Abstract
One caveat to current loss-of-function approaches in zebrafish is that they typically disrupt gene function from the beginning of development. This can be problematic when attempting to study later developmental events. In vivo electroporation is a method that has been shown to be effective at incorporating reagents into the developing nervous system at multiple later developmental stages. The temporal and spatial characteristics of in vivo electroporation that have been previously demonstrated suggest that this could be a powerful approach for time-resolved loss-of-function analysis. Here, in an attempt to demonstrate the efficacy of this approach for analysis of a specific developmental timeframe—that of initial development of the zebrafish visual system—we have done a systematic characterization of the efficiency of in vivo electroporation in zebrafish across multiple developmental stages, from 24 to 96 h postfertilization. We show that electroporation is efficient at delivering expression plasmids to large numbers of neurons at multiple developmental steps, including 24, 48, or 96 h postfertilization. Expression from electroporated plasmids is maximal within 24 h, and significant and useful expression is seen within 6 h. Electroporation can be used to deliver two separate expression plasmids (green fluorescent protein and mCherry), resulting in coexpression in 97% of cells. Most importantly, electroporation can be used to incorporate siRNA reagents, resulting in 84% knockdown of a target protein (green fluorescent protein). In conclusion, in vivo electroporation is an effective method for delivering both DNA-based expression plasmids and RNA interference-based loss-of-function reagents, and exhibits the appropriate characteristics to be useful as a time-resolved genetic approach to investigate the molecular mechanisms of visual system development.
Introduction
Zebrafish (Danio rerio) has emerged as a powerful model system to study the molecular mechanisms of development.1 In addition to being a well-characterized genetic system, zebrafish is particularly well suited to in vivo imaging because the embryos are small and essentially transparent through the early stages of development. Thus, developmental events such as cell shape changes, cell migration, and tissue formation can be directly observed in live embryos by expressing a fluorescent protein—such as the green fluorescent protein (GFP)—in early differentiating cells or their precursors. This ability to assess developmental events in a live (unfixed) embryo is particularly important when investigating neural system development because the complex extracellular environment and elaborate spatial cues that are required to appropriately wire the vertebrate brain cannot be reliably reproduced in vitro.2
Identifying the molecular mechanisms that control zebrafish neural development requires two methods: (1) a technique that can assess the developmental event in vivo in live embryos; (2) a loss-of-function approach that can target specific genes within the target tissue or cell type. As stated above, in vivo imaging of fluorescent proteins is a good approach for monitoring developmental events in live embryos because it allows the assessment of several different cellular parameters, including differentiation, migration, and axonal/dendritic pathfinding, without having to fix or otherwise disturb the embryonic tissues. Of particular importance to an in vivo imaging approach is how the fluorescent protein expression construct will be targeted to the cells or tissues of interest. Ideally, one would be able to reproducibly target a specific cell type or tissue with both spatial and temporal resolution.
Historically, the most widely used loss-of-function approach has been mutagenesis and mutant analysis, which has yielded a wealth of knowledge about what genes are necessary for development of model organisms. More recently, molecular-targeted approaches such as RNA interference (RNAi) have greatly facilitated the feasibility of genetic loss of function, while maintaining specificity.3 In zebrafish, an analogous approach using antisense morpholino oligonucleotides has been widely used because of the ease of incorporating morpholinos by intracellular microinjection at the one- or two-cell embryo stage.4,5 A major caveat to both mutant analysis and injection of RNAi or morpholino reagents at the one-cell stage is that the loss of function is initiated at the beginning of development. This is a major problem when attempting to analyze later development events, such as development of the nervous system, because many of the genes involved in neural development are also required for earlier developmental steps. Loss of function for these genes is predicted to lead to dysfunctional early development, compromising the analysis of later events. Thus, a time-resolved loss-of-function method, which allows for the disruption of gene function at specific developmental stages, would be an ideal approach for the genetic analysis of later development events.
In vivo electroporation is a method that can be used for intracellular delivery of oligonucleotides to developing embryos in multiple model organisms,6 which offers excellent spatial and temporal resolution. This technique has become a very powerful method for gain-of-function and loss-of-function analysis in the developing chick system.7–9 In Xenopus, in vivo electroporation has been shown to be particularly well suited for targeting oligonucleotide reagents to the developing nervous systems.10–13 Although currently not as widely used, in vivo electroporation has long been known to be an effective method for incorporating reagents into developing zebrafish embryos.14 The efficacy of using in vivo electroporation for targeting later developmental stages was first demonstrated by targeting the neural tube for injection and electroporation.15 In vivo electroporation has now has been shown to be an effective method for delivering dyes and expression plasmids to large numbers of cells in different regions of the developing nervous system in zebrafish embryos15–18 and adults,19,20 and a modified version of the method can be used to target single cells.21,22 Also, in vivo electroporation has been used successfully to incorporate RNAi and morpholino loss-of-function reagents.17,23,24 However, if in vivo electroporation is to become a primary method for loss-of-function analysis in zebrafish (as it is in chick), it is important to first quantitatively assess the efficacy of the method, and, most importantly, to determine the temporal resolution of the technique as it relates to the timeframe of the developmental events of interest.
In vivo electroporation works by delivering brief (5–50 ms) pulses of an electric field across an embryo, which leads to the opening of very-short-lived (∼1s) pores in the plasma membrane, allowing for oligonucleotides injected outside of the cell to cross the plasma membrane.22 Charged reagents, such as DNA expression plasmids or RNAi oligonucleotides, are further facilitated in entering cells due to an ionophoresis effect, which facilitates movement of the negatively charged oligonucleotides toward the positive electrode. Thus, by localizing the injection of charged reagents, and orientating the electric field to control the direction of ionophoresis, this technique can be used to target different regions of the developing nervous system. In zebrafish, several studies have demonstrated that the method has excellent spatial resolution, while still being able to target large numbers developing neurons in the target tissue.15,16,18 For even more precise spatial resolution, a modified version of the method can even target single cells.21,22
Given the spatial and temporal resolution previously shown for in vivo electroporation, the method should provide an excellent approach for temporally controlled gene loss-of-function experiments. Through incorporation of charged loss-of-function reagents, such as RNAi oligonucleotides, shRNA-based plasmids, or modified morpholino oligonucleotides,26,27 in vivo electroporation can be used to induce loss of function at specific developmental stages. Further, by localized injection and appropriate orientation of the electric field, loss-of-function oligonucleotides can be delivered to specific tissues or cell types. Given these spatial and temporal characteristics, in vivo electroporation provides an excellent method for the analysis of the molecular mechanism controlling later developmental events.
Here we have quantified the efficacy and temporal parameters of in vivo electroporation in zebrafish as they relate to the analysis of neural development. We have shown that in vivo electroporation is a robust method for incorporating GFP expression plasmids and RNAi oligonucleotides into cells of the developing nervous system. We have shown that in vivo electroporation is effective for incorporation of these reagents in embryos at multiple developmental stages, spanning those important for initial development of the zebrafish visual system. Finally, we have shown that delivery of RNAi oligonucleotides via in vivo electroporation is an effective method for gene knockdown.
Materials and Methods
DNA expression plasmids and RNAi oligonucleotides
All expression constructs were provided by R. Koster (Institute of Developmental Genetics, Neuherberg, Germany). To maximize GFP and mCherry transient expression, we used a Gal4-VP16 activator/effector expression system.28 This system requires two plasmids: (1) a plasmid including the coding sequence for a fusion protein of the Gal4 DNA binding domain and the VP16 transcriptional activation domain, under control of the ubiquitous elongation factor-1 alpha (EF-1α) promoter; (2) a plasmid including the coding sequence of either enhanced GFP (EGFP) or mCherry under the control of 14 tandem upstream activating sequences (UAS) (Gal4 binding) sequences and the fish basal promoter E1b.28 Both plasmids include the beta globin polyadenylation sequence. For experiments monitoring the efficacy of RNAi-mediated knockdown of GFP, expression vectors driven by the human cytomegalovirus promoter (CMV) were used to reduce the amount of mRNA produced as compared to the high levels mediated by the Gal4-VP16 system. The coding sequence for EGFP or mCherry was cloned into the pCS2+ Xenopus expression plasmid.28 Plasmids were purified by standard maxi prep column protocols (Qiagen, Valencia, CA).
Previously characterized short interfering RNA (siRNA) oligonucleotides were used to target EGFP expression.31 Duplex siRNA oligonucleotides targeting the EGFP sequence, 5′- GAC GUA AAC GGC CAC AAG U -3′, were purchased from Thermo Fisher Scientific (Waltham, MA). For control electroporations, siGENOME nontargeting siRNA #2 oligonucleotides were used (Thermo Fisher Scientific). Duplex siRNA oligonucleotides were dissolved in siRNA buffer (Thermo Fisher Scientific) at a concentration of 200 μM for injection and electroporation.
Embryo preparation and in vivo electroporation
Embryonic and adult zebrafish were maintained by standard protocols.29 Briefly, embryos were grown at 28°C in egg water plus 0.0001% methylene blue. For embryos to be imaged, pigment formation was inhibited by inclusion of 100 μM N-phenylthiourea.
For electroporations, 24 h postfertilization (hpf ) embryos were manually dechorionated using fine forceps (not required for later stage embryos). Embryos were then transferred to electroporation buffer (180 mM NaCl, 5 mM KCl, 1.8 mM CaCl2, and 5 mM HEPES, pH 7.2).17 For injections, tricaine (0.017%) was included in the electroporation buffer to prevent embryo movements. Embryos were trapped in 0.2% low-melting-point agarose (Sigma-Aldrich, St. Louis, MO) and positioned manually with forceps. DNA expression plasmids were dissolved in electroporation buffer at a concentration of 0.1–1.0 mg/mL with the addition of 0.03% phenol red to observe injections. Glass micropipettes were fabricated to a fine point using a Sutter P-30 Pipette Puller using glass capillary tubes (inner diameter = 0.5 mm; outer diameter = 1.0 mm with filament). To obtain a sharp injection tip, the sealed fine point of the pulled pipet was broken back manually using a submerged kimwipe. Embryos were injected with DNA solution using a pressure injection apparatus (MPPI-2 Pressure Injector; Applied Scientific Instrumentation; Eugene, OR). Electroporation was initiated as soon after injection as possible. Hand-held electrodes were positioned outside the embryo, spanning the DNA-injected ventricular space, and square-shaped electroporation pulses were applied using a Grasss SD-9 Stimulator (Grass-Telefactor, West Warwick, RI). A typical electroporation protocol consisted of seven 5 ms pulses, initiated manually, space by approximately 1 s. Voltages of electroporation pulses ranged from 60 to 100 V. It should be noted that the biphasic or “bi” setting on the SD-9 stimulator was used to decrease production of bubbles from the platinum electrodes. This setting produces a biphasic output by placing a capacitor in series with the output. The useful voltage range when using the mono setting will be significantly lower (10–50 V). Electrodes were custom built using Grass E2 Platinum Subdermal Electrodes (Grass-Telefactor). Embryos were then allowed to recover for at least 5 min before being manually released from the agarose using fine forceps, placed in normal embryo media, and returned to 28°C.
Fluorescent imaging and image processing
Embryos to be imaged were immobilized in 0.2% low-melting-point agarose. Fluorescent and bright-field images were acquired using either a Zeiss Axioplan (Oberkochen, Germany) fluorescent microscope or an Olympus (Center Valley, PA) BF60 fluorescent microscope. In both cases images were acquired using a computer-controlled digital monochrome camera. Higher-resolution confocal images were acquired using an Olympus Fluoview 300 confocal microscope.
Digital image files were converted from 16 bit pixel depth to 8 bit pixel depth using ImageJ image-processing software (ImageJ 1.37v; National Institutes of Health, Bethesda, MD). In some cases gray-scale fluorescent images were pseudocolored green or red using the lookup table function in ImageJ, and adjusted for brightness and contrast. Bright-field and fluorescent images were combined in ImageJ using the Z-project function. For figure presentation, image size and resolution was adjusted using Adobe Photoshop CS3 (Adobe Systems Incorporated, San Jose, CA).
ImageJ was also used to quantify the level of fluorescence in individual cells on saved image files. GFP and mCherry images of the same cell were combined in an image stack, and then the brightness within a box drawn to be totally enclosed within the cell was processed using the measure-stack function. This measurement function allows us to determine the total brightness of all pixels within the box, which could be used to determine the ratio of GFP to mCherry fluorescence. A box of the same size was also placed adjacent to the cells to determine background fluorescence, which was subtracted from the cell levels before the GFP/mCherry ratio was determined. The median pixel value within a cell was also determined using the measure-stack function.
Results
As discussed in the Introduction, in vivo electroporation has been shown to be effective for delivering reagents to the developing nervous system at multiple developmental stages,15–18 thus offering excellent temporal resolution. The method has also been shown to have excellent spatial resolution, allowing for the targeting of large numbers of neurons in different regions of the developing brain,15–18 or even the targeting of single neurons.21,22 Given these characteristics, and the use of RNAi or morpholino-based loss-of-function reagents,17,23,24 in vivo electroporation should provide a very powerful approach for time-resolved loss-of-function analysis. For this to become a widely used loss-of-function approach, the method should be efficient at inducing the knockdown of target genes, and should be time-resolved with respect to the timeframe of the developmental event of interest. Thus, the goal of this study was twofold: (1) to quantitatively assess the efficacy of using in vivo electroporation for knockdown of a target gene; (2) to quantitatively assess the temporal resolution of in vivo electroporation across the timeframe for a particular developmental event—in this case, from 24 to 96 hpf, which spans the major developmental events for the initial differentiation of the visual system.
We used expression of GFP as a way to monitor the efficiency of in vivo electroporation. The sensitivity of this characterization is dependent upon the efficiency of the expression plasmid used, and for this reason we utilized a Gal4-VP16 system that had been optimized for expression in the zebrafish nervous system28 (provided by R. Koster). This system requires the introduction of two expression plasmids: one with the coding sequence of GFP under the control of multiple tandem UAS sequences, and a second plasmid encoding the Gal4-VP16 transcription factor under the control of the ubiquitous EF-1α promoter. Although this system requires the simultaneous introduction of two plasmids, previous studies have shown that simply including both plasmids in the solution to be electroporated leads to very efficient GFP expression,18 and we also found this to be the case in our hands. This expression system gave us a very sensitive assay for successful incorporation of DNA.
Targeting GFP expression to the nervous system using in vivo electroporation
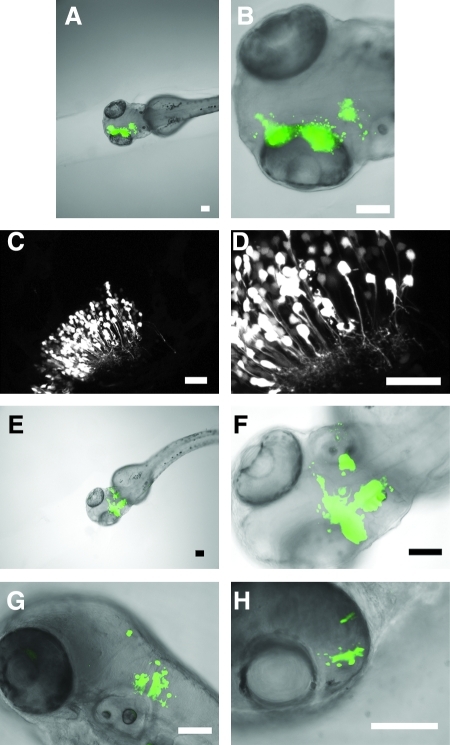
We first determined the efficacy of in vivo electroporation for delivering GFP expression plasmids to large areas of the developing central nervous system (CNS) in zebrafish embryos. To target large regions, a solution of the GFP expression plasmid was pressure-injected into the midbrain ventricle of 24 hpf embryos. Multiple injection pulses were used such that the DNA solution (observed with phenol red) spreads throughout the ventricles of the early developing brain. Electroporation was then initiated using hand-held platinum wire electrodes (electrodes separated by 1 mm). Electrodes were positioned on either side of the head, outside of the embryo, such that the electric filed was oriented across the eyes. Electroporation pulses were applied as soon after injection as possible. The typical voltage protocol used was seven consecutive square pulses (75 V) initiated manually, each lasting 5 ms. Images of embryos 24 h after electroporation (at 48 hpf ) revealed that large numbers of cells were expressing GFP throughout the early brain (Fig. 1A, B). Orientation of the electric field across the eyes at this stage seems to preferentially target cells in the midbrain, and expression is typically seen only on one side of the brain. This preferential targeting of cells on one side of the midline is likely due to the ionophoretic movement of DNA from the ventricle into adjacent tissue in the direction of the positive electrode. Higher-resolution confocal imaging of a similarly electroporated embryo showed that a majority of the GFP-expressing cells have morphology typical of that for early developing neurons, with extended and branched neurites (Fig. 1C, D). The number of neurons labeled is somewhat variable, but is typically more than 100 cells. Supplemental Movies made from a Z-series of confocal sections give a more accurate impression of the number of cells labeled through a section of midbrain (Supplemental Movies S1 and S2, available online at www.liebertonline.com).
FIG. 1.
Delivering green fluorescent protein (GFP) expression plasmids to large regions of the developing brain by in vivo electroporation. (A, B) Embryos were electroporated at 24 h postfertilization (hpf ). Combined fluorescence (green) and bright-field (gray scale) images of embryos 24 h after electroporation show GFP expression in large regions of the developing brain. (C, D) Confocal images demonstrate the typical neurite morphology of GFP-expressing neurons of the optic tectum. These images represent a z-projection of a series of confocal slices acquired by focusing dorsal to ventral through the area of labeled neurons. Supplemental Movies S1 and S2 are animated versions of the stacks of confocal images through this section of optic tectum, which demonstrate the number of cells expressing GFP within a given section of brain, and also reveal the elaborated structure of neurites. (E, F) Combined fluorescence (green) and bright-field (gray scale) images of embryos 24 h after electroporation showing GFP expression in the early developing cerebellum. (G, H) Combined fluorescence (green) and bright-field (gray scale) images of embryos 24 h after electroporation showing GFP expression in the hindbrain (G) and retina (H). Scale bars equal 100 μm (A, B, E–H) and 50 μm (C, D).
While characterizing in vivo electroporation for zebrafish, we found that there were two critical modifications of the method that vastly improved the efficacy of the technique. First, we found that the use of the Gal4-VP16 system that had been optimized for zebrafish expression greatly increased the sensitivity of our assay for successful incorporation of DNA. Mammalian vectors and even some Xenopus vectors proved to be of low efficiency for gaining expression in zebrafish. Second, as has been previously reported,17 the embryo medium used for electroporation needs to be much higher in salt concentration than what is typically used for the zebrafish embryo medium. This is likely because a certain level of conductivity in the medium is required for the electric field to have its full effect. We used an electroporation buffer previously described (180 mM NaCl, 5 mM KCl, 1.8 mM CaCl2, and 5 mM HEPES, pH 7.2).17
We next determined whether other regions of the developing CNS could be targeted by this electroporation approach. Spatial targeting was achieved by localizing the injections to the appropriate ventricle or intercellular space and by positioning the positive electrode such that the DNA will be drawn toward the appropriate cells. As has been previously shown,16 the cerebellum can be targeted by injecting the hindbrain ventricle and then positioning the positive electrode at the anterior end of the embryo. Thus, when an electric field is applied the DNA travels in the direction of the positive electrode, from the ventricular space into the cells of the anterior wall of the hindbrain ventricle, where early cerebellar neurons are localized. This approach allowed us to incorporate GFP expression plasmids into large numbers of cells in the developing cerebellum (Fig. 1E, F). Using the same injection protocol, hindbrain neurons can be targeted by orienting the electrodes across the medial–lateral axis, which draws the DNA into cells along the lateral walls of the ventricles (Fig. 1G). Cells in the developing retina can be targeted by direct injection of DNA into the developing eye. Subsequent orientation of the electrodes across the eyes, with the positive electrode outside the embryo adjacent to the opposite eye, draws the DNA from the center of the eye into the retina (Fig. 1H). Thus, large injections of DNA combined with careful placement of the positive electrode can be used to target large numbers of neurons in specific regions of the developing CNS.
Measuring the temporal resolution of in vivo electroporation
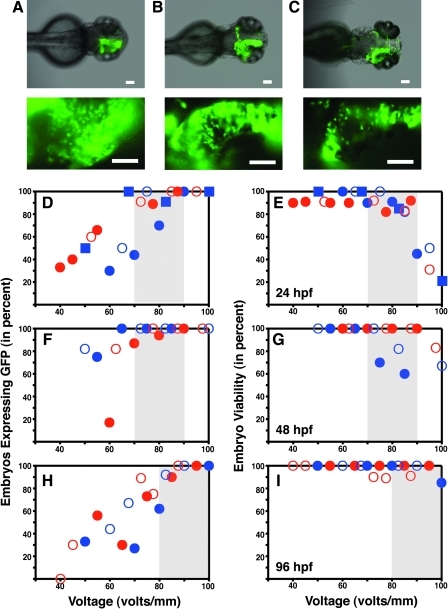
Several previous studies have demonstrated that zebrafish embryos at different developmental stages can be targeted by in vivo electroporation.15–20 Here, our goal was to systematically compare the efficiency of electroporation across several developmental stages, from 24 to 96 hpf. We chose this range of developmental stages because it covers the time required for initial development of the visual system. Embryos at 24, 48, and 96 hpf can all be successfully targeted by in vivo electroporation as demonstrated by robust GFP expression 24 h after electroporation (Fig. 2A–C). Although GFP-expressing cells become somewhat more dispersed in 96 hpf embryos (Fig. 2C), we saw no systematic difference in the ability to incorporate GFP expression plasmids into embryos from 24 to 96 hpf.
FIG. 2.
Temporal resolution of in vivo electroporation. (A–C) Images of embryos 24 h after electroporation for embryos electroporated at 24 hpf (A), 48 hpf (B), and 96 hpf (C). Voltages used were 75, 80, and 95 V, respectively. Scale bars represent 100 μm top panels and 50 μm bottom panels. To characterize the temporal resolution of in vivo electroporation, embryos were electroporated at 24, 48, or 96 hpf. Twenty-four hours after electroporation the embryos were screened for expression of GFP and for viability. Voltage was varied to assess the efficiency of electroporation at that stage of development. (D, F, H) The percentage of embryos expressing GFP was determined for each voltage. Embryos were considered positive if at least 10 cells with neuronal morphology were expressing GFP; however, the vast majority of positive embryos had 100-plus cells showing fluorescence. (E, G, I) Viability was assessed by embryo morphology and the presence of a strong heartbeat. Gray boxes indicate a range of voltages for which there is high expression and very high viability. Data points are color-coded to investigator to further demonstrate the robustness of the technique (blue: S.A.; red: S.K.). Common data symbols (squares/circles and closed/open) represent all of the data points for one experiment.
For in vivo electroporation to be a useful time-resolved genetic approach, it must be efficient at delivering reagents to early neurons at multiple developmental steps, and it should not damage the embryos or disrupt developmental processes. One of the advantages of electroporation, as opposed to microinjection, is that the size and duration of the holes induced in the plasma membrane are a function of the voltage applied.25 Thus, by adjusting the voltage protocol it should be possible to achieve efficient delivery of reagents without significant cell death.
To characterize the efficiency of the in vivo electroporation technique, and its effects on embryo viability, we determined the voltage dependence of GFP expression and embryo viability. As a measure of the efficiency of GFP expression, we determined the percentage of embryos expressing GFP 24 h after electroporation. We found that this was a more appropriate measure of the efficacy of the technique as compared to the number of cells expressing in each embryo because it assesses the number of successful electroporations—a more direct measurement of the usefulness of the technique. Also, we found that if an embryo was expressing GFP it was typically expressed in hundreds of cells; thus, expression in an embryo was to a large extent all or none. To compensate for false-positives from spurious expression in a few cells, we set a threshold of at least 10 GFP-expressing cells to score an embryo as positive. Still, the vast majority of the positive embryos were expressing GFP in at least 100 cells.
The voltage dependence of GFP expression can be seen in Figure 2D. For this experiment embryos were electroporated at 24 hpf, and the number of embryos expressing were determined 24 h after electroporation. To assess electroporation at many different voltages, nonoverlapping voltages were chosen for each experiment. This did not allow us to average the data at individual voltages; however, the noise in the data can be seen in the scatter plot, and by plotting all of the data the robustness of the technique can be directly observed (note: each data point represents an experiment consisting of 6–12 electroporated embryos). In addition, to further demonstrate the robustness of the technique, we have color-coded experiments according to the investigator doing the electroporations (red: S.K.; blue: S.A.). As can be seen, there does not seem to be any bias due to investigator, further demonstrating that the method is consistent.
The same embryos were also scored for viability (Fig. 2E) to demonstrate that electroporation can deliver DNA reagents without disrupting developmental processes. Embryos were assessed for normal morphology 24 h after electroporation, and any embryos displaying altered tissue morphology were scored as dead (typical aberrations included curved tails or distended ventral epidermis). We found that embryos displaying normal morphology at 24 h after electroporation survived and developed normally at least until 96 hpf, the longest time point assessed. Swimming behavior was also the same as in un-electroporated controls, and all embryos scored as viable displayed normal heartbeat. Another indication that electroporated cells maintained normal function is their ability to transcribe and translate GFP (continuing to at least 96 hpf, the longest time point assessed), and GFP-expressing cells in the developing CNS showed typical neuronal morphology with elaborated axonal and dendritic processes.
As expected, the percentage of embryos successfully electroporated—as measured by expression of GFP—increases as you increase the voltage of the electroporation pulse. The percentage of embryos expressing GFP increases roughly linearly from 40 to 80 V (Fig. 2D). On the other hand, the viability of embryos begins to decrease at the upper range of voltages. When electroporating 24 hpf embryos, viability begins to decrease at electroporation voltages greater than 90 V (Fig. 2E). If in vivo electroporation is to be a useful technique for delivering reagents to early developing neurons, there should be a voltage range for which there is a high percentage of successful incorporation and also high viability of the embryos. For 24 hpf embryos, electroporation voltages between 70 and 90 V fulfill this requirement of high expression and high viability (see range highlighted in gray, Fig. 2D, E). Thus, in vivo electroporation is an efficient and robust technique for targeting developing neurons in 24 hpf embryos.
For in vivo electroporation to be a useful method for time-resolved genetic analysis, it must be an effective method for delivering reagents at any time point across the timeframe of the developmental events of interest. As a proof of principle, we chose to assess whether in vivo electroporation was an effective method for delivering reagents across the developmental period for which the zebrafish visual system is initially formed.30 Thus, we determined the efficacy of in vivo electroporation at 24, 48, and 96 hpf. As discussed above, in 24 hpf embryos electroporation voltages between 70 and 90 V lead to a high percentage of GFP-expressing embryos, while also maintaining close to 100% viability (Fig. 2D, E). A similar voltage range is also effective when electroporating 48 hpf embryos. The shaded regions in Figure 2F and G highlight the high percentage of embryos expressing and the high viability. When electroporating 96 hpf embryos, higher voltages are required to achieve the same percentage of embryos expressing, which is likely due to the further development of the skin, increasing its integrity, and somewhat reducing the actual voltage experienced by cells in the CNS. To obtain maximal expression when electroporating, 96 hpf embryos required a voltage range of 80–100 V while the viability remained essentially 100% at these voltages (Fig. 2H, I). These experiments demonstrate that in vivo electroporation is an effective method for delivering reagents to embryos from 24 to 96 hpf. Thus, the temporal resolution of the approach is sufficient for assessing gene function throughout initial development of the nervous systems in zebrafish.
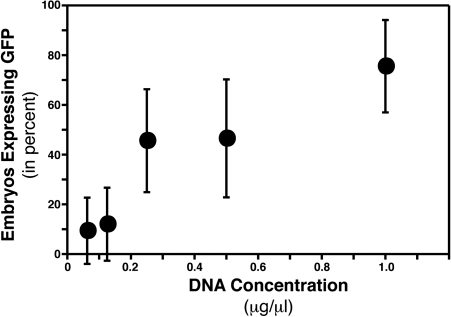
Another parameter that is important to define is the concentration of DNA necessary to achieve good expression of the transgene. Again, using the percentage of embryos expressing GFP as a measure of plasmid delivery, we found that the steepest dependence on DNA concentration in the injection pipet was from 0.1 to 0.3 μg/μL (Fig. 3). Thus, to ensure good expression a concentration of approximately 0.5 μg/μL should suffice. It should be noted that this range of effective DNA concentrations is higher than what is typically used for direct microinjection at the one-cell stage (0.05–0.3 μg/μL),29 and is 10-fold higher than what has been shown to be sufficient for these particular Gal4-VP16 vectors (0.03 μg/μL).28 As would be expected, this difference suggests that the percentage of the injection solution getting into the cells during in vivo electroporation is much lower than that for direct injection. It is important to take this into consideration when determining the appropriate concentration to be used.
FIG. 3.
DNA concentration dependence of in vivo electroporation. Zebrafish embryos were electroporated at 24 hpf with different concentrations of DNA in the injection pipet. The percentage of embryos expressing GFP was determined 24 h after electroporation, and the mean percentage of embryos expressing GFP was plotted as a function of DNA concentration in the injection pipet. Embryos were considered positive if at least 10 cells with neuronal morphology were expressing GFP; however, the vast majority of positive embryos had 100-plus cells showing fluorescence. Error bars display standard deviation (n = 4–6).
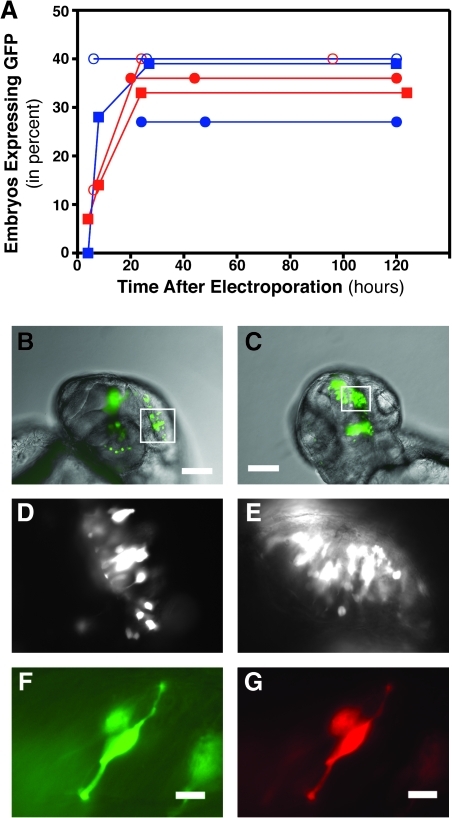
If in vivo electroporation is to be used as a time-resolved loss-of-function approach, coelectroporation of a GFP-based expression plasmid (or other color fluorescent protein) will likely be required to identify which cells have received the loss-of-function reagent. Thus, the timeframe of GFP expression after electroporation is another aspect of the temporal resolution of the technique. We measured the time course of GFP expression by determining the percentage of embryos expressing GFP at different time intervals after electroporation. Figure 4A shows the time course of expression for six different experiments, each of which followed the same group of electroporated embryos for multiple time points. Experiments following the same group of electroporated embryos are connected by lines, and the data are color-coded as to the investigator doing the electroporations (blue: S.A.; red: S.K., another way to monitor the robustness of the technique). The maximal number of embryos expressing GFP is seen by 24 h (Fig. 4A). We chose a voltage range (70–75 V) that does not typically yield 100% of the embryos expressing to have sufficient sensitivity to follow changes in expression at the early time points. Although maximal expression is seen by 24 h for all experiments, significant and useful expression can be seen by 6 h after electroporation (Fig. 4B–G).
FIG. 4.
Temporal resolution of GFP expression. Zebrafish embryos were electroporated at 24 hpf. (A) The percentage of embryos expressing GFP was determined at various times after electroporation. Embryos were considered positive if at least 10 cells with neuronal morphology were expressing GFP; however, the vast majority of positive embryos had 100-plus cells showing fluorescence. Data points connected by a line are from the same group of embryos followed over time. Each point represents the percentage expression for 6–15 embryos. Data points are color-coded to investigator (blue: S.A.; red: S.K.). Data points connected by a line (and having the same symbol) are from the same group of embryos followed over time. (B, D, F) Embryos electroporated at 24 hpf displayed significant GFP expression at 6 h after electroporation. (C, E, G) Embryos electroporated at 48 hpf also displayed significant GFP expression at 6 h after electroporation. White boxes in the combined fluorescent/bright-field images in (B) and (C) represent the area of the higher magnification fluorescent images shown in (D) and (E), respectively. (F, G) Coelectroporation of a GFP expression plasmid and an mCherry expression plasmid resulted in coexpression of GFP (F) and mCherry (G) 6 h after electroporation. Scale bars equal 100 μm (B, C) and 25 μm (F, G).
To confirm that the method is efficient at multiple stages of development, we determined whether GFP expression occurs within 6 h after electroporation for embryos electroporated at 24 hpf (Fig. 4B, D), and for embryos electroporated at 48 hpf (Fig. 4C, E). Embryos at both developmental stages could be targeted by in vivo electroporation, and large areas of the developing brain could be imaged within 6 h. Further, expression at 6 h is robust: coelectroporation of two expression plasmids (one for GFP and one for mCherry) yields expression of both proteins at 6 h after electroporation (Fig. 4F, G). Thus, expression of fluorescent proteins can be used as a marker to identify electroporated cells at least as early as 6 h after electroporation.
Using in vivo electroporation for RNAi-based loss-of-function analysis
We have thus far shown that in vivo electroporation can be used to introduce reagents into cells of the early developing nervous system for embryos from 24 to 96 hpf, and that expression plasmids introduced by electroporation can yield GFP expression within 6 h. Thus, the technique has sufficient temporal resolution to target multiple different stages in the development of the nervous system. The next step is to demonstrate that in vivo electroporation can be used to incorporate loss-of-function reagents, and that this leads to efficient knockdown of the target protein. To use in vivo electroporation as an acute loss-of-function approach requires that two reagents are coelectroporated into target cells: (1) the loss-of-function reagent; (2) a GFP expression plasmid such that the electroporated cells can be identified. Thus, to determine the efficacy of using in vivo electroporation as a loss-of-function method, we must first determine the efficiency of delivering two reagents simultaneously by coelectroporation.
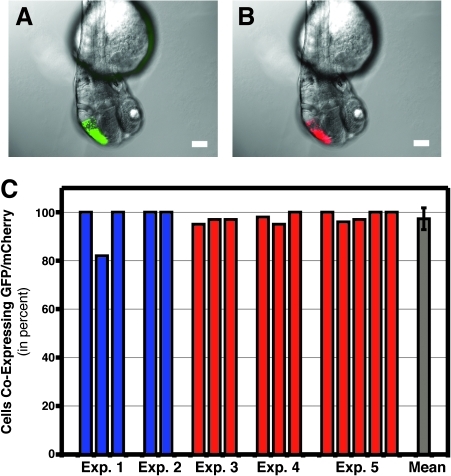
To determine the efficiency of using in vivo electroporation to coelectroporate two different reagents, we used two different expression plasmids—one coding for GFP and one coding for the red fluorescent protein, mCherry. Using the Gal4-VP16 system actually requires that we include three plasmids in the injection pipet: a UAS-GFP plasmid, a UAS-mCherry plasmid, and a constitutive Gal4-VP16 expression plasmid. Electroporation of the mixture including all three plasmids (each at 0.5 μg/μL) leads to coexpression of both GFP (Fig. 5A) and mCherry (Fig. 5B). To quantify the degree of coexpression, we analyzed embryos cell by cell using a fluorescent microscope to determine if both fluorescent proteins were expressed in individual cells. All of the data for five different experiments are plotted as colored bars in Figure 5C (color coded to investigator: blue, S.A.; red, S.K.). Each bar represents a single electroporated embryo, and 20–50 cells were analyzed per embryo. As can be seen, the vast majority of embryos had greater than 95% of the cells coexpressing both GFP and mCherry. The mean of all experiments showed that 97.3% of all cells showed coexpression of both proteins (standard deviation = 4.5%). Thus, in vivo electroporation is quite efficient for coelectroporation of two different reagents.
FIG. 5.
Coelectroporation of GFP and mCherry expression plasmids. (A, B) Mixing together equal concentrations of two different expression plasmids—one for GFP and one for mCherry—leads to coexpression of GFP and mCherry in large regions of the early developing brain. Combined bright-field and fluorescent images show overlapping expression of GFP (A) and mCherry (B) 24 h after electroporation. Scale bars equal 100 μm. (C) Quantification of the degree of coexpression. Single cells were examined for GFP and mCherry expression. Colored bars represent the percentage of cells coexpressing both GFP and mCherry in a single embryo (20–50 cells/embryo). Bars are color-coded to investigator (blue: S.A.; red: S.K.). Gray bar represents the mean number of cells coexpressing both GFP and mCherry (97.3%; n = 16 embryos; standard deviation = 4.5%).
Next, to actually measure the efficiency of using in vivo electroporation for loss-of-function analysis, we used a previously characterized anti-EGFP RNAi oligonucleotide31 to target expression of GFP. By targeting GFP, we can directly monitor the loss of protein expression by fluorescent microscopy. The experimental design was to coelectroporate the anti-EGFP RNAi oligonucleotide, the GFP expression plasmid, and an mCherry expression plasmid that could be used to identify electroporated cells even after GFP knockdown. A nontargeting RNA oligonucleotide was used for control electroporations. Given that RNAi functions by stoichiometric block of mRNA, we decided to use a GFP expression plasmid driven by the CMV promoter that would produce somewhat lower levels of GFP mRNA than the extremely high levels of message produced by the Gal4-VP16 system.
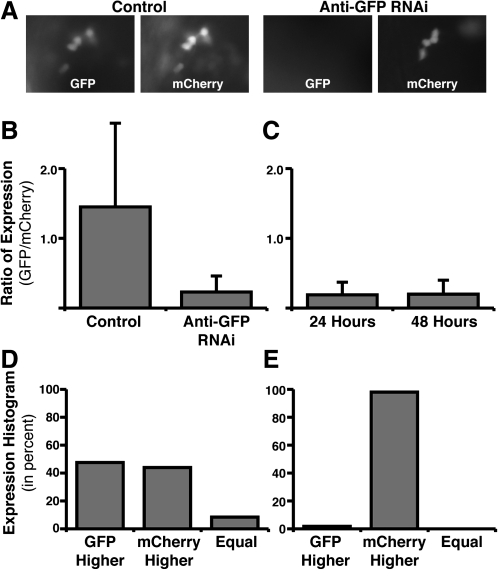
Embryos were electroporated at 24 hpf with either a control mixture (nontargeting RNAi oligo, pCS GFP, and pCS mCherry) or the anti-GFP mixture (anti-EGFP RNAi oligo, pCS GFP, and pCS mCherry). As can be seen in Figure 6A, 24 h after electroporation both GFP and mCherry are expressed in control cells. However, although cells receiving the anti-GFP RNAi showed robust mCherry expression, GFP expression was frequently not visible (Fig. 6A). The degree of GFP knockdown was then quantified by determining the ratio of GFP fluorescence to mCherry fluorescence in single cells (analysis done using Image J software; see Materials and Methods). Thus, if GFP and mCherry are equally expressed in control embryos we would expect a ratio of 1.0, and knockdown of GFP expression should lead to ratios of less than 1.0 depending of the degree of knockdown. The combined results of four experiments are shown in Figure 6B (n = 82 cells for control; n = 108 cells for anti-GFP RNAi). In control embryos, the ratio of GFP to mCherry fluorescence is quite variable (see error bar, Fig. 6B), which is to be expected given the nonlinear relationship between the amount of expression plasmid and the amount of protein produced. Nonetheless, for embryos receiving the anti-GFP RNAi, the amount of GFP protein is dramatically reduced as compared to control. The mean ratio for anti-GFP RNAi cells is 0.23 as compared to a ratio of 1.45 for control cells, indicating that anti-GFP RNAi oligos lead to an 84% knockdown of GFP expression. This demonstrates that in vivo electroporation can be used to efficiently knockdown a target protein. Further, this 84% reduction in GFP expression is particularly striking given that the levels of mRNA produced from these CMV-based expression plasmids are likely much greater than that for endogenous genes.
FIG. 6.
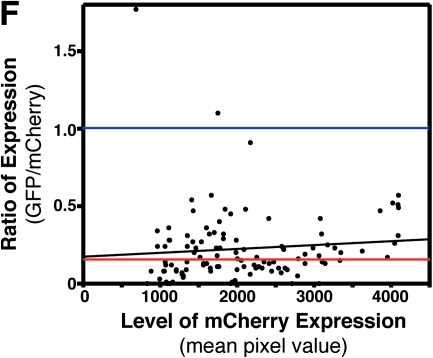
Using in vivo electroporation for RNA interference (RNAi)–based loss of function. (A) Fluorescent images showing that coelectroporation of an anti-GFP-targeted siRNA along with an expression plasmid for GFP leads to decreased GFP fluorescence as compared to cells electroporated with a nontargeting siRNA. A plasmid coding for mCherry was also included to identify the cells successfully electroporated. (B) The degree to which GFP expression was blocked was quantified by ratioing GFP to mCherry fluorescence in single cells (see Materials and Methods). Control electroporations included a nontargeting siRNA, a GFP expression plasmid, and an mCherry expression plasmid. Anti-GFP RNAi electroporations included an siRNA targeting GFP, a GFP expression plasmid, and an mCherry expression plasmid. (C) The degree to which GFP expression was blocked was analyzed in the same embryos 24 and 48 h after electroporation. (D) Histogram of all control cells analyzed. The cells have been categorized by the GFP/mCherry ratio for three different categories: (i) GFP higher expressed, (ii) mCherry higher expressed, or (iii) equal expression of GFP and mCherry (within 5%). (E) Histogram for all anti-GFP RNAi cells analyzed. (F) The GFP/mCherry ratio for all analyzed anti-GFP RNAi cells plotted against the total mCherry fluorescence (mean pixel value). This graph represents the degree of knockdown (ratio) as a function of total expression via the plasmids (mCherry expression). Black line represents a linear regression of all RNAi data. Red line represents the median ratio for RNAi cells. Blue line represents the median ratio for all control cells (note: control cells are not plotted on the graph).
What is the temporal resolution of RNAi-mediated knockdown using in vivo electroporation? RNAi works by direct binding of the RNAi oligo to the target mRNA. Thus, RNAi would be expected to have an immediate effect on all mRNA produced after the introduction of the RNAi oligo. Given that we coelectroporate the GFP expression plasmid along with the anti-GFP oligonucleotide, we would expect that there is no delay in the action of the RNAi. To confirm that no further knockdown occurs at later time points, we analyzed the same embryos at both 24 and 48 h after electroporation. Figure 6C shows that there is no further knockdown at 48 h, demonstrating that there is no delay in the action of anti-GFP RNAi when delivered by in vivo electroporation.
Given the unavoidable variability in the ratio of GFP to mCherry fluorescence in control cells (Fig. 6B), we performed some further analysis to demonstrate the robustness of RNA-mediated knockdown using in vivo electroporation. As suggested by the averaged data (Fig. 6B), whether GFP or mCherry is more highly expressed in individual control cells appears to be random. This is confirmed by plotting a histogram of individual cells and binning the data for GFP higher expressed, mCherry higher expressed, or GFP and mCherry equally expressed (within 5%). Figure 6D shows that which fluorescent protein is higher expressed in control cells is essentially random, with about half of the cells with higher GFP and the other half with higher mCherry. In contrast, for cells that received the anti-GFP RNAi oligo, essentially all of the cells had higher expression of mCherry (98%), confirming successful knockdown of GFP in the vast majority of cells electroporated with the anti-GFP RNAi oligo (Fig. 6E).
One potential caveat to using in vivo electroporation for delivery of RNAi oligonucleotides is whether sufficient amounts of the oligo are incorporated into the target cells. We have shown that a somewhat greater concentration of plasmid DNA is necessary for electroporation as compared to direct microinjection at the one-cell stage (Fig. 4). Given that RNAi works by stoichiometric block of mRNA, it is important to assess whether the amount of RNAi oligos that we are delivering by electroporation is sufficient for efficient knockdown of the target protein or whether the amount of oligo is borderline for cells expressing higher amounts of mRNA. We addressed this by plotting the level of knockdown (using the ratio of GFP to mCherry fluorescence) versus the relative amount of mRNA being produced. As an estimate of the level of mRNA, we used the average pixel intensity of mCherry fluorescence. Although this is not directly measuring the amount of GFP mRNA being produced, it is likely that cells receiving higher levels of the mCherry plasmid during electroporation also received higher levels of the GFP plasmid. If the level of anti-GFP RNAi oligos was borderline for stoichiometric block of the GFP mRNA being produced in the cells, then we would expect to see a decrease in the amount of knockdown at higher mRNA expression levels. Figure 6F is a plot of all 108 cells receiving the anti-GFP RNAi oligo and shows that there is relatively little decrease in the knockdown of GFP at higher levels of mRNA expression. The black line represents a linear regression of the data, showing that there is very little decrease in the level of knockdown (84–74%) across the whole range of mRNA levels observed. Given this, it seems that the levels of siRNA oligos delivered by in vivo electroporation in these experiments are far from being saturated, even by the high levels of mRNA produced from these CMV-driven expression plasmids.
Because Figure 6F represents all of the RNAi data in a single plot, it is also useful for observing the robustness of knockdown by electroporation. This graph makes it apparent that the vast majority of RNAi cells have significant knockdown of GFP expression. The red line represents the median ratio for RNAi cells, highlighting that half of the cells had a GFP/mCherry ratio of less that 0.16. Thus, half the cells showed more than an 84% decrease in GFP expression as compared to the control median (blue line; median ratio = 1.01). This plot also shows that the vast majority of cells cluster around the median line, demonstrating that knockdown induced through in vivo electroporation is consistent and robust.
Discussion
Although mutant analysis and morpholino injections at the one-cell stage are well-established genetic approaches in zebrafish, there is a paucity of methods for which gene knockout or knockdown can be specifically induced at later developmental stages. Such an approach is critically required to analyze the function of genes that are important for multiple developmental events. Previous studies characterizing the temporal resolution of in vivo electroporation15–18 have suggested that it could provide a powerful approach for time-resolved loss of function. As a proof of principle, in this study we have quantitatively assessed the efficacy of using in vivo electroporation to deliver siRNA oligonucleotides and induce knockdown of a specific target protein (GFP). In addition, we have directly measured the temporal resolution of in vivo electroporation, as it relates to the time frame of development of the zebrafish visual system. The results of this characterization further confirm that in vivo electroporation is a powerful method for time-resolved loss-of-function analysis in zebrafish.
Using expression of GFP from DNA plasmids as a way to monitor the efficiency of incorporation of reagents, we have shown that in vivo electroporation can target large regions of the developing brain (Fig. 1; Supplemental Movies S1 and S2), and this is achieved at voltages for which there is very high viability (Fig. 2). The concentration of DNA in the injection pipet required for maximal expression is 2- to 10-fold higher than what is typically used for microinjection of constructs at the one-cell stage,29 suggesting that the holes induced via electroporation are much smaller in diameter or much shorter lived in time (Fig. 3). Different regions of the developing CNS, including midbrain, hindbrain, cerebellum, and retina, can be targeted by adjusting the site of DNA injections and by appropriately orienting the electric field (Fig. 1).
We have shown that in vivo electroporation can effectively deliver DNA expression plasmids to the developing nervous system of embryos at 24, 48, and 96 hpf (Fig. 2). For each stage, a significant range of voltages could be found for which there was a high percentage of expressing embryos and very high viability. Thus, in vivo electroporation can effectively deliver reagents at any developmental stage, from 1 to 5 days after fertilization. As many experiments utilizing in vivo electroporation will also require a means to identify the cells targeted, we also measured the time required for GFP expression after incorporation of the GFP-coding plasmid. The maximal percentage of embryos expressing GFP was reached by 24 h after electroporation, and significant and useful expression can be observed by 6 h after electroporation (Fig. 4A–E). This early expression is robust as demonstrated by consistent coexpression of two coelectroporated plasmids (Fig. 4F–4G).
Finally, if in vivo electroporation is to be used as a genetic loss-of-function approach, it must be effective for delivering loss-of-function reagents, and these loss-of-function reagents must be efficient at inducing knockdown of the target protein. To determine the efficiency of RNAi-based knockdown by electroporation, we used coelectroporation of two expression plasmids, one for GFP and one for mCherry. This allowed us to directly monitor the loss of the target, GFP in this case, by directly observing GFP fluorescence. Using this approach we showed that coelectroporation of an anti-GFP duplex RNAi oligo along with the GFP expression plasmid resulted in the inhibition of GFP expression by at least 84% (Fig. 6A–B). This is significant block for a highly expressed mRNA being driven by the strong CMV promoter. Block of expression was immediate (Fig. 6C), as would be expected given that the mRNA was not made until after RNAi incorporation. Thus, with this particular experimental approach, the temporal resolution of knockdown is immediate. This should also hold true for endogenous target genes if the RNAi reagent can be delivered at a developmental stage before expression of the target gene is turned on. For other target genes that are already being expressed at the time of RNAi delivery, the temporal resolution of knockdown will be determined by the half-life of the protein. This can be vastly different depending upon the specific target protein, and will need to be determined on a case-by-case basis.
Our choice to use short duplex siRNA oligos instead of morpholino-based antisense oligos was strictly for technical reasons relating to this particular experiment. Because the ionophoresis effect of charged reagents is thought to facilitate incorporation by electroporation, it is likely that labeled morpholinos will be required for in vivo electroporation experiments.26,27 The experimental approach we used here to quantify the level of knockdown required that we use both a GFP to quantify the level of knockdown and a red fluorescent protein to identify cells that were successfully electroporated. Thus, to use a labeled morpholino, it would be required to go to a third, nonoverlapping fluorophore. This is possible with confocal microscopy, but was not as simple with our in-house fluorescent scope. Other experiments aimed at endogenous genes will typically not require expression of two colored proteins. Thus, for endogenous genes, fluorescently labeled morpholinos should be amenable to loss-of-function analysis by in vivo electroporation in zebrafish, as has been previously shown.17,23,24 Given this, the temporal and spatial characteristic of in vivo electroporation that we have characterized here should also apply for future experiments using morpholino-based reagents.
Although RNAi technology has revolutionized loss-of-function analysis for most model organisms,31–33 and initial experiments were promising in zebrafish,34 the method is not generally used for the zebrafish system. This is likely due to early work showing that some forms of RNAi reagents lead to nonspecific and deleterious effects.35 Subsequent studies have not yet been conclusive as to the potential of RNAi technology for studying zebrafish development.36 That debate is clearly beyond the scope of this study, and the reasons for our using siRNA oligos were strictly due to the technical specifics of our experiment. However, it is worth noting here that short-duplex siRNA oligos delivered by in vivo electroporation worked very efficiently for target knockdown in our hands (Fig. 6). Further, we saw no change in embryo viability or gross morphology, nor did we see any effect on cellular morphology of mCherry expressing neurons targeted with either control or anti-GFF siRNA oligos. One possibility is that by targeting embryos at later developmental stages we are avoiding complications due to inhibiting the processing of microRNAs at the maternal-to-zygotic transition, which has been shown to be one of the off-target effects of siRNA in zebrafish.37,38 Also, it is possible that electroporation results in much lower concentrations of siRNA oligos in cells, thus lessening the stoichiometric block of microRNA processing machinery.
Supplementary Material
Acknowledgments
We would like to thank Drs. Nancy Krucher, Reinhard Koster, Jeff Gross, and Scott Fraser for discussions and helpful suggestions. All wild-type zebrafish stocks used in this study were provided by Dr. Jeff Gross (University of Texas, Austin, TX). All GFP expression plasmids were obtained from Dr. Reinhard Koster (Institute of Developmental Genetics, Neuherberg, Germany). We would also like to thank Dr. Todd Evans (Albert Einstein College of Medicine, New York, NY) for advice on zebrafish techniques and for the use of his Zeiss Axioplan Fluorescence Microscope, and Dr. Diana Petit (Albert Einstein College of Medicine, New York, NY) for imaging expertise and use of her Olympus Fluoview 300 confocal microscope. This work was funded through an NIH R15 grant to J.H. (NIMH; R15MH083221), and through Pace University Scholarly Research Funds (J.H.) and a Pace University Lang Fellowship (J.H. and S.A.).
Disclosure Statement
No competing financial interests exist.
References
- 1.Grunwald DJ. Eisen JS. Headwaters of the zebrafish—emergence of a new model vertebrate. Nat Rev Genet. 2002;3:717–724. doi: 10.1038/nrg892. [DOI] [PubMed] [Google Scholar]
- 2.Lichtman JW. Fraser SE. The neuronal naturalist: watching neurons in their native habitat. Nat Neurosci. 2001;4(Suppl):1215–1220. doi: 10.1038/nn754. [DOI] [PubMed] [Google Scholar]
- 3.Boutros M. Ahriner J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9:554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- 4.Heasman J. Mopholino oligos: making sense of antisense. Dev Biol. 2002;243:209–214. doi: 10.1006/dbio.2001.0565. [DOI] [PubMed] [Google Scholar]
- 5.Eisen JS. Smith JC. Controlling morpholino experiments: don't stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- 6.Swartz M. Eberhart J. Mastick G. Krull CE. Sparking new frontiers: using in vivo electroporation for genetic manipulations. Dev Biol. 2001;233:13–21. doi: 10.1006/dbio.2001.0181. [DOI] [PubMed] [Google Scholar]
- 7.Krull CE. A primer on using in ovo electroporation to analyze gene function. Dev Dyn. 2004;229:433–439. doi: 10.1002/dvdy.10473. [DOI] [PubMed] [Google Scholar]
- 8.Stern CD. The chick; a great model system becomes even greater. Dev Cell. 2005;8:9–17. doi: 10.1016/j.devcel.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 9.Sauka-Spengler T. Barembaum M. Gain- and loss-of-function approaches in the chick embryo. Methods Cell Biol. 2008;87:237–256. doi: 10.1016/S0091-679X(08)00212-4. [DOI] [PubMed] [Google Scholar]
- 10.Haas K. Sin WC. Javaherian A. Li Z. Cline HT. Single-cell electroporation for gene transfer in vivo. Neuron. 2001;229:583–591. doi: 10.1016/s0896-6273(01)00235-5. [DOI] [PubMed] [Google Scholar]
- 11.Haas K. Jensen K. Sin WC. Foa L. Cline HT. Targeted electroporation in Xenopus tadpoles in vivo—from single cells to the entire brain. Differentiation. 2002;70:148–154. doi: 10.1046/j.1432-0436.2002.700404.x. [DOI] [PubMed] [Google Scholar]
- 12.Bestman JE. Ewald RC. Chiu S. Cline HT. In vivo single-cell electroporation for transfer of DNA and macromolecules. Nat Protoc. 2006;1:1267–1272. doi: 10.1038/nprot.2006.186. [DOI] [PubMed] [Google Scholar]
- 13.Falk J. Drinjakovic J. Leung KM. Dwivedy A. Regan AG. Piper M. Holt CE. Electroporation of cDNA/Morpholinos to targeted areas of the embryonic CNS in Xenopus. BMC Dev Biol. 2007;7:107–115. doi: 10.1186/1471-213X-7-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller F. Lele Z. Varadi L. Menczel L. Orban L. Efficient transient expression system based on square pulse electroporation and in vivo luciferase assay of fertilized fish eggs. FEBS Lett. 1993;324:27–32. doi: 10.1016/0014-5793(93)81525-5. [DOI] [PubMed] [Google Scholar]
- 15.Teh C. Chong SW. Korzh V. DNA delivery into anterior neural tube of zebrafish embryos by electroporation. Biotechniques. 2003;35:950–954. doi: 10.2144/03355st02. [DOI] [PubMed] [Google Scholar]
- 16.Teh C. Parinov S. Korzh V. New ways to admire zebrafish: progress in functional genomics research methodology. Biotechniques. 2005;38:897–906. doi: 10.2144/05386RV01. [DOI] [PubMed] [Google Scholar]
- 17.Cerda GA. Thomas JE. Allende ML. Karlstrom RO. Palma V. Electroporation of DNA, RNA, and morpholinos into zebrafish embryos. Methods. 2006;39:207–211. doi: 10.1016/j.ymeth.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Hendricks M. Jesuthasan S. Electroporation-based methods for in vivo, whole mount and primary culture analysis of zebrafish brain development. Neural Dev. 2007;15:2–6. doi: 10.1186/1749-8104-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rambabu KM. Rao SH. Rao NM. Efficient expression of transgenes in adult zebrafish by electroporation. BMC Biotechnol. 2005;13(5):29. doi: 10.1186/1472-6750-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao NM. Rambabu KM. Rao SH. Electroporation of adult zebrafish. Methods Mol Biol. 2008;423:289–298. doi: 10.1007/978-1-59745-194-9_21. [DOI] [PubMed] [Google Scholar]
- 21.Bhatt DH. Otto SJ. Depoister B. Fetcho JR. Cyclic AMP-induced repair of zebrafish spinal circuits. Science. 2004;305:254–258. doi: 10.1126/science.1098439. [DOI] [PubMed] [Google Scholar]
- 22.Tawk M. Bianco IH. Clarke JD. Focal electroporation in zebrafish embryos and larvae. Methods Mol Biol. 2009;546:145–151. doi: 10.1007/978-1-60327-977-2_10. [DOI] [PubMed] [Google Scholar]
- 23.Thummel R. Bai S. Sarras MP. Song P. McDermott J. Brewer J, et al. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev Dyn. 2006;235:336–346. doi: 10.1002/dvdy.20630. [DOI] [PubMed] [Google Scholar]
- 24.Thummel R. Kassen SC. Montgomery JE. Enright JM. Hyde DR. Inhibition of Muller glial cell division blocks regeneration of the light-damaged zebrafish retina. Dev Neurobiol. 2008;68:392–408. doi: 10.1002/dneu.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teruel MN. Meyer T. Electroporation-induced formation of individual calcium entry sites in the cell body and processes of adherent cells. Biophys J. 1997;73:1785–1796. doi: 10.1016/S0006-3495(97)78209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCauley DW. Bronner-Fraser M. Importance of SoxE in neural crest development and the evolution of the pharynx. Nature. 2006;441:750–752. doi: 10.1038/nature04691. [DOI] [PubMed] [Google Scholar]
- 27.Taneyhill LA. Coles EG. Bronner-Fraser M. Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development. 2007;134:1481–1490. doi: 10.1242/dev.02834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koster RW. Fraser SE. Tracing transgene expression in living zebrafish embryos. Dev Biol. 2001;233:329–346. doi: 10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
- 29.Nusslein-Volhard C. Dahm R. Zebrafish. first. Oxford, UK: Oxford University Press; 2002. [Google Scholar]
- 30.Campbell DS. Stringham SA. Timm A. Xiao T. Law MY. Baier H, et al. Slit1a inhibits retinal ganglion cell arborization and synaptogenesis via Robo2-dependent and—independent pathways. Neuron. 2007;55:231–245. doi: 10.1016/j.neuron.2007.06.034. [DOI] [PubMed] [Google Scholar]
- 31.Lewis DL. Hagstrom JE. Loomis AG. Wolff JA. Herweijer H. Efficient delivery of siRNA for inhibition of gene expression in post natal mice. Nat Genet. 2002;32:17–108. doi: 10.1038/ng944. [DOI] [PubMed] [Google Scholar]
- 32.Martin SE. Caplen NJ. Application of RNA interference in mammalian systems. Annu Rev Genomics Hum Genet. 2007;8:81–108. doi: 10.1146/annurev.genom.8.080706.092424. [DOI] [PubMed] [Google Scholar]
- 33.Boutros M. Ahringer J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9:554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- 34.Li YX. Farrell MJ. Liu R. Mohanty N. Kirby ML. Double-stranded RNA injection produces null phenotypes in zebraifish. Dev Biol. 2000;217:394–405. doi: 10.1006/dbio.1999.9540. [DOI] [PubMed] [Google Scholar]
- 35.Oates AC. Bruce AE. Ho RK. Too much interference: injection of double-stranded RNA as nonspecific effects in the zebrafish embryo. Dev Biol. 2000;224:20–28. doi: 10.1006/dbio.2000.9761. [DOI] [PubMed] [Google Scholar]
- 36.Skromne I. Prince VE. Current perspectives in zebrafish reverse genetics: moving forward. Dev Dyn. 2008;237:861–882. doi: 10.1002/dvdy.21484. [DOI] [PubMed] [Google Scholar]
- 37.Zhao XF. Fjose A. Larsen N. Helvik JV. Drivenes O. Treatment with small interfering RNA affects the microRNA pathway and causes unspecific defects in zebrafish embryos. FEBS J. 2008;275:2177–2184. doi: 10.1111/j.1742-4658.2008.06371.x. [DOI] [PubMed] [Google Scholar]
- 38.Giraldez AJ. Cinalli RM. Glasner ME. Enright AJ. Thomson JM. Baskerville S, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.