Abstract
The ‘rate of living’ theory predicts that longevity should be inversely correlated with the rate of mitochondrial respiration. However, recent studies in a number of model organisms, including mice, have reported that interventions that retard the aging process are, in fact, associated with an increase in mitochondrial activity. To better understand the relationship between energy metabolism and longevity, we supplemented the endogenous respiratory chain machinery of the fruit fly Drosophila melanogaster with the alternative single-subunit NADH–ubiquinone oxidoreductase (Ndi1) of the baker's yeast Saccharomyces cerevisiae. Here, we report that expression of Ndi1 in fly mitochondria leads to an increase in NADH–ubiquinone oxidoreductase activity, oxygen consumption and ATP levels. In addition, exogenous Ndi1 expression results in increased CO2 production in living flies. Using an inducible gene expression system, we expressed Ndi1 in different cells and tissues and examined the impact on longevity. In doing so, we discovered that targeted expression of Ndi1 in fly neurons significantly increases lifespan without compromising fertility or physical activity. These findings are consistent with the idea that enhanced respiratory chain activity in neuronal tissue can prolong fly lifespan.
Keywords: Aging, fly, longevity, mitochondria, respiration
Introduction
Defects in mitochondrial energy metabolism and their effects on aging and age-related disease are widely reported (Wallace 2005; Lin & Beal 2006). Aged mammalian tissues show a decreased capacity to produce ATP and the impairment of mitochondrial function is due to decreased rates of electron transfer by the selectively diminished activities of complexes I and IV (Navarro & Boveris 2007). Dysfunctional mitochondria in aged rodents are characterized, besides decreased electron transfer and oxygen uptake, by an increased content of oxidation products of phospholipids, proteins and DNA, a decreased membrane potential, and altered morphology (Shigenaga et al. 1994). These features of the aging process appear to be conserved across the animal kingdom. For example, in Drosophila aging is also associated with changes in mitochondrial structure (Burch et al. 1970; Walker & Benzer 2004) and defects in mitochondrial respiratory chain function (Ferguson et al. 2005).
Although it is widely accepted that aging is associated with a decline in mitochondrial activity, an understanding of the causal relationship between the rate of respiratory chain activity and animal aging remains elusive. The ‘rate of living’ theory (Pearl 1928) predicts an inverse relationship between the rate of mitochondrial activity and longevity. However, inactivation of genes important for respiratory chain function has been associated with both increased (Feng et al. 2001; Dillin et al. 2002; Lee et al. 2003; Liu et al. 2005; Dell'agnello et al. 2007; Copeland et al. 2009) and decreased longevity (Ishii et al. 1998; Walker et al. 2006) and it is not yet known whether reduced mitochondrial activity is important for changes in longevity mediated by reduced expression of ETC genes. For example, long-lived flies with reduced expression of ETC genes do not consistently show energetic or physiological trade-offs (Copeland et al. 2009).
In recent years, a number of studies have reported that interventions that retard the aging process may, in fact, lead to an increase in mitochondrial respiratory chain activity (Guarente 2008). Moderate dietary restriction (DR) slows aging and delays the onset of age-related pathologies in a wide range of species (Mair & Dillin 2008). Recent studies have reported that DR is associated with an increase in mitochondrial activity in yeast (Lin et al. 2002), C. elegans (Bishop & Guarente 2007), Drosophila (Zid et al. 2009) and mice (Nisoli et al. 2005). In addition, long-lived mice with a fat-specific insulin receptor knockout (FIRKO) (Bluher et al. 2003) display an increase in basal metabolic rate and respiratory exchange ratio (Katic et al. 2007). The TOR (target of rapamycin) pathway modulates longevity in yeast, worms, flies and rodents (Vellai et al. 2003; Kapahi et al. 2004; Kaeberlein et al. 2005; Harrison et al. 2009). In a recent mechanistic study, it was shown that reduced TOR signaling extends chronological lifespan in yeast via increased respiration (Bonawitz et al. 2007). In both yeast (Barros et al. 2004) and mice (Caldeira da Silva et al. 2008), uncoupling agents such as 2,4-dinitrophenol (DNP) have been reported to increase both respiratory activity and longevity. In Drosophila, neuronal expression of human uncoupling protein 2 (hUCP2) was reported to increase longevity (Fridell et al. 2005). However, in an independent study, expression of human UCP3 (hUCP3) at moderate levels in adult neurons resulted in a marginal lifespan-extension in male flies and high expression of hUCP3 in neuronal tissue shortened lifespan (Humphrey et al. 2009). Interpretation of these findings is hampered by a lack of detailed understanding of the physiological function of individual UCPs. Interestingly, overexpression of neurofibromatosis-1 (NF1) was reported to increase both respiration and lifespan in the fly (Tong et al. 2007). It has been suggested that increased respiration is sufficient to prolong lifespan in yeast: overexpression of Hap4, a transcription factor expected to activate many genes involved in respiration, extends lifespan in yeast (Lin et al. 2002). Taken together, these studies raise an intriguing question: Is increased mitochondrial respiratory chain activity sufficient to delay animal aging? And, if so, which tissues are important in mediating longevity?
Progress towards developing strategies to directly increase the activity of mitochondrial respiratory chain enzymes in animals has been confounded by technical difficulties associated with their coordinated assembly. For example, mitochondrial complex I (NADH–ubiquinone oxidoreductase), the major entry point for electrons into the respiratory chain, is comprised of at least 45 subunits encoded by both the nuclear and mitochondrial genomes (Carroll et al. 2006). In contrast, the alternative NADH-ubiquinone oxidoreductase (Ndi1; GeneID: 854919 Entrez Gene) of Saccharomyces cerevisiae mitochondria is composed of a single nuclear-encoded polypeptide (Luttik et al. 1998). Previously, Ndi1 expression has been shown to restore mitochondrial activity in animal and cell-based models of respiratory chain disorders (DeCorby et al. 2007; Perales-Clemente et al. 2008) (Marella et al. 2008). Ndi1 expression is also sufficient to restore mitochondrial activity in a fly mutant with impaired endogenous complex I activity (JC & DWW, manuscript in preparation). We reasoned that expression of Ndi1 in wild-type Drosophila may be an effective approach to directly increase respiratory chain activity in an animal model. Here, we demonstrate that supplementation of the endogenous fly respiratory chain with the NDI1 enzyme leads to an increase in NADH–ubiquinone oxidoreductase activity, oxygen consumption and ATP levels. In addition, Ndi1-expressing flies display an increased metabolic rate. Using our transgenic fly model, we report the consequences of tissue-specific expression of Ndi1 on longevity: expression in adipose tissue shortens lifespan, while neuronal expression leads to increased longevity. Long-lived flies display increased NADH–ubiquinone oxidoreductase activity and ATP levels in tissue isolated from heads.
Results
Expression of the yeast Ndi1 gene in Drosophila mitochondria leads to an increase in respiratory chain activity
To investigate whether expression of the alternative, rotenone-insensitive NADH-ubiquinone oxidoreductase (Ndi1) of Saccharomyces cerevisiae was sufficient to enhance mitochondrial activity in Drosophila, we expressed Ndi1 in flies using the GAL4/UAS system (Brand & Perrimon 1993). We transformed flies with UAS-constructs containing the Ndi1 gene and performed seven rounds of backcrossing into a w1118 background. In all subsequent experiments, w1118 was used as a control strain. Except for the insertion of the UAS-Ndi1 transgene, these control flies were genetically very similar to the experimental flies. Using a ubiquitous expression GAL4 driver line, daughterless (da)-GAL4, we confirmed the mitochondrial-specific expression of the NDI1 protein by Western blot (Figure 1A). To determine whether expression of an exogenous inner membrane protein (Ndi1) may affect the structure of the endogenous respiratory chain enzymes, we examined their assembly by Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE). Expression of Ndi1 in fly mitochondria did not affect the expression or assembly of the endogenous respiratory chain machinery (Figure 1B). To determine whether NDI1 could function to promote respiration in fly mitochondria, we examined oxygen consumption in the presence of various substrate/inhibitor combinations. To distinguish the NADH–ubiquinone oxidoreductase activites of the endogenous Drosophila complex I and of the yeast NDI1 enzyme, we monitored oxygen consumption in the presence of the endogenous complex I inhibitor rotenone. In contrast to control flies, mitochondria from Ndi1-expressing flies supported a substantial rotenone-insensitive substrate oxidation (Figure 1C). We confirmed that this rotenone-insensitive respiration was due to the yeast enzyme, using the Ndi1-specific inhibitor flavone. Moreover, we observed that expression of Ndi1 in fly mitochondria leads to a 30% increase in glutamate/malate-stimulated (complex I-specific) respiration, while complex II (succinate-stimulated)-specific respiration was unaffected by Ndi1 expression (Figure 1C). Our results indicate that NDI1 can function in fly mitochondria to promote respiration without affecting the structure or function of the endogenous respiratory chain.
Figure 1. NDI1 can function in Drosophila mitochondria.
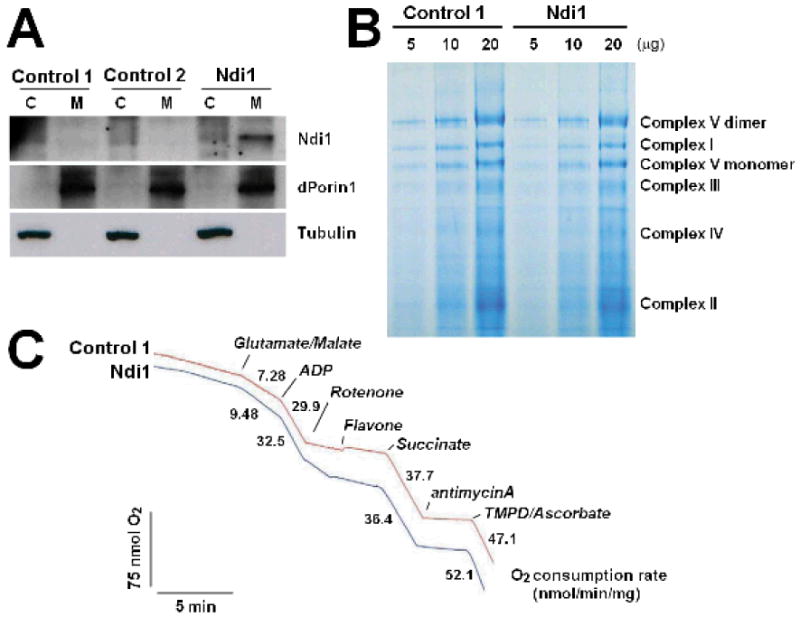
All assays were performed on female flies between 6-10 days of age.
A Targeted expression of NDI1 in fly mitochondria. The purity of cytoplasmic (C) and mitochondrial (M) fractions was confirmed using antibodies against Drosophila Porin 1 (dPorin1) and α-tubulin as mitochondrial and cytoplasmic markers, respectively. The expression of NDI1 protein in each fraction was examined by immunoblotting with polyclonal rabbit antisera against NDI1. Genotypes: Control 1: +/da-GAL4, Control 2: UAS-Ndi1/+; Ndi1: UAS-Ndi1/da-GAL4. B Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE) was performed on mitochondrial proteins isolated from Ndi1-expressing flies and controls. Each lane represents increasing amount (5, 10, 20μg) of protein. Expression of NDI1 does not affect the assembly of the endogenous respiratory chain enzymes. Genotypes: Control: +/da-GAL4, Ndi1: UAS-Ndi1/da-GAL4. C. Representative polarographic trace showing rates of oxygen consumption in mitochondria isolated from Ndi1-expressing flies and controls. Ndi1 confers a 30% increase in glutamate/malate-stimulated (complex I-specific) respiration and promotes rotenone-insensitive respiration, without affecting succinate-stimulated (complex II-specific) respiration. Genotypes: Control: +/da-GAL4, Ndi1: UAS-Ndi1/da-GAL4.
We observe that expression of NDI1 in fly mitochondria leads to an increase in complex I-specific oxygen consumption. Next, we assayed NADH-ubiquinone oxidoreductase (complex I) enzymatic activity and confirmed that mitochondria from Ndi1-expressing flies display a 28% increase in this activity (Figure 2A). In addition, Ndi1 conferred a 2.6-fold increase in rotenone-insensitive NADH-ubiquinone oxidoreductase activity. To determine whether this increase in NADH-ubiquinone oxidoreductase activity affects overall energy production, we took two independent approaches. Firstly, we examined ATP levels in Ndi1-expressing flies and controls. Expression of Ndi1 conferred a 24% increase in ATP levels (Figure 2B). Next, we set out to determine whether expression of Ndi1 affects respiratory chain activity in living flies. To do so, we assayed CO2 production in Ndi1-expressing flies and controls. Flies expressing Ndi1 display a 16% increase in metabolic rate compared to controls (Figure 2C). This result indicates that Ndi1 can promote an increase in respiratory rate in the intracellular environment in which mitochondria respire in vivo. Unlike the endogenous fly complex I enzyme, NDI1 cannot promote proton pumping. Therefore, the NDI1-mediated increase in CO2 production and ATP levels likely results from an increase in electron flow through the ETC. In Figure S1, we depict a schematic drawing of the putative mode of Ndi1 function in fly mitochondria.
Figure 2. Expression of Ndi1 can increase respiratory chain activity in Drosophila.
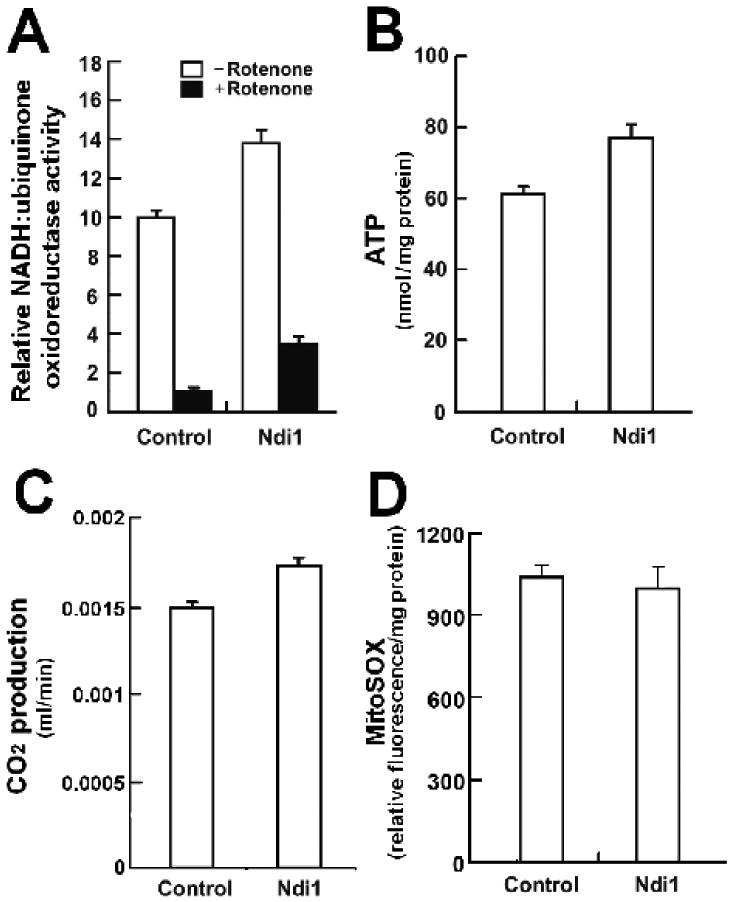
Genotypes: Control: +/da-GAL4, Ndi1: UAS-Ndi1/da-GAL4. All assays were performed on female flies between 6-10 days of age.
A Ndi1 confers a significant increase in NADH-ubiquinone oxidoreductase (complex I) enzymatic activity compared to controls (p=0.0007). Rotenone insensitive activity was measured after addition of 1 μM rotenone. B Ndi1-expressing flies display a significant increase in mean ATP levels compared to controls (p=0.004). C. Ndi1-expressing flies display a significant increase in CO2 production compared to controls (P < 0.001). D. Expression of Ndi1 in fly mitochondria does not affect ROS production (p>0.05). A, B, D n = 3, C n=8. Error bars indicate standard deviation. Student's t-test was used.
The relationship between the rate of respiratory chain activity and the production of reactive oxygen species (ROS) is complicated and controversial (Balaban et al. 2005). We examined whether Ndi1-mediated increased respiratory chain activity affects ROS production in fly mitochondria using the MitoSOX fluorescent indicator to monitor superoxide anion production. We observe that increased NADH-ubiquinone oxidoreductase activity, resulting from Ndi1 expression, did not affect mitochondrial ROS production (Figure 2D). Our data indicate that Ndi1 expression in fly mitochondria can increase respiratory chain activity without affecting ROS production.
Targeted expression of Ndi1 in adipose tissue shortens lifespan, while expression in neurons increases lifespan
Having established that expression of Ndi1 in fly mitochondria can increase respiratory chain activity, we next set out to examine the impact on longevity of targeted expression of Ndi1 in different cells and tissues. To do so, we used the mifepristone (RU486) inducible-GAL4 system (annotated P[Switch] or Gene-Switch (Osterwalder et al. 2001; Roman et al. 2001)). This system eliminates genetic background effects since all flies share the same genetic background and only differ with respect to the presence of the inducing agent (RU486) in the food. To eliminate the possible contribution of induction of endogenous genes at the site of insertion of the Ndi1 transgene, we tested two independent insertions of the UAS-Ndi1 transgene in all longevity experiments. Firstly, we set out to investigate the impact of expression of Ndi1 in all cells and tissues. Ubiquitous induction of Ndi1 expression during both development and adulthood using tubulin (tub)-Gene-Switch (GS) had no robust positive effects on longevity in male (Figures S2A & S2B; Table S1) or female flies (Figures 3A & 3B; Table S1). Next, we set out to investigate the effects of targeted expression of Ndi1 in specific tissues. Adipose tissue has been reported to play an important role in insulin/IGF-1-mediated longevity across the animal kingdom (Russell & Kahn 2007). To examine the impact of Ndi1 expression in adipose tissue, we used the Gene-Switch driver S1106, which is predominately expressed in the fat-body (Poirier et al. 2008). Expression of Ndi1 in adipose tissue resulted in shortened lifespan in both male (Figures S2C & S2D; Table S2) and female flies (Figure 3C & 3D; Table S2).
Figure 3. Effects of tissue-specific expression of Ndi1 on fly longevity.
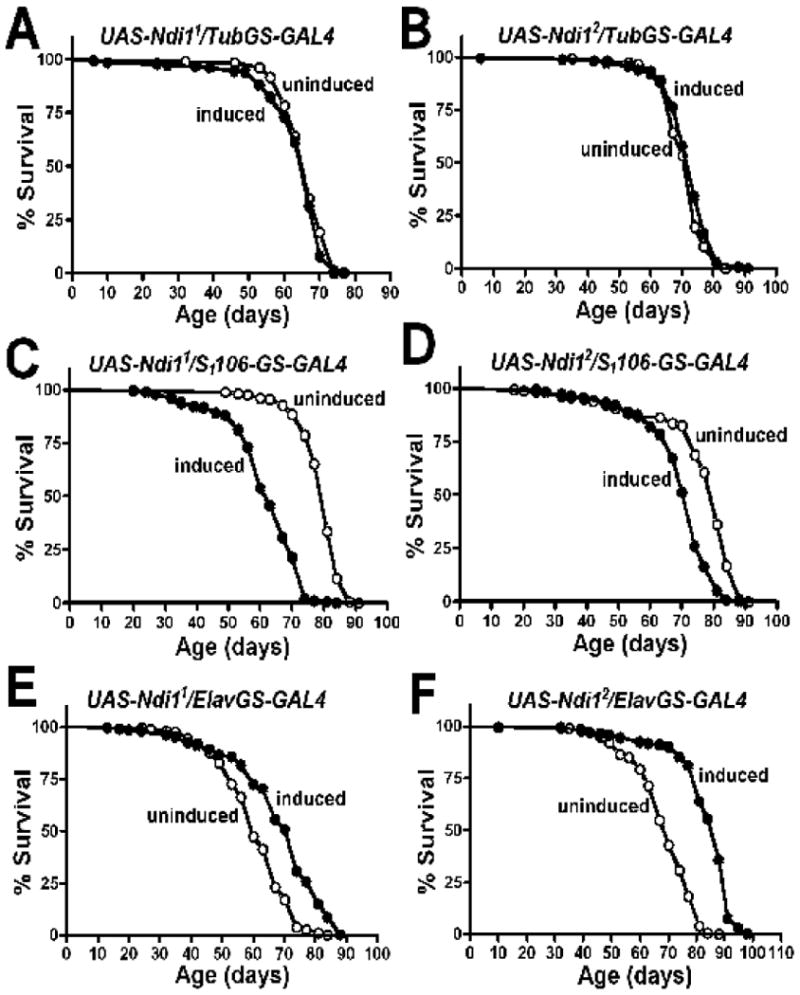
Two independent UAS-Ndi1 lines (UAS-Ndi11 & UAS-Ndi12), were crossed to GeneSwitch driver lines (A-B the ubiquitous Tubulin (tub)-GS driver, C-D the fat-body driver S1106, E-F the pan-neuronal driver Elav-GS) and lifespan curves are shown as induced (2 mg/ml RU486 during development and 10 mg/ml RU486 from the onset of adulthood (black circles) or uninduced (−RU486, open circles).
A. Lifespan curves of UAS-Ndi11/tub-GS females. A moderate decrease in survival was observed in response to RU486 (P=0.0441). B. Lifespan curves of UAS-Ndi12/tub-GS females. A moderate increase in survival was observed in response to RU486 (P=0.018). C. Lifespan curves of UAS-Ndi11/S1106 females. A 22% decrease in survival was observed in response to RU486 (p<0.0001). D. Lifespan curves of UAS-Ndi12/S1106 females. A 9% decrease in survival was observed in response to RU486 (p<0.0001). E. Lifespan curves of UAS-Ndi11/Elav-GS females. A 12% increase in survival was observed in response to RU486 (p<0.0001). F. Lifespan curves of UAS-Ndi12/Elav-GS females. A 21% increase in survival was observed in response to RU486 (p<0.0001). Complete survival data on different RU486 concentrations are given in Tables S1, S2, S3. The significance of the difference between survival curves was analyzed using log-rank statistical test.
Previous studies have reported that the nervous system is an important target tissue in mediating longevity in flies (Bauer et al. 2005; Fridell et al. 2005; Copeland et al. 2009). Therefore, we used the pan-neuronal Elav-Gene-Switch (Elav-GS) driver line to induce Ndi1 expression specifically in neurons during both development and adulthood. In contrast to the effects we observed using both the ubiquitous and the fat-body driver lines, we observed life extension when Ndi1 was expressed in neurons in both male (Figures S2E, S2F; Table S3) and female flies (Figures 3E & 3F; S3A & S3B; Table S3). Specifically, neuronal expression of Ndi1 increased lifespan by up to 21% in females and by up to 7% in males. We investigated the possibility that RU486 itself may increase longevity in our fly strains by simultaneously feeding RU486 or diluent alone to the progeny of Elav-GS crossed to w1118, the genetic background of the two Ndi1 transgenic lines. RU486 had an overall negative impact on longevity in control flies (Table S3). Therefore, it is possible that the negative effects of RU486 on lifespan may partially ‘mask’ the beneficial effects of NDI1.
To examine the impact of Ndi1 expression in muscle cells on longevity, we used the constitutive muscle specific driver MHC-GAL4. We used this constitutive driver line because MHC-GeneSwitch displays very high-level inducer-independent (‘leaky’) expression in muscle, but does not induce in a broad set of muscles, but instead induces in digestive tissue (Poirier et al. 2008). Expression of two independent UAS-Ndi1 transgenes mediated by MHC-GAL4 had no major impact on longevity in male or female flies (Figure S4; Table S4).
Neuronal expression of Ndi1 can increase respiratory chain activity
To investigate the mechanism by which neuronal Ndi1 expression extends lifespan, we examined the physiology of mitochondria from heads of Ndi1-expressing flies. First, using quantitative real-time PCR (qRT-PCR), we confirmed that the Ndi1 transcript was induced in heads, which are greatly enriched for neuronal tissue (Figure 4A). Next, we examined whether neuron-specific expression of Ndi1 leads to an increase in NADH-ubiquinone oxidoreductase activity. We observe that induced neuronal expression of Ndi1 leads to a 16% increase in complex I enzymatic activity in mitochondria isolated from heads (Figure 4B). In addition, Ndi1 conferred a 65% increase in rotenone-insensitive NADH-ubiquinone oxidoreductase activity in head tissue. RU486 had no effect on in NADH-ubiquinone oxidoreductase activity in control flies (w1118/Elav-GS; Figure S5A).
Figure 4. Neuronal expression of Ndi1 increases complex I enzymatic activity and ATP levels in heads of long-lived flies.
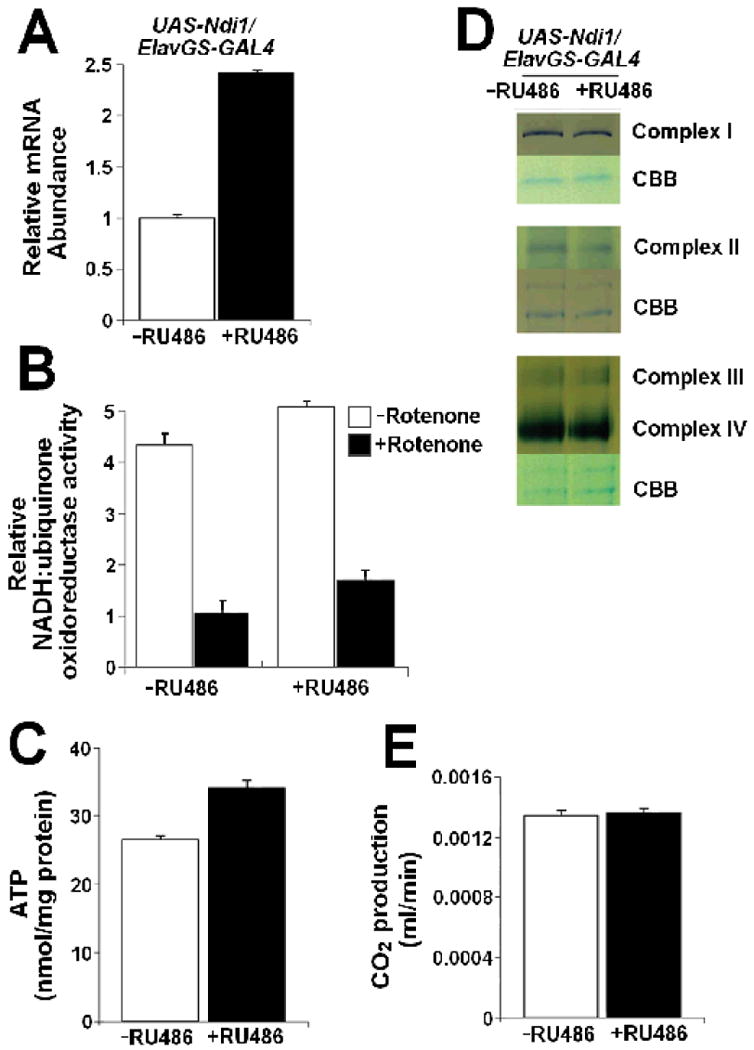
A-E. UAS-Ndi1/Elav-GeneSwitch flies were fed with 2 μg/ml RU486 during development and 10 μg/ml RU486 during the adulthood stage; uninduced flies were fed with diluent. All assays were performed on female flies between 6-10 days of age.
A. Increased Ndi1 mRNA in heads of long-lived flies (p<0.0001). B. Increased NADH-ubiquinone oxidoreductase (complex I) enzymatic activity in heads of long-lived flies compared to uninduced controls (p = 0.0029). Rotenone insensitive activity was measured after addition of 1μM rotenone. C. Increased ATP levels in heads of long-lived flies (p=0.0002). D. The expression and activities of endogenous respiratory chain enzymes are unchanged in heads of long-lived flies as assayed by Blue native-polyacrylamide gel electrophoresis (BN-PAGE) followed by in-gel activity staining. Coomasie Brilliant Blue (CBB) was used for estimating the amounts of the separated protein complexes. E. Neuronal expression of Ndi1 does not affect CO2 production in living flies (p>0.05). A-D n = 3, E n=8. Error bars indicate standard deviation. Student's t-test was used.
To determine whether this increase in NADH-ubiquinone oxidoreductase activity affects overall energy production in neuronal tissue, we examined ATP levels in heads of Ndi1-expressing flies and controls. Expression of Ndi1 in fly neurons resulted in a 28% increase in ATP levels in the heads of long-lived flies (Figure 4C). RU486 had no effect on ATP levels in control flies (w1118/Elav-GS; Figure S5B). To investigate whether this increase in energy production was due to an increase in the activity of the endogenous Drosophila respiratory chain machinery, we assayed the activities of each of the individual fly respiratory chain enzymes using BN-PAGE followed by in-gel enzyme activity staining. As Figure 4D illustrates, the activities of the endogenous respiratory enzymes were unaffected in the heads of long-lived flies. To determine whether neuron-specific expression of Ndi1 could affect metabolism at the whole-animal level, we examined the metabolic rate of Ndi1 flies and controls. Neuron-specific expression of Ndi1 did not lead to an increase in CO2 production in living flies (Figure 4E). RU486 had no effect on CO2 production in control flies either (w1118/Elav-GS; Figure S5C).
Neuronal expression of Ndi1 protects against oxidative stress and rotenone toxicity
To investigate whether neuronal expression of Ndi1 leads to a reduction in oxidative stress, we assayed ROS levels in dissected brains of long-lived flies. Dihydroethidium staining revealed that targeted expression of Ndi1 conferred a 51% decrease in ROS levels in the brains of 45-day old flies compared to uninduced controls (Figures 5A & 5B). Inducer has no impact on ROS levels in control flies (w1118/Elav-GS; S.B & D.W.W, unpublished observations). We observe that expression of Ndi1 in fly mitochondria supports rotenone-insensitive respiration (Figure 1C). To determine whether Ndi1 can function in vivo to support respiration in the presence of an endogenous complex I inhibitor, we tested flies for resistance to rotenone. Induced expression of Ndi1 in fly neurons results in an improved tolerance to rotenone poisoning (Figure 6A), with RU486 having no effect on rotenone resistance in control flies (w1118/Elav-GS; Figure S6A). We observe that targeted neuronal expression of NdiI leads to reduced ROS production in fly brains (Figure 5A & 5B). Next, we investigated whether neuronal Ndi1 expression can protect against acute oxidative stress at the whole-animal level. To do so, we examined whether long-lived flies, with targeted neuronal expression of Ndi1, were more resistant to dietary paraquat which has been shown to generate ROS in vivo (Hassan & Fridovich 1979). Expression of Ndi1 in fly neurons resulted in improved tolerance to paraquat poisoning relative to uninduced cohorts (Figure 6B), while RU486 had no effect on paraquat tolerance in control flies (w1118/Elav-GS; Figure S6B). To determine whether Ndi1-expressing flies display a general resistance to extrinsic stressors, we examined their resistance to starvation. As opposed to the increase in resistance to both dietary paraquat and rotenone, expression of Ndi1 in fly neurons did not improve their ability to withstand starvation (Figure 6C). RU486 had no effect on starvation resistance in control flies (w1118/Elav-GS; Figure S6C).
Figure 5. Reduced levels of reactive oxygen species (ROS) in brains of Ndi1-expressing flies.
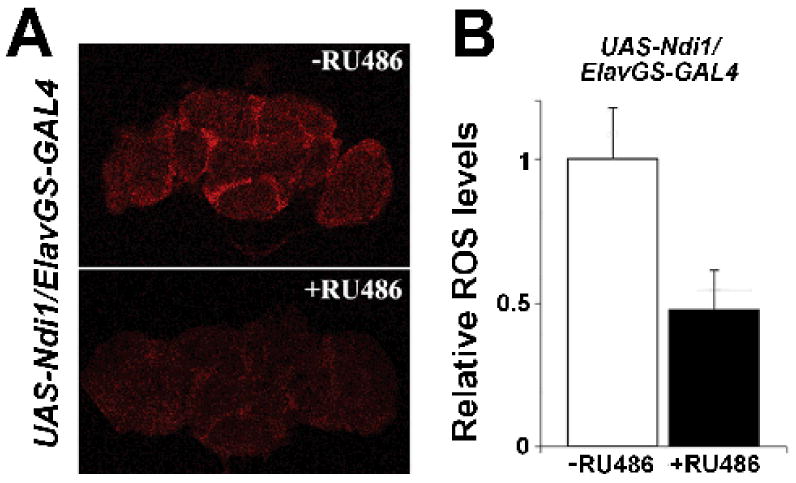
A-B. UAS-Ndi1/Elav-GeneSwitch flies were fed with 2 μg/ml RU486 during development and 10 μg/ml RU486 during the adulthood stage; uninduced flies were fed with diluent. Assays were performed on 45 day-old female flies.
A. Neuronal expression of Ndi1 reduces ROS abundance in fly brains detected by dihydroethidium. B. Quantification of the relative ROS levels in brains of Ndi1-expressing flies normalized to uninduced control brains; Ndi1 expression in neurons reduces ROS levels by 51% (p=0.001). n = 5 brains per treatment. Error bars indicate standard deviation. Student's t-test was used.
Figure 6. Ndi1 expressing flies are resistant to the endogenous complex I inhibitor rotenone and the free radical generator paraquat.
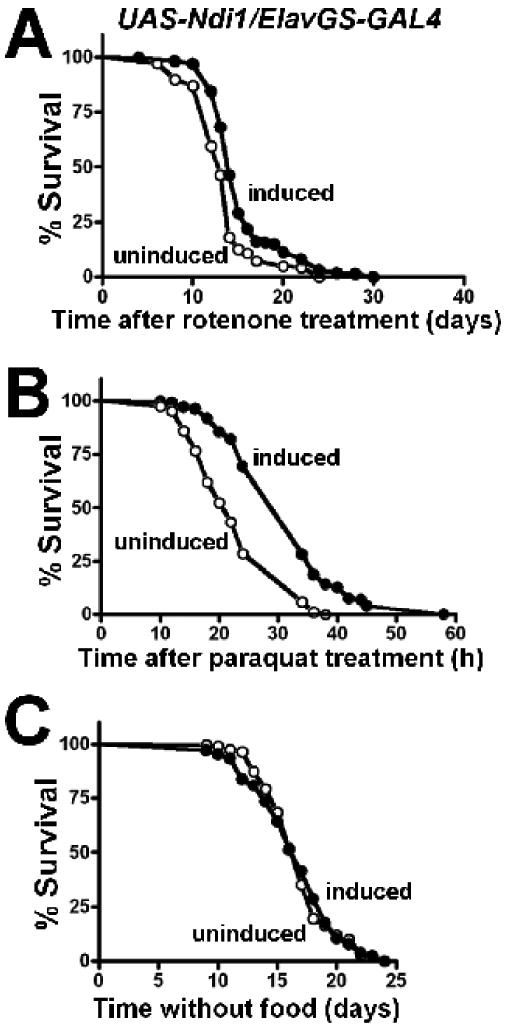
A-C, UAS-Ndi1/Elav-GeneSwitch flies were fed with 2 μg/ml RU486 during development and 10 μg/ml RU486 during the adulthood stage; uninduced flies were fed with diluent. Survival curves are shown as induced (+RU486, solid circles) or uninduced (-RU486, open circles). All assays were performed on female flies between 6-10 days of age. A Ndi1 confers tolerance to 250 μM rotenone (p< 0.0001) and B 30 mM paraquat (p< 0.0001), but does not affect C starvation resistance (p=0.9278). The significance of the difference between survival curves was analyzed using the log-rank statistical test.
Long-lived Ndi1-expressing flies display normal physical activity and fertility
In many cases, interventions that extend longevity result in reproductive trade-offs (Partridge et al. 2005). Therefore, we examined female fertility over the first 30 days of adulthood in Ndi1-expressing flies and uninduced controls. As has been previously reported (Fridell et al. 2005), we observed that RU486 had a negative impact on fertility in control flies (w1118/Elav-GS; Figure S7A). In contrast, RU486-induced expression of Ndi1 in fly neurons did not decrease fertility relative to uninduced cohorts (Figure 7A). Decreased physical activity has also been associated with extended lifespan in flies (Helfand & Rogina 2003). To determine whether a decline in physical activity contributes to the lifespan extension of the Ndi1-expressing flies, we monitored the physical activity of flies with a 24 hr Drosophila activity monitor and found that long-lived Ndi1-expressing flies exhibited spontaneous physical activity equal to uninduced controls (Figure 7B). RU486 had no effect on physical activity in control flies (w1118/Elav-GS; Figure S7B). Thus, Ndi1-mediated life extension is not a result of a decrease in reproduction or physical activity.
Figure 7. Long-lived Ndi1-expressing flies display normal fertility and physical activity.
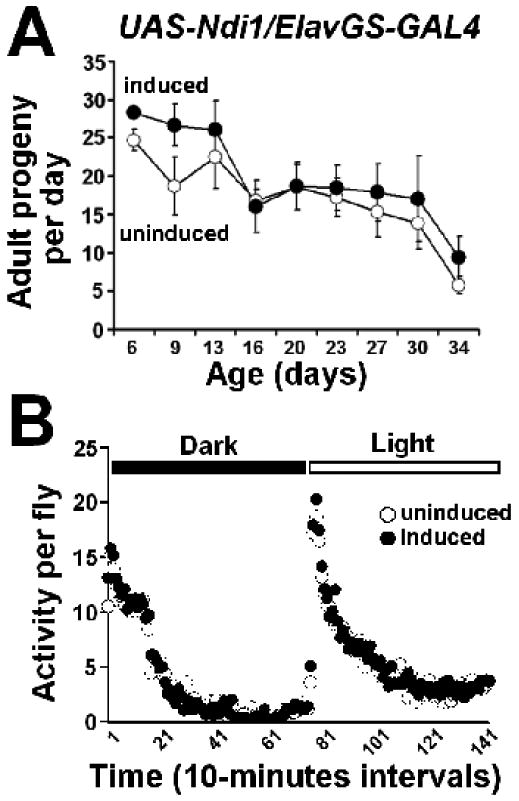
UAS-Ndi1/Elav-GeneSwitch flies were fed with 2 μg/ml RU486 during development and 10 μg/ml RU486 during the adulthood stage; uninduced flies were fed with diluent.
A. Average number of progeny produced from crosses between 5 wild-type males and 5 UAS-Ndi1/Elav-GeneSwitch-GAL4 females fed RU486 or diluent. Long-lived Ndi1 expressing flies display normal fertility compared to uninduced controls (p>0.05; two-way Analysis of Variance). B. Activity of 6- 10 day-old flies recorded with Drosophila Activity Monitor over a 24 hour period is shown. Long-lived Ndi1 expressing flies display normal physical activity compared to uninduced controls (p>0.05; two-way Analysis of Variance).
Discussion
The mechanisms that cause the deterioration of cellular functions in aging animals remain poorly understood. One attractive hypothesis, supported by a growing body of correlative data (Shigenaga et al. 1994; Wallace 2005), is that impaired mitochondrial respiratory chain function may play an important role in the aging process. However, the causal relationship between the rate of mitochondrial activity and animal aging remains poorly understood. The ‘rate of living – free-radical damage’ theory (Pearl 1928; Harman 1956; Sohal 2002) suggests that higher rates of metabolism should be linked to increased ROS and, as a result, decreased longevity. In recent years, however, this idea has been challenged by a number of studies reporting a positive correlation between respiratory activity and longevity (Speakman et al. 2004; Guarente 2008).
In this study, we report that targeted expression of the single subunit alternative NADH-ubiquinone oxidoreductase (Ndi1) of Saccharomyces cerevisiae can increase respiratory chain activity in fly mitochondria and in living flies. While expression of Ndi1 in fat tissue leads to detrimental effects on longevity, expression in neurons can promote life extension. These contradictory effects in different tissues may explain our observation that ubiquitous expression of Ndi1 had no major effect on longevity. Flies expressing Ndi1 in neurons displayed increased NADH-ubiquinone oxidoreductase activity and ATP levels in tissue prepared from heads. However, expression of Ndi1 in neurons did not result in an increase in CO2 production in living flies. This result implies that increased respiration in neurons did not lead to an increase in energy production in other tissues of the animal.
Our study suggests that reduced oxidative stress in neurons may play a role in Ndi1-mediated longevity. In cell culture, Ndi1 has been reported to reduce ROS production under conditions of endogenous complex I deficiency (Seo et al. 2006; Park et al. 2007). Here, we report that expression of Ndi1 in wild-type neurons leads to reduced ROS levels in the brains of long-lived flies. In addition, neuronal expression of Ndi1 results in improved tolerance to the free radical generator paraquat. However, at present, the underlying mechanism by which neuronal expression of Ndi1 results in reduced oxidative stress remains unknown. We observed that Ndi1 expression does not affect ROS production in mitochondria isolated from whole flies. It is possible that expression of Ndi1 may lead to different effects on ROS production in different cells and tissues. Another possibility is that Ndi1 may reduce oxidative stress indirectly via altered cell signaling, resulting in an up-regulation of genes important for ROS detoxification.
Previous studies in mammalian model systems have reported a positive correlation between increased respiratory chain activity and longevity (Speakman et al. 2004; Nisoli et al. 2005; Katic et al. 2007; Caldeira da Silva et al. 2008). Our Ndi1 fly model builds upon these studies. However, as yet, we have not demonstrated that the observed changes in mitochondrial function in response to Ndi1 expression are a cause of increased longevity. Future work will focus on examining the molecular and cellular correlates of NDI1-mediated longevity and establishing causal relationships between rates of energy metabolism and rates of aging in different tissues.
Experimental Procedures
Drosophila strains
Tubulin-Gene-Switch was provided by S. Pletcher. Elav-Gene-Switch was provided by H. Keshishian. S1106 was provided by L. Seroude. We transformed flies with pUAST plasmids containing Ndi1 (amplified by PCR from yeast genomic DNA) and performed seven rounds of backcrossing into a w1118 background.
Survivorship
Flies were collected under light nitrogen-induced anesthesia and housed at a density of 25-30 flies per vial. At least 6 vials were assayed per treatment. All flies were kept in a humidified, temperature-controlled incubator with 12 h on/off light cycle at 25 °C in vials containing standard cornmeal medium (Lewis 1960). RU486 was administered by adding 200 μl of 0, 2, 10, or 50 μg/ml RU486 dissolved in ethanol to the top of the fly food, as previously described (Poirier et al. 2008; Copeland et al. 2009). Flies were flipped to fresh vials twice per week and scored for death.
Purification of Mitochondria
Mitochondria were isolated from adult flies as previously described (Walker et al. 2006). Fifty flies were gently crushed in 1 ml chilled mitochondrial isolation medium (MIM: 250 mM sucrose, 10 mM Tris (pH 7.4), 0.15 mM MgCl2) by using a glass-on-glass homogenizer, then spun twice at 1,000 × g for 5 min at 4°C to remove debris. The supernatant was then spun at 13,000 × g, for 5 min at 4°C. The pellet, containing the mitochondria, was washed with 1 ml MIM and resuspended in 50 μl MIM.
NADH-ubiquinone oxidoreductase Activity assay
Mitochondrial complex I (NADH-ubiquinone oxidoreductase) activity was measured spectrophotometrically at 600 nm in an incubation volume of 1 ml containing 25 mM potassium phosphate, 3.5 g/l BSA, 60 μM DCIP, 70 μM decylubiquinone, 1 μM antimycin A, and 0.2 mM NADH, pH 7.8. We preincubated an aliquot of 10 μl mitochondrial suspension, prepared as described above, at 25°C in 960 μl incubation mixture without NADH. After 3 minutes, 20 μl of 10 mM NADH was added, and the absorbance was measured at 30 sec intervals for 4 minutes at 25°C. After 4 minutes, we added 1 μl rotenone (1 mM) and measured the absorbance again at 30 sec intervals for 4 minutes. Complex I activity was expressed as a reduction rate of DCIP.
Analysis of CO2 release
The rate of CO2 emission was used to determine the metabolic rate of 8 groups of flies (10 individuals per group) from each treatment. We used flow through respirometry to measure CO2 release with Sable Systems (Las Vegas, NV, USA) data acquisition software logging data from an infrared CO2 analyzer (Li-Cor model 6251; Lincoln, NE, USA). During an experimental run, room air was pumped through two silica and one ascarite/drierite column to be scrubbed of water and CO2. Air then flowed through one of five 30 ml chambers attached to the multiplexor and subsequently passed into the CO2 analyzer. The chambers connected to the multiplexor included an empty control chamber used to collect baseline values, and four chambers containing flies from a particular treatment. The order in which each chamber was measured was controlled with SABLE system data acquisition software (Las Vegas, NV, USA). When a chamber was not being measured, it was still perfused with H20- and CO2 -free air at a rate equal to the regulated flow entering the measured chamber. Flow rate was adjusted using a mass flow controller (Sierra Instruments, Monterey, CA, USA) and was held at 100 ml/min.
An experimental run lasted a total of 75 minutes. Three five-minute baseline readings were recorded at the beginning, middle and end of the run and used to provide accurate zero values. The rate of CO2 emission of four fly groups was measured for 15 minutes within each run. The room temperature was maintained at 25 ± 1°C. The CO2 levels (ppm) were averaged and recorded once/second using Sable Systems data acquisition software. Sable Systems (Las Vegas, NV, USA) Expedata analysis software was used to process VCO2 measurements. CO2 levels were recorded in parts per million and, after data were zeroed using baseline values, converted to milliliters/minute.
Dihydroethidium (DHE) staining
Fly brains were dissected in PBS buffer (8 mM Na2HPO4, 150 mM NaCl, 2 mM KH2PO4, 3 mM KCl) and stained with 30 μM DHE solution for 10-15 minutes. Brains were washed 3 times in PBS buffer and visualized by confocal microscopy. The intensity of the brains was measured using the Scion Image Software (Scion Corp., Frederick, MD). To minimize variations in signal intensity, brains were mounted on the same slide and fluorescence intensities of five different brains of each treatment were averaged to give one data point.
Immunoblot assay
Polyclonal rabbit antisera against NDI1 (1:2000), polyclonal rabbit antisera against dPorin1 (1:1000), and monoclonal mouse antibody against α-tubulin (1:5000 dilution, Cell Signalling). The primary antibodies were detected by HRP-conjugated anti-mouse or anti-rabbit IgG antibodies (1:10000 dilution, Sigma) and SuperSignal West Pico Chemiluminescent reagents (Thermo Scientific).
Mitochondrial respiration assay
Rate of oxygen consumption was measured polarographically using a Clark-type oxygen electrode connected to a computer-operated Oxygraph control unit (Hansatech Instruments) at 25°C. Freshly isolated mitochondria was prepared from 50 flies and then suspended in a respiration medium containing 20 mM Hepes-KOH (pH 7.1), 110 mM sucrose, 10 mM KH2PO4, 40mM KCl, 3 mM MgCl2 and 0.5 mM EGTA. The rate was measured continuously, with the following sequential additions: 5 mM glutamate plus 5 mM malate, 200 μM ADP, 0.5 μM rotenone, 5 mM succinate, 2.5 μM antimycin A, 1 mM ascorbate plus 0.25 mM N′-tetramethyl-1,4-phenylenediamine (TMPD).
MitoSox assay
Superoxide production was assayed, in isolated mitochondria, in respiration medium with 5 mM glutamate, 5 mM malate and 50 μM MitoSox (Invitrogen) at 510 nm excitation/580 nm emission.
ATP assay
Five flies were homogenized in 100 μl of 6 M guanidine-HCl in extraction buffer (100 mM Tris and 4 mM EDTA, pH 7.8) and boiled for 3 minutes. The samples were then centrifuged to collect the supernatant, which was then diluted (1/750) with extraction buffer. The luminescence was measured by a luminometer using autoinjection of the luminescent solution (Invitrogen), and the results were compared to the standards. The ATP level was then calculated by dividing the luminescence by the total protein concentration, which was determined by the Bradford method.
Quantitative Real-Time PCR
Total RNA was extracted from heads of female flies using Trizol reagent. RNA concentration was measured with a Nanodrop spectrophotometer, and 1 μg RNA was used for qRT-PCR measurements. Actin5C was the reference gene used to normalize amplicon amounts. The following primer pairs were used: Act57B: GTGCTATGTTGCCCTGGACT, GCTGGAAGGTGGACAGAGAG; Ndi1: GGTGGGCCTACTGGTGTAGA, CAATGGCGAAAATGTTGTTG. cDNA synthesis and qRT-PCR were combined into one step using Power SYBR Green RNA-to-CT 1-Step kit (Applied Biosystems) and DNA amount was monitored during the 40-cycle PCR by using a 7300 thermal cycler (Applied Biosystems).
Blue native-polyacrylamide gel electrophoresis (BN-PAGE) and In-gel enzyme activity staining
BN-PAGE was performed using Novex Native PAGE Bis-Tris Gel System followed the manufacturer's (Invitrogen) procedure. In brief, 80 μg of the purified mitochondria were resuspended in 25 μl of 1× Native PAGE Sample buffer (Invitrogen) with 1% digitonin and protease inhibitors (Roche) and incubated on ice for 15 min. After centrifugation at 20,000 × g for 30 min, the 25 μl of supernatant was resuspended with 1.25 μl of 5% G-250 sample additive and 10 μl of 4× Native PAGE Sample Buffer (Invitrogen). The final volume was adjusted to 40 μl by addition of double-distilled H2O. Samples were loaded on 3-12% Bis-Tris Native PAGE gels and electrophoresed using 1× Native PAGE Running buffer system (Invitrogen). The upper buffer included 1× Cathode Buffer Additive (Invitrogen). NativeMark Protein standard (Invitrogen) was used as the molecular weight marker.
For in-gel activity assay, the gels were rinsed briefly and equilibrated in the appropriate reaction buffer without chromogenic reagents for 10 minutes. The gels were then incubated in fresh reaction buffer plus chromogenic reagents for varying lengths of time. For complex I, the reaction buffer contains 5 mM Tris-HCl (pH 7.4), 2.5 mg/ml NTB (nitrotetrazolium blue) and 0.1 mg/ml NADH. For Complex II, the reaction buffer contains 5 mM Tris-HCl (pH 7.4), 20 mM sodium succinate, 0.2 mM phenazine methosulfate, and 2.5 mg/ml NTB. For complex III and IV, the reaction buffer contains 50mM sodium phosphate (pH 7.2), 0.05% 3,3′-diaminobenzidine tetrahydrochloride, 50 mM horse heart cytochrome c. All activity staining was carried out at room temperature. The reactions were stopped by fixing the gels in 45% (v/v) methanol and 10% (v/v) acetic acid after color development. The band intensities of both protein contents and activity staining were estimated by optical densitometry.
Stress resistance
Paraquat resistance
Female flies were aged on ethanol (diluent) or RU486-supplemented media for 10 days, starved for 6 h and then transferred to vials containing two 2.4cm glass fiber filter circles (Whatman) wetted with 150μl of 30 mM paraquat (Sigma) in 5% sucrose solution. Flies were kept in the dark at all times, except for scoring death and the daily addition of 150μl 30mM paraquat solution.
Rotenone resistance
female flies were aged on ethanol (diluent) or RU486-supplemented media for 10 days and then transferred to 250 μM rotenone-supplemented medium with diluent or RU486. Survival rate was measured every 2 days, with survivors transferred into fresh vials every 4 days.
Starvation resistance
Female flies were aged on ethanol (diluent) or RU486-supplemented medium for 10 days and then transferred to 1% agar solution with 200 μl diluent or RU486. Percentage survival was measured every day, with survivors transferred to fresh vials every 3-4 days.
Fertility
Four individual crosses were established each with five virgin females that developed on either 0 μg/ml or 2 μg/ml RU486 and five wild-type (Canton-S) males. Crosses were transferred twice per week to fresh food containing either 0 μg/ml or 10 μg/ml RU486. Adult progeny from each vial were recorded and averaged to generate the fertility over time.
Physical Activity
30 adult female flies were placed in a Drosophila activity meter (TriKinetics Inc., Massachusetts). Movements were recorded continuously under normal culturing conditions for 24 hours on a 12:12-hour dark:light cycle. Triplicate samples were used for each activity measurements.
Supplementary Material
Acknowledgments
The authors would like to thank J. Hur for helpful comments, H. Keshishian, S. Pletcher, L. Seroude and the Drosophila Stock Center (Bloomington) for fly stocks and T. Yagi and V. de Pinto for antisera against NDI1 and dPORIN1 respectively. We are indebted to the late Seymour Benzer for providing laboratory resources to J.C to generate Ndi1 transgenic flies. We thank Kevin Vu for help with fly work. We are grateful to the Phelps, Lin and Jacobsen labs for generously allowing us to use their equipment. T.J.B received support from the National Science Foundation (NSF IOS-0920683). D.W.W received support from the UCLA Older Americans Independence Center, NIH/NIA Grant P30-AG028748, and the content does not necessarily represent the official views of the National Institute on Aging or the National Institutes of Health. D.W.W also received support from the Ellison Medical Foundation, the American Federation for Aging Research, the UCLA Center for gene environment in Parkinson's Disease, and the Muscular Dystrophy Association. D.W.W is an Ellison Medical Foundation New Scholar in Aging.
Footnotes
Author Contributions: S.B, J.C and T.L in the Walker laboratory designed and performed the experiments and analyzed the data. H.C. and T.J.B in the Bradley lab assayed CO2 production in living flies. H.O.K and D.E.K in the Krantz laboratory assisted with experiments involving dihydroethidium staining of fly brains; D.W.W designed experiments, supervised the work, analyzed the data and wrote the manuscript with helpful comments from S.B. and J.C.
References
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Barros MH, Bandy B, Tahara EB, Kowaltowski AJ. Higher respiratory activity decreases mitochondrial reactive oxygen release and increases life span in Saccharomyces cerevisiae. J Biol Chem. 2004;279:49883–49888. doi: 10.1074/jbc.M408918200. [DOI] [PubMed] [Google Scholar]
- Bauer JH, Poon PC, Glatt-Deeley H, Abrams JM, Helfand SL. Neuronal expression of p53 dominant-negative proteins in adult Drosophila melanogaster extends life span. Curr Biol. 2005;15:2063–2068. doi: 10.1016/j.cub.2005.10.051. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Bluher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Burch GE, Sohal R, Fairbanks LD. Ultrastructural changes in Drosophila heart with age. Arch Pathol. 1970;89:128–136. [PubMed] [Google Scholar]
- Caldeira da Silva CC, Cerqueira FM, Barbosa LF, Medeiros MH, Kowaltowski AJ. Mild mitochondrial uncoupling in mice affects energy metabolism, redox balance and longevity. Aging Cell. 2008;7:552–560. doi: 10.1111/j.1474-9726.2008.00407.x. [DOI] [PubMed] [Google Scholar]
- Carroll J, Fearnley IM, Skehel JM, Shannon RJ, Hirst J, Walker JE. Bovine complex I is a complex of 45 different subunits. J Biol Chem. 2006;281:32724–32727. doi: 10.1074/jbc.M607135200. [DOI] [PubMed] [Google Scholar]
- Copeland JM, Cho J, Lo T, Jr, Hur JH, Bahadorani S, Arabyan T, Rabie J, Soh J, Walker DW. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol. 2009;19:1591–1598. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- DeCorby A, Gaskova D, Sayles LC, Lemire BD. Expression of Ndi1p, an alternative NADH:ubiquinone oxidoreductase, increases mitochondrial membrane potential in a C. elegans model of mitochondrial disease. Biochim Biophys Acta. 2007;1767:1157–1163. doi: 10.1016/j.bbabio.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Dell'agnello C, Leo S, Agostino A, Szabadkai G, Tiveron C, Zulian A, Prelle A, Roubertoux P, Rizzuto R, Zeviani M. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum Mol Genet. 2007;16:431–444. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- Ferguson M, Mockett RJ, Shen Y, Orr WC, Sohal RS. Age-associated decline in mitochondrial respiration and electron transport in Drosophila melanogaster. Biochem J. 2005;390:501–511. doi: 10.1042/BJ20042130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridell YW, Sanchez-Blanco A, Silvia BA, Helfand SL. Targeted expression of the human uncoupling protein 2 (hUCP2) to adult neurons extends life span in the fly. Cell Metab. 2005;1:145–152. doi: 10.1016/j.cmet.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Guarente L. Mitochondria--a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171–176. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan HM, Fridovich I. Intracellular production of superoxide radical and of hydrogen peroxide by redox active compounds. Arch Biochem Biophys. 1979;196:385–395. doi: 10.1016/0003-9861(79)90289-3. [DOI] [PubMed] [Google Scholar]
- Helfand SL, Rogina B. From genes to aging in Drosophila. Adv Genet. 2003;49:67–109. doi: 10.1016/s0065-2660(03)01002-2. [DOI] [PubMed] [Google Scholar]
- Humphrey DM, Toivonen JM, Giannakou M, Partridge L, Brand MD. Expression of human uncoupling protein-3 in Drosophila insulin-producing cells increases insulin-like peptide (DILP) levels and shortens lifespan. Exp Gerontol. 2009;44:316–327. doi: 10.1016/j.exger.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Fujii M, Hartman PS, Tsuda M, Yasuda K, Senoo-Matsuda N, Yanase S, Ayusawa D, Suzuki K. A mutation in succinate dehydrogenase cytochrome b causes oxidative stress and ageing in nematodes. Nature. 1998;394:694–697. doi: 10.1038/29331. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katic M, Kennedy AR, Leykin I, Norris A, McGettrick A, Gesta S, Russell SJ, Bluher M, Maratos-Flier E, Kahn CR. Mitochondrial gene expression and increased oxidative metabolism: role in increased lifespan of fat-specific insulin receptor knock-out mice. Aging Cell. 2007;6:827–839. doi: 10.1111/j.1474-9726.2007.00346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Lewis EB. A new standard food medium. Drosophila Information service. 1960;34:118–119. [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttik MA, Overkamp KM, Kotter P, de Vries S, van Dijken JP, Pronk JT. The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J Biol Chem. 1998;273:24529–24534. doi: 10.1074/jbc.273.38.24529. [DOI] [PubMed] [Google Scholar]
- Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- Marella M, Seo BB, Nakamaru-Ogiso E, Greenamyre JT, Matsuno-Yagi A, Yagi T. Protection by the NDI1 gene against neurodegeneration in a rotenone rat model of Parkinson's disease. PLoS One. 2008;3:e1433. doi: 10.1371/journal.pone.0001433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Boveris A. The mitochondrial energy transduction system and the aging process. Am J Physiol Cell Physiol. 2007;292:C670–686. doi: 10.1152/ajpcell.00213.2006. [DOI] [PubMed] [Google Scholar]
- Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, Carruba MO. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Li YF, Bai Y. Yeast NDI1 improves oxidative phosphorylation capacity and increases protection against oxidative stress and cell death in cells carrying a Leber's hereditary optic neuropathy mutation. Biochim Biophys Acta. 2007;1772:533–542. doi: 10.1016/j.bbadis.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Gems D, Withers DJ. Sex and death: what is the connection. Cell. 2005;120:461–472. doi: 10.1016/j.cell.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Pearl R. Rate of Living. University of London Press; 1928. [Google Scholar]
- Perales-Clemente E, Bayona-Bafaluy MP, Perez-Martos A, Barrientos A, Fernandez-Silva P, Enriquez JA. Restoration of electron transport without proton pumping in mammalian mitochondria. Proc Natl Acad Sci U S A. 2008;105:18735–18739. doi: 10.1073/pnas.0810518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier L, Shane A, Zheng J, Seroude L. Characterization of the Drosophila Gene-Switch system in aging studies: a cautionary tale. Aging Cell. 2008;7:758–770. doi: 10.1111/j.1474-9726.2008.00421.x. [DOI] [PubMed] [Google Scholar]
- Roman G, Endo K, Zong L, Davis RL. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2001;98:12602–12607. doi: 10.1073/pnas.221303998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SJ, Kahn CR. Endocrine regulation of ageing. Nat Rev Mol Cell Biol. 2007;8:681–691. doi: 10.1038/nrm2234. [DOI] [PubMed] [Google Scholar]
- Seo BB, Marella M, Yagi T, Matsuno-Yagi A. The single subunit NADH dehydrogenase reduces generation of reactive oxygen species from complex I. FEBS Lett. 2006;580:6105–6108. doi: 10.1016/j.febslet.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Shigenaga MK, Hagen TM, Ames BN. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci U S A. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal RS. Oxidative stress hypothesis of aging. Free Radic Biol Med. 2002;33:573–574. doi: 10.1016/s0891-5849(02)00885-7. [DOI] [PubMed] [Google Scholar]
- Speakman JR, Talbot DA, Selman C, Snart S, McLaren JS, Redman P, Krol E, Jackson DM, Johnson MS, Brand MD. Uncoupled and surviving: individual mice with high metabolism have greater mitochondrial uncoupling and live longer. Aging Cell. 2004;3:87–95. doi: 10.1111/j.1474-9728.2004.00097.x. [DOI] [PubMed] [Google Scholar]
- Tong JJ, Schriner SE, McCleary D, Day BJ, Wallace DC. Life extension through neurofibromin mitochondrial regulation and antioxidant therapy for neurofibromatosis-1 in Drosophila melanogaster. Nat Genet. 2007;39:476–485. doi: 10.1038/ng2004. [DOI] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Walker DW, Benzer S. Mitochondrial “swirls” induced by oxygen stress and in the Drosophila mutant hyperswirl. Proc Natl Acad Sci U S A. 2004;101:10290–10295. doi: 10.1073/pnas.0403767101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DW, Hajek P, Muffat J, Knoepfle D, Cornelison S, Attardi G, Benzer S. Hypersensitivity to oxygen and shortened lifespan in a Drosophila mitochondrial complex II mutant. Proc Natl Acad Sci U S A. 2006;103:16382–16387. doi: 10.1073/pnas.0607918103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


