Abstract
Background & Aims
Approximately half of the families that fulfill Amsterdam criteria for Lynch syndrome or hereditary non-polyposis colorectal cancer (HNPCC) do not have evidence of the germline mismatch repair (MMR) gene mutations that define this syndrome and result in microsatellite instability. The carcinogenic pathways and the best diagnostic approaches to detect microsatellite stable (MSS) HNPCC tumors are unclear. We investigated the contribution of epigenetic alterations to development of MSS HNPCC tumors.
Methods
Colorectal cancers were divided in four groups: 1. Microsatellite stable, Amsterdam positive (MSS HNPCC) (N=22); 2. Lynch syndrome cancers (identified mismatch repair mutations) (N=21); 3. Sporadic MSS (N=92); 4. Sporadic MSI (N=46). Methylation status was evaluated for CACNAG1, SOCS1, RUNX3, NEUROG1, MLH1, and LINE-1. KRAS and BRAF mutations status was analyzed.
Results
MSS HNPCC tumors displayed a significantly lower degree of LINE-1 methylation, marker for global methylation, than any other group. Whereas most MSS HNPCC tumors had some degree of CpG island methylation, none presented a high index of methylation. MSS HNPCC tumors had KRAS mutations exclusively in codon 12, but none harbored V600E BRAF mutations.
Conclusions
Tumors from Amsterdam-positive patients without mismatch repair deficiency (MSS HNPCC) have certain molecular features, including global hypomethylation that distinguish them from all other colorectal cancers. These characteristics could have an important impact on tumor behavior or treatment response. Studies are underway to further assess the cause and effects of these features.
Keywords: Colorectal cancer, Microsatellite stable Hereditary Non-Polyposis Colorectal Cancer, Non-Lynch HNPCC, DNA Methylation, hypomethylation
Background and Aims
Hereditary nonpolyposis colorectal cancer (HNPCC) is the most common colorectal cancer syndrome (∼3% of all colorectal cancers)1 and it has long been defined clinically through the Amsterdam I criteria2. These criteria reveal a syndrome without polyposis, an autosomal pattern of inheritance and a penetrance for cancer close to 80%. Subsequently, the Amsterdam II criteria were drafted in order to include other tumors that often associate with this syndrome3. Germline mutations in mismatch repair (MMR) genes are responsible for approximately half of the families that fulfill Amsterdam criteria of HNPCC and these tumors are now defined as Lynch syndrome colorectal cancers1. Mutations in the MMR genes MLH1 and MSH2 account for almost 90% of patients with Lynch syndrome4, 5. The remaining are located in the MSH66 or PMS27 genes. These mutations result in the absence of the corresponding protein's expression and cause a loss of DNA mismatch repair activity. This results in microsatellite instability (MSI) and a hypermutable phenotype, which is believed to underlie the rapid neoplastic progression of these tumors8. This molecular phenotype is also present in approximately 15% of sporadic colorectal cancers but, in contrast to Lynch syndrome patients, the underlying defect is somatic inactivation by biallelic promoter hypermethylation of MLH19.
Approximately half of the families that fulfill Amsterdam criteria of HNPCC do not have evidence of MMR deficiency and therefore their tumors are microsatellite stable (MSS)10. The observation that MSS HNPCC tumors constitute a distinct entity from Lynch syndrome was reported almost simultaneously in three different studies, one population based10, one clinic-based11 and one with mixed patients12. Lindor and colleagues proposed the term Familial Colorectal Cancer Type X for these MSS HNPCC families12. These studies established that these families have a lower risk of CRC cancer than Lynch syndrome families; the average age at diagnosis is older than Lynch patients, but about 10 years younger than sporadic cases; tumors are mostly on the left side of the colon and they do not have the characteristic lymphocytic infiltrate seen in MSI CRC. These families have a lower incidence of extra colonic malignancies. Because MSS HNPCC families fulfill Amsterdam criteria, they all display an autosomal dominant pattern of inheritance. However, further studies are still needed to clarify some aspects of these families due to the distinct findings among these three publications. Specifically, the risk of extracolonic malignancies was not found to be increased in Lindor's article as opposed to the other studies that showed slight increases in the risk of endometrial13 or both gastric and endometrial cancers11. Some of the discrepancies could be explained by selection bias as Lindor's study only included families fulfilling Amsterdam I criteria, or differences in ascertainment and classification between studies.
The carcinogenic pathways involved in the development of CRC in MSS HNPCC families are poorly understood. Abdel-Rahman et al.14 reported active Wnt signaling in a third of the cases, and no beta-catenin gene (CTNNB1) mutations in the MSS CRC. The frequency of P53 and KRAS mutations was also low in tumors without active Wnt signaling. A more recent study confirmed a low percentage of active Wnt signaling in MSS HNPCC tumors, and the frequency of APC mutations was reported as low as in Lynch tumors, with similar mutation profiles in both groups15. Taken together there is little evidence of the involvement of the common CRC carcinogenic pathways in MSS HNPCC cases.
As many as 30-40% of sporadic CRCs and a subset of Lynch syndrome tumors have been shown to harbor an epigenetic DNA methylation defect that causes transcriptional inactivation of candidate growth regulatory genes in the colon16,17. Alterations in the KRAS/BRAF signaling pathway have been reported to associate with a CpG island methylator phenotype or CIMP17. DNA methylation changes can be influenced by dietary and lifestyle factors, and can manifest either as biallelic events (in sporadic CRCs) or as mono-allelic events (in Lynch syndrome patients)18,17. On the other hand, global hypomethylation has also been implicated in the early steps of CRC development19,20 as it leads to reactivation of otherwise silenced proto-oncogenes21.
In view of the paucity of mutations observed in MSS HNPCC patients, and the fact that epigenetic changes are frequent mechanisms of gene silencing or activation of normally silenced genes in CRC, we hypothesized that epigenetic alterations may constitute a key mechanism in the evolution of MSS HNPCC cancers. In support of our hypothesis, we provide novel evidence that hypomethylation of LINE-1 retrotransposable elements, surrogate markers for global methylation, is frequently observed in MSS HNPCC tumors. The majority of these tumors also display some degree of hypermethylation at specific methylation markers but none show a high CpG island methylation index. Additionally, these epigenetic events occur independently of mutations in the KRAS/BRAF pathway.
Materials and Methods
Study sample
This study included patients from gastrointestinal cancer high-risk units at Baylor University Medical Center (Dallas, TX), The Dana-Farber Cancer Institute (Boston, MA), Hospital Comarcal Inca (Mallorca, Spain), Hospital Vírgen del Rocio (Sevilla, Spain), The University of Illinois at Chicago (Chicago, IL), and from the Epicolon I cohort13. Patients were included under the Institutional Review Board (IRB) approved protocol in the respective institutions and after written informed consent was obtained. Demographic, clinical, and tumor-related features of the proband, as well as a detailed family history of cancer were obtained expanding backward and laterally at least two generations. Samples were characterized for MSI status, MMR protein expression, BRAF and KRAS mutations, and methylation of 5 CIMP-related promoter markers and LINE-1 sequence. Patients with MSI tumors and/or tumors with absent expression of a MMR protein underwent germline mutation analysis for MLH1, and MSH2 genes. Samples were divided into 4 groups: 1) and 2) sporadic CRC cases with no family history of CRC or any other HNPCC-related tumors3, with and without MSI; 3) Lynch syndrome patients (with identified MMR gene mutations) and 4) MSS HNPCC patients (probands from families fulfilling Amsterdam I or II criteria and with no evidence of MMR deficiency: MSS, normal MMR protein expression, no MMR gene mutations.
Microsatellite instability analysis
MSI status was assessed as previously described22. Briefly, a single amplification of the quasi-monomorphic mononucleotide markers BAT26 and NR 24 was performed from tumor DNA. Each forward primer had a fluorescent tag, 6FAM™ or VIC™ (Applied Biosystems, Foster City, CA) at the 5′ end, to allow amplicon detection by the ABI 3730 DNA Analyzer (Applied Biosystems). Polymerase chain reaction (PCR) conditions were the following: 10μL of HotStartTaq Master Mix (Qiagen Science, Germantown, MD), including 1.5mM MgCl2, 2.5 units of HotStart Taq DNA polymerase, 0.2mM dNTPs, 0.5μM of each primer, and 30ng of DNA per reaction with a final volume of 20μL. Cycling steps were: initial Taq activation (95°C) for 15 min, and 36 cycles as follows: 94°C 20 sec, 55°C annealing 20 sec, 72°C 30 sec and final extension 72°C 10 min. PCRs were performed on a Veriti PCR system (Applied Biosystems). PCR products (≤ 2μL) were mixed with 9 μL of Hi-Di formamide (Applied Biosystems) and 0.5 μL of the size standard LIZ (Applied Biosystems). Samples were denatured at 95°C for 3 min and analyzed with GeneMapper (Applied Biosystems). Microsatellites were considered unstable if PCR fragments showed alterations ≥3bp at a given locus. Tumors were classified as MSI when either of the two markers was unstable.
MMR protein expression
MMR protein expression was assessed by immunohistochemical staining of MLH1, MSH2, and MSH6 as previously described22. Four micron-thick tumor sections were de-paraffinized and rehydrated using xylene and alcohol. Before immunostaining, antigen retrieval was performed by immersing sections in boiling citrate buffer (10 mmol/L, pH 6.0) in a pressure cooker for 5 minutes. Sections were then incubated for 20 minutes at room temperature with mouse monoclonal antibodies against MLH1 (clone G168-15, dilution 1:30; BD PharMingen, San Diego, CA), MSH2 (clone FE11, dilution 1:30; Oncogene Research Products, Boston, MA), or MSH6 (clone 44 MSH6-GTBP, dilution 1:100; BD Biosciences, San Jose, CA). The Ultra-Vision streptavidin–biotin peroxidase detection kit (Dako, Carpinteria, CA) was used as the secondary detection system according to manufacturer's instructions. Loss of MLH1, MSH2, or MSH6 expression was recorded when there was a complete absence of nuclear staining in neoplastic cells but there was nuclear staining in normal epithelial cells, lymphocytes, and stromal cells. When there was no nuclear staining of the internal control in three separate experiments, the results were considered inconclusive.
BRAF and KRAS gene mutation analysis
The most common mutations of BRAF (V600E)23 and KRAS (codons 12 and 13)24 were analyzed through direct sequencing. PCR conditions were: 10μL of HotStartTaq Master Mix (Qiagen Science), including 1.5mM MgCl2, 2.5 units of HotStart Taq DNA polymerase, 0.2mM dNTPs, 0.5μM of each primer, and 30ng of DNA per reaction with a final volume of 20μL. Cycling steps were: initial Taq activation (95°C) for 15 min, 36 cycles as follows: 94°C 20 sec, annealing for 20 sec at 50°C for BRAF and 55°C for KRAS, 72°C 30 sec and final extension 72°C 10 min. Primers are shown in the Supplementary table 1. The PCR products were purified using a 96-well vacuum filter system from Millipore (Billerica, MA) and the QIAquick PCR purification Kit (Qiagen Science). Amplified fragments went through a PCR sequencing amplification. Briefly, the PCR product was amplified using ABI Big Dye Terminator Sequencing Kit, v3.1 (Applied Biosystems). The resulting PCR product was purified with the ABI BigDye XTerminator Purification kit (Applied Biosystems). The cleaned product was loaded into a 3730 DNA Analyzer also from ABI. Sequence histograms were analyzed searching for heterozygous and homozygous substitutions.
Mutation Analysis of MMR genes
Lynch syndrome patients had pathogenic mutations in MLH1 or MSH2 MMR genes, diagnosed either in the research laboratory or at Myriad Genetic Laboratories Inc. (Salt Lake City, UT). There were no cases of unique loss of MSH6 protein expression and therefore MSH6 mutation analysis was not performed in this series. In the former case, genes underwent germline genetic testing by both multiple ligation probe amplification (MLPA) analysis and sequencing. MLPA was performed using the MLH1/MSH2 exon deletion assay (MRCHolland, Amsterdam, the Netherlands), which allows the detection of genomic rearrangements in these genes. Thirty-four ligation products were amplified using a fluorescently labeled primer and analyzed on an ABI 3100 sequencer using GeneScan and Genotyper Analysis software (Applied Biosystems). Control DNA samples with known MSH2 or MLH1 genomic rearrangements were included in each batch of experiments. MLPA results were confirmed by RT-PCR encompassing contiguous exons of the suspected deleted fragment. Germline mutations in the MSH2 and MLH1 genes were also sought by direct exon-by-exon sequencing. Amplification products were generated with primers located in the flanking introns approximately 50 base pairs from the respective intron/exon borders to detect all possible splice junction mutations. The sequences were determined on the genetic analyzer (ABI 3100, Applied Biosystems) using fluorescently labeled primers and protocols supplied by the manufacturer.
Analysis of tumor methylation status
As reported previously, the following five markers were used to assess the methylation status of the tumor tissue: CACNAG1, SOCS1, RUNX3, NEUROG1, MLH125. Long interspersed nucleotide element-1 (LINE-1) methylation was used as a surrogate marker for genome-wide methylation. Previous studies have shown that LINE-1 methylation correlates with global DNA methylation26,27. DNA was modified with sodium-bisulfite using the EZ Methylation Gold Kit (Zymo Research, Orange, CA) and markers were amplified by PCR containing bisulfite DNA, Hotstartaq, forward primer, biotinylated reverse primers and water. Each marker has a different primer set as well as sequencing primer (Supplementary table 1). Four microliters of PCR product were added to 38 μl of binding buffer (Biotage, Uppsala, Sweden), 2 μl streptavidin sepharose high-performance beads (GE Healthcare, Buckinghamshire, England) and 36 μl of sterile water. Single-stranded biotinylated templates were isolated using the PyroMark Vacuum Prep WorkStation (Biotage). The products were dispensed into 96 well plates containing 0.36 μl 10 μM sequencing primer and 11.64 μl annealing buffer (Biotage) at 80°C for 3 min, and then placed at room temperature for 10 min. Pyrosequencing reactions were carried out in the Pyro-Mark MD (Qiagen, Hilden, Germany) using PyroGold reagents and results were analyzed using pyro Q-CpG Software (Biotage). To assess the total methylation status for each marker we calculated the mean percentage of methylation for all these CpG sites. We classified each marker as methylated when the mean percentage was higher than 5% for the following markers: CACNAG1, SOCS1, RUNX3 and MLH1; and 10% for NEUROG1 marker. Cut offs were chosen after determining the average methylation level of each marker in healthy mucosa samples plus two standard deviations. Final results were represented as Methylation index (MI), a simple representation of total number of CIMP markers methylated in each tumor. A tumor was considered to have a “Low MI” if it had up to 3 methylated markers and a “High MI” if it had 4 or 5 methylated markers.
Statistical analysis
The χ2 or Fisher exact test was applied for comparison between categorical variables. To compare continuous variables, when equal variances were assumed, we used the T-test. However, when variances were not equal, we used the Kruskal-Wallis one-way analysis of variance followed or the non-parametric Mann-Witney U test. To perform pairwise comparisons, we applied the Mann-Witney U test when the Kruskal-Wallis test showed a significant difference. In this case, we set the alpha error rate to the usual (.05) divided by the total number of comparisons to control for Type I error. All calculations were performed using the 17.0 SPSS software package (SPSS Inc., Chicago, IL).
Results
CRC cases were divided in 4 groups according to molecular features and family history of cancer. Group 1 included 22 probands from families fulfilling Amsterdam criteria for HNPCC but with no evidence of MMR deficiency (MSS HNPCC). Group 2 included 21 Lynch syndrome probands with identified pathogenic MMR gene mutations. Group 3 included 92 sporadic CRC patients with no family history of CRC or any Lynch-related cancers, and no evidence of MMR deficiency (MSS sporadic). Group 4 was comprised of 46 mismatch repair deficient tumors and no family history for CRC or any Lynch-related cancers (MSI sporadic).
Clinical and molecular features of Group 1 (MSS HNPCC)
Patients were included in this group according to the previously reported features describing it as a distinct entity13. These families display an autosomal dominant pattern of inheritance, but they have a lower number of affected family members than in Lynch syndrome; they do not have a significant number of polyps; and their cancers are MMR-proficient. Table 1 summarizes the clinical and molecular features of the cases included in this study. Fourteen families fulfilled Amsterdam I criteria for HNPCC, and 5 fulfilled Amsterdam II. The mean age for diagnosis of CRC was 59 years in the probands and 57 considering all affected family members. The majority of the tumors (80%) were located in the left side of the colon. All tumors were wild type for BRAF V600E mutation (one was not evaluated due to the lack of available DNA) and 32% had KRAS mutations (all in codon 12) (Table 1).
Table 1. Molecular and clinical features of the 22 MSS HNPCC patients.
Tumor location: R: right side of the colon including cecum, ascending colon, transverse colon up to splenic flexure. L: left side of the colon including descending colon and recto-sigmoid. wt: wild type. mut: mutated
| BRAF V600E | KRAS Codon 12 | KRAS Codon 13 | Gender | Tumor location | Amsterdam I or II | Age of the proband | Mean age of affected family members | |
|---|---|---|---|---|---|---|---|---|
| 1 | wt | wt | wt | F | R | I | 79 | 61 |
| 2 | wt | mut | wt | F | L | II | 89 | 60 |
| 3 | wt | wt | wt | M | L | II | 64 | 59 |
| 4 | wt | wt | wt | M | R | I | 49 | 51 |
| 5 | wt | wt | wt | M | L | I | 52 | 47 |
| 6 | wt | wt | wt | M | L | I | 47 | 67 |
| 7 | mut | wt | M | L | I | 50 | 63 | |
| 8 | wt | wt | wt | M | L | I | 63 | 59 |
| 9 | wt | mut | wt | M | L | I | 55 | 53 |
| 10 | wt | wt | wt | M | L | I | 51 | 58 |
| 11 | wt | mut | wt | M | L | I | 73 | 49 |
| 12 | wt | wt | wt | M | R | I | 66 | 58 |
| 13 | wt | wt | wt | M | L | I | 52 | 54 |
| 14 | wt | wt | wt | F | R | I | 67 | 59 |
| 15 | wt | wt | wt | M | L | I | 48 | 59 |
| 16 | wt | wt | wt | M | L | II | 79 | 63 |
| 17 | wt | wt | wt | F | L | II | 80 | 62 |
| 18 | wt | wt | wt | M | I | 47 | 61 | |
| 19 | wt | mut | wt | F | L | I | 54 | 52 |
| 20 | wt | wt | wt | M | L | II | 44 | 54 |
| 21 | wt | mut | wt | M | L | I | 38 | 57 |
| 22 | wt | mut | wt | M | L | I | 40 | 55 |
Presence of CpG island hypermethylation in hereditary and sporadic colorectal tumors
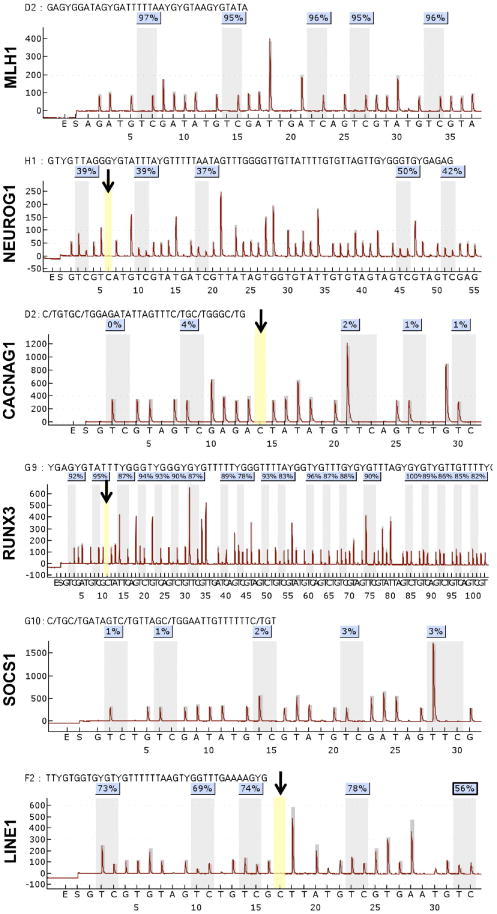
Herein, we first determined as to the total number of CIMP markers (CACNAG1, SOCS1, RUNX3, NEUROG1, and MLH1) required to be methylated for a tumor to clearly correlate with the clinical and molecular phenotype commonly associated with CIMP; such as presence of MSI, mutated BRAF, less often mutated KRAS, females, right sided CRC, and older age. After studying all potential combinations, we determined that when 4 of the 5 markers were methylated, tumors showed statistically significant differences in all the above-mentioned features associated with CIMP in comparison with tumors with 3 or less methylated markers. Accordingly, we categorized tumors as “Low MI” in the samples that had up to 3 methylated markers and “High MI” where at least 4 markers were methylated (Table 2). When methylation status was studied in the four different cancer groups, the overwhelming majority of tumors with a high MI came from the MSI sporadic group. Over two thirds of all MSI sporadic cancers had a high MI. Only a small proportion of Lynch syndrome and MSS sporadic tumors had a high MI, while none of the MSS HNPCC tumors did. On the other hand, some degree of promoter methylation was not only seen in sporadic cases and Lynch syndrome tumors as previously reported16,17, but also in most MSS HNPCC tumors (Table 3). Figure 1 depicts a representative pyrogram of all methylation markers.
Table 2. Association of methylation index (MI) with other molecular features and clinical data.
| Low MI | High MI | ||
|---|---|---|---|
| MSI status | P<0.001 | ||
| MSI | 50.7% (34) | 49.3% (33) | |
| MSS | 96.5% (109) | 3.5% (4) | |
| KRAS | P = 0.006 | ||
| wild type | 74.6% (100) | 25.4% (34) | |
| mutant | 93.5% (43) | 6.5% (3) | |
| BRAF | P<0.001 | ||
| wild type | 85.4% (140) | 14.6% (24) | |
| mutant | 13.3% (2) | 86.7% (13) | |
| Gender | P = 0.010 | ||
| Male | 86.7% (85) | 13.3% (13) | |
| Female | 70.7% (58) | 29.3% (24) | |
| Tumor Location | P<0.001 | ||
| Right side | 64.7% (55) | 35.5% (30) | |
| Left side | 92.2% (83) | 7.8% (7) | |
| Age | 64.6 (143) | 73.0 (37) | P = 0.004 |
Table 3. Methylation index (MI) in hereditary and sporadic CRC tumors.
| Low MI | High MI | |
|---|---|---|
| MSS HNPCC | 100% (22) | 0% (0) |
| Lynch syndrome | 90.5% (19) | 9.5% (2) |
| MSI sporadic | 32.6% (15) | 67.4% (31) |
| MSS sporadic | 95.6% (87) | 4.4% (4) |
Figure 1. Pyrograms of methylation markers MLH1, NEUROG1, CACNAG1, RUNX3, SOCS1, and LINE-1.
The black arrow points the internal control cytosine residues that check for the adequacy of bisulfite treatment in the pyrosequencing reactions. No signal amplification was seen for the internal control suggesting that 100% of the DNA samples used for various CIMP markers were satisfactorily converted by bisulfite treatment.
Involvement of the RAS/RAF signaling pathway in hereditary and sporadic colorectal tumors
We next studied the implication of KRAS and BRAF mutations in the different CRC groups. As expected, MSI sporadic tumors were frequently mutated at BRAF (28.3%; Table 4). However, neither Lynch syndrome, nor MSS HNPCC tumors exhibited V600E BRAF mutations. In addition, only a small proportion of MSS sporadic cancers (2/92; ∼2%) carried BRAF mutations. As the overwhelming majority of BRAF mutated tumors were MSI sporadic cancers and BRAF mutations were clearly associated with a high MI (Table 2), a mechanism implicating BRAF and CIMP seems to be exclusively implicated in a significant proportion of MSI sporadic tumors.
Table 4. Mutation in the RAS-RAF pathway in hereditary and sporadic CRC tumors.
P < 0.001 for BRAF and KRAS comparisons among CRC groups. P = 0.001 for codon 12 and P = 0.033 for codon 13 comparisons among groups.
| BRAF mutation | KRAS mutation | codon 12 | codon 13 | |
|---|---|---|---|---|
| MSS HNPCC | 0% | 31.8% (7) | 31.8% (7) | 0% |
| Lynch syndrome | 0% | 9.5% (2) | 9.5% (2) | 0% |
| MSI sporadic | 28.3% (13) | 4.4% (2) | 2.2% (1) | 2.2% (1) |
| MSS sporadic | 2.2% (2) | 39.2% (36) | 27.2% (25) | 12% (11) |
On the other hand, KRAS mutations were present in 9.5% of Lynch syndrome tumors, 31.8% of MSS HNPCC, 39.2% of MSS sporadic cases and 4.4% of MSI sporadic cancers (Table 4). Although the frequency of KRAS mutations was not significantly different between sporadic MSS and MSS HNPCC cancers, the latter showed mutations solely in codon 12. KRAS codon 13 mutations were only seen in sporadic cancers. Not surprisingly, concomitant KRAS and BRAF mutations were not observed in any tumor subtypes.
LINE-1 hypomethylation in hereditary and sporadic colorectal tumors
As none of the MSS HNPCC tumors displayed a high index of candidate tumor suppressor locus hypermethylation, we hypothesized that these tumors might evolve through the acquisition of CIN due to genome-wide hypomethylation. LINE-1 methylation is a surrogate marker for genome-wide methylation of intronic CpG dinucleotides, and hypomethylation of these CpG sequences has been associated with chromosomal instability (CIN)28.
We performed quantitative pyrosequencing for LINE-1 sequences (Figure 1) and observed that LINE-1 methylation was positively associated with the presence of MSI, high MI, and right-sided CRCs (Table 5). Interestingly, when we analyzed LINE-1 methylation in the 4 subtypes of colonic tumors, we identified that MSS HNPCC cancers had the lowest degree of LINE-1 methylation (60.08%) and this was significantly different from the other three groups of CRCs (Table 6). These findings clearly suggest that a higher degree of genome-wide hypomethylation distinguishes MSS HNPCC from other CRCs and is indicative of increased CIN in these neoplasms.
Table 5. LINE-1 methylation according to other molecular features and clinical data.
Mean Rank value when applying the Mann-Witney U test. *T test applied
| Mean % LINE-1 methylation | Mean Rank | ||
|---|---|---|---|
| MI | P = 0.003 | ||
| high | 68.17 (138) | 109.66 | |
| low | 64.42 (37) | 82.19 | |
| MS status | P = 0.007 | ||
| MSI | 66.97 (65) | 102.15 | |
| MSS | 64.18 (111) | 80.51 | |
| KRAS | P = 0.441 | ||
| wild type | 65.22 (130) | 90.26 | |
| mutant | 65.18 (46) | 83.52 | |
| BRAF* | P = 0.494 | ||
| wild type | 65.09 (160) | ||
| mutant | 66.32 (15) | ||
| Gender* | P = 0.272 | ||
| male | 64.70 (95) | ||
| female | 65.80 (81) | ||
| Location | P = 0.001 | ||
| right | 67.10 (81) | 99.69 | |
| left | 63.53 (90) | 73.68 | |
| Age | P = 0.064 | ||
| ≤50 | 64.77 (32) | 85.86 | |
| 50-70 | 63.35 (51) | 75.77 | |
| ≥70 | 66.37 (93) | 96.39 |
Table 6. LINE-1 methylation in hereditary and sporadic CRC tumors.
Mean Rank value when applying the Kruskal-Wallis test. P values for comparison between MSS HNPCC and the other types of CRC, using the Mann-Witney U test. We performed a total of 3 comparisons, therefore differences were statistically significant when be P<(.05/3)=<.017.
| Mean % LINE1 methylation | Mean Rank | P | |
|---|---|---|---|
| MSS HNPCC | 60.08 (21) | 56.05 | |
| Lynch syndrome | 66.29 (20) | 94.80 | 0.015 |
| MSI sporadic | 67.27 (45) | 105.41 | 0.001 |
| MSS sporadic | 65.13 (90) | 86.22 | 0.009 |
Discussion
The main objectives of this study were to elucidate the potential role of epigenetic events in the carcinogenic process of CRC from patients with MSS HNPCC tumors. Furthermore, we asked if those could be related to activating mutations of the RAS/RAF signaling pathway.
Two epigenetic changes have been found to lead to distinct carcinogenic pathways in the mammalian genome29. The first one consists of hypermethylation of CpG islands in gene promoter regions that results in transcriptional silencing of tumor suppressor genes30. The second consists of global hypomethylation that leads to reactivation of otherwise silenced proto-oncogenes21.
CpG island methylator phenotype or CIMP is present in as many as 30-40% of all CRCs16. CIMP tumors have a distinct clinical, pathological, and molecular profile, such as associations with proximal tumor location, female sex, poor differentiation, MSI as a consequence of hypermethylation of MLH1 gene, high BRAF, and low TP53 mutation rates25, 31-33. Presently there is no consensus regarding the best markers for the evaluation of methylation status, and how many methylated markers are needed in order to classify a tumor as CIMP. The markers we used included CACNAG1, SOCS1, RUNX3, and NEUROG1, which were found to be excellent for the detection of CIMP in CRC in two independent series25,34, along with MLH1. In our study, when we used a cut off of 4 or 5 methylated markers, the positive samples shared the clinical and molecular features that commonly associate with a CIMP phenotype. However, this issue is not definitely settled and we are using methylation index as a mean of distinguishing the two groups: high MI and low MI. Using these criteria, the majority of highly methylated tumors were sporadic cancers with MSI. Only a very small percentage of sporadic MSS and Lynch syndrome tumors had a high MI, and none of the MSS HNPCC did. In contrast, using a different set of markers, Nagasaka et al. reported that a subset of Lynch Syndrome CRC was highly methylated and in that case they were associated with mutated KRAS rather than BRAF35. A significant percentage of methylated tumors from MSS HNPCC was also recently reported36. The difference between these studies and our findings could presumably be explained by the use of different markers, but it is more likely due to the threshold used to define methylation. For instance, Bettstetter et al.37 established a cutoff value of 18% methylation for MLH1 to distinguish sporadic MSI cases from Lynch and sporadic MSS. In any case, there is some degree of methylation in hereditary CRC but the significance of it is far from clear.
As none of the MSS HNPCC tumors in our study had a high MI we questioned if global hypomethylation might be an important mechanism underlying the carcinogenic process in these MSS HNPCC tumors. In order to do so, we used the retrotransposable element LINE-1 as a surrogate marker, as this has been found to correlate consistently with global methylation levels. LINE-1 hypomethylation is inversely associated with microsatellite instability and the CIMP phenotype in CRC38. In our series, MSS HNPCC tumors had a significantly lower degree of LINE-1 methylation than any other group of cancers. This is remarkable as DNA hypomethylation has been shown to precede genomic instability in gastrointestinal cancers, and it has been suggested that DNA hypomethylation may have more oncogenic importance than DNA hypermethylation39. The degree of genome-wide demethylation seems to correlate with the level of chromosomal alterations and it may affect the stability of all chromosomes40, imprinting mechanisms, and the activation of normally silenced genes41. The genomic instability related to global hypomethylation seems to be independent of P53 status40.
LINE-1 encodes an antisense promoter that has been shown to provide an alternative transcription start site for several genes that can lead to transformation and tumorigenicity, and LINE-1 hypermethylation would be a major defense mechanism to repress these genes and their mechanisms42
The implications of our findings can be remarkable as LINE-1 hypomethylation is so far one of the few distinguishing features of MSS HNPCC tumors, and this opens new venues to explore potentially heritable defects that can result in this molecular phenotype. Furthermore, other aspects such as cancer surveillance or response to chemotherapy may need to be evaluated in this group as LINE-1 hypomethylation has been associated with a shorter survival, independent of clinical (sex, age, tumor location, and stage) or molecular features (CIMP, MSI, KRAS, BRAF, p53 and chromosomal instability status)43. It will be important to find out if the widespread low hypermethylation index seen in MSS HNPCC tumors plays a significant role in their development, or if it is merely a background phenomenon, unrelated to causation.
Finally, the significant absence of codon 13 KRAS mutations in MSS HNPCC cannot be explained at this point. KRAS codon 12 mutations have been found to associate with increased tumor aggressiveness as they seem to confer a higher resistance to apoptosis and predisposition to anchorage-independent growth44.
In summary, an epigenetic event based on genome-wide hypomethylation is a distinctive feature of the tumors from MSS HNPCC patients. These tumors do not have V600E BRAF mutations and KRAS mutations are limited to codon 12. Further studies will need to address the underlying mechanism and the potentially heritable causes that result in the described phenotype.
Supplementary Material
primer sequences and location of CpG sites
mean/average methylation across all CpG sites in the MSS HNPCC patients
Acknowledgments
Grant Support: The Sirazi Foundation, Raymond Cole Memorial Foundation and Internal Grant from the Department of Medicine and the Cancer Center of the University of Illinois at Chicago to X. Llor. NIH grants CA72851 and CA129286 and funds from the Baylor Research Institute to C.R. Boland and A. Goel. Grant from the Spanish Ministerio de Ciencia e Innovación (SAF 07-64873) to A. Castells.
Abbreviations
- CIN
Chromosomal instability
- CIMP
CpG island methylation phenotype
- CRC
Colorectal cancer
- HNPCC
Hereditary nonpolyposis colorectal cancer
- LINE-1
Long interspersed nucleotide element-1
- MI
Methylation index
- MSI
Microsatellite instability
- MSS
Microsatellite stable
- MSS HNPCC
Microsatellite stable hereditary nonpolyposis colorectal cancer
- MLPA
Multiple ligation probe amplification
Footnotes
Authors' roles: A. Goel, R.M. Xicola, and X. Llor, developed the study concept and design, analyzed and interpreted the data, and drafted the manuscript.
R.M Xicola, T-P. Nguyen, and B.J. Doyle acquired the data.
R.M Xicola performed the statistical analysis.
A. Goel, and X. Llor supervised the study.
B.J. Doyle, V. R. Sohn, F. Balaguer, P. Bandipalliam, J. Reyes, C. Cordero, A. Castells, R. Jover, M. Andreu, S. Syngal, and C.R. Boland critically reviewed the manuscript and provided suggestions.
The authors do not have any potential conflicts of interest to disclose that are relevant to the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Xicola RM, Llor X. Hereditary Colorectal Cancer. In: Kim K, editor. Colorectal Cancer: Early detection and prevention. New Jersey: Slack Inc.; 2009. pp. 149–166. [Google Scholar]
- 2.Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC) Dis Colon Rectum. 1991;34:424–5. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 3.Vasen HFA, Watson P, Mecklin JP, Lynch HT, HNPCC I. New Clinical Criteria for Hereditary Nonpolyposis Colorectal Cancer (HNPCC, Lynch Syndrome) Proposed by the International Collaborative Group on HNPCC. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 4.Beck NE, Tomlinson IP, Homfray T, Hodgson SV, Harocopos CJ, Bodmer WF. Genetic testing is important in families with a history suggestive of hereditary non-polyposis colorectal cancer even if the Amsterdam criteria are not fulfilled. Br J Surg. 1997;84:233–7. [PubMed] [Google Scholar]
- 5.Chung DC, Rustgi AK. The hereditary nonpolyposis colorectal cancer syndrome: genetics and clinical implications. Ann Intern Med. 2003;138:560–70. doi: 10.7326/0003-4819-138-7-200304010-00012. [DOI] [PubMed] [Google Scholar]
- 6.Kolodner RD, Tytell JD, Schmeits JL, Kane MF, Gupta RD, Weger J, Wahlberg S, Fox EA, Peel D, Ziogas A, Garber JE, Syngal S, Anton-Culver H, Li FP. Germ-line msh6 Mutations in Colorectal Cancer. Cancer Res. 1999;59:5068–5074. [PubMed] [Google Scholar]
- 7.Nakagawa H, Lockman JC, Frankel WL, Hampel H, Steenblock K, Burgart LJ, Thibodeau SN, de la Chapelle A. Mismatch repair gene PMS2: disease-causing germline mutations are frequent in patients whose tumors stain negative for PMS2 protein, but paralogous genes obscure mutation detection and interpretation. Cancer Res. 2004;64:4721–7. doi: 10.1158/0008-5472.CAN-03-2879. [DOI] [PubMed] [Google Scholar]
- 8.Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Powell SM, Jen J, Hamilton SR, et al. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–6. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 9.Fishel R. Signaling mismatch repair in cancer. Nature Medicine. 1999;5:1239–1241. doi: 10.1038/15191. [DOI] [PubMed] [Google Scholar]
- 10.Llor X, Pons E, Xicola RM, Castells A, Alenda C, Pinol V, Andreu M, Castellvi-Bel S, Paya A, Jover R, Bessa X, Giros A, Roca A, Gassull MA. Differential features of colorectal cancers fulfilling Amsterdam criteria without involvement of the mutator pathway. Clin Cancer Res. 2005;11:7304–10. doi: 10.1158/1078-0432.CCR-05-0965. [DOI] [PubMed] [Google Scholar]
- 11.Mueller-Koch Y, Vogelsang H, Kopp R, Lohse P, Keller G, Aust D, Muders M, Gross M, Daum J, Schiemann U, Grabowski M, Scholz M, Kerker B, Becker I, Henke G, Holinski-Feder E. HNPCC - clinical and molecular evidence for a new entity of hereditary colorectal cancer. Gut. 2005;14:14. doi: 10.1136/gut.2004.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindor NM, Rabe K, Petersen GM, Haile R, Casey G, Baron J, Gallinger S, Bapat B, Aronson M, Hopper J, Jass J, LeMarchand L, Grove J, Potter J, Newcomb P, Terdiman JP, Conrad P, Moslein G, Goldberg R, Ziogas A, Anton-Culver H, de Andrade M, Siegmund K, Thibodeau SN, Boardman LA, Seminara D. Lower cancer incidence in Amsterdam-I criteria families without mismatch repair deficiency: familial colorectal cancer type X. Jama. 2005;293:1979–85. doi: 10.1001/jama.293.16.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llor X, Pons E, Xicola RM, Castells A, Alenda C, Pinol V, Andreu M, Castellvi-Bel S, Paya A, Jover R, Bessa X, Giros A, Roca A, Gassull MA. Differential features of colorectal cancers fulfilling Amsterdam criteria without involvement of the mutator pathway. Clin Cancer Res. 2005;11:7304–10. doi: 10.1158/1078-0432.CCR-05-0965. [DOI] [PubMed] [Google Scholar]
- 14.Abdel-Rahman WM, Ollikainen M, Kariola R, Jarvinen HJ, Mecklin JP, Nystrom-Lahti M, Knuutila S, Peltomaki P. Comprehensive characterization of HNPCC-related colorectal cancers reveals striking molecular features in families with no germline mismatch repair gene mutations. Oncogene. 2005;17:17. doi: 10.1038/sj.onc.1208387. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez-de-Abajo A, de la Hoya M, van Puijenbroek M, Tosar A, Lopez-Asenjo JA, Diaz-Rubio E, Morreau H, Caldes T. Molecular analysis of colorectal cancer tumors from patients with mismatch repair proficient hereditary nonpolyposis colorectal cancer suggests novel carcinogenic pathways. Clin Cancer Res. 2007;13:5729–35. doi: 10.1158/1078-0432.CCR-06-2996. [DOI] [PubMed] [Google Scholar]
- 16.Goel A, Nagasaka T, Arnold CN, Inoue T, Hamilton C, Niedzwiecki D, Compton C, Mayer RJ, Goldberg R, Bertagnolli MM, Boland CR. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127–38. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 17.Nagasaka T, Koi M, Kloor M, Gebert J, Vilkin A, Nishida N, Shin SK, Sasamoto H, Tanaka N, Matsubara N, Boland CR, Goel A. Mutations in both KRAS and BRAF may contribute to the methylator phenotype in colon cancer. Gastroenterology. 2008;134:1950–60. 1960 e1. doi: 10.1053/j.gastro.2008.02.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gylling A, Ridanpaa M, Vierimaa O, Aittomaki K, Avela K, Kaariainen H, Laivuori H, Poyhonen M, Sallinen SL, Wallgren-Pettersson C, Jarvinen HJ, Mecklin JP, Peltomaki P. Large genomic rearrangements and germline epimutations in Lynch syndrome. Int J Cancer. 2009;124:2333–40. doi: 10.1002/ijc.24230. [DOI] [PubMed] [Google Scholar]
- 19.Feinberg AP, Vogelstein B. Alterations in DNA methylation in human colon neoplasia. Semin Surg Oncol. 1987;3:149–51. doi: 10.1002/ssu.2980030304. [DOI] [PubMed] [Google Scholar]
- 20.Goelz SE, Vogelstein B, Hamilton SR, Feinberg AP. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science. 1985;228:187–90. doi: 10.1126/science.2579435. [DOI] [PubMed] [Google Scholar]
- 21.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632–42. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 22.Xicola RM, Llor X, Pons E, Castells A, Alenda C, Pinol V, Andreu M, Castellvi-Bel S, Paya A, Jover R, Bessa X, Giros A, Duque JM, Nicolas-Perez D, Garcia AM, Rigau J, Gassull MA. Performance of different microsatellite marker panels for detection of mismatch repair-deficient colorectal tumors. J Natl Cancer Inst. 2007;99:244–52. doi: 10.1093/jnci/djk033. [DOI] [PubMed] [Google Scholar]
- 23.Kumar R, Angelini S, Czene K, Sauroja I, Hahka-Kemppinen M, Pyrhonen S, Hemminki K. BRAF mutations in metastatic melanoma: a possible association with clinical outcome. Clin Cancer Res. 2003;9:3362–8. [PubMed] [Google Scholar]
- 24.Maruta H, Holden J, Sizeland A, D'Abaco G. The residues of Ras and Rap proteins that determine their GAP specificities. J Biol Chem. 1991;266:11661–8. [PubMed] [Google Scholar]
- 25.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93. doi: 10.1038/ng1834. Epub 2006 Jun 25. [DOI] [PubMed] [Google Scholar]
- 26.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang AS, Doshi KD, Choi SW, Mason JB, Mannari RK, Gharybian V, Luna R, Rashid A, Shen L, Estecio MR, Kantarjian HM, Garcia-Manero G, Issa JP. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006;66:5495–503. doi: 10.1158/0008-5472.CAN-05-2385. [DOI] [PubMed] [Google Scholar]
- 28.Matsuzaki K, Deng G, Tanaka H, Kakar S, Miura S, Kim YS. The relationship between global methylation level, loss of heterozygosity, and microsatellite instability in sporadic colorectal cancer. Clin Cancer Res. 2005;11:8564–9. doi: 10.1158/1078-0432.CCR-05-0859. [DOI] [PubMed] [Google Scholar]
- 29.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 30.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–6. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogino S, Cantor M, Kawasaki T, Brahmandam M, Kirkner GJ, Weisenberger DJ, Campan M, Laird PW, Loda M, Fuchs CS. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000–6. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–93. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 33.Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci U S A. 2000;97:710–5. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–14. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagasaka T, Koi M, Kloor M, Gebert J, Vilkin A, Nishida N, Shin SK, Sasamoto H, Tanaka N, Matsubara N, Boland CR, Goel A. Mutations in both KRAS and BRAF may contribute to the methylator phenotype in colon cancer. Gastroenterology. 1960;2008 Jun;134:1950–60. doi: 10.1053/j.gastro.2008.02.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joensuu EI, Abdel-Rahman WM, Ollikainen M, Ruosaari S, Knuutila S, Peltomaki P. Epigenetic signatures of familial cancer are characteristic of tumor type and family category. Cancer Res. 2008;68:4597–605. doi: 10.1158/0008-5472.CAN-07-6645. [DOI] [PubMed] [Google Scholar]
- 37.Bettstetter M, Dechant S, Ruemmele P, Grabowski M, Keller G, Holinski-Feder E, Hartmann A, Hofstaedter F, Dietmaier W. Distinction of hereditary nonpolyposis colorectal cancer and sporadic microsatellite-unstable colorectal cancer through quantification of MLH1 methylation by real-time PCR. Clin Cancer Res. 2007;13:3221–8. doi: 10.1158/1078-0432.CCR-06-3064. [DOI] [PubMed] [Google Scholar]
- 38.Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–73. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki K, Suzuki I, Leodolter A, Alonso S, Horiuchi S, Yamashita K, Perucho M. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell. 2006;9:199–207. doi: 10.1016/j.ccr.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C, Moreno V, Esteller M, Capella G, Ribas M, Peinado MA. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462–9468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 41.Pogribny IP, Beland FA. DNA hypomethylation in the origin and pathogenesis of human diseases. Cell Mol Life Sci. 2009;66:2249–61. doi: 10.1007/s00018-009-0015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervantes F, Sanchez J, Garate L, Barrios M, Castillejo JA, Navarro G, Colomer D, Prosper F, Heiniger A, Torres A. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–23. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- 43.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Chan AT, Schernhammer ES, Giovannucci EL, Fuchs CS. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734–8. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guerrero S, Casanova I, Farre L, Mazo A, Capella G, Mangues R. K-ras codon 12 mutation induces higher level of resistance to apoptosis and predisposition to anchorage-independent growth than codon 13 mutation or proto-oncogene overexpression. Cancer Res. 2000;60:6750–6. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
primer sequences and location of CpG sites
mean/average methylation across all CpG sites in the MSS HNPCC patients