Summary
Paramutation involves trans interactions between alleles or homologous sequences that establish distinct gene expression states that are heritable for generations. It was first described in maize by Alexander Brink in the 1950's, with his studies of the r1 (red1) locus. Since that time, paramutation-like phenomena have been reported in other maize genes, other plants, fungi and animals. Paramutation can occur between endogenous genes, two transgenes or an endogenous gene and transgene. Recent results indicate that paramutation involves RNA-mediated heritable chromatin changes and a number of genes implicated in RNAi pathways. However, not all aspects of paramutation can be explained by known mechanisms of RNAi-mediated transcriptional silencing.
Introduction
Paramutation was first described by Alexander Brink in 1956 for the maize red1 (r1) gene [1] and subsequently for the b1 (Booster 1) gene by Ed Coe Jr. in 1959 [2], both genes that encode transcription factors that activate the anthocyanin biosynthetic pathway. Subsequently paramutation has been demonstrated at other genes in maize, pl1 [3], p1 [4] and lpa1-241 [5]. The classic definition of paramutation is an allele interaction that leads to a heritable change in gene expression. The nature of the interaction between alleles is not known, but several possibilities will be discussed in this review. When an allele's expression is changed (i.e. sensitive to paramutation), it is referred to as paramutable. The allele that induces the change in expression is referred to as paramutagenic. Following paramutation, previously paramutable (sensitive alleles) are termed paramutant (or paramutated). Paramutated alleles are capable of inducing paramutation; they become paramutagenic. Most alleles do not participate in paramutation (they neither induce a change nor are they changed in the presence of a paramutagenic allele); these types of alleles will be referred to as neutral in this review. Using the criteria of trans interactions that induce gene expression changes or activities that are heritable in subsequent generations, a number of related phenomena have been reported in multiple species [6-8].
In summary, the classic maize systems share three key features: (1) the newly silenced expression state efficiently transmitted to subsequent generations even though the allele originally issuing the instructions (i.e. the paramutagenic allele) is not transmitted; (2) the paramutated allele continues to issue similar instructions to homologous sequences in subsequent generations (it is paramutagenic); and (3) where examined, there are no associated DNA sequence changes; thus, the altered expression states are mediated by epigenetic mechanisms [6].
This review is not meant to be comprehensive, but instead focuses on data from studies of several paramutation systems in maize, reviewing the key sequence requirements and the genes that have been identified that are required for paramutation. These studies demonstrate a role for RNA and small interfering RNA (siRNA) biogenesis in paramutation and that mutations in the maize siRNA biogenesis pathway can lead to developmental defects. The most extensively characterized system, b1, is used to present a model for paramutation and open questions to stimulate future research directions are discussed.
Sequence Requirements for Maize Paramutation
Key sequences mediating paramutation have been identified for two plant paramutation systems, b1 [9,10] and p1 [4]. The b1 locus encodes a transcription factor required to activate the anthocyanin biosynthetic pathway [11], resulting in purple pigmentation of most mature plant tissues. The amount of transcription of b1 quantitatively correlates with the pigment levels in plants providing a powerful system for investigating paramutation. When the highly transcribed, darkly pigmented B-I allele is crossed with B′, which is lightly pigmented and has much lower transcription, paramutation always occurs: B-I is always changed into B′. This new B′ allele (designated as B′*) is as paramutagenic as the parental B′ allele.
The key sequences required for b1 paramutation are seven direct tandem repeats of noncoding DNA located ∼100 Kb upstream of the b1 transcription start site [9]. Each of the repeat units is 853 bp and both B-I and B′ carry seven tandem repeats, whereas neutral alleles have a single copy of the repeat unit [9]. Characterization of an allelic series that were identical except for the number of repeat units (one, three and five), revealed that multiple repeat units are required for paramutation, and the strength of paramutation correlated with the number of repeats [9]. In addition to sharing identical numbers of the tandem repeats, B-I and B′ have identical sequences throughout the 150 kb surrounding the gene. B-I and B′ do show distinct patterns of DNA methylation and chromatin structure within the tandem repeats and are thus epialleles [9,12,13].
Spontaneous changes of B-I to B′ can occur at high frequency (0.1-10%), but B′ is extremely stable, with no changes to B-I observed over 50 years and tens of thousands of plants [14]. It is intriguing that the stability of B′ and B-I is dramatically different. One possibility for why B′ is so stable might be that the default chromatin state for the seven tandem repeats is the silent paramutagenic B′ state. Thus, once it is formed, B′ is faithfully inherited. This view suggests that B-I is the “mutant” form requiring regulatory mechanisms at each cell division to maintain the chromatin state mediating its high transcription. These postulated mechanisms must be relatively reproducible, but clearly not foolproof, such that B-I is changed to B′ occasionally when maintained homozygous.
Paramutation at the maize p1 gene, which encodes a transcription factor mediating phlobaphene biogenesis, is associated with silencing of the endogenous P1-rr allele that normally has red ear and cob pigmentation. Upon exposure to a transgene carrying specific P1-rr regulatory sequences, P1-rr becomes paramutated and this newly established transcriptionally silenced state, termed P1-rr′, is paramutagenic in subsequent generations [4,15]. Analyses of transgenes containing different regions of the p1 regulatory region, have demonstrated that a specific 1.2 kb enhancer fragment is sufficient to establish paramutation, even with a heterologous promoter or no promoter [15]. The 1.2 kb fragment is part of a direct repeat that flanks the p1 coding region and this region functions as an enhancer [16].
It is intriguing that the sequences mediating b1 paramutation are tandem repeats that also mediate enhancer activities [9]. In addition, tandem repeats are also required for paramutation at the r1 locus, but with major differences when compared to b1 and p1. At r1 the tandem repeats span and therefore contain the r1 coding regions and are of unknown length [17]. Recombination mapping generated a series of alleles with differing numbers of tandem repeats [18-20]; and unlike the b1 and p1 studies, the r1 results revealed that no specific region within the repeat is required, it is the number of repeats that matters with the strength of paramutation correlating with the repeat numbers. While an important theme emerging is that repeat sequences are involved in paramutation, studies in both maize and Arabidopsis thaliana (Arabidopsis) demonstrate that being epigenetically silenced and carrying tandem repeats is not always sufficient for paramutation. In maize, the P1-wr locus is composed of multiple tandem repeats, which are large and span the coding region and it is epigenetically silenced [21], yet it does not participate in paramutation with P1-rr or P1-rr′ [15]. Transcription of two tandem repeats and generation of siRNAs are also not sufficient for trans silencing of the FWA locus in Arabidopsis [22]. For further discussion of the similarities and differences between b1 paramutation and FWA silencing see Chandler, 2007 [6] and Henderson and Jacobsen, 2007 [23].
Paramutation is associated with siRNA biogenesis in maize
Several genes required for paramutation, have been identified through forward genetic screens, and to date all encode genes that have been associated with siRNA biogenesis in other species. The mediator of paramutation (mop) genes [24,25] and the required to maintain repression (rmr) genes [26-28] were isolated using the b1 and pl1 systems, respectively. The first gene cloned was mop1, which encodes an RNA-dependent RNA polymerase (RDR), which is the predicted ortholog of RDR2 in Arabidopsis [29]. The mop1 gene is required for paramutation at multiple loci [15,24], is involved in Mutator transposon silencing [30,31] and is required for the accumulation of the vast majority of 24 nt siRNAs [32]; RDR2 in Arabidopsis is also required for 24 nt siRNA biogenesis [33].
Multiple studies in Arabidopsis have revealed that plant specific RNA Polymerases (Pol IV and Pol V) are required for siRNA biogenesis and transcriptional gene silencing [34-36]. Three mutants in maize have identified two genes required for maize paramutation that are highly similar to subunits of these plant-specific RNA polymerases, which when mutated reduce maize siRNAs. The rmr6 gene encodes a large subunit most similar to NRPD1 in Arabidopsis, which is the large subunit of Pol IV [37]. The mop2/rmr7 gene encodes a second largest subunit most similar to NRPD2/E2 [25,28], which in Arabidopsis, functions in both Pol IV and Pol V complexes [36]. Maize has three such subunits [25,28] and it remains unclear how many Pol IV/Pol V-related complexes maize contains [25,28,38]. The rmr1 gene is also required for the accumulation of siRNAs [39]. It encodes a putative chromatin-remodeling protein with a Snf2 domain that shares similarity to the helicase domain of DRD1, but it is likely not the ortholog of either DRD1 or CLSY1 in Arabidopsis and it falls into a distinct clade [39]. In Arabidopsis, RDR2, NRPD1, NRPD2/E2 and DRD1 are all components of an RNA-dependent DNA methylation (RdDM) pathway that mediates sequence-specific, chromatin based transcriptional gene silencing [40,41].
While there are clearly multiple genetic components in common between paramutation in maize and RdDM in Arabidopsis (Table 1), there are unique and distinct properties of paramutation in maize that are not shared with the RdDM pathway in Arabidopsis. For example, RdDM gene silencing is much less heritable than paramutation, as RdDM is rarely heritable if separated from the locus producing the siRNAs. Most dramatically, genes silenced by RdDM are not paramutagenic; they cannot silence active alleles [6,23].
Table 1.
All of the genes indicated in the model (Figure 1A) are listed. To simplify the Figure 1A diagram we have focused on the most extensively characterized Arabidopsis genes that have been recovered in multiple screens. A full description of genes identified through mutational analysis to prevent RdDM is found in [41].
| Arabidopsis RdDM Mutations | Maize Paramutation Mutants | Shared Protein Domains | Hypothesized Function |
|---|---|---|---|
| nrpd1 | rmr6 | Largest subunit of RNA polymerase IV | Biogenesis of 24 nt heterochromatic siRNAs |
| nrpd2/nrpe2 | Mop2/rmr7 | Second largest subunit of Pol IV and Pol V | Biogenesis of 24 nt heterochromatic siRNAs |
| dcl3 | NYIa | Rnase-III | Processing of double-stranded RNA to 24 nt heterochromatic siRNAs |
| rdr2 | mop1 | RNA dependent RNA polymerase | Biogenesis of 24 nt heterochromatic siRNAs and synthesis of double stranded RNA |
| ago4 | NYIa | PAZ/PIWI | RNA cleavage, 24 nt siRNA binding, de novo DNA methylation |
| hen1 | NYIa | RNA methyltransferase | Methylation of 21–24 nt small RNAs |
| drm2 | NYIa | DNA methyltransferase | De novo methylation (CG, CNG, CNN) |
| drd1 | NYIb | SNF2 | Chromatin remodeling |
NYI: “Not Yet Identified”: maize genes with strong homology exist (www.chromdb.org) but no mutants have been either identified or tested for their involvement in paramutation.
RMR1 shares similarity with the helicase domain of DRD1 but phylogenetic analysis put it into a different clade relative to either DRD1 or CLSY1 [39].
RNA as a signaling molecule that establishes trans-interactions
The requirement for all the genes described above strongly indicates that an RNA-dependent mechanism is critical for paramutation. In addition, the tandem repeats absolutely required for b1 paramutation are transcribed on both strands [29]. Recent results indicate that siRNAs are produced from the b1 tandem repeats in multiple alleles: B-I and B′, the alleles carrying seven tandem repeats and undergoing paramutation and, a neutral allele (b) not involved in paramutation that has a single copy of the repeat (M. Arteaga-Vazquez and Chandler, unpublished). The b1 tandem repeat siRNAs are reduced in both mop2 [25] and mop1 mutants (M. Arteaga-Vazquez and Chandler, unpublished), consistent with siRNA involvement in paramutation. However, the presence of b1 tandem repeat siRNAs in alleles that cannot undergo paramutation, suggests that the b1 tandem repeat siRNAs may not be sufficient for paramutation, similar to what is observed with FWA in Arabidopsis [22]. One explanation is that while siRNAs are produced from all alleles, it is a longer transcript that is actually the trigger for paramutation and this hypothetical trigger RNA is differentially produced in distinct alleles. While we cannot eliminate this model, we favor a direct role for b1 repeat siRNAs given the multiple mutants that are required for siRNA biogenesis that have been isolated in paramutation screens. One model is that the b1 tandem repeat siRNAs are required early in development when paramutation is established [14]; these are tissues that are not amenable to biochemical analyses of siRNAs. Recent work has shown that the b1 repeat siRNAs are not required to maintain the silencing state associated with B′, as a mop2 mutant that has a severe reduction in siRNAs maintains the silenced state. However the same mutation completely prevents the establishment of paramutation [25]. These results indicate that the steps involved in establishing the silent state and in maintaining it can be separated, suggesting different mechanisms. These results are similar to the observations with FWA, where siRNAs are necessary but not sufficient for silencing and are required to establish, but not maintain silencing [20].
Further evidence for a role for the b1 tandem repeat siRNAs in establishing paramutation comes from recent results artificially generating b1 tandem repeat siRNAs (L. Sidorenko and Chandler, unpublished). The experiment used transgenic plants that produced b1 tandem repeat siRNAs by expressing an inverted repeat of a b1 repeat unit under a strong constitutive promoter (35S CaMV); the type of construct used to induce RdDM in Arabidopsis. These transgenic plants were able to recapitulate all key features of paramutation, i.e. a naïve B-I allele was silenced and that silenced state was fully heritable and paramutagenic when the inducing transgene was segregated away (L. Sidorenko and Chandler, unpublished).
Model
We will focus our discussion of a model for paramutation (Table 1, Figure 1) using the maize paramutation system, b1, which has the most data on both the key sequences and proteins required. The key sequences required for b1 paramutation are multiple tandem repeats located ∼100 kb upstream, which are also required for high expression in B-I. Thus, our model discusses two roles for the tandem repeats. First, the tandem repeats contain an enhancer that functions in cis to upregulate the b1 gene [9]. We hypothesize that it is the chromatin structure differences within the b1 tandem repeats in B-I and B′ that determines whether the enhancer can function to increase b1 transcription (Figure 1A). Furthermore as discussed below and shown in Figure 1B, the repeats are necessary to mediate the trans-interactions that establish and maintain paramutation.
Figure 1. Model for paramutation at the b1 locus in maize.
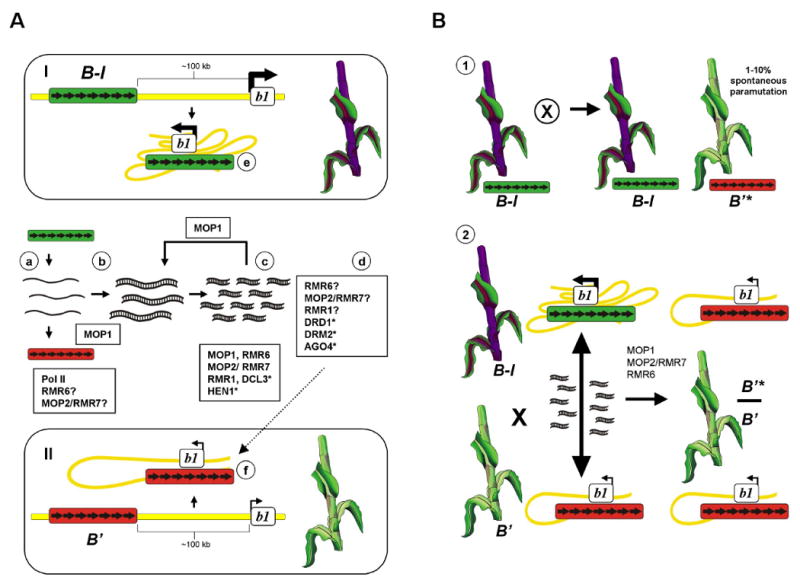
(A) Description of the two alleles and model for long distance cis regulation of b1 transcription. The two epialleles involved in paramutation, B-I (darkly pigmented) and B′ (lightly pigmented) exhibit different b1 transcription levels. The seven tandem repeats (depicted as black arrows) required to induce paramutation in maize are located ∼100 kb upstream of the transcription start site (TSS) of the b1 coding region. These tandem repeats also contain enhancer activity and interaction between the tandem repeats and the TSS is proposed to mediate high b1 transcription by a multi-loop structure in B-I (I) in contrast to a simple loop structure in B′ (II). a) B-I and B′ tandem repeats are bi-directionally transcribed at similar rates, with the majority of transcription by Pol II (Pol IV/V-like enzymes may also play a role, see text). b) transcripts derived from the tandem repeats are used to generate dsRNA either by pairing of opposite strands or through the activity of MOP1 (RDR), which may function in a reinforcement loop to maintain and amplify dsRNA. c) accumulation of short interfering siRNAs depends on the activity of MOP1, RMR6, MOP2/RMR7, RMR1 and hypothesized proteins (indicated by an asterisk) such as DCL3* and HEN1*. d) siRNA-guided chromatin based modification may involve the activity of RMR6, MOP2/RMR7, RMR1, DRD1*, DRM2* and AGO4*, and other RdDM related components. Experimental data indicate the alleles have two contrasting chromatin states with B-I (e) in an active conformation (green box) and B′ (f) in a silent conformation (red box). One model to explain how B-I can remain active in spite of producing siRNAs is that its chromatin structure is immune to silencing, potentially because RNA silencing complexes cannot assemble or the B-I repeats are not localized in the same subnuclear compartment as B′. (B) Model for paramutation and RNA-mediated trans-interactions between alleles. (1) B-I can spontaneously turn into B′ at a relatively high frequency (1-10%), indicating that while heritable, the B-I state is unstable. B′* symbolizes a B′ allele that was B-I in the previous generation. B′ and B′* are indistinguishable in terms of their paramutation activity and B′ (B′*) is extremely stable; no changes from B′ to B-I have ever been observed in wild type backgrounds. (2) When B-I and B′ are crossed paramutation always occurs; B-I is always changed into B′* (asterisk indicates that the newly generated B′ state was B-I in the previous generation). The genes required for paramutation are indicated and repeat siRNAs are assumed to be generated using a pathway similar to that diagramed in Panel A. Experimental evidence strongly suggests RNA plays a key role in mediating these trans-interactions. However, RNA may not be sufficient. One possibility is that B-I is efficiently silenced in the presence of B′ through chromatin associated pairing with the B′ repeats that allows the RNA silencing complexes to assemble on B-I and establish a B′* state.
In a previous review multiple models have been discussed for the long distance cis regulation of high expression, drawing parallels with models deriving from studies on the globin loci and imprinted genes in mammals [42]. Recent evidence supports a looping model as proposed by Louwers et al., 2009 [13]. They used chromosome conformation capture (3C) analyses to demonstrate that the b1 tandem repeats interact with the transcription start site of the b1 gene differentially in B-I versus B′. There were more interactions potentially generating more loops in B-I relative to B′ (diagramed in Figure 1A). There is no evidence that the b1 repeat siRNAs are involved in enhancer activity, as there is no sequence homology between the b1 promoter region and the b1 tandem repeat siRNAs. Thus, we favor a model where these interactions are mediated by proteins bound to the repeats and the promoter region.
The model for generation of b1 repeat siRNAs (Figure 1A) is a modification of models for RdDM in Arabidopsis [40,41]. Current data indicate that bi-directional transcription of the seven tandem repeats occurs at similar rates in both B-I and B′ [29]. The bulk of this transcription is most likely mediated by RNA Pol II, as b1 repeat transcription is blocked by alpha-amanitin concentrations that block Pol II (L. Sidorenko and Chandler, unpublished) and mutants in mop2 and mop1 show no reduction in repeat transcription as measured by nuclear run-on assays (L. Sidorenko and Chandler, unpublished) [25]. Clearly Pol IV/Pol V-like enzymes (MOP2/RMR7 and RMR6) are absolutely required for paramutation and production of b1 repeat siRNAs, but their exact role(s) remains to be determined as these enzymes are not contributing to the majority of transcription from the b1 repeats. It is possible that Pol IV/V-like enzymes in maize are sensitive to low levels of alpha-amanatin, and that they do contribute to generation of some primary transcripts. It is also possible that the Pol IV/V-like enzymes function further downstream at one or more of the steps indicated in Figure 1A. Subsequent amplification by RDR (mop1) leads to the production of double stranded RNA (dsRNA) and this dsRNA is processed into siRNAs by multiple enzymes not yet shown to be required for paramutation in maize (Figure 1A). The siRNAs are then hypothesized to induce chromatin modifications within the b1 repeats, through mechanisms that have not been elucidated in any system. Potentially RMR1, MOP2/RMR7 and RMR6 and many other proteins yet to be described, contribute to inducing and/or maintaining the chromatin modifications in addition to their roles in generation of siRNAs. We hypothesize that the chromatin state of B-I makes it relatively immune to the RNA-directed chromatin modifications, such that most of the time it remains in an actively transcribed state in spite of producing similar levels of siRNAs as B′.
The second role of the b1 tandem repeats is to mediate the communication in trans between B-I and B′ that turns B-I into B′. We hypothesize the trans communication is mediated at least in part through b1 tandem repeat siRNAs, which could provide the homology target to guide the transfer of the B′ chromatin structure to B-I (Figure 1B). While RNA clearly is involved, DNA/DNA or DNA/Protein/DNA interactions between the B′ and B-I repeats may also be occurring. Potentially, proteins that bind to the b1 tandem repeats could form higher order complexes and bring the two alleles in close proximity, much like what has been proposed for the role of zeste in trans-communication in Drosophila [43]. These types of postulated pairing interactions have been observed with transgene repeat arrays in Drosophila [44]. Close proximity of the two alleles could mediate localization to a “silencing” nuclear compartment where siRNAs can mediate silencing through establishment of heritable chromatin states. Chromatin structure differences between B-I and B′ could influence the binding of these postulated proteins, with stochastic changes at B-I enabling binding that leads to a B′ state.
Maize siRNA Biogenesis and Development
Mutations in three genes that prevent paramutation [mop1, mop2 and rmr6] show pleiotropic developmental phenotypes including reduced height, compromised sex determination, altered leaf polarity, late flowering and reduced fertility [24,25,27]. One possible explanation for the maize phenotypes is that miss regulation of siRNAs leads to miss regulation of key genes required for normal maize development. Consistent with this hypothesis, a decrease in the levels of miR156 and an increase in transcript levels of many putative mir156-targets was observed in tassels exhibiting a tasselseed phenotype in both mop1 and tasselseed 1 (ts1) mutants [45]. Interestingly, the decrease in miR156 levels in mop1 mutants correlates with a decrease in pri-miRNA transcript levels, which raises the possibility that loss of MOP1 influences miRNA biogenesis at early steps either during transcription or stability of pri-miRNAs (M. Arteaga-Vazquez and Chandler, unpublished). Other data are also consistent with an indirect role for siRNAs in maize development. Recent results show that transcript levels of a large proportion of chromatin modification genes and almost all of the genes implicated by homology to the Arabidopsis RdDM pathway are reduced in shoot apical meristems of the mop1 mutant [46]. In unpublished work, we have shown that levels of several miRNAs, including miR156, miR160, miR168 and miR169 are reduced in immature ears from mop1 and mop2 mutants (M. Arteaga-Vazquez and Chandler, unpublished).
As maize has nearly 80% of its genome composed of transposons [47], which are targeted by the majority of siRNAs [32,48], another contributing factor could be that transposon silencing is relieved when siRNAs are dramatically decreased, which could result in the generation of epimutations or mutations with developmental phenotypes. Consistent with this idea, Arabidopsis mutants defective in DNA methylation, such as ddm1 and met1 [49-52], show activation of transposable elements that can lead to mutations due to transposon insertions and epimutations due to miss regulation of nearby genes.
Conclusions
Paramutation is a fascinating process in which alleles or homologous sequences can communicate in trans to establish distinct expression states that are heritable over multiple generations. In the past few years a number of studies have revealed that paramutation in maize involves the establishment and heritable transmission of changes in chromatin and that the siRNA biogenesis pathway plays a critical role as mutations in multiple genes in that pathway prevent paramutation. However, multiple questions remain. Two questions are discussed below as a context for future directions.
Why are tandem repeats required for b1 paramutation?
Focusing on the b1 system, experiments examining an allelic series with differing numbers of repeats demonstrated that seven or five copies of the tandem repeat were highly paramutagenic, while alleles with a single repeat unit were completely inert [9]. Interestingly, an allele with three copies of the repeat was paramutagenic, but unstable and less penetrant, suggesting a mechanism for sensing the number of repeats exits [9]. One possibility is that repeats are needed to generate a large pool of either dsRNA or siRNAs. This model predicts that alleles with more repeats would show higher transcription rates and larger amounts of siRNAs, potentially proportional to the number of repeats. While theoretically attractive, current data are not consistent in that transcription is similar between alleles with different numbers of repeats [29] and there are only modest differences in the levels of siRNAs (M. Arteaga-Vaquez and Chandler, unpublished) among alleles, at least in the tissues examined to date.
Another possibility is that repeats are necessary to mediate some type of pairing interaction that is also required for paramutation. Tandem repeats of transgenes have been shown to mediate trans interactions in Drosophila through pairing [44,53]. It is possible that the b1 tandem repeats increase the local concentration of key proteins bound to those sequences resulting in distinct chromatin properties relative to the same sequence when unique. Distinct chromatin states could influence whether or not RNA silencing complexes assemble or target the sequences to distinct subnuclear compartments, with more repeats leading to more efficient complex assembly or subnuclear localization. RNA silencing complexes have been observed in distinct compartments in Arabidopsis [54] and subnuclear localization of repeats has been observed in Drosophila [55,56].
Cytological experiments to examine potential pairing interactions and RNAs present in different alleles, especially during early embryogenesis when paramutation is established [14] would be one approach to investigate these possibilities.
What is the function of paramutation?
Previous reviews have discussed at some length potential roles for paramutation [6,7], so herein we will only briefly summarize potential functions. One highly speculative role, given that in plants somatic tissues give rise to meiotic cell precursors and eventually gametes, is that paramutation provides a mechanism for transferring environmentally adapted expression states to offspring. Another possibility arising from the observation that paramutation requires repeats that are structurally similar to repeats at centromeres, which are also regulated by RNAi processes [57,58], is that paramutation derived from regulatory systems that evolved to defend against invasive viruses and transposons [59,60]. Paramutation, which involves trans interactions among homologous sequences may also provide a mechanism for generating functional homozygosity in polyploids [61].
Characterization of additional genes required for paramutation may shed light on potential functions for paramutation. There are a number of mutations defective in paramutation that have not yet been cloned and screens are far from saturated, so much remains to be learned. Another promising direction will be to identify other genes undergoing paramutation in maize. To date five genes have been shown to undergo paramutation in maize [1-5]. Four of these five genes encode transcription factors that regulate purple and red flavonoid pigments, which provide exceptional visual markers. The ease of simply looking at plant or seed color to identify changes in gene expression of these regulatory genes is likely to have facilitated the discovery of paramutation. Thus, we suspect paramutation may be much more common, at least in maize, than previously appreciated. Approaches to examine global expression patterns in multiple paramutation, especially of genes associated with tandem repeats might identify other genes that can undergo paramutation. Knowledge of other genes subject to the paramutation regulatory system could contribute toward understanding how widespread paramutation is and may also provide insight into potential functions.
Acknowledgments
We are grateful to past and present members of the Chandler laboratory. We thank Lyudmila Sidorenko for sharing unpublished results and Jocelyn E. Arteaga-Vazquez for her assistance with artwork. The work from the Chandler laboratory was supported by grants from the National Science Foundation (NSF) and the National Institutes of Health (NIH).
Footnotes
Annotations
** Stam M, Belele C, Dorweiler JE, Chandler VL: Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev 2002, 16:1906-1918.
This study shows that the key sequences required for paramutation are seven tandem repeats located ∼100 Kb upstream of the transcription start site of the b1 gene and demonstrates that the chromatin structure in the epialleles, B-I and B′, is different.
* Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler JE, White J, Sikkink K, Chandler VL: An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 2006, 442:295-298.
The authors report the cloning of the mop1 gene, whose molecular nature revealed that an RNAi-based mechanism is involved in paramutation in maize and suggested a role for RNA as a signaling molecule mediating trans-interactions and trans-generational silencing.
* Erhard KF, Jr., Stonaker JL, Parkinson SE, Lim JP, Hale CJ, Hollick JB: RNA polymerase IV functions in paramutation in Zea mays. Science 2009, 323:1201-1205.
This study show evidence in support of an RNAi-based mechanism involved in paramutation by demonstrating that rmr6 encodes the largest subunit of a predicted Pol IV RNA polymerase required to mediate heritable epigenetic changes and proper maize development.
** Chan SW, Zhang X, Bernatavichute YV, Jacobsen SE: Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol 2006, 4:e363.
This study shows that tandem repeats in Arabidopsis thaliana are involved but not sufficient to establish heritable trans-interactions as those observed in paramutation in maize.
* Suter CM, Martin DI: Paramutation: the tip of an epigenetic iceberg? Trends Genet 2009.
The authors present a current view of the diversity of paramutation and paramutation-like phenomena and discuss the possibility of a broad distribution of paramutation in eukaryotes.
** Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al.: The B73 maize genome: complexity, diversity, and dynamics. Science 2009, 326:1112-1115.
This study represents a milestone in plant biology. The sequence of the highly repetitive maize genome, one of the most important crop plants for humanity and a classical biological research model.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brink RA. A Genetic Change Associated with the R Locus in Maize Which Is Directed and Potentially Reversible. Genetics. 1956;41:872–889. doi: 10.1093/genetics/41.6.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coe EH. A Regular and Continuing Conversion-Type Phenomenon at the B Locus in Maize. Proc Natl Acad Sci U S A. 1959;45:828–832. doi: 10.1073/pnas.45.6.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hollick JB, Patterson GI, Coe EH, Jr, Cone KC, Chandler VL. Allelic interactions heritably alter the activity of a metastable maize pl allele. Genetics. 1995;141:709–719. doi: 10.1093/genetics/141.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sidorenko LV, Peterson T. Transgene-induced silencing identifies sequences involved in the establishment of paramutation of the maize p1 gene. Plant Cell. 2001;13:319–335. doi: 10.1105/tpc.13.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pilu R, Panzeri D, Cassani E, Cerino Badone F, Landoni M, Nielsen E. A paramutation phenomenon is involved in the genetics of maize low phytic acid1-241 (lpa1-241) trait. Heredity. 2009;102:236–245. doi: 10.1038/hdy.2008.96. [DOI] [PubMed] [Google Scholar]
- 6.Chandler VL. Paramutation: from maize to mice. Cell. 2007;128:641–645. doi: 10.1016/j.cell.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Chandler VL, Stam M. Chromatin conversations: mechanisms and implications of paramutation. Nat Rev Genet. 2004;5:532–544. doi: 10.1038/nrg1378. [DOI] [PubMed] [Google Scholar]
- 8.Suter CM, Martin DI. Paramutation: the tip of an epigenetic iceberg? Trends Genet. 2009 doi: 10.1016/j.tig.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stam M, Belele C, Dorweiler JE, Chandler VL. Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 2002;16:1906–1918. doi: 10.1101/gad.1006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stam M, Belele C, Ramakrishna W, Dorweiler JE, Bennetzen JL, Chandler VL. The regulatory regions required for B′ paramutation and expression are located far upstream of the maize b1 transcribed sequences. Genetics. 2002;162:917–930. doi: 10.1093/genetics/162.2.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandler VL, Radicella JP, Robbins TP, Chen J, Turks D. Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R genomic sequences. Plant Cell. 1989;1:1175–1183. doi: 10.1105/tpc.1.12.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandler VL, Eggleston WB, Dorweiler JE. Paramutation in maize. Plant Mol Biol. 2000;43:121–145. doi: 10.1023/a:1006499808317. [DOI] [PubMed] [Google Scholar]
- 13.Louwers M, Bader R, Haring M, van Driel R, de Laat W, Stam M. Tissue- and expression level-specific chromatin looping at maize b1 epialleles. Plant Cell. 2009;21:832–842. doi: 10.1105/tpc.108.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coe EH. The Properties, Origin, and Mechanism of Conversion-Type Inheritance at the B Locus in Maize. Genetics. 1966;53:1035–1063. doi: 10.1093/genetics/53.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sidorenko L, Chandler V. RNA-dependent RNA polymerase is required for enhancer-mediated transcriptional silencing associated with paramutation at the maize p1 gene. Genetics. 2008;180:1983–1993. doi: 10.1534/genetics.108.095281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sidorenko LV, Li X, Cocciolone SM, Chopra S, Tagliani L, Bowen B, Daniels M, Peterson T. Complex structure of a maize Myb gene promoter: functional analysis in transgenic plants. Plant J. 2000;22:471–482. doi: 10.1046/j.1365-313x.2000.00750.x. [DOI] [PubMed] [Google Scholar]
- 17.Eggleston WB, Alleman M, Kermicle JL. Molecular organization and germinal instability of R-stippled maize. Genetics. 1995;141:347–360. doi: 10.1093/genetics/141.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kermicle JL, Eggleston WB, Alleman M. Organization of paramutagenicity in R-stippled maize. Genetics. 1995;141:361–372. doi: 10.1093/genetics/141.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kermicle JL. Epigenetic silencing and activation of a maize r gene. In: Russo VEA, Martienssen RA, Riggs AD, editors. Epigenetic mechanisms of gene expression. Cold Spring Harbor Laboratory Press; 1996. pp. 267–287. [Google Scholar]
- 20.Panavas T, Weir J, Walker EL. The structure and paramutagenicity of the R-marbled haplotype of Zea mays. Genetics. 1999;153:979–991. doi: 10.1093/genetics/153.2.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chopra S, Athma P, Li XG, Peterson T. A maize Myb homolog is encoded by a multicopy gene complex. Mol Gen Genet. 1998;260:372–380. doi: 10.1007/s004380050906. [DOI] [PubMed] [Google Scholar]
- 22.Chan SW, Zhang X, Bernatavichute YV, Jacobsen SE. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol. 2006;4:e363. doi: 10.1371/journal.pbio.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 24.Dorweiler JE, Carey CC, Kubo KM, Hollick JB, Kermicle JL, Chandler VL. mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell. 2000;12:2101–2118. doi: 10.1105/tpc.12.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sidorenko L, Dorweiler JE, Cigan AM, Arteaga-Vazquez M, Vyas M, Kermicle J, Jurcin D, Brzeski J, Cai Y, Chandler VL. A dominant mutation in mediator of paramutation2, one of three second-largest subunits of a plant-specific RNA polymerase, disrupts multiple siRNA silencing processes. PLoS Genet. 2009;5:e1000725. doi: 10.1371/journal.pgen.1000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollick JB, Chandler VL. Genetic factors required to maintain repression of a paramutagenic maize pl1 allele. Genetics. 2001;157:369–378. doi: 10.1093/genetics/157.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkinson SE, Gross SM, Hollick JB. Maize sex determination and abaxial leaf fates are canalized by a factor that maintains repressed epigenetic states. Dev Biol. 2007;308:462–473. doi: 10.1016/j.ydbio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Stonaker JL, Lim JP, Erhard KF, Jr, Hollick JB. Diversity of Pol IV function is defined by mutations at the maize rmr7 locus. PLoS Genet. 2009;5:e1000706. doi: 10.1371/journal.pgen.1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler JE, White J, Sikkink K, Chandler VL. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- 30.Lisch D, Carey CC, Dorweiler JE, Chandler VL. A mutation that prevents paramutation in maize also reverses Mutator transposon methylation and silencing. Proc Natl Acad Sci U S A. 2002;99:6130–6135. doi: 10.1073/pnas.052152199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woodhouse MR, Freeling M, Lisch D. Initiation, establishment, and maintenance of heritable MuDR transposon silencing in maize are mediated by distinct factors. PLoS Biol. 2006;4:e339. doi: 10.1371/journal.pbio.0040339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nobuta K, Lu C, Shrivastava R, Pillay M, De Paoli E, Accerbi M, Arteaga-Vazquez M, Sidorenko L, Jeong DH, Yen Y, et al. Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc Natl Acad Sci U S A. 2008;105:14958–14963. doi: 10.1073/pnas.0808066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu C, Kulkarni K, Souret FF, MuthuValliappan R, Tej SS, Poethig RS, Henderson IR, Jacobsen SE, Wang W, Green PJ, et al. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006;16:1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 35.Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 36.Pikaard CS, Haag JR, Ream T, Wierzbicki AT. Roles of RNA polymerase IV in gene silencing. Trends Plant Sci. 2008;13:390–397. doi: 10.1016/j.tplants.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erhard KF, Jr, Stonaker JL, Parkinson SE, Lim JP, Hale CJ, Hollick JB. RNA polymerase IV functions in paramutation in Zea mays. Science. 2009;323:1201–1205. doi: 10.1126/science.1164508. [DOI] [PubMed] [Google Scholar]
- 38.Pikaard CS, Tucker S. RNA-silencing enzymes Pol IV and Pol V in maize: more than one flavor? PLoS Genet. 2009;5:e1000736. doi: 10.1371/journal.pgen.1000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hale CJ, Stonaker JL, Gross SM, Hollick JB. A novel Snf2 protein maintains trans-generational regulatory states established by paramutation in maize. PLoS Biol. 2007;5:e275. doi: 10.1371/journal.pbio.0050275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 41.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 42.Chandler VL, Stam M, Sidorenko LV. Long-distance cis and trans interactions mediate paramutation. Adv Genet. 2002;46:215–234. doi: 10.1016/s0065-2660(02)46008-7. [DOI] [PubMed] [Google Scholar]
- 43.Bickel S, Pirrotta V. Self-association of the Drosophila zeste protein is responsible for transvection effects. Embo J. 1990;9:2959–2967. doi: 10.1002/j.1460-2075.1990.tb07488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dorer DR, Henikoff S. Transgene repeat arrays interact with distant heterochromatin and cause silencing in cis and trans. Genetics. 1997;147:1181–1190. doi: 10.1093/genetics/147.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hultquist JF, Dorweiler JE. Feminized tassels of maize mop1 and ts1 mutants exhibit altered levels of miR156 and specific SBP-box genes. Planta. 2008;229:99–113. doi: 10.1007/s00425-008-0813-2. [DOI] [PubMed] [Google Scholar]
- 46.Jia Y, Lisch DR, Ohtsu K, Scanlon MJ, Nettleton D, Schnable PS. Loss of RNA-dependent RNA polymerase 2 (RDR2) function causes widespread and unexpected changes in the expression of transposons, genes, and 24-nt small RNAs. PLoS Genet. 2009;5:e1000737. doi: 10.1371/journal.pgen.1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 48.Hale CJ, Erhard KF, Jr, Lisch D, Hollick JB. Production and processing of siRNA precursor transcripts from the highly repetitive maize genome. PLoS Genet. 2009;5:e1000598. doi: 10.1371/journal.pgen.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johannes F, Porcher E, Teixeira FK, Saliba-Colombani V, Simon M, Agier N, Bulski A, Albuisson J, Heredia F, Audigier P, et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009;5:e1000530. doi: 10.1371/journal.pgen.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature. 2001;411:212–214. doi: 10.1038/35075612. [DOI] [PubMed] [Google Scholar]
- 51.Reinders J, Wulff BB, Mirouze M, Mari-Ordonez A, Dapp M, Rozhon W, Bucher E, Theiler G, Paszkowski J. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 2009;23:939–950. doi: 10.1101/gad.524609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stokes TL, Kunkel BN, Richards EJ. Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 2002;16:171–182. doi: 10.1101/gad.952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Csink AK, Bounoutas A, Griffith ML, Sabl JF, Sage BT. Differential gene silencing by trans-heterochromatin in Drosophila melanogaster. Genetics. 2002;160:257–269. doi: 10.1093/genetics/160.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pontes O, Pikaard CS. siRNA and miRNA processing: new functions for Cajal bodies. Curr Opin Genet Dev. 2008;18:197–203. doi: 10.1016/j.gde.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Csink AK, Henikoff S. Genetic modification of heterochromatic association and nuclear organization in Drosophila. Nature. 1996;381:529–531. doi: 10.1038/381529a0. [DOI] [PubMed] [Google Scholar]
- 56.Dernburg AF, Broman KW, Fung JC, Marshall WF, Philips J, Agard DA, Sedat JW. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell. 1996;85:745–759. doi: 10.1016/s0092-8674(00)81240-4. [DOI] [PubMed] [Google Scholar]
- 57.Martienssen RA, Kloc A, Slotkin RK, Tanurdzic M. Epigenetic inheritance and reprogramming in plants and fission yeast. Cold Spring Harb Symp Quant Biol. 2008;73:265–271. doi: 10.1101/sqb.2008.73.062. [DOI] [PubMed] [Google Scholar]
- 58.Morris CA, Moazed D. Centromere assembly and propagation. Cell. 2007;128:647–650. doi: 10.1016/j.cell.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 59.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 61.Mittelsten Scheid O, Afsar K, Paszkowski J. Formation of stable epialleles and their paramutation-like interaction in tetraploid Arabidopsis thaliana. Nat Genet. 2003;34:450–454. doi: 10.1038/ng1210. [DOI] [PubMed] [Google Scholar]


