Abstract
The Hepatitis B Virus (HBV) core gene codes for two closely related antigens: a 21-kDa protein which forms dimers that assemble as multi-megadalton capsids, and a 17-kDa protein which also forms dimers but that do not assemble. The proteins, respectively referred to as core and e-antigen, share a sequence of 149 residues but have different amino- and carboxy-termini. Their structural and serological relationship has long been unclear. With insights gained from recent structural studies on immune complexes of the capsids, the relationship was reassessed using recombinant forms of the antigens and a panel of monoclonal antibodies (mab) commonly believed to discriminate between core and e-antigen. Surface plasmon resonance (SPR) was used to measure the affinities, in contrast to previous studies that used more error prone and less sensitive plate-type assays. Four of the six mabs did not discriminate between core and e-antigen, nor did they discriminate between e-antigen and dimers of dissociated core antigen capsids. One mab (3120) was specific for assembled capsids and one (e6) was specific for unassembled dimers. Epitope valency of the e-antigen was also studied, using a sandwich SPR assay where e-antigen was captured with one mab and probed with a second. The e-antigen is often considered to be a monomeric protein on the basis of monovalent reactivity with antibody pairs specific for either an α or β epitope (in a prior nomenclature for e-antigen specificity). This model, however, is incorrect as recombinant e-antigen is a stable dimer and its apparent monovalency is due to steric blockage. This was proven by the formation of a 2:1 Fab e6:e-antigen complex. These results suggest new approaches for the isolation of the authentic e-antigen, its biological assay, and its stabilization as an immune complex for structural studies.
Keywords: hepatitis B virus, surface plasmon resonance, monoclonal antibody, e-antigen, core antigen
Introduction
Hepatitis B virus (HBV) causes 350 - 400 million chronic infections and approximately 1 million deaths annually.1; 2 Infection results in the expression of three protein antigens known as surface-antigen (HBsAg), core-antigen (HBcAg), and e-antigen (HBeAg).3; 4 HBsAg occurs in three forms (S, M, L) in the viral envelope and large quantities of it are found in aggregated form in the serum of infected individuals. HBcAg refers to the viral nucleocapsid which is seldom found in non-enveloped form outside of infected cells. HBeAg is a soluble protein that is secreted into the circulation and is thought to promote chronic infection.5; 6; 7
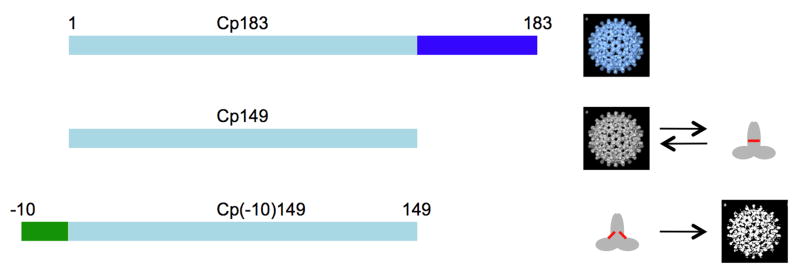
The full-length HBcAg polypeptide is 183 residues long; however, the amino-terminal 149 residues are fully competent to form dimers that can assemble to form capsids. The dimers have a central four-helix bundle flanked on either side by an α-helical domain8. Capsids are assembled from either 90 or 120 such dimers, with the four-helix bundles projecting as spikes.9 HBeAg differs from the 149-residue amino-terminal portion of HBcAg only by the presence of an additional 10 residues at its amino terminus.10; 11; 12 No atomic structure is available for HBeAg, but the extensive sequence identity with HBcAg predicts that their structures are similar.13
Despite this close sequence similarity, HBcAg and HBeAg differ in solubility and in their assembly properties,14 in their B cell and T cell responses,7 in the antibodies they are recognized by and in the kinetics they exhibit during infection,15 and in having different functions.6 On the other hand, the sequence similarity poses problems for discriminating between the two proteins in diagnostic immunoassays.16 Accordingly, a great effort has been made to identify determinants specific for each of these two antigens, and at least 70 monoclonal antibodies (Mabs) have been generated for this purpose.13; 16; 17; 18; 19; 20; 21 The results, although often unclear, can be approximately summarized as follows: (a) determinants recognized on HBcAg are primarily conformational whereas those on HBeAg are mostly linear;16 (b) there is one group of similar determinants clustered around residue 80 that comprise the principal antigenic aspect of HBcAg and another near the carboxy-terminus, and this latter can be divided into two subsets;13; 17; 18 (c) HBcAg and HBeAg share the first group of determinants but differ with regard to the second, which may become masked during assembly of capsids;13 and (d) there are a number of less-well characterized epitopes at other locations on the dimer.16; 22 However, any review of the literature quickly reveals numerous counter-examples and ambiguities. Fortunately, since the time that these studies were done the structure of HBcAg has been determined at high resolution8 and the epitopes of seven HBcAg-specific Mabs have been identified by cryoelectron microscopy and image reconstruction.23; 24; 25; 26; 27
The goal of the present study was to reassess and clarify the serological relationship between HBcAg and HBeAg using surface plasmon resonance. This technique is not only very sensitive but also less error prone than the previously employed plate-based assays and it was therefore used to measure the affinities of a panel of six historically well-documented monoclonal antibodies for a set of highly-purified and well-characterized recombinant forms of HBcAg and HBeAg. Mabs 904, 905, 3105, and 3120 are the original monoclonal antibodies from the Mayumi group that define the four primary HBcAg and HBeAg determinants. Mabs 904 and 905 define the e-antigen determinants HBeAg/a and HBeAg/b,21 and Mabs 3105 and 3120 define the core-antigen determinants HBcAg/α and HBcAg/β.20 Mab e6 defines the e-antigen determinant HBe-β19 and Mab F11A4 recognizes the “dominant” core-antigen determinant.18 The epitopes of Mabs 3105, 3120, and F11A4 have been determined structurally.9 HBcAg and HBeAg were produced as recombinant proteins in E. coli (see Table 1 and Fig. 1). For HBcAg, we used capsids of the full-length 183-residue protein (Cp183) that contain bacterial nucleic acid (substituting for the viral pregenome in authentic HBcAg) and capsids composed of the 149-residue protein (Cp149). The latter are free of nucleic acid and, unlike Cp183 capsids, may be disassembled in vitro to dimers. For HBeAg, we expressed in E. coli a construct corresponding to the same polypeptide chain as native HBeAg, i.e. Cp149 extended at its amino-terminus by a 10-residue peptide corresponding to the residual propeptide viz. Cp(-10)149.10; 11; 12 We assumed that this highly purified and well-defined dimeric protein closely resembles native (serum-derived) HBeAg. In order to distinguish assembly-dependent properties from conformation-dependent properties of these proteins, we also determined conditions to form novel capsid-like structures from Cp(-10)149, and exploited a point mutation that greatly reduces the propensity of both core- and e-antigen dimers to assemble into capsids, and included these reagents in the SPR assays. The suffices “c” and “d” are used to denote, respectively, the capsid and dimer forms of a given protein, e.g. Cp149c, and Cp149d.
Table 1.
Recombinant HBcAg and HBeAg-related proteins
| HBV Protein | Residues | Mutations | Assembly State |
|---|---|---|---|
| Cp183ca, b | 1-183 | None | Capsid |
| Cp149c | 1-149 | C48A, C107A | Capsid |
| Cp149dc | 1-149 | C48A, C107A, G123A | Dimer |
| Cp(-10)149c | (-10)-149 | C48A, C107A | Capsid |
| Cp(-10)149dd | (-10)-149 | C48A, C107A, G123A | Dimer |
Cp183c contains encapsulated nucleic acid derived from E. coli 14 and corresponds to serum-derived HBV core antigen (HBcAg).
The subscripts c and d refer to capsids and dimer proteins, respectively.
Cp149c does not contain encapsulated nucleic acid, is immunologically similar to HBcAg, and has been used extensively for structural studies.
Fig. 1.
Schematic representation of the recombinant HBV proteins used in this study. The three capsid-related proteins (Cp) correspond to those listed in Table 1 and in greater detail in Fig. 2. Cp183 is expressed in E. coli as nucleic acid-filled capsids (Cp183c) with a mass of ∼6 MDa39 (represented by the blue fenestrated structure). Such capsids are very stable and cannot be dissociated into subunits without protein denaturation. In Cp149 the arginine rich carboxy-terminal domain has been deleted and when this protein is expressed in E. coli it forms empty capsids (Cp149c) with masses of 3 and 4 MDa39 (represented by a grey fenestrated structure). These capsids can be reversibly dissociated into dimeric subunits with a mass of ∼35 kDa (grey object). The red line indicates an intermolecular disulfide bond [C61-C61]. The Cp(-10)149 or e-antigen is expressed in E. coli in a form that is neither assembled capsid nor an aggregated inclusion body-type protein. The protein can be extracted with 2 - 3 M urea at pH 9.5 to give soluble folded dimers. Purified dimers can be induced to form capsids (white fenestrated structure). The reversibility of this has not been fully explored. The red lines indicate intramolecular disulfide bonds [C(-7) – C61]. See Figs. S1 and S2 for analytical data on these various proteins.
Results and Discussion
Protein Expression and Purification
Unlike the very stable Cp183c, Cp149c can be dissociated into dimers under conditions which do not denature the constituent subunits.14 The resulting Cp149d is not very soluble, readily re-associating into capsids in the presence of salt at neutral pH;14 however, the solubility is dramatically enhanced by introducing either the single (G123A) or double (G123A, R127A) mutations (unpublished observations). Cp(-10)149d is more soluble than Cp149d but can be induced to form capsids by elevating the temperature (see Materials and Methods). The mutation G123A also enhances the solubility of Cp(-10)149d. The two cysteine 61 residues in Cp149d form an intermolecular disulfide bond (Fig. S1, a) whereas in Cp(-10)149d they form two intramolecular disulfides with the cysteine (-7) residues (Fig. S1, a)11; 12 and for this reason, cysteine 61 was retained in all constructs. However, cysteines 48 and 104 are not involved in disulfide bond formation8 and were routinely substituted with alanine. The capsid proteins and the various mutants used in this study are listed in Table 1 and schematically represented in Fig. 1. The assembly state of capsids under native conditions (i.e. in buffered saline), was confirmed either by gel filtration using Sepharose 4B or by sedimentation velocity ultracentrifugation.14 SDS-PAGE of freshly prepared Cp183 capsids under reducing conditions gave one main band corresponding to the monomer (Fig. S1, b) and under non-reducing condition, two major bands corresponding to monomers and dimers (Fig. S1, b) and lesser amounts of higher order multimers28. The native molecular weights of Cp149d (32,744 ± 500 Da) and Cp(-10)149d (36,834 ± 177 Da) were directly determined by sedimentation equilibrium ultracentrifugation (Fig. S2), indicating that the proteins are dimeric at neutral pH.
Monoclonal Antibodies
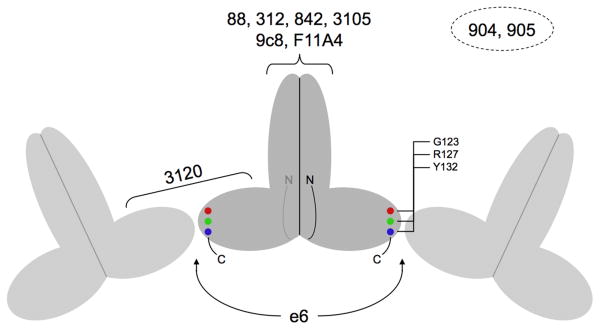
Supplementary Table 1 summarizes the principal antigenic regions on HBcAg and HBeAg reported in the literature, and Table 2 summarizes the epitopes of the antibodies employed in this study. The locations of the epitopes are also shown in Fig. 2.
Table 2.
Antibodies characterized in this study
| Mab | Immunogen | Specificity | Epitope Site | Residues | Ref |
|---|---|---|---|---|---|
| 904 | HBeAg/serum | HBeAg/a | NDa | - | |
| 905 | HBeAg/serum | HBeAg/b | ND | - | |
| 3105 | Virions/serum | HBcAg/α | Spike tip | (77-80)+(77-80)b | 25 |
| 3120 | Virions/serum | HBcAg/β | Capsid; inter-dimer | (20-22)+(25-29)+(126-127) + (20-22)+(129-132)c | 26 |
| F11A4 | rHBcAg/E. coli | HBcAg | Spike tip | (74-77)+(78-80)+(83-84)d | 25 |
| e6 | dHBcAge | HBeAg/β | Dimer; C-terminus | (124-132)f |
Not determined.
Mab 3105 can bind to the epitope in either of two symmetrical orientations.
Mab 3120 binds at the interface between two dimers, involving similar sets of residues on the two subunits.
Mab F11A4 binds to both loops and one helix of the four-helix bundle.
Reduced and denatured recombinant HBcAg.
Approximate extent, from residues identified in this study.
Fig. 2.
Locations of the epitopes on HBV capsids and subunits. The cartoon represents three adjacent dimeric subunits on a capsid with the protruding spikes corresponding to the four-helix bundles. The epitopes for Mabs 88, 312, 842, 3105, 9c8, and F11A4 are all located on the apices of the spikes whereas the epitope for Mab 3120 is located between the spikes and involves residues from two adjacent subunits.23; 24; 25; 26; 27 The two copies of the Mab e6 epitope are located near the C-termini of the dimer and are accessible only on free subunits. The locations of the epitopes for Mabs 904 and 905 are uncertain, but epitope 904 may be on the spike apex and 905 near the C-terminus (see text). The positions of the three carboxy-terminal mutations employed to map the e6 epitope, and which affect capsid assembly, are also indicated.
Direct Binding of Capsids and Dimers to the Antibodies
The binding kinetics of various protein preparations to the panel of Mabs are summarized in Table 3. From a cursory inspection the affinities shown in this table do not appear to be consistent with the published specificities of the antibodies used (Table 2). One might expect that HBeAg-specific and HBcAg-specific Mabs would, by definition, have higher affinities for their respective antigens. For example, the HBeAg-specific Mab 905 exhibits an affinity for Cp183c almost as high (Kd = 8.4 × 10-10 M) as for Cp(-10)149d (Kd = 2.0 × 10-10 M). At the same time, the affinity of this Mab for Cp183c is much higher than the affinity of the HBcAg-specific Mab 3105 for Cp183c (Kd = 3.4 × 10-8 M). How these results can be explained is discussed below.
Table 3.
Affinities of HBcAg and HBeAg binding to Mabs
| Ligand | Analyte | ka (1/Ms) | kd (1/s) | Kd (M) |
|---|---|---|---|---|
| Mab 904 | Cp183c | 2.5 × 104 | 4.3 × 10-4 | 1.7 × 10-8 |
| Cp149c | 5.3 × 104 | 8.2 × 10-4 | 1.5 × 10-8 | |
| Cp(-10)149c | 1.2 × 104 | 3.0 × 10-4 | 2.6 × 10-8 | |
| Cp149d | 2.5 × 104 | 8.4 × 10-5 | 3.4 × 10-9 | |
| Cp(-10)149d | 6.9 × 104 | 2.4 × 10-4 | 3.5 × 10-9 | |
| Mab 905 | Cp183c | 5.2 × 104 | 4.3 × 10-5 | 8.4 × 10-10 |
| Cp149c | 2.3 × 105 | 2.3 × 10-4 | 1.0 × 10-9 | |
| Cp(-10)149c | 4.0 × 104 | 5.5 × 10-4 | 1.4 × 10-8 | |
| Cp149d | 3.7 × 103 | 1.6 × 10-3 | 4.4 × 10-7 | |
| Cp(-10)149d | 1.9 × 105 | 3.8 × 10-5 | 2.0 × 10-10 | |
| Mab 3105 | Cp183c | 4.1 × 104 | 1.4 × 10-3 | 3.4 × 10-8 |
| Cp149c | 2.2 × 105 | 1.1 × 10-3 | 4.9 × 10-9 | |
| Cp(-10)149c | 1.4 × 104 | 4.6 × 10-5 | 3.2 × 10-9 | |
| Cp149d | 5.1 × 104 | 1.7 × 10-3 | 3.3 × 10-8 | |
| Cp(-10)149d | 3.5 × 104 | 1.2 × 10-2 | 3.4 × 10-7 | |
| Mab 3120 | Cp183c | 1.5 × 104 | 3.6 × 10-7 | 2.3 × 10-11 |
| Cp149c | 3.0 × 104 | 3.8 × 10-7 | 1.2 × 10-11 | |
| Cp(-10)149c | 1.2 × 104 | 2.4 × 10-7 | 1.9 × 10-11 | |
| Mab F11A4 | Cp183c | 2.9 × 104 | 2.7 × 10-5 | 9.4 × 10-10 |
| Cp149c | 1.8 × 106 | 1.8 × 10-4 | 1.0 × 10-10 | |
| Cp(-10)149c | 1.0 × 104 | 1.4 × 10-6 | 1.3 × 10-10 | |
| Cp149d | 5.7 × 105 | 3.9 × 10-3 | 6.9 × 10-9 | |
| Cp(-10)149d | 5.3 × 104 | 7.5 × 10-3 | 1.4 × 10-7 | |
| Mab e6 | Cp149d | 6.5 × 104 | 2.2 × 10-4 | 3.3 × 10-9 |
| Cp(-10)149d | 5.9 × 103 | 6.9 × 10-4 | 1.2 × 10-7 |
Capsids formed from the constructs Cp183, Cp149 and Cp(-10)149 all bound with moderate (Kd ∼ 10-8 to 10-9 M) to high (Kd < 10-9 M) affinity to all of the antibodies except Mab e6, which only bound dimeric protein. The dimers Cp149d and Cp(-10)149d also bound with moderate to high affinity to all members of the same panel, except Mab 3120. Thus four of the six Mabs designated as either HBcAg- or HBeAg-specific exhibit little discrimination between capsids and dimers. For example, the respective bindings of Cp149c and Cp149d to Mab 3105 (nominally HBcAg-specific) show fairly similar kinetics and affinity (Fig. 3). Thus - with the exceptions noted above - these Mabs are largely insensitive to the assembly state of the capsid protein or, for a given assembly state, to the sequences appended at either terminus.
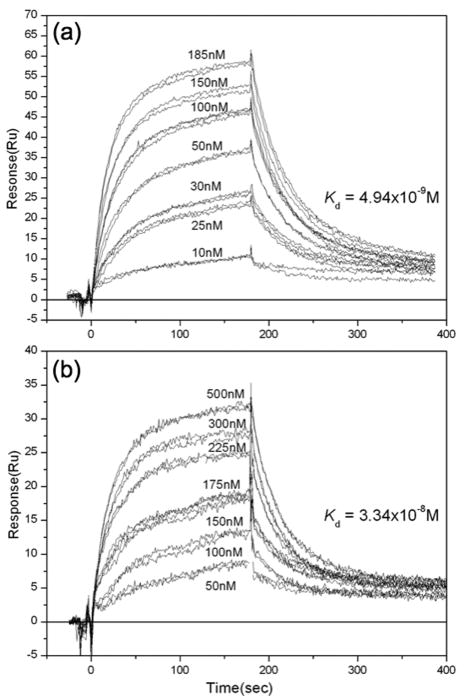
Fig. 3.
Typical sensograms illustrating the binding of antigen to Mab 3105. Antigen was allowed to bind to immobilized antibody. Duplicate, and in some cases triplicate, injections of antigen were done. The concentrations refer to the concentration of antigen, in terms of dimers. The kinetics and affinities of Cp149c (A) and Cp149d (B) for Mab 3105 are similar, indicating that the epitope is not affected significantly by the assembly state of the antigen. This is in keeping with its known location on the apex of the spike(s).
The failure of Cp149d and Cp(-10)149d to bind Mab 3120 is consistent with the identification of its epitope as bridging two subunits on adjacent dimers.26 For the same reason, this Mab exhibits little difference in its affinities for the three capsids (Cp183c, Cp149c, and Cp(-10)149c): all are characterized by very slow off-rates (kd) indicative of high affinity binding (very low Kd) (Table 3). The other nominally HBcAg-specific Mabs, 3105 and F11A4, bind to the two so-called immunodominant loops located at the apex of the capsid spike.25 The four-helix bundles in capsid subunits and free dimers are expected to have similar, though not necessarily identical,29 conformations. Thus, the binding of these spike-specific antibodies is unaffected by the assembly state of the capsid protein: they all bind strongly to both capsids and dimers (Table 3).
Valency Considerations
We have assumed that the interactions in Table 3 represent 1:1 interactions between immobilized antibodies and antigens, and furthermore, that these interactions represent affinity rather than avidity, despite the fact that the antibody is bivalent and the antigens are either bi- or polyvalent. To promote monovalent interactions between antibody and antigen we employed the minimum density of immobilized antibody consistent with reliable measurements (densities of 2000, 500, and 50 RU were tested). The equilibrium constants reported in Table 3 were essentially unaltered over this range of densities. Hence, at the routinely used density of 500 RU, the contribution to the data of single antigens bound to more than one antibody was considered to be minimal.
With regard as to whether the interactions represent affinity or avidity, we considered the dimers to be too small to engage with both antigen-binding domains of a given IgG simultaneously. This was confirmed by experiment, for example, Cp(-10)149d bound to immobilized Fab 904 and Mab 904 with Kd values of 6.8 and 3.5 × 10-9 M, respectively. Similar experiments with capsids were not possible because capsids failed to bind to immobilized Fabs, probably due to restricted access. However, the binding of Mabs 3105, 3120, and F11A4 to capsids has been visualized:9 in all cases the Fabs were oriented essentially perpendicular to the capsid surface, suggesting that the two antigen binding domains of a given IgG would probably not be able to interact with the same capsid,30 although a bivalent interaction has been observed with a different viral capsid.31 That all these Mabs readily aggregate capsids, as evidenced by light scattering, precipitation, and electron microscopy (data not shown) supports the interpretation that the values obtained represent affinity and not avidity.
The e-antigen is Both Physically and Antigenically Dimeric
The number of antibody molecules that a single dimer can bind depends on the positioning of the epitopes in question; binding of one Mab or Fab to its epitope should allow another, different, Mab/Fab to bind only if its epitope is not occluded. Similarly, two copies of the same Mab/Fab may be expected to bind to a dimer only if the two copies of its epitope are suitably spaced apart. To study epitope accessibility of dimers with respect to the HBeAg-specific Mabs, we used three-layer sandwich assays in which one Mab (the ligand) is immobilized on the Biacore chip and, after capture of either Cp149d or Cp(-10)149d, the binding of a second Mab is measured (Supplementary Fig. 3).
The results with Mab e6 as the ligand are the most clear; captured HBeAg dimers are detected with a second e6 antibody, indicating the presence of two accessible epitopes (Table 4). Mabs 904 and 905 also bind to Mab e6-captured protein, indicating that, in both cases, as least one copy of their epitope is still accessible on Mab e6-bound dimer.
Table 4.
SPR sandwich assay of capsid proteinsa
| Ligand (Mab) |
Analyte-1 (Antigen) |
Analyte-2 (Mab) |
Binding | Commentb |
|---|---|---|---|---|
| 904 | Cp149d | e6 | + | |
| 904 | - | Binding to single epitope | ||
| 905 | + | |||
| 3105 | + | |||
| 905 | Cp149d | e6 | + | |
| 904 | + | |||
| 905 | - | Binding to single epitope | ||
| 3105 | + | |||
| 3120 | - | No binding to dimers | ||
| Cp149c | e6 | - | No binding to capsids | |
| 904 | + | |||
| 905 | + | Multiple epitopes available | ||
| 3120 | + | |||
| e6 | Cp149d | e6 | + | Binding to two epitopes |
| 904 | + | |||
| 905 | + | |||
| 3105 | + | |||
| F11A4 (Fab) | + | |||
| 4B5c | - | Negative control | ||
Ligand (Mab) is immobilized on Biacore chip; capsid protein (Analyte 1) is captured by ligand and used to monitor binding of second Mab (Analyte 2). See Fig. 3 for typical sensogram tracing.
Comments refer to binding of Analyte-2.
The Mab 4B5 is anti-HBsAg.
The situation is more complicated when the other HBeAg-specific Mabs, 904 and 905, are used to capture antigen because, in three-layer assays with either of these Mabs, Cp149d behaves as if only one epitope is accessible. Thus, dimer captured with Mab 905 only binds Mab 904 and not Mab 905, and vice versa. Presumably, antibody bound to one epitope on the dimer occludes the second copy of the same epitope. We have described previously how proximal epitopes can introduce steric barriers to antibody binding to HBV capsids.9 In contrast, when capsids are captured with immobilized Mab 905, they can be detected with Mab 905. This was as predicted because capsids present multiple copies of each epitope, distributed over a large surface. All the other Mabs tested, including 3120, also detect capsids captured with Mab 905 (and Mab 904) (Table 4). The exception, Mab e6, also as predicted, does not bind to the capsids and this is consistent with the direct binding studies (Table 3).
The apparently restricted binding of Mab 904 was further tested by binding to Cp149d and then probing with Fab 904 (rather than Mab 904). Weak binding was observed characterized by fast on- and off-rates (data not shown) supporting the view that steric blockage is responsible for an apparently single epitope on a dimeric protein. This point is important as historically it has guided the development of assays for the e-antigen and is based on the long-held view that the e-antigen is monomeric, for example, see model for p17 (e-antigen).12 This physical picture of the e-antigen was not based on an accurate mass determination of serum-isolated antigen, not to date carried out, but on its antigenic properties. According to this model, Mabs 904 and 905, for example, do not bind in a bivalent mode because each is directed to a different epitope (α or β, respectively) located on a monomeric protein. We have described elsewhere9 that recombinant e-antigen is a very stable dimer (Table 1). Furthermore, the binding properties of Mab e-6 (Table 4) and direct measurement of binding stoichiometry described below, clearly show the dimeric nature of e-antigen both in terms of structure and antigenicity. It should be pointed out that although the direct determination of the physical state of serum e-antigen has not been reported, the high molecular weight forms mentioned in previous studies, for example,19 are most likely complexes of dimeric e-antigen with specific anti-HBe IgG 32; 33
Location of Anti-HBeAg Mab Epitopes
Cryo-EM and molecular modeling of Fab-labeled capsids has been used to localize and define the epitopes of a total of seven anti-HBcAg Mabs,23; 24; 25; 26; 27 including three involved in this study (Fig. 2 and Table 2). Attempts to use this approach with Mabs 904 and 905 were unsuccessful because we were unable to achieve stable decoration of capsids with these Fabs under the conditions used for electron microscopy. This outcome was surprising given the SPR result that their affinities for capsids appear to be as strong as that of Mab 3105, which decorates capsids well. We performed many control experiments to confirm that capsids do, in fact, bind these antibodies: for example, when capsids are titrated with either Mab 904 or Mab 905, their binding to immobilized Mab 904 or Mab 905 is completely blocked (Fab 904 or Fab 905 also block the binding).
We also studied the binding of Mab e6 to dimers with various point mutations (Supplementary Table 2). The R127A substitution in both Cp149d and Cp(-10)149d effectively abrogated binding to Mab e6. The Y132A substitution also blocked binding of this mab. In the crystal structure of T = 4 capsids,8 R127 is located at the end of the helix-5 (residues 112–127) and Y132 is in the following loop (see Fig. 2). These motifs pack together in adjacent dimers in capsids and therefore should be accessible on dimers but occluded in capsids. This localization places the two copies of the e6 epitope at opposite ends of the crosspiece of the T-shaped dimer, providing a ready explanation for its observed ability to bind two copies of Mab e6. In this context, it is noteworthy that the G123A mutation, also located in helix-5, affects dimer solubility but has no effect on Mab e6 binding.
The same set of Cp149d mutants was tested with Mabs 904 and 905. R127A had no effect on the binding of Mab 904 but that of Mab 905 was greatly reduced (not shown). Y132A did not affect the binding of either Mab. These observations suggest that part of the 905 epitope is located in helix-5; however, it clearly differs from that of Mab e6 as indicated by its reactivity with Y132A dimer and because this Mab binds to capsids whereas Mab e6 does not. In two-layer competition experiments using immobilized Mab e6, Cp149d was mixed separately with Mabs e6, 904, and 905 prior to injection; as expected, dimer binding was blocked completely by Mab e6. Binding of Mab 905 was blocked to a lesser degree, whereas Mab 904 had no effect on binding. Taken together, these results support assigning the 905 epitope to the carboxy-terminal region of Cp149 and its partial overlap with the e6 epitope.
Electron Microscopy
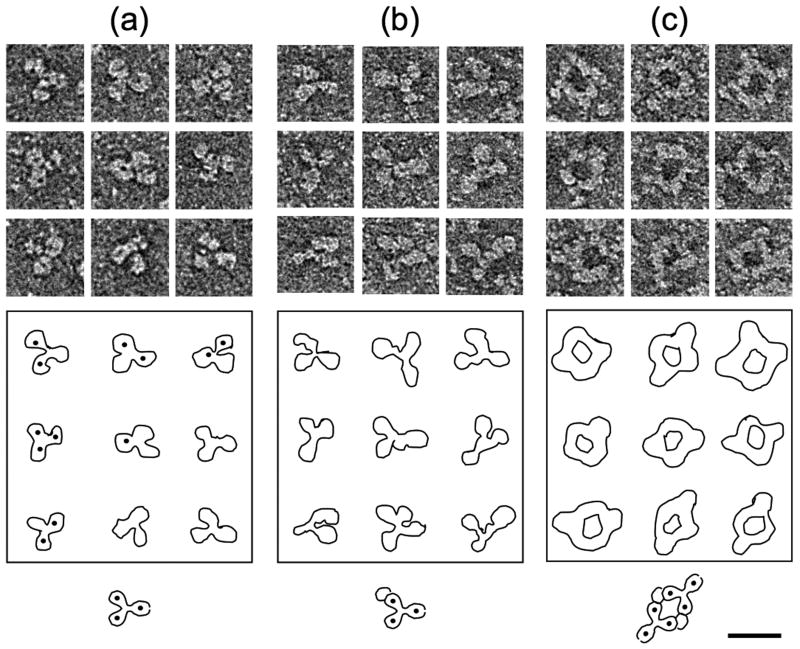
The locations of the binding sites of Mabs 3105, 3120, and F11A4 are known9 and those of Mab e6 and Mab 905 can be assigned to the vicinity of the carboxy-termini of the HBc/eAg dimer. To obtain additional information about the locations of the binding sites of Mabs 904, 905, and e6, complexes of these mabs with Cp(-10)149d were examined by negative stain electron microscopy (Fig. 4). Mab 904 appears to form a 1:1 antibody:dimer complex consistent with the SPR results (Table 3). Complexes with 1:2 stoichiometry probably also formed but were either rare or, more probably, not recognized as such, however no ring-like complexes or linear polymers were ever observed with this antibody. The two-fold symmetry of the dimer implies the presence of two unique sites, namely at the exact apex and underside of the four-helix bundle. The interaction of Mab 904 with Cp149d in an apparently monovalent manner, together with its affinity for both dimers and capsids, suggests that the 904 epitope may be located at the unique site on the apex of the four-helix bundle (the underside of the dimer being inaccessible in capsids).
Fig. 4.
Immune complexes visualized by electron microscopy. Cp(-10)149d were mixed with antibodies in solution and visualized in negative stain. (A) Mab alone, (B) Dimer + Mab 904, (C) Dimer + Mab 905. Outlines and an interpretative diagram are shown in each case. The formation of the circular complexes in (C) suggests the presence of two copies of the 905 epitope per Cp(-10)149d molecule. Very similar complexes were observed when Mab e6 was employed (not shown). Bar = 200 Å.
Unlike Mab 904, Mab 905 forms ring-like antibody:dimer complexes which are consistent with dimers crosslinked by antibody binding to the carboxy-terminal regions (Fig. 4). However, this interpretation is not completely consistent with the biochemical data because, as we have seen, the binding of Mab 905 to dimeric protein appears monomeric in terms of epitope valency. A rationale analogous to that given above for the monovalent nature of the Mab 904 interaction with dimers is incompatible with the likely location of the 905 epitope(s) at the carboxy-termini. As may always be possible, the immobilization and binding of proteins can restrict access by steric hindrance and this would not be an issue with freely interacting proteins in solution. Complexes of Mab e6 with Cp(-10)149d also formed rings similar to those formed by Mab 905; however, despite repeated attempts, the structural integrity of these complexes was not as well preserved (not shown).
Specific Immuno-assays for HBcAg and HBeAg
In practical terms, the use of Mab 904 and Mab 905 to distinguish between HBcAg and HBeAg is problematical, as both antibodies will capture capsids as well as dimers. A more specific assay system would consist of Mab 904 or Mab 905 as the primary ligand for capture of both HBcAg and HBeAg followed by Mab e6 for the specific detection of HBeAg and Mab 3120 for the specific detection of HBcAg. Based on the specificity of Mab e6 for dimer, this assay provides a bona fide assay for HBeAg when Mab e6 is used for ligation and any of Mabs e6, 904, 905, or 3105 for detection.
A Complex of Fab e6 and HBc/eAg Dimers Suitable For Structural Studies
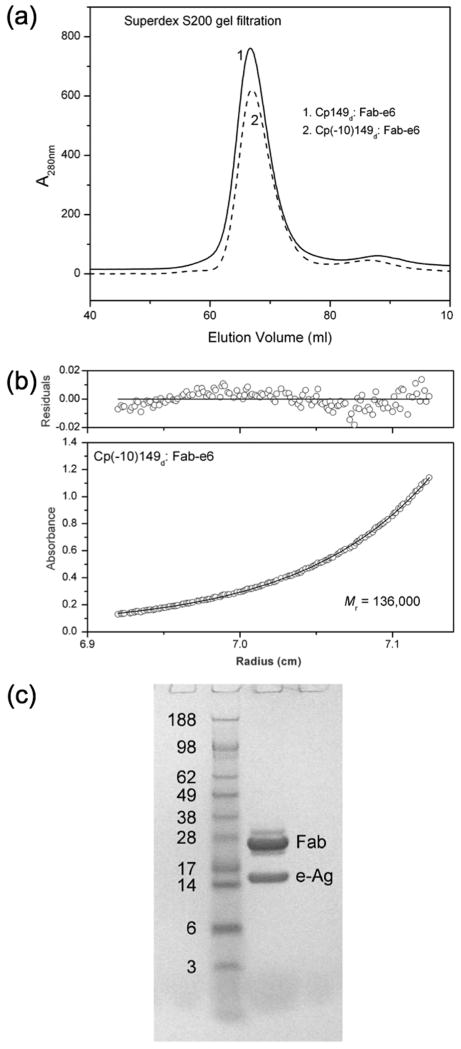
Direct crystallization of the dimeric proteins Cp149d and Cp(-10)149d has so far proven unsuccessful, in part due to their tendency to self-associate at higher protein concentrations despite the G123A mutation. Antibody fragments have long been used to effect the co-crystallization of proteins.34 Based on our results, Mab e6 appears to be a useful candidate to prepare immune complexes with these dimers for screening crystallization conditions. The SPR data indicate that Mab e6 binds to dimeric protein with good affinity (Kd = 3.3×10-9 M for Cp149d; 1.2×10-7 M for Cp(-10)149d) and that the carboxy-terminal location of the epitopes allows unrestricted access, i.e. binding can occur independently at both sites. Further, the G123A mutation, important in enhancing the solubility of dimeric protein, has little effect on the binding of the antibody (Supplementary Table 2). The preparation and crystallization of the immune complexes Cp149d:Fab e6 and Cp(-10)149d:Fab e6, both with a subunit stoichiometry of 1:2 (Fig. 5), will be described elsewhere.
Fig. 5.
Characterization of Cp149d:Fab e6 and Cp(-10)149d:Fab e6 immune complexes. Analysis of the complexes by (a) gel filtration chromatography, (b) analytical ultracentrifugation, and (c) reducing SDS-PAGE indicates a 1:2 molar ratio in both cases.
Conclusion
There is a large yet inconclusive literature regarding the immunochemical detection of HBV core- and e-antigens (for reviews see 16; 35). There are four problems that have contributed to this situation. First, HBcAg and HBeAg are composed of almost identical polypeptides yet the folded proteins are distinct in several ways. Second, there has been a lack of well-characterized antigens available for study. The HBeAg in particular has been problematical as it has not been isolated from serum as a native protein and well characterized. It has been derived from various sources (e.g. serum, liver, and even bacterially-expressed and denatured HBcAg), with various molecular weights and degrees of purity, and with uncertainties regarding its polymeric state. Third, until recently8 there has been no structural knowledge about either of the antigens. Fourth, the earlier work was hampered by the cumbersome and involved nature of the immunoassays. Here we have employed precisely defined antigens, antibodies that recognize the major antigenic sites on these antigens, and a sensitive means to measure the affinities.
The main observation is that most of the antibodies, regardless of reported specificity, and regardless of the assembly state of the antigen, bind with relatively high affinity. The exceptions are the anti-HBeAg Mab e6, which detects an epitope accessible only on the non-polymerized HBc/eAg dimer, and the anti-HBcAg Mab 3120, which detects an epitope formed only when dimers associate to form capsids. The conclusion is that an antigenic distinction between HBcAg and HBeAg can be partially made on the basis of the assembly state of the protein. The 10-residue amino-terminal precore region in HBeAg positions the cysteine at position (-7) close to cysteine 61 at the dimer interface which forms an intramolecular disulfide [C(-7) – C61] in preference to a intermolecular one [C61-C61] as in core antigen. This both enhances the solubility of dimeric e-antigen compared to the dimeric subunits of core antigen and may also promote local conformation changes. We have not yet identified antibodies that are truly specific for e-antigen and which could distinguish between dimeric core capsid subunits and e-antigen. However, Mab e6 promises to be a useful tool for the isolation and characterization of the authentic e-antigen, which has surprisingly eluded isolation and characterization. Furthermore, we have found that the antigen-binding domains of Mab e6 form soluble complexes with dimeric HBc/eAg that hold promise for a high-resolution structure determination. This would allow a direct comparison of the core-antigen and e-antigen dimer structures and open a new avenue to structure-function studies of both of these proteins. It will then still remain to be seen how the interactions of the recombinant antigens with monoclonal antibodies relate to those of the authentic antigens with the immune system.
Materials and Methods
Purification of HBV antigens
The capsids and capsid-related proteins used in this study were expressed in E. coli. Site-specific mutations (see Table 1 and Fig. 1) were introduced by standard procedures. The proteins were prepared essentially as described previously.14 Briefly, the cells were resuspended in 100 mM Tris-HCl (pH 7.0) containing protease inhibitors (Sigma) and lysed in a French press. Cp183c, which always forms intact RNA-containing capsids, was purified by a combination of gel filtration and centrifugation. For Cp149 and Cp(-10)149 constructs containing the G123A solubilizing mutation, lysates were centrifuged at 25,000 ×g for 60 min. Cp(-10)149 constructs were all located in the pellets whereas Cp149 constructs were located in the supernatants. Cp(-10)149 protein was solubilized from the pellets with 100 mM Na2C03-NaHC03 (pH 9.5) (Buffer A) containing 3.0 M urea and, following clarification of the solution by centrifugation, concentrated by the addition of 40% (w/v) (NH4)2SO4. The precipitated proteins were dissolved in Buffer A plus 3 M urea and then applied to a Superdex 200 column (GE Healthcare) equilibrated in Buffer A containing 2 M urea. Cp(-10)149d proteins (eluting with a mass of ∼30 kDa) were resolved from aggregates and other contaminants and then further purified using a Q Sepharose column (GE Healthcare) equilibrated with 50 mM Tris-HCl (pH 8.0) containing 2 M urea. The protein was eluted with a 0 - 0.5 M NaCl gradient and then dialyzed against 50 mM Tris-HCl (pH 7.8). Occasionally, an additional Superdex 200 chromatography step was performed to ensure removal of any aggregated protein and to exchange buffer. The Cp149d located in the supernatant of the initial low speed centrifugation were concentrated by precipitation with 40% (NH4)2SO4 and then purified as described for Cp(-10)149d. All proteins were stored in small aliquots at -80 °C in the absence of reducing agents. Wild-type Cp149d was assembled into Cp149c at 0.5 mg/ml by dilution with 100 mM HEPES (pH 7.0), containing 350 mM NaCl (assembly buffer). The Cp(-10)149d was assembled into Cp(-10)149c similarly to Cp149d except that the protein was warmed to 37 °C for 1 hour following dilution into assembly buffer.
Monoclonal Antibodies
Mabs 904, 905, 3105, and 3120 were purchased from the Institute of Immunology, Tokyo. Mabs e6 and F1 1A4 have been described previously.18; 19 Mab 4B5 recognizes HBsAg (unpublished observations). Fabs were prepared with immobilized papain and Protein A (Pierce) using standard procedures.
Surface Plasmon Resonance
The kinetics of the capsid-related proteins binding to immobilized Mabs (two-layer configuration) was studied by surface plasmon resonance (Biacore). Mabs were immobilized on the surface of CM5 sensor chips by the EDC-NHS [N-ethyl-N ″ (dimethylaminopropyl) carbodiimide-N-hydroxysuccinimide] coupling method. Antibody was immobilized on chips at densities of 2000, 500 and 50 RU (1000 RU ∼ 1 ng bound protein/mm2 36), with 500 RU used for routine measurements. HBS-EP (100 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20) was used as running buffer and samples in running buffer (90 μl) were passed over the Mab surface at a flow rate of 30 ul/min. At the end of the association phase, the dissociation was monitored for at least 30 min (very slow off-rates were monitored up to 120 min). Analytes were injected at concentrations between 0.5 – 500 nM. For each injection, the reference surface data (Fc1) was subtracted from the reaction surface data (Fc2) and the resulting data (Fc2-Fc1) provided the actual binding. For all assays at least two replicate injections at the same concentrations were employed to calculate the kinetic data. In some experiments a three-layer configuration was employed. Immobilized Mab was overlaid with capsid protein (analyte-1) by injection of 100 μl sample at 20 μl/min and after the end of injection (∼320 seconds) the second Mab (analyte-2) was injected (100 μl). In this assay, protein captured with one antibody is probed with a second antibody.
The data were analyzed with BIAevalution software, version 3.1. The interactions of the capsid proteins with the Mabs were globally fitted to a 1:1 interaction model with the association and dissociation phases of the interaction fitted simultaneously. The goodness of fit between the fitted curves and the experimental curves was assessed by visual comparison. The rate constants (ka and kd) and the equilibrium constants (Kd) of each interaction were calculated from the best-fit kinetic parameters. After each fitting, the function ln(abs(dY/dX)) during the association phase was evaluated, and when this produced a straight line with a negative slope it was taken to indicate that there was no mass transfer limitation (BIAevaluation handbook BR 1002-29, Biacore AB). When necessary, a lower level of Mab was employed to reduce the mass transfer effects. The Langmuir model was used in most instances unless there was need to use either the Langmuir with mass transfer or the Langmuir with drifting baseline models. Nevertheless, in all instances, rate constants for each kinetic set were checked with the three different models to determine whether ka and kd deviated significantly between models.
Electron Microscopy Of Immune Complexes
Immune complexes with dimers were formed in solution. Mabs 904, 905, and e6 were mixed with Cp(-10)149d. G123A in 1:1 molar ratio, incubated overnight at room temperature, and then diluted to 0.01 mg/ml with respect to Mab with 20 mM ammonium acetate. A thin carbon film deposited on freshly cleft mica was floated onto a drop containing the immune complexes and allowed to adsorb for 60 seconds, then floated onto 1% uranyl formate, before being picked up from above with a lacy carbon film supported on a 400 mesh copper grid. Micrographs were recorded at 60,000× magnification on a Philips CM-120 electron microscope (FEI) equipped with a CCD camera.
Preparation of Fab e6 Immune Complexes
The Cp149d or Cp(-10)149d (both with the G123A mutation) were mixed with Fab e6 in a 1:2 molar ratio. The complexes (12 mg/ml) were gel filtrated by applying 1 ml to a Superdex 200 column (1.6 cm × 60 cm) equilibrated with 50 mM HEPES (pH 7.0).
Analytical Ultracentrifugation
A Beckman Optima XL-A analytical ultracentrifuge with an An-60 Ti rotor and standard double-sector centerpiece cells was used. Sedimentation equilibrium measurements of the Cp149d and Cp(-10)149d constructs were performed at 18,000 and 17,000 rpm, respectively, and the immune complexes at 11,500 rpm. Centrifugations were done at 20 °C for 15 - 20 h with an additional 3 h at 45,000 rpm to establish baselines. The buffers were 50 mM Tris-HCl (pH 7.5) for Cp149d and Cp(-10)149d and 50 mM HEPES (pH 7.0) for the immune complexes. The data were analyzed with Beckman-Origin software. Protein partial specific volumes were calculated from the amino acid compositions37 and solvent densities as previously described.38
Supplementary Material
Acknowledgments
We thank Joshua Kaufman and Ira Palmer (NIAMS) for expert technical assistance. This research was supported by the NIAMS Intramural Research Program of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lesmana LA, Leung NW, Mahachai V, Phiet PH, Suh DJ, Yao G, Zhuang H. Hepatitis B: overview of the burden of disease in the Asia-Pacific region. Liver Int. 2006;26 2:3–10. [Google Scholar]
- 2.Liaw YF. Management of chronic hepatitis B: an evolving issue. Liver Int. 2006;26 2:1–2. [Google Scholar]
- 3.Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fields BN, Knipe DM, Howley PM, Chanock RM, Melnick JL, Monath TP, Roizman B, Straus SE. Fields Virology. 3. Lippincott-Raven Publishers; Philadelphia: 1996. [Google Scholar]
- 5.Ou JH. Molecular biology of hepatitis B virus e antigen. J Gastroenterol Hepatol. 1997;12:S178–87. doi: 10.1111/j.1440-1746.1997.tb00499.x. [DOI] [PubMed] [Google Scholar]
- 6.Yang CY, Kuo TH, Ting LP. Human hepatitis B viral e antigen interacts with cellular interleukin-1 receptor accessory protein and triggers interleukin-1 response. J Biol Chem. 2006;281:34525–36. doi: 10.1074/jbc.M510981200. [DOI] [PubMed] [Google Scholar]
- 7.Milich D, Liang TJ. Exploring the biological basis of hepatitis B e antigen in hepatitis B virus infection. Hepatology. 2003;38:1075–86. doi: 10.1053/jhep.2003.50453. [DOI] [PubMed] [Google Scholar]
- 8.Wynne SA, Crowther RA, Leslie AG. The crystal structure of the human hepatitis B virus capsid. Mol Cell. 1999;3:771–80. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- 9.Steven AC, Conway JF, Cheng N, Watts NR, Belnap DM, Harris A, Stahl SJ, Wingfield PT. Structure, assembly, and antigenicity of hepatitis B virus capsid proteins. Adv Virus Res. 2005;64:127–165. doi: 10.1016/S0065-3527(05)64005-5. [DOI] [PubMed] [Google Scholar]
- 10.Schödel F, Peterson D, Zheng J, Jones JE, Hughes JL, Milich DR. Structure of hepatitis B virus core and e-antigen. A single precore amino acid prevents nucleocapsid assembly. J Biol Chem. 1993;268:1332–7. [PubMed] [Google Scholar]
- 11.Wasenauer G, Kock J, Schlicht HJ. A cysteine and a hydrophobic sequence in the noncleaved portion of the pre-C leader peptide determine the biophysical properties of the secretory core protein (HBe protein) of human hepatitis B virus. J Virol. 1993;66:5338–5346. doi: 10.1128/jvi.66.9.5338-5346.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nassal M, Rieger A. An intramolecular disulfide bridge between Cys-7 and Cys61 determines the structure of the secretory core gene product (e antigen) of hepatitis B virus. J Virol. 1993;67:4307–4315. doi: 10.1128/jvi.67.7.4307-4315.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salfeld J, Pfaff E, Noah M, Schaller H. Antigenic determinants and functional domains in core antigen and e antigen from hepatitis B virus. J Virol. 1989;63:798–808. doi: 10.1128/jvi.63.2.798-808.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wingfield PT, Stahl SJ, Williams RW, Steven AC. Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis, and in vitro capsid assembly. Biochemistry. 1995;34:4919–4932. doi: 10.1021/bi00015a003. [DOI] [PubMed] [Google Scholar]
- 15.Milich DR, Sällberg M, Maruyama T. The humoral immune response in acute and chronic hepatitis B virus infection. Springer Semin Immunopathol. 1995;17:149–66. doi: 10.1007/BF00196163. [DOI] [PubMed] [Google Scholar]
- 16.Baumeister MA, Medina-Selby A, Coit D, Nguyen S, George-Nascimento C, Gyenes A, Valenzuela P, Kuo G, Chien DY. Hepatitis B virus e antigen specific epitopes and limitations of commercial anti-HBe immunoassays. J Med Virol. 2000;60:256–63. [PubMed] [Google Scholar]
- 17.Sällberg M, Pushko P, Berzinsh I, Bichko V, Sillekens P, Noah M, Pumpens P, Grens E, Wahren B, Magnius LO. Immunochemical structure of the carboxy-terminal part of hepatitis B e antigen: identification of internal and surface-exposed sequences. J gen Virol. 1993;74:1335–40. doi: 10.1099/0022-1317-74-7-1335. [DOI] [PubMed] [Google Scholar]
- 18.Ferns RB, Tedder RS. Human and monoclonal antibodies to hepatitis B core antigen recognise a single immunodominant epitope. J Med Virol. 1986;19:193–203. doi: 10.1002/jmv.1890190213. [DOI] [PubMed] [Google Scholar]
- 19.Ferns RB, Tedder RS. Monoclonal antibodies to hepatitis Be antigen (HBeAg) derived from hepatitis B core antigen (HBcAg): their use in characterization and detection of HBeAg. J Gen Virol. 1984;65:899–908. doi: 10.1099/0022-1317-65-5-899. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi K, Machida A, Funatsu G, Nomura M, Usuda S, Aoyagi S, Tachibana K, Miyamoto H, Imai M, Nakamura T, Miyakawa Y, Mayumi M. Immunochemical structure of hepatitis B e antigen in the serum. J Immunol. 1983;130:2903–2907. [PubMed] [Google Scholar]
- 21.Imai M, Nomura M, Gotanda T, Sano T, Tachibana K, Miyamoto H, Takahashi K, Toyama S, Miyakawa Y, Mayumi M. Demonstration of two distinct antigenic determinants on hepatitis B e antigen by monoclonal antibodies. J Immunol. 1982;128:69–72. [PubMed] [Google Scholar]
- 22.Usuda S, Okamoto H, Ohnuma H, Tanaka T, Machida A, Miyakawa Y, Mayumi M. A monoclonal antibody against a hepatitis B e antigen epitope borne by six amino acids encoded by the precore region. J Virol Methods. 1997;68:207–15. doi: 10.1016/s0166-0934(97)00125-0. [DOI] [PubMed] [Google Scholar]
- 23.Watts NR, Cardone G, Vethanayagam JG, Cheng N, Hultgren C, Stahl SJ, Steven AC, Sallberg M, Wingfield PT. Non-canonical binding of an antibody resembling a naive B cell receptor immunoglobulin to hepatitis B virus capsids. J Mol Biol. 2008;379:1119–29. doi: 10.1016/j.jmb.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris A, Belnap DM, Watts NR, Conway JF, Cheng N, Stahl SJ, Vethanayagam JG, Wingfield PT, Steven AC. Epitope diversity of hepatitis B virus capsids: quasi-equivalent variations in spike epitopes and binding of different antibodies to the same epitope. J Mol Biol. 2006;355:562–76. doi: 10.1016/j.jmb.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 25.Belnap DM, Watts NR, Conway JF, Cheng N, Stahl SJ, Wingfield PT, Steven AC. Diversity of core antigen epitopes of hepatitis B virus. Proc Nat'l Acad Sci USA. 2003;100:10884–9. doi: 10.1073/pnas.1834404100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conway JF, Watts NR, Belnap DM, Cheng N, Stahl SJ, Wingfield PT, Steven AC. Characterization of a conformational epitope on hepatitis B virus core antigen and quasi-equivalent variations in antibody binding. J Virol. 2003;77:6466–6473. doi: 10.1128/JVI.77.11.6466-6473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conway JF, Cheng N, Zlotnick A, Stahl SJ, Wingfield PT, Belnap DM, Kanngiesser U, Noah M, Steven AC. Hepatitis B virus capsid: localization of the putative immunodominant loop (residues 78 to 83) on the capsid surface, and implications for the distinction between c and e-antigens. J Mol Biol. 1998;279:1111–21. doi: 10.1006/jmbi.1998.1845. [DOI] [PubMed] [Google Scholar]
- 28.Zheng J, Schödel F, Peterson DL. The structure of hepadnaviral core antigens: identification of free thiols and determination of the disulfide bonding pattern. J Biol Chem. 1992;267:9422–9429. [PubMed] [Google Scholar]
- 29.Packianathan C, Katen SP, Zlotnick A. Conformational changes in the Hepatitis B virus core protein are consistent with a role for allostery in virus assembly. Journal of Virology. 2009 doi: 10.1128/JVI.02033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dimmock NJ, Hardy SA. Valency of antibody binding to virions and its determination by surface plasmon resonance. Rev Med Virol. 2004;14:123–35. doi: 10.1002/rmv.419. [DOI] [PubMed] [Google Scholar]
- 31.Hewat EA, Blaas D. Structure of a neutralizing antibody bound bivalently to human rhinovirus 2. EMBO J. 1996;15:1515–1523. [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi K, Imai M, Miyakawa Y, Iwakiri S, Mayumi M. Duality of hepatitis B e antigen in serum of persons infected with hepatitis B virus: evidence for the nonidentity of e antigen with immunoglobulins. Proc Natl Acad Sci U S A. 1978;75:1952–6. doi: 10.1073/pnas.75.4.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sallberg M, Norder H, Lindh G, Magnius LO. IgG subclasses in circulating immune complexes with hepatitis B e antigen in chronic hepatitis B. Clin Exp Immunol. 1991;84:116–21. [PMC free article] [PubMed] [Google Scholar]
- 34.Hunte C, Michel H. Crystallisation of membrane proteins mediated by antibody fragments. Curr Opin Struct Biol. 2002;12:503–8. doi: 10.1016/s0959-440x(02)00354-8. [DOI] [PubMed] [Google Scholar]
- 35.Seifer M, Standring DN. Assembly and antigenicity of hepatitis B virus core particles. Intervirology. 1995;38:47–62. doi: 10.1159/000150414. [DOI] [PubMed] [Google Scholar]
- 36.Stenberg E, Persson B, Roos H, Urbaniczky C. Quantitative determination of surface concentration of protein with surface plasmon resonance using radiolabeled proteins. Journal of Colloid and Interface Science. 1991;143:513–526. [Google Scholar]
- 37.Cohn EJ, Edsall JT. Proteins, amino acids and peptides. Van Nostrand-Reinhold; Princeton, NJ: 1943. [Google Scholar]
- 38.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Centrifugation in Biochemistry and Polymer Science. Royal Society for Chemistry; Cambridge, U.K.: 1992. pp. 90–125. [Google Scholar]
- 39.Uetrecht C, Versluis C, Watts NR, Roos WH, Wuite GJ, Wingfield PT, Steven AC, Heck AJ. High-resolution mass spectrometry of viral assemblies: molecular composition and stability of dimorphic hepatitis B virus capsids. Proc Natl Acad Sci U S A. 2008;105:9216–20. doi: 10.1073/pnas.0800406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Standring DN, Ou JH, Masiarz FR, Rutter WJ. A signal peptide encoded within the precore region of hepatitis B virus directs the secretion of a heterogeneous population of e antigens in Xenopus oocytes. Proc Nat'l Acad Sci USA. 1988;85:8405–9. doi: 10.1073/pnas.85.22.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schödel F, Moriarty AM, Peterson DL, Zheng JA, Hughes JL, Will H, Leturcq DJ, McGee JS, Milich DR. The position of heterologous epitopes inserted in hepatitis B virus core particles determines their immunogenicity. J Virol. 1992;66:106–114. doi: 10.1128/jvi.66.1.106-114.1992. published erratum appears in. [DOI] [PMC free article] [PubMed] [Google Scholar]; J Virol. 1992 Jun;66(6):3977. [Google Scholar]
- 42.Tordjeman M, Fontan G, Rabillon V, Martin J, Trepo C, Hoffenbach A, Mabrouk K, Sabatier JM, Van Rietschoten J, Somme G. Characterization of minor and major antigenic regions within the hepatitis B virus nucleocapsid. J Med Virol. 1993;41:221–9. doi: 10.1002/jmv.1890410310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.