Abstract
OBJECTIVE
In the United States, the rate of preterm delivery (PTD) is higher in African Americans (17.8%) than non-Hispanic whites (11.5%). Such disparity cannot be fully explained by differences in socioenvironmental factors.
STUDY DESIGN
We genotyped 812 mothers in a case-control PTD study at Boston Medical Center who self-reported their ethnicity as “black.” Regression analysis and Wilcoxon rank-sum test were applied to evaluate ancestral distribution and the association between genetic ancestry and PTD-related traits, as well as the potential confounding effect of population stratification.
RESULTS
The estimated African ancestral proportion was 0.90 ± 0.13. We found significant associations of ancestral proportion with PTD as a whole and PTD subgrouped by the presence of maternal hypertensive disorders. We did not observe significant confounding as a result of population stratification in this case-control PTD study.
CONCLUSION
Our data underline the need for more intensive investigation of genetic admixture in African Americans to identify novel susceptibility genes of PTD.
Keywords: admixture, genetic ancestry, population stratification
Preterm delivery (PTD) (< 37 weeks of gestation) remains a significant clinical and public health problem. The rate of PTD is high in the United States at about 12.5%1 and has increased in the past 2 decades.2 PTD threatens all ethnic groups, particularly African American women (17.8% compared with 11.5% in non-Hispanic whites).1 However, socio-demographic risk factors cannot fully explain the racial disparity found in PTD. Compelling evidence suggests that PTD is a complex disease influenced by multiple genetic and environmental factors and gene-environment interactions.1,3,4 However, the role of the genetic influence in PTD remains largely unexplored.
The contemporary African American population is recognized as an admixed population with genetic admixture between Africans and Europeans.5 The ethnic definition of African Americans in most PTD studies is based on self-reported ethnicity. Recently, genetic ancestry among African Americans can be estimated by applying advanced statistical methods using a set of ancestry informative markers (AIMs).6–9 The degree of estimated European admixture among African Americans has previously been reported between 3.5–25%.5,10
Accurate assessment of genetic ancestry is important for several reasons. First, self-reported ethnicity, which has been a surrogate of individual’s ancestral background, may not sufficiently capture genetic variations. This may lead to inadequate adjustment for potential population stratification confounding when studying genetic association of PTD in admixed populations. Alternatively, the use of individual ancestral estimates (IAEs) may more precisely predict individual genetic ancestral variation to disease outcomes.11 Specifically, ancestral information cannot only provide a validation on how well self-reported ethnicity corresponds to ancestral proportion as estimated by AIMs, but it also can be used to evaluate differences in ancestral proportion between cases and controls in genetic association studies. It may also offer the opportunity to assess whether different ethnic definitions (self-report vs ancestral proportion estimate) alter the pattern or associations in epidemiologic and/or genetic studies.
Second, although racial disparities in PTD have been observed for decades, the role of genetic ancestry in PTD among African Americans has remained under investigation. Ancestral information provides a potential opportunity for mapping novel susceptibility genes in diseases with marked ethnic disparity that cannot be fully explained by socioeconomic and/or environmental factors.12 This approach, known as admixture mapping, has successfully identified candidate regions related to disease of interest, including multiple sclerosis and prostate cancer.13–15
Third, population stratification confounding has been a main concern in case-control genetic association studies in admixed populations, such as African Americans and Latino Americans.16,17 Population stratification occurs when there are different allele frequencies between cases and controls as a result of heterogeneity in ancestry, which is unrelated to disease’s affection status. Ignoring population stratification in association tests may lead to a potential excess of false-positive and/or false-negative results.18,19 With ancestral information, we can evaluate the degree of confounding effect caused by population stratification and apply such information to detect and control for the potential confounding effect.
In this study, we genotyped a panel of 61 AIMs and used 57 of the 61 AIMs to estimate genetic ancestry in 812 mothers enrolled in an ongoing case-control study of PTD at the Boston Medical Center (BMC), who self-reported their ethnicity as “black.” We sought to address 3 issues. First, we estimated genetic ancestral proportion by using AIMs data and evaluated whether there was a difference in ancestral proportion between cases and controls. Second, we investigated whether genetic ancestral background was associated with PTD and with pre-term subgroups. Finally, we assessed whether ancestral proportion confounded the association tests between 3 candidate genes (F5 factor, CYP1A1, and GSTT1) and PTD.
Materials and Methods
Study population and data collection
In this study, we included the first 812 black mothers (326 preterm cases and 486 matched controls) enrolled in an ongoing case-control study of preterm birth at the BMC.20 Case mothers were those who delivered singleton, live births occurring at < 37 weeks of gestation, and controls were defined as mothers delivering at ≥ 37 weeks of gestation with birthweight appropriate for gestational age as defined by the National Center for Health Statistics/Centers for Disease Control and Prevention guidelines (birthweight 2500–4000 g).21 Pregnancies resulting in multiple births and newborns with major birth defects were excluded. Control subjects were matched by race, maternal age, and date of delivery for each case delivery. Detailed description of the study population was published previously.20 Briefly, we collected epidemiologic data, clinical data, and maternal venous blood samples. In addition, placentae were sent for histopathology based on routine indications, including preterm birth.22 The institutional review boards of the BMC, the Massachusetts Department of Public Health, the Chicago Children’s Memorial Hospital, and the Harvard School of Public Health approved the study protocol. All participants gave written informed consent.
Definition of phenotypes
PTD was evaluated as both binary (< 37 weeks of gestation vs ≥ 37 weeks of gestation) and continuous (gestational age) variables. Gestational age was assessed using an algorithm based on last menstrual period and the result of early ultrasound (< 20 weeks of gestation). The last menstrual period estimate was used only if confirmed by ultrasound within 7 days or if no ultrasound estimate was obtained; otherwise, the ultrasound estimate was used. This approach has been used in previous studies.20,23
PTD subgroups investigated in this study were defined as follows. (1) By mode of delivery: we categorized PTD cases as spontaneous PTD (vaginally or by cesarean section) that occurred secondary to documented active preterm labor (uterine contractions with cervical effacement and dilation at < 37 weeks), preterm premature rupture of membranes (< 37 weeks without uterine contractions), or by both uterine contractions and preterm premature rupture of membranes occurring simultaneously or as medically induced PTD defined as delivery (vaginally or by cesarean section) that was not preceded by the presence of uterine contractions and/or rupture of membranes. (2) By degree of prematurity: in this study, we used a cut point of < 32 weeks to define very PTD and 34.0–36.86 weeks to define late PTD, which has been used by other groups.24,25 (3) By major pregnancy complications: because of sample size constraints, we focused on 2 relatively common pregnancy complications: (a) PTD with intrauterine infection/inflammation using placental histologic chorioamnionitis as a proxy—detailed description on placental collection, pathological methods, definition of maternal and fetal inflammatory responses, and quality control was published previously22 and (b) PTD complicated by maternal hypertensive disorders—this group consisted of PTD cases with an accompanying diagnosis of maternal preeclampsia, eclampsia, gestational hypertension, or HELLP syndrome as defined in our previous publication,26 with or without a history of chronic hypertension.
Selection and genotyping of AIMs
Selection
We selected and genotyped a panel of 61 AIMs with averaged δ (difference of allele frequencies between African and European ancestral populations) equal to 0.54 in 812 black mothers. Detailed information of the 61 AIMs regarding chromosomal location, allele frequencies among different ancestral populations, and difference of allele frequencies between different ancestral populations is provided in Supplemental Table S1. Polymorphic site, flanking sequence, and other relevant information of these 61 AIMs are available elsewhere.27
Genotyping
DNA was extracted from samples of venous whole blood using a standard protocol.28 Genotyping was performed using the TaqMan genotyping assay (Applied Biosystems, Foster City, CA). Polymerase chain reaction was performed in a 5-μL reaction containing 10 ng of genomic DNA, 1 × master mix, 900 nmol/L primers, and two 250 nmol/L TaqMan MGB probes by use of 384-well plates on a PTC-225 Tetrad Thermal Cycler (MJ Research, Waltham, MA) under the following conditions: 95°C for 10 minutes, 50 cycles of 92°C for 15 seconds, and 60°C for 1 minute. After polymerase chain reaction, an endpoint plate read of the fluorescence intensity of each well was performed on an ABI Primer 7900 (Applied Biosystems). Genotypes were called automatically using SDS, version 2.1 (Applied Biosystems) and were inspected visually on the plot.
Statistical analyses
Before data analysis, we computed genotype frequencies of the 61 AIMs and examined whether genotype frequencies of each marker were under Hardy-Weinberg equilibrium (HWE) by using exact tests implemented in the single nucleotide polymorphism-HWE program (http://www.sph.umich.edu/csg/abecasis/Exact/index.html).29
To estimate individual admixture, we first retrieved the available data of these 57 AIMs of subjects inherited from African and European ancestral populations (90 Yoruba subjects and 90 CEPH subjects) from the International HapMap Project (http://www.hapmap.org/). We then applied an admixture model implemented in the Structure program (http://pritch.bsd.uchicago.edu/software.html) using the AIMs data obtained from Hap-Map Project and the 812 African American mothers.30,31 Specifically, the admixture model in Structure assumes that each individual inherits some proportion of his or her ancestry from each ancestral population. To compute IAEs, we input genotyping data from both ancestral populations (Africans and Europeans), specified as known populations, and admixed subjects, specified as an unknown population; we assumed an admixture model and used default values for other parameters by Structure with 5000 burn-in and 50,000 further iterations through the Markov chain Monte Carlo algorithm implemented in the Structure program. In addition, to examine whether the distribution of genetic ancestry was similar between cases and controls, a nonparametric approach, the Wilcoxon rank-sum test, was applied because of the skewness of the distribution of individual ancestral proportions.
To examine the impact of genetic ancestry on PTD and related traits, we applied multiple logistic and linear regression models, separately, to test the association between IAEs and PTD-related phenotypes after adjusting covariates. Specifically, we recoded genetic ancestry into 4 groups (first quartile, second plus third quartiles, and fourth quartile of African ancestral proportion) and treated genetic ancestry as an ordinary variable in the model. Trend tests were performed to examine the associations between genetic ancestry and PTD and related traits as well. To evaluate a potential excess of false-positive and false-negative results, multiple logistic and linear regressions were carried out, separately, to test the association of 3 susceptibility genetic polymorphisms (F5, CYP1A1, and GSTT1) with PTD and gestational age, individually, with and without the adjustment of genetic ancestry—specifically, African IAEs.
The covariates adjusted in the analyses included: maternal age (< 20, 20–24, 25–29, 30–34, and ≥ 35 years), education (≤ middle school, high school, and > high school), parity (0, 1, and ≥ 2), marital status (married, other), maternal prepregnant body mass index (< 20, 20–24, 25–29, and ≥ 30), maternal smoking during pregnancy (never, quitter, and persistent), alcohol use (yes/no), illicit drug use (yes/no), and infant sex. Data analyses were performed using statistical packages R 2.3.1 (http://www.r-project.org) and Intercooled STATA 8.0 (Stata Corp, College Station, TX).
Results
Demographic characteristics and genetic ancestral estimation in BMC cohort
A total of 812 African American mothers (326 with PTD, and 486 with full-term delivery) were included in this study. The demographic characteristics and clinical data are shown in Table 1. The distributions of age, maternal prepregnant body mass index, education, parity, marital status, passive smoking during pregnancy, alcohol use, and infant sex were similar in both groups (Table 1). The rates of maternal smoking during pregnancy and illicit drug use were higher among the PTD cases.
TABLE 1.
Demographic and clinical characteristics of African American mothers, by preterm status
| Characteristic | PTD (n = 326) | Control (n = 486) |
|---|---|---|
| Demographic characteristics | ||
| Age (y) | 28.8 ± 6.8 | 27.9 ± 6.7 |
| BMI (kg/m2) | 26.7 ± 6.7 | 26.0 ± 6.1 |
| Education (%) | ||
| ≤ Middle school | 27.3 | 29.0 |
| High school | 40.5 | 32.5 |
| > High school | 32.2 | 38.5 |
| Infant sex (%) | ||
| Male | 51.5 | 54.3 |
| Female | 48.5 | 45.7 |
| Parity (%) | ||
| 0 | 37.7 | 36.2 |
| 1 | 26.4 | 32.3 |
| ≥ 2 | 35.9 | 31.5 |
| Marital status (%) | ||
| Married | 29.8 | 36.8 |
| Other | 70.2 | 63.2 |
| Maternal smoking during pregnancy (%) | ||
| Never | 75.3 | 82.7 |
| Quitter | 5.6 | 4.8 |
| Persistent | 19.1 | 12.5 |
| Passive smoking during pregnancy (%) | ||
| No | 71.6 | 77.9 |
| Yes | 28.4 | 22.1 |
| Alcohol use (%) | ||
| No | 95.7 | 96.1 |
| Yes | 4.3 | 3.9 |
| Illicit drug use (%) | ||
| No | 84.1 | 88.3 |
| Yes | 15.9 | 11.7 |
| Clinical characteristics | ||
| Gestational age (wk) | 33.1 ± 3.7 | 39.7 ± 1.0 |
| Infant birthweight (g) | 2024.2 ± 762.7 | 3159.6 ± 551.4 |
BMI, body mass index; PTD, preterm delivery.
Tsai. Association of genetic ancestry with preterm delivery and related traits among African American mothers. Am J Obstet Gynecol 2009.
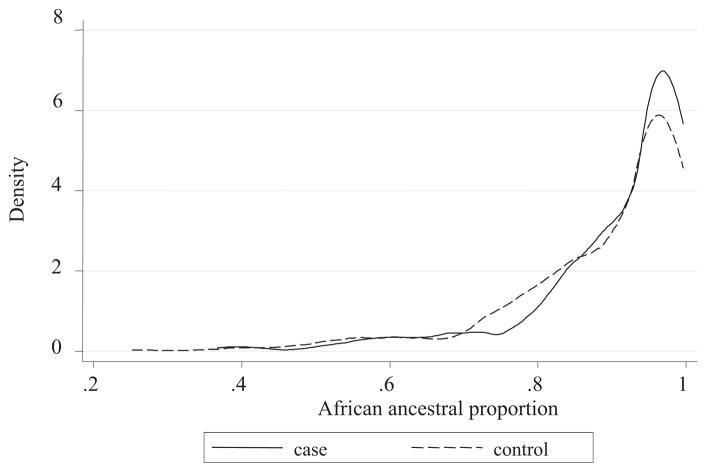
In all, 61 AIMs were genotyped in this study; 4 of 61 AIMs deviated from HWE (Supplemental Table S1) and were excluded in the subsequent analysis. We next applied the Structure program to estimate individual ancestry using the remaining 57 AIMs data. We observed that the average and corresponding SE of African proportion were 0.91 ± 0.13, 0.91 ± 0.12, and 0.90 ± 0.13 for all subjects, PTD cases, and controls, respectively. In addition, the results from the Wilcoxon rank-sum test indicated that the distributions of ancestral proportions were only slightly different between PTD cases and controls (P = .05; Figure).
FIGURE.
Distribution of African ancestral proportion in cases and controls
Tsai. Association of genetic ancestry with preterm delivery and related traits among African American mothers. Am J Obstet Gynecol 2009.
Associations between genetic ancestry and PTD and related traits
We examined the associations between IAEs and PTD and related traits with and without the adjustment of covariates. As shown in Table 2, significant associations were found for PTD and related traits, including spontaneous PTD, very PTD, and PTD with maternal hypertensive disorders, respectively. In detail, when comparing subjects in the first quartile of African ancestral proportion (bottom 25%), subjects in the second or third quartile had an increased risk of PTD (odds ratio [OR], 1.5; 95% confidence interval [CI], 1.1–2.2), medically induced PTD (OR, 2.1; 95% CI, 1.1–3.8), near-term PTD (OR, 1.6; 95% CI, 1.0–2.5), and PTD with maternal hypertensive disorders (OR, 2.4; 95% CI, 1.3–4.6). Likewise, subjects in the top 25% distribution of African ancestral proportion had an increased risk of PTD (OR, 1.7; 95% CI, 1.1–2.7), spontaneous PTD (OR, 1.7; 95% CI, 1.1–2.8), very PTD (OR, 2.1; 95% CI, 1.1–4.2), and PTD with maternal hypertensive disorders (OR, 2.2; 95% CI, 1.1–4.5). Moreover, the results from trend test in Table 2 also suggested significant associations of ancestral proportion with PTD (P = .03) and PTD with maternal hypertensive disorders (P = .01).
TABLE 2.
Associations between African ancestry and preterm delivery and preterm subgroups
| Preterm traits | Crude |
Adjusteda |
||
|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | |
| PTD | ||||
| First quartile | Ref | Ref | ||
| Second + third quartile | 1.4 (1.0–2.0) | .05 | 1.5 (1.1–2.2) | .04b |
| Fourth quartile | 1.6 (1.1–2.3) | .03b | 1.7 (1.1–2.7) | .01b |
| Trend test | .03b | |||
|
SPONTANEOUS PTD | ||||
| First quartile | Ref | Ref | ||
| Second + third quartile | 1.3 (0.9–2.0) | .15 | 1.3 (0.9–2.0) | .22 |
| Fourth quartile | 1.5 (1.0–2.4) | .07 | 1.7 (1.1–2.8) | .03b |
| Trend test | .07 | |||
|
MEDICALLY INDICATED PTD | ||||
| First quartile | Ref | Ref | ||
| Second + third quartile | 2.0 (1.1–3.7) | .03b | 2.1 (1.1–3.8) | .02b |
| Fourth quartile | 1.9 (1.0–3.9) | .06 | 1.9 (0.9–3.8) | .09 |
| Trend test | .06 | |||
|
VERY PTD | ||||
| First quartile | Ref | Ref | ||
| Second + third quartile | 1.3 (0.8–2.3) | .33 | 1.4 (0.8–2.6) | .29 |
| Fourth quartile | 1.7 (0.9–3.2) | .08 | 2.1 (1.1–4.2) | .03b |
| Trend test | .08 | |||
|
NEAR-TERM PTD | ||||
| First quartile | Ref | Ref | ||
| Second + third quartile | 1.6 (1.1–2.5) | .03b | 1.6 (1.0–2.5) | .04b |
| Fourth quartile | 1.5 (0.9–2.4) | .12 | 1.5 (0.9–2.6) | .13 |
| Trend test | .12 | |||
|
PTD WITH MATERNAL HYPERTENSIVE DISORDER | ||||
| First quartile | Ref | Ref | ||
| Second + third quartile | 2.4 (1.3–4.5) | .006b | 2.4 (1.3–4.6) | .007b |
| Fourth quartile | 2.5 (1.2–4.9) | .009b | 2.2 (1.1–4.5) | .03b |
| Trend test | .01b | |||
|
PTD ACCOMPANIED BY HISTOLOGIC CHORIOAMNIONITIS | ||||
| First quartile | Ref | Ref | ||
| Second + third quartile | 1.0 (0.6–1.6) | .89 | 1.1 (0.6–1.9) | .72 |
| Fourth quartile | 1.2 (0.7–2.1) | .55 | 1.2 (0.6–2.2) | .65 |
| Trend test | .54 | |||
| β (SE) | P | β (SE) | P | |
|
GEAA (WK) | ||||
| First quartile | Ref | Ref | ||
| Second + third quartile | −0.40 (0.35) | .24 | −0.44 (0.38) | .24 |
| Fourth quartile | −0.60 (0.40) | .13 | −0.86 (0.45) | .05 |
| Trend test | .10 | |||
CI, confidence interval; GEAA, gestational age; OR, odds ratio; PTD, preterm delivery; Ref, reference group.
Adjusted for maternal age, education, parity, marital status, prepregnant body mass index, maternal smoking during pregnancy, drug use, alcohol use, and infant sex;
P < .05.
Tsai. Association of genetic ancestry with preterm delivery and related traits among African American mothers. Am J Obstet Gynecol 2009.
Evaluation of potential population stratification confounding effect
To evaluate potential confounding effect caused by population stratification, we examined the associations between 3 PTD candidate genes (F5, CYP1A1, and GSTT1) and PTD and gestational age, separately, with the adjustment of covariates only, and with the adjustment of both covariates and genetic ancestry, respectively. Our data showed that the association results were almost identical with or without adjustment for genetic ancestry (Table 3). In other words, we did not observe an excess in false-positive or false-negative results and did not find significant confounding effect caused by population stratification.
TABLE 3.
Associations of genetic polymorphisms with preterm delivery and length of gestation in 812 African Americans, with and without adjustment of African ancestral proportion
| Variable | PTD (< 37 wk) |
Gestational age |
||||||
|---|---|---|---|---|---|---|---|---|
| Unadjusted AIMs |
Adjusted AIMs |
Unadjusted AIMs |
Adjusted AIMs |
|||||
| OR (95% CI) | P | OR (95% CI) | P | β (SE) | P | β (SE) | P | |
| F5 gene (rs6019) | ||||||||
| GG | 1.0 | — | 1.0 | — | Ref | — | Ref | — |
| GC | 1.3 (1.0–1.8) | .10 | 1.3 (0.9–1.8) | .11 | −0.5 (0.3) | .14 | −0.4 (0.3) | .16 |
| CC | 1.8 (1.2–2.8) | .01a | 1.7 (1.1–2.7) | .02a | −1.5 (0.4) | .001a | −1.4 (0.4) | .001a |
|
CYP1A1 Msp I | ||||||||
| AA | 1.0 | — | 1.0 | — | Ref | — | Ref | — |
| Aa/aa | 0.9 (0.7–1.3) | .68 | 0.9 (0.7–1.3) | .73 | 0.4 (0.3) | .21 | 0.4 (0.3) | .23 |
|
GSTT1 | ||||||||
| Present | 1.0 | — | 1.0 | — | Ref | — | Ref | — |
| Delete | 0.8 (0.6–1.1) | .18 | 0.8 (0.6–1.1) | .19 | 0.6 (0.4) | .13 | 0.6 (0.4) | .12 |
All analyses were adjusted for maternal age, education, parity, marital status, prepregnant body mass index, maternal smoking during pregnancy, drug use, alcohol use, and infant sex. AIM, ancestry informative markers; CI, confidence interval; OR, odds ratio; PTD, preterm delivery; Ref, reference group.
P < .05.
Tsai. Association of genetic ancestry with preterm delivery and related traits among African American mothers. Am J Obstet Gynecol 2009.
Comment
It has been long observed that the highest rates of PTD in the United States occur among racial minorities, especially African Americans (17.8%). Although the impact of genetic ancestry and its potential confounding effect have been examined in several common complex diseases, such as asthma, breast cancer, and cardiovascular disease, none of the previous studies has assessed the influence of genetic ancestry in PTD and related traits. This study is the first to evaluate whether genetic ancestry is associated with PTD and related traits and whether genetic ancestry may confound association tests of candidate loci with PTD and related traits. Several important findings have been observed in this study.
First, the average estimates of European ancestry were 9.5% in this sample of African American mothers recruited from BMC. The estimates in the study sample were comparable with those reported in previous studies, ranging from 3.5–25%.5,32 For instance, Parra et al5 in 1998 reported 12% European ancestry estimated from African Americans residing in Charleston, SC, whereas Wassel Fyr et al33 reported 25% European ancestry estimated from African Americans residing in Pittsburgh, PA. Our results have supported that genetic admixture occurs among contemporary African American populations across different regions of the United States.11,34 On the other hand, our results have also suggested high consistency between self-reported ethnicity as “black” and African ancestral background as estimated by AIMs in this sample.
Of note, the accuracy of estimating individual ancestry is affected by the number of AIMs, and the informativeness of AIMs. In this study, we estimated individual ancestry using a set of 57 AIMs with averaged δ equal to 0.54. To assess whether the number and corresponding informativeness of AIMs were sufficient for ancestral estimation, we calculated IAEs by using the first 30, the first 45, and all 57 AIMs, separately. The results in Supplemental Table S2 indicated that IAEs obtained from using ≥ 45 AIMs in estimation were highly similar. This suggested that the potential computing inaccuracy in estimating genetic ancestry may be avoided while genotyping and incorporating a set of ≥ 45 AIMs with averaged δ ≥ 0.5. However, previous studies have reported that when sample size of admixed subjects increases, more numbers and more informativeness of AIMs are required for precisely estimating individual ancestry and efficiently controlling for excess of type I error.35,36 Therefore, the number of AIMs for precise ancestral estimation should be carefully selected.
Our results suggested that African ancestry was associated with an increased risk of PTD and related traits in African American mothers. Specifically, subjects with higher African ancestry had a higher risk in developing PTD. Furthermore, African ancestry was also associated with PTD subgroups, such as spontaneous PTD, very PTD, and PTD complicated by maternal hypertensive disorders. Consistently, previous studies also reported higher prevalence of spontaneous PTD, very PTD, and PTD complicated by maternal hypertensive disorders among African American mothers than other ethnic groups.37,38 Taken together, our findings that subjects carrying higher African ancestral background had a higher risk of developing PTD and certain preterm subgroups lend further support for future studies to conduct admixture mapping of PTD in African Americans.
Admixture mapping has been considered to be a promising method to search for susceptibility genes in a genome scale for complex diseases in admixed populations such as African Americans.39 Suitable diseases for admixture mapping approach are those with marked ethnic disparity that cannot be fully explained by socioeconomic and/or environmental factors. As such, this approach may be a useful tool in determining novel susceptibility genes associated to PTD and related traits.
From an epidemiologic standpoint, great attention has been drawn to whether genetic ancestral estimates can more precisely capture genetic variation than self-reported race.40,41 We performed association tests in African American subjects with and without adjusting genetic ancestral proportion. The results were highly consistent in the association tests for 3 candidate genes of PTD. In addition, we also performed the association tests between 57 AIMs and PTD. Similarly, inflation of type I error was not observed in those results (data not shown). Our data supported the findings reported previously that population stratification confounding could be overcome by well-matched case-control study design when conducting genetic association studies in admixed populations.16,42,43 Although we did not observe detectable population stratification confounding effect in this study, as mentioned above, when sample size of admixed subjects increases, more numbers and more informativeness of AIMs are required to make the appropriate correction.36
In summary, the estimated average African ancestral proportion was approximately 90% in this BMC sample of African American mothers who self-reported their ethnicity as “black.” Our data did not show a substantial confounding effect caused by population stratification in genetic association tests with PTD. Novel associations were found between African ancestry and PTD as a whole and certain preterm subgroups. The findings have provided strong support for further conducting more intensive investigation of genetic admixture in African Americans, including genome-wide admixture mapping to dissect PTD-related susceptibility genes.
Supplementary Material
Acknowledgments
The study was supported in part by Grants from the National Institute of Child Health and Human Development (R01 HD41702; K24 HD 042489), National Institute of Environmental Health Sciences (R01ES11682, R21ES11666), March of Dimes Birth Defects Foundation (20-FY98-0701, 20-FY02-56 and #21-FY07-605), and Food Allergy Project.
We thank the nursing staff of Labor and Delivery at BMC for their continuous support and assistance to the study, Lingling Fu for data management, and Ann Ramsay for administrative support. We would like to particularly thank the outstanding expert consultants of the BMC Pre-term Study team: Drs Paul Wise, Jerome Klein, Phillip G. Stubblefield, John M. Kasznica, and Milton Kotelchuck. Finally, we thank all of the participating mothers and their families.
References
- 1.Behrman RE, Butler AS, editors. Preterm birth: cause, consequences, and prevention. Washington, DC: Institute of Medicine of the National Academies (committee on understanding premature birth and assuring healthy outcomes board on health sciences policy); 2006. [Google Scholar]
- 2.Preterm birth: crisis and opportunity. Lancet. 2006;368:339. doi: 10.1016/S0140-6736(06)69080-6. [DOI] [PubMed] [Google Scholar]
- 3.Macones GA, Parry S, Elkousy M, Clothier B, Ural SH, Strauss JF., III A polymorphism in the promoter region of TNF and bacterial vaginosis: preliminary evidence of gene-environment interaction in the etiology of spontaneous preterm birth. Am J Obstet Gynecol. 2004;190:1504–8. doi: 10.1016/j.ajog.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Crider KS, Whitehead N, Buus RM. Genetic variation associated with preterm birth: a HuGE review. Genet Med. 2005;7:593–604. doi: 10.1097/01.gim.0000187223.69947.db. [DOI] [PubMed] [Google Scholar]
- 5.Parra EJ, Marcini A, Akey J, et al. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63:1839–51. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfaff CL, Parra EJ, Bonilla C, et al. Population structure in admixed populations: effect of admixture dynamics on the pattern of linkage disequilibrium. Am J Hum Genet. 2001;68:198–207. doi: 10.1086/316935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–14. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shriver MD, Parra EJ, Dios S, et al. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet. 2003;112:387–99. doi: 10.1007/s00439-002-0896-y. [DOI] [PubMed] [Google Scholar]
- 9.Shriver MD, Mei R, Parra EJ, et al. Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Hum Genomics. 2005;2:81–9. doi: 10.1186/1479-7364-2-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parra EJ, Kittles RA, Argyropoulos G, et al. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am J Phys Anthropol. 2001;114:18–29. doi: 10.1002/1096-8644(200101)114:1<18::AID-AJPA1002>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 11.Reiner AP, Ziv E, Lind DL, et al. Population structure, admixture, and aging-related phenotypes in African American adults: the cardiovascular health study. Am J Hum Genet. 2005;76:463–77. doi: 10.1086/428654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith MW, O’Brien SJ. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nat Rev Genet. 2005;6:623–32. doi: 10.1038/nrg1657. [DOI] [PubMed] [Google Scholar]
- 13.Reich D, Patterson N, De Jager PL, et al. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet. 2005;37:1113–8. doi: 10.1038/ng1646. [DOI] [PubMed] [Google Scholar]
- 14.Freedman ML, Haiman CA, Patterson N, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103:14068–73. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haiman CA, Patterson N, Freedman ML, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–44. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- 17.Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2:91–9. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- 18.Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–48. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 19.Burchard EG, Ziv E, Coyle N, et al. The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med. 2003;348:1170–5. doi: 10.1056/NEJMsb025007. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Zuckerman B, Pearson C, et al. Maternal cigarette smoking, metabolic gene polymorphism, and infant birth weight. JAMA. 2002;287:195–202. doi: 10.1001/jama.287.2.195. [DOI] [PubMed] [Google Scholar]
- 21.Hamill PV, Drizd TA, Johnson CL, Reed RB, Roche AF, Moore WM. Physical growth: National Center for Health Statistics percentiles. Am J Clin Nutr. 1979;32:607–29. doi: 10.1093/ajcn/32.3.607. [DOI] [PubMed] [Google Scholar]
- 22.Gupta M, Mestan KK, Martin CR, et al. Impact of clinical and histologic correlates of maternal and fetal inflammatory response on gestational age in preterm births. J Matern Fetal Neonatal Med. 2007;20:39–46. doi: 10.1080/14767050601156861. [DOI] [PubMed] [Google Scholar]
- 23.Kramer MS, Platt R, Yang H, et al. Secular trends in preterm birth: a hospital-based cohort study. JAMA. 1998;280:1849–54. doi: 10.1001/jama.280.21.1849. [DOI] [PubMed] [Google Scholar]
- 24.Engle WA, Tomashek KM, Wallman C. “Late-preterm” infants: a population at risk. Pediatrics. 2007;120:1390–401. doi: 10.1542/peds.2007-2952. [DOI] [PubMed] [Google Scholar]
- 25.Mathews TJ, Menacker F, MacDorman MF. Infant mortality statistics from the 2002 period: linked birth/infant death data set. Natl Vital Stat Rep. 2004;53:1–29. [PubMed] [Google Scholar]
- 26.Wang L, Feng Y, Zhang Y, et al. Prolylcarboxypeptidase gene, chronic hypertension, and risk of preeclampsia. Am J Obstet Gynecol. 2006;195:162–71. doi: 10.1016/j.ajog.2006.01.079. [DOI] [PubMed] [Google Scholar]
- 27.Yang N, Li H, Criswell LA, et al. Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Hum Genet. 2005;118:382–92. doi: 10.1007/s00439-005-0012-1. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–93. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–87. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reiner AP, Carlson CS, Ziv E, Iribarren C, Jaquish CE, Nickerson DA. Genetic ancestry, population sub-structure, and cardiovascular disease-related traits among African-American participants in the CARDIA Study. Hum Genet. 2007;121:565–75. doi: 10.1007/s00439-007-0350-2. [DOI] [PubMed] [Google Scholar]
- 33.Wassel Fyr CL, Kanaya AM, Cummings SR, et al. Genetic admixture, adipocytokines, and adiposity in Black Americans: the Health, Aging, and Body Composition study. Hum Genet. 2007;121:615–24. doi: 10.1007/s00439-007-0353-z. [DOI] [PubMed] [Google Scholar]
- 34.Kittles RA, Weiss KM. Race, ancestry, and genes: implications for defining disease risk. Annu Rev Genomics Hum Genet. 2003;4:33–67. doi: 10.1146/annurev.genom.4.070802.110356. [DOI] [PubMed] [Google Scholar]
- 35.Rosenberg NA, Li LM, Ward R, Pritchard JK. Informativeness of genetic markers for inference of ancestry. Am J Hum Genet. 2003;73:1402–22. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai HJ, Choudhry S, Naqvi M, Rodriguez-Cintron W, Burchard EG, Ziv E. Comparison of three methods to estimate genetic ancestry and control for stratification in genetic association studies among admixed populations. Hum Genet. 2005;118:424–33. doi: 10.1007/s00439-005-0067-z. [DOI] [PubMed] [Google Scholar]
- 37.Samadi AR, Mayberry RM, Zaidi AA, Pleasant JC, McGhee N, Jr, Rice RJ. Maternal hypertension and associated pregnancy complications among African-American and other women in the United States. Obstet Gynecol. 1996;87:557–63. doi: 10.1016/0029-7844(95)00480-7. [DOI] [PubMed] [Google Scholar]
- 38.Blackmore-Prince C, Kieke B, Jr, Kugaraj KA, et al. Racial differences in the patterns of singleton preterm delivery in the 1988 national maternal and infant health survey. Matern Child Health J. 1999;3:189–97. doi: 10.1023/a:1022373205005. [DOI] [PubMed] [Google Scholar]
- 39.Patterson N, Hattangadi N, Lane B, et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang H, Quertermous T, Rodriguez B, et al. Genetic structure, self-identified race/ethnicity, and confounding in case-control association studies. Am J Hum Genet. 2005;76:268–75. doi: 10.1086/427888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Risch N. Dissecting racial and ethnic differences. N Engl J Med. 2006;354:408–11. doi: 10.1056/NEJMe058265. [DOI] [PubMed] [Google Scholar]
- 42.Pennell CE, Jacobsson B, Williams SM, et al. Genetic epidemiologic studies of preterm birth: guidelines for research. Am J Obstet Gynecol. 2007;196:107–18. doi: 10.1016/j.ajog.2006.03.109. [DOI] [PubMed] [Google Scholar]
- 43.Tsai HJ, Kho JY, Shaikh N, et al. Admixture-matched case-control study: a practical approach for genetic association studies in admixed populations. Hum Genet. 2006;118:626–39. doi: 10.1007/s00439-005-0080-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.