking PNGase activity in vivo, we demonstrated that all of these intermediates were glycosylated, and their appearance was also linked to the presence in the cytosol of full-length tyrosinase molecules that had been previously glycosylated. The accumulation of these substrates was not altered by addition of inhibitors of calpain (z-val-phe-cho), caspase (Qvd-oph, z-vad-fmk), serine proteases (TPCK, amino ethyl benzene sulfonyl fluoride, TPPII (butabindide), endosomal enzymes (NH4Cl), or nitric oxide synthase (L-NG-monomethyl arginine citrate), suggesting they are produced by the proteasome itself. The presence of these intermediates is difficult to explain by simple models of processive processing by the proteasome (20). However, it has been shown that the proteasome opening allows up to 5 polypeptide chains to pass through it at the same time (21), enabling degradation of substrates at internal peptide bonds even when they lacked accessible termini (22). Partial degradation by the proteasome has demonstrated in several protein processing reactions (23-28) and has been explained by presence of stably folded domains that are protease resistant. In some cases, it has been established that the most proximal proteolytic cleavage occurs 80-90 amino acids from the resistant domain (29, 30). Thus, a possible mechanism leading to the discrete glycosylated intermediates we observe might be that the tyrosinase polypeptide enters the catalytic chamber in a hairpin conformation at some distance from any glycosylation site. The proteasome would degrade these molecules until it encounters an N-glycan chain, and would then release the remainder of the molecule. While this remnant could be subsequently degraded by the same or other proteasomes in the same manner, this mechanism would give rise to a set of discrete intermediates with N-glycosylated residues in proximity to one terminus or the other. Consistent with this model, when the molecular weights of the intermediates we observe are aligned to the tyrosinase sequence, their N-termini lie within 29+/- 15 residues upstream of a glycosylation site.
Given that melanoma cells and transfectants express unglycosylated tyrosinase, a significant question is why the Tyr369(N) peptide epitope is not presented. Using tyrosinase with a mutation that abrogates N371 glycosylation, we previously found that this sequence can undergo deamidation to Tyr369(D) by a mechanism independent of glycosylation (11). Similarly, we have also shown that short N-terminally extended precursors containing the Tyr369(N) sequence can undergo glycosylation when they are transported into the ER from the cytosol, and undergo deglycosylation and N-terminal trimming upon reverse translocation into the cytosol (11). However, in both cases, these substrates also led to a significant level of Tyr369(N) presentation. The failure to present Tyr369(N) from completely unglycosylated tyrosinase may be because the latter molecule is more rapidly or more thoroughly destroyed by the proteasome. This hypothesis is consistent with the observation that smaller molecular weight proteasomal degradation intermediates are all derived from glycosylated tyrosinase. It is also consistent with our inability our inability to rescue presentation of Tyr369(N) using the protease inhibitors described above (data not shown). It is also consistent with the observation that the presentation of Tyr369(N) in cells expressing minigene encoded epitopes is not proteasome dependent (11). This latter result also suggests the possibility that the peptide fragments generated when glycosylation blocks proteasomal cleavage may be further trimmed in a proteasome-independent manner.
Other studies have examined the influence of PNGase on protein degradation by the proteasome. Inhibition of PNGase reduced the rate of degradation of the MHC class I heavy chain (15), RNaseB (31) and synthetic glycopeptides (32), but not TCRα (15). In addition, the presence of a glycosylation site near a peptide epitope was shown to diminish, but not abolish, its presentation in a PNGase dependent manner. While our observation that glycosylation is associated with enhanced epitope presentation is seemingly at odds with these observations, an important variable is likely to be the location of glycosylation in relation to the epitope, and the relative stability of glycosylated and unglycosylated forms of the proteins. We propose that the lack of presentation of the Tyr369(N) epitope derived from fully unglycosylated tyrosinase is due to either more rapid or more non-selective degradation of the entire protein. This is in keeping with other work demonstrating that more rapid degradation sometimes results in diminished, rather than enhanced, epitope presentation (33). We also propose that glycosylation either slows this degradation, particularly in proximity to the glycosylation sites, or alters the pattern of proteasome cleavage to protect epitope precursors, leading to enhanced presentation of Tyr369(D). This is in keeping with the studies cited above, particularly those of Kario et al (31), who demonstrated that glycosylation alters the pattern of proteasomal cleavage in vitro.
In sum, our results have revealed an important role for glycosylation in augmenting the generation of a class I MHC restricted epitope derived from tyrosinase. This is separate from the previously described role of glycosylation in enabling deamidation of the epitope, and instead seems tied to alteration of the pattern of proteasomal degradation. While demonstrated specifically in this instance for tyrosinase, there are many other examples of both deamidated epitopes, and epitopes derived from presumptively glycosylated but misfolded precursors. It will be useful to determine whether unglycosylated precursors also exist in these latter situations, and whether the spectrum or efficiency of epitope production from glycosylated and unglycosylated species is altered.
Footnotes
This work was supported by USPHS grants AI20963 and AI33134 to VHE.
Abbreviations used in this paper: Wild Type (WT), endoplasmic reticulum (ER), peptide N-glycanase (PNGase)
References
- 1.Petrescu SM, Branza-Nichita N, Negroiu G, Petrescu AJ, Dwek RA. Tyrosinase and glycoprotein folding: roles of chaperones that recognize glycans. Biochemistry. 2000;39:5229–5237. doi: 10.1021/bi000107z. [DOI] [PubMed] [Google Scholar]
- 2.Jimbow K, Park JS, Kato F, Hirosaki K, Toyofuku K, Hua C, Yamashita T. Assembly, target-signaling and intracellular transport of tyrosinase gene family proteins in the initial stage of melanosome biogenesis. Pigment Cell Res. 2000;13:222–229. doi: 10.1034/j.1600-0749.2000.130403.x. [DOI] [PubMed] [Google Scholar]
- 3.Wolfel T, Van Pel A, Brichard V, Schneider J, Seliger B, zum Buschenfelde KHM, Boon T. Two tyrosinase nonapeptides recognized on HLA-A2 melanomas by autologous cytolytic T lymphocytes. Eur J Immunol. 1994;24:759–764. doi: 10.1002/eji.1830240340. [DOI] [PubMed] [Google Scholar]
- 4.Skipper JCA, Hendrickson RC, Gulden PH, Brichard V, Van Pel A, Chen Y, Shabanowitz J, Wolfel T, Slingluff CL, Boon T, Hunt DF, Engelhard VH. An HLA-A2 restricted tyrosinase antigen on melanoma cells results from post-translational modification and suggests a novel processing pathway for membrane proteins. J Exp Med. 1996;183:527–534. doi: 10.1084/jem.183.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kittlesen DJ, Thompson LW, Gulden PH, Skipper JC, Colella TA, Shabanowitz JA, Hunt DF, Engelhard VH, Slingluff CL., Jr Human melanoma patients recognize an HLA-A1-restricted CTL epitope from tyrosinase containing two cysteine residues: implications for tumor vaccine development. J Immunol. 1998;160:2099–2106. [PubMed] [Google Scholar]
- 6.Topalian SL, Rivoltini L, Mancini M, Markus NR, Robbins PF, Kawakami Y, Rosenberg SA. Human CD4+ T cells specifically recognize a shared melanoma- associated antigen encoded by the tyrosinase gene. Proc Natl Acad Sci USA. 1994;91:9461–9465. doi: 10.1073/pnas.91.20.9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halaban R, Cheng E, Zhang Y, Moellmann G, Hanlon D, Michalak M, Setaluri V, Hebert DN. Aberrant retention of tyrosinase in the endoplasmic reticulum mediates accelerated degradation of the enzyme and contributes to the dedifferentiated phenotype of amelanotic melanoma cells. Proc Natl Acad Sci USA. 1997;94:6210–6215. doi: 10.1073/pnas.94.12.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosse CA, Meadows L, Luckey CJ, Kittlesen DJ, Huczko EL, Slingluff CL, Jr, Shabanowitz J, Hunt DF, Engelhard VH. The class I antigen processing pathway for the membrane protein tyrosinase involves translation in the endoplasmic reticulum and processing in the cytosol. J Exp Med. 1998;187:37–48. doi: 10.1084/jem.187.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luckey CJ, King GM, Marto JA, Venketeswaran S, Maier BF, Crotzer VL, Colella TA, Shabanowitz J, Hunt DF, Engelhard VH. Proteasomes can either generate or destroy MHC class I epitopes: evidence for nonproteasomal epitope generation in the cytosol. J Immunol. 1998;161:112–121. [PubMed] [Google Scholar]
- 10.Ostankovitch M, Robila V, Engelhard VH. Regulated folding of tyrosinase in the endoplasmic reticulum demonstrates that misfolded full-length proteins are efficient substrates for class I processing and presentation. J Immunol. 2005;174:2544–2551. doi: 10.4049/jimmunol.174.5.2544. [DOI] [PubMed] [Google Scholar]
- 11.Altrich-VanLith ML, Ostankovitch M, Polefrone JM, Mosse CA, Shabanowitz J, Hunt DF, Engelhard VH. Processing of a class I-restricted epitope from tyrosinase requires peptide N-glycanase and the cooperative action of endoplasmic reticulum aminopeptidase 1 and cytosolic proteases. J Immunol. 2006;177:5440–5450. doi: 10.4049/jimmunol.177.8.5440. [DOI] [PubMed] [Google Scholar]
- 12.Halaban R, Cheng E, Hebert DN. Coexpression of wild-type tyrosinase enhances maturation of temperature-sensitive tyrosinase mutants. J Invest Dermatol. 2002;119:481–488. doi: 10.1046/j.1523-1747.2002.01824.x. [DOI] [PubMed] [Google Scholar]
- 13.Blom D, Hirsch C, Stern P, Tortorella D, Ploegh HL. A glycosylated type I membrane protein becomes cytosolic when peptide: N-glycanase is compromised. EMBO J. 2004;23:650–658. doi: 10.1038/sj.emboj.7600090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang W, Man S, Gulden PH, Hunt DF, Engelhard VH. Class I-restricted alloreactive cytotoxic T lymphocytes recognize a complex array of specific MHC-associated peptides. J Immunol. 1998;160:1091–1097. [PubMed] [Google Scholar]
- 15.Misaghi S, Pacold ME, Blom D, Ploegh HL, Korbel GA. Using a small molecule inhibitor of peptide: N-glycanase to probe its role in glycoprotein turnover. Chem Biol. 2004;11:1677–1687. doi: 10.1016/j.chembiol.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Kwon BS, Haq AK, Pomerantz SH, Halaban R. Isolation and sequence of a cDNA clone for human tyrosinase that maps at the mouse c-albino locus [published erratum appears in Proc Natl Acad Sci U S A 1988 Sep;85(17):6352] Proc Natl Acad Sci USA. 1987;84:7473–7477. doi: 10.1073/pnas.84.21.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ujvari A, Aron R, Eisenhaure T, Cheng E, Parag HA, Smicun Y, Halaban R, Hebert DN. Translation rate of human tyrosinase determines its N-linked glycosylation level. J Biol Chem. 2001;276:5924–5931. doi: 10.1074/jbc.M009203200. [DOI] [PubMed] [Google Scholar]
- 18.Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 19.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 20.Nussbaum AK, Dick TP, Keilholz W, Schirle M, Stevanovic S, Dietz K, Heinemeyer W, Groll M, Wolf DH, Huber R, Rammensee HG, Schild H. Cleavage motifs of the yeast 20S proteasome beta subunits deduced from digests of enolase 1. Proc Natl Acad Sci USA. 1998;95:12504–12509. doi: 10.1073/pnas.95.21.12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee C, Prakash S, Matouschek A. Concurrent translocation of multiple polypeptide chains through the proteasomal degradation channel. J Biol Chem. 2002;277:34760–34765. doi: 10.1074/jbc.M204750200. [DOI] [PubMed] [Google Scholar]
- 22.Liu CW, Corboy MJ, DeMartino GN, Thomas PJ. Endoproteolytic activity of the proteasome. Science. 2003;299:408–411. doi: 10.1126/science.1079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 24.Chen CH, von Kessler DP, Park W, Wang B, Ma Y, Beachy PA. Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell. 1999;98:305–316. doi: 10.1016/s0092-8674(00)81960-1. [DOI] [PubMed] [Google Scholar]
- 25.Jia J, Amanai K, Wang G, Tang J, Wang B, Jiang J. Shaggy/GSK3 antagonizes Hedgehog signalling by regulating Cubitus interruptus. Nature. 2002;416:548–552. doi: 10.1038/nature733. [DOI] [PubMed] [Google Scholar]
- 26.Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature. 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- 27.Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 28.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 29.Lin L, DeMartino GN, Greene WC. Cotranslational dimerization of the Rel homology domain of NF-kappaB1 generates p50-p105 heterodimers and is required for effective p50 production. EMBO J. 2000;19:4712–4722. doi: 10.1093/emboj/19.17.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 31.Kario E, Tirosh B, Ploegh HL, Navon A. N-linked glycosylation does not impair proteasomal degradation but affects class I major histocompatibility complex presentation. J Biol Chem. 2008;283:244–254. doi: 10.1074/jbc.M706237200. [DOI] [PubMed] [Google Scholar]
- 32.Hagihara S, Goda K, Matsuo I, Ito Y. Analysis of ER-associated glycoprotein degradation using synthetic glycopeptide probes. Biochem Biophys Res Commun. 2007;360:357–362. doi: 10.1016/j.bbrc.2007.06.053. [DOI] [PubMed] [Google Scholar]
- 33.Golovina TN, Morrison SE, Eisenlohr LC. The impact of misfolding versus targeted degradation on the efficiency of the MHC class I-restricted antigen processing. J Immunol. 2005;174:2763–2769. doi: 10.4049/jimmunol.174.5.2763. [DOI] [PubMed] [Google Scholar]


