Abstract
Enterotoxigenic Escherichia coli (ETEC) strains are leading causes of childhood diarrhea in developing countries. ETEC pili and non-pili adherence factors designated colonization surface antigens (CSA) are believed to be important in the pathogenesis of diarrhea. Longus, a type IV pilus identified as the CSA21, is expressed in up to one-third of ETEC strains, and share similarities to the toxin-coregulated pilus of Vibrio cholerae, and the bundle-forming pilus of enteropathogenic E. coli. To identify longus phenotype and possible function, a site-directed mutation of the lngA major subunit gene in the E9034A wild type ETEC strain was constructed. Lack of longus expression from the lngA mutant was demonstrated by immunoblot analysis and electron microscopy using specific anti-LngA antibody. Formation of self-aggregates by ETEC was shown to be dependent on longus expression as the lngA mutant or wild type grown under poor longus-expression conditions was unable to express this phenotype. Longus-expressing ETEC were also associated with improved survival when exposed to antibacterial factors including lysozyme and antibiotics. This suggests that longus-mediated bacterial self-aggregates protect bacteria against antimicrobial environmental agents and may promote gut colonization.
Keywords: Enterotoxigenic E. coli, diarrhea, pili, longus, mutagenesis
1. INTRODUCTION
Enterotoxigenic Escherichia coli (ETEC) strains are leading causes of diarrhea in children living in developing countries and the most common cause of traveler’s diarrhea (19). Among all colonization surface antigens (CSA) described in ETEC, the colonization factor antigen-I (CFA-I) and longus (CS21) are the most prevalent based on screening of ETEC isolates from different geographic regions of the world (11, 14, 22, 24). Longus, a type IV pili (10) expressed at 37°C when grown in blood agar plates, consist of long intertwining filaments similar to the bundle-forming pili of enteropathogenic E. coli (EPEC) (9). The 28-kDa LngA major subunit of longus is processed by a prepilin peptidase to a mature 22-kDa major subunit. Longus filaments result from linear polymers of a single structural subunit designated LngA. Longus pilin has sequence homology to the well characterized toxin-coregulated pilus (TCP) of V. cholerae, N. gonorrheae pilin, the bundle-forming pilus (BFP) of EPEC, the Pseudomonas aeruginosa strain K (PAK) pilin, the colonization factor III A (CofA) of ETEC, among others (13). Type IV pili are divided in subclasses type IVa, and type IVb. Type IVb pilin, expressed among enteropathogens that colonize human intestine, including longus, have a longer leader peptide, a variable methylated N-terminal residue, and minimal similarity a the C-terminal domain, except for a common C-terminal disulfide bond (8, 28). High-resolution X-ray crystal structures of the type IVb TCP pilin and the type IVa, PAK pilin indicate that virtually all type IV pilin subunits assemble as alpha helices (5). Conserved structural features of type IV pilins lead to the characteristic long hair-like structure and variable features result in unique assembly domains with well defined roles in adherence, self-aggregation phenotypes, and mucosal colonization (5).
A DNA cluster present in the virulence plasmid of some ETEC strains carries the biosynthetic genes necessary for expression of longus (12). The cluster is 14 kb in size and carries 16 putative biosynthetic genes with 57 to 95% homology to CFA/III type IV pili genes and identical topology. CFA/III is a less prevalent CSA of ETEC that was reported to mediate ETEC adherence to CaCo-2 cells (29, 30). The cluster has also significant homology and topology to the colonization factor Citrobacter (CFC) cluster of Citrobacter rodentium, a murine pathogen that cause diarrhea and colonic hyperplasia and typical attaching and effacing lesions. C. rodentium is an in vivo model system for clinically significant enteric pathogens (21).
The purpose of this study is to characterize the longus structural subunit, its phenotype, and the possible role it may have in ETEC pathogenesis. First, we will construct a tridimensional longus structure, derived by automated comparison from known type IV pili, to evaluate structural and functional components. A site-directed mutant at the lngA major subunit gene will be constructed and compared with the wild type strains to identify possible phenotypes. We report that longus major subunits structure is similar to other type IVb major subunits with a C-terminus likely involved in pili-pili, or bacterial-pili interactions. Longus induces ETEC self-agglutination, or microcolony formation as other type IV pili and this phenotype is believed to protect longus-expressing ETEC against antimicrobial agents in vitro.
2. RESULTS
2.1. LngA secondary and tertiary protein structure
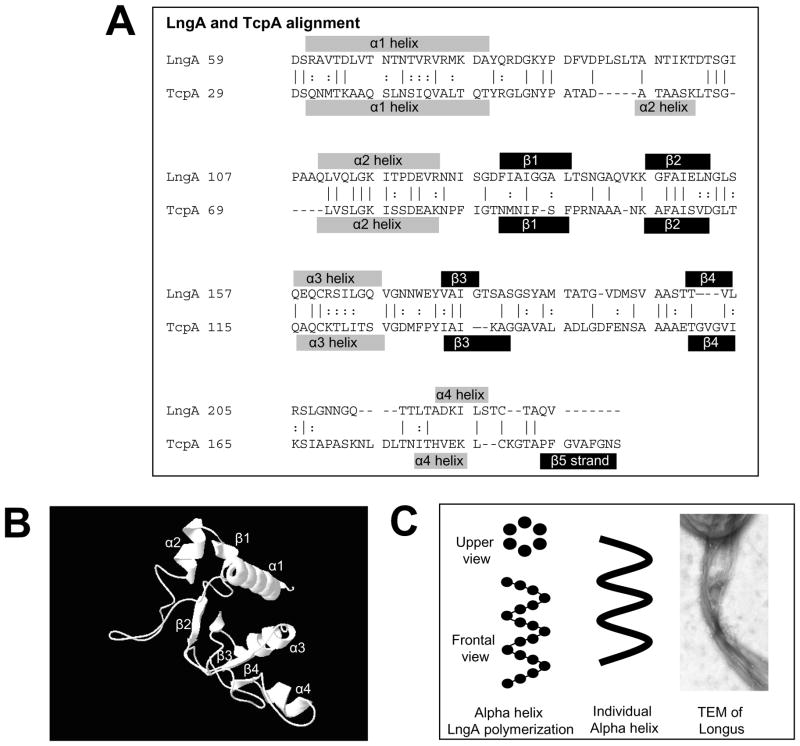
Molecular characterization of TCP and PAK pili structures indicate that type IV pili assemble as a conserved N-terminal alpha helix wrapped by beta strands that anchors a variable globular C-terminus domain. To determine if longus pilin shares the same secondary and tertiary structure, an automated comparison using Swissprotein web-based software was performed. As shown in Figure 1 it is predicted that the basic elements of the secondary and tertiary structure of TcpA are also present in LngA. The LngA residues analyzed were 59 to 228, which correspond to the mature pilin. This fragment excludes the leader peptide that is cleaved by prepilin peptidase. LngA has the same four alpha helices and four out of the five beta-sheets present in TcpA (Figure 1, panel A). The alpha1 and alpha2 helixes conform the N-terminus domain, while the alpha3 and alpha4 helices conform the C-terminus domain (Figure 1, panel B). Alpha 3 and 4 helices are linked through a disulfide bond between a cystein residue present at each helix at positions 162 and 224 of the complete LngA protein. The topology of the alpha and beta sheet structures is also identical in both TcpA and LngA (Figure 1, panel B). This suggests that LngA assembles and polymerizes in similar fashion as TcpA, where the N-terminus domain helps assemble the alpha helix polymer, whereas the C-terminus domain located external to the alpha helix polymer axis may participate in pili-pili and bacterial-pili interactions. Individual LngA subunits would arrange to form alpha helices, with six subunits for every helix turn (Figure 1, panel C), and three parallel alpha helices may assemble simultaneously to form longus pili. Individual longus pilus structures also form pili-pili interactions or bundles as described for TCP, PAK, and BFP of enteropathogenic E. coli (Figure 1, panel C), which may result in bacterial aggregate formation or microcolonies.
Figure 1. Predicted protein LngA model based on automated comparison with TcpA.
Panel A. Protein sequence and structural alignment of LngA and TcpA. The alpha helices or beta strands are shown above or below each LngA or TcpA sequences, respectively. Panel B. Frontal view of the tertiary structure of LngA based on automated comparison with TcpA showing alpha spins and beta strands. Alpha1 and 2 helices N-terminus LngA regions are predicted to face the inner core of the alpha helix while the C-terminus alpha 3 and 4 are predicted to face the outer surface of the helix. Panel C. Diagram of the LngA subunit arrangement into an alpha helix, showing six subunits per helix turn. Three alpha helix is predicted to form the longus filaments as described for TCP of V. cholerae. Longus filaments may also group in bundles as shown by TEM.
2.2. Isogenic lngA mutant of ETEC E9034A strain do not express LngA
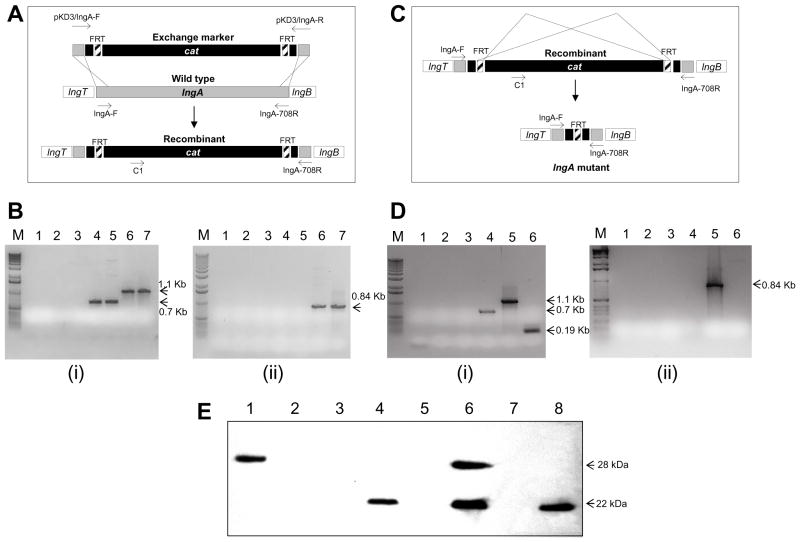
To evaluate the role of the lngA gene in pilin expression, longus bundle formation and to test the possible function of longus, an lngA gene deletion mutation was constructed. The lambda Red system was used to construct an isogenic site-directed mutation in the lngA gene of the ETEC wild-type E9034A strain as described by Datsenko and Warner (6). The construction of the isogenic mutant involved two independent recombination events, one mediated by the lambda Red recombinase and the second by the FLP recombinase. The first homologous recombination in the ETEC E9034A(pKD46) strain exchanged the wild-type lngA gene for the ΔlngA::FRT-cat linear DNA fragment (Figure 1, Panel A) that was transferred by electroporation. The resulting chloramphenicol-resistant strain carrying the ΔlngA::FRT-cat DNA fragment was designated E9034AΔlngA::FRT-cat(pKD46). PCR amplification of lngA with lngA-specific primers resulted in a 708bp DNA band in the E9034A(pKD46) strain and a 1,107bp in the E9034AΔlngA::FRT-cat(pKD46) strain (See Figure 1 panels A and B). In addition, the presence of the cat gene in the E9034A ΔlngA::FRT-cat was confirmed by the generation of a 864 bp PCR fragment with primers C1 and LngA-R. No DNA amplification with the same primers was observed from the wild type E9034A strain (Figure 1, panels B). The isolation of the isogenic mutant containing a deletion mutation of lngA without the cat cassette was accomplished by a second homologous recombination event mediated by the Flp recombinase (Figure 1, Panel C). A single chloramphenicol-sensitive colony was evaluated by PCR analysis to confirm the creation of the truncated lngA deletion mutant. Amplification of the truncated lngA gene with lngA-specific primers resulted in a single 190-bp DNA fragment from E9034AΔlngA(pCP20), whereas no DNA fragment was amplified when primers C1 and lngA708R were used (Figure 1, panels C and D), indicating the FRT-cat cassette was absent.
Except for the lngA gene, both wild-type E9034A and mutant E9034AΔlngA strains were identical, including colony morphology, antibiotic resistance markers, time growth curves, and protein binding patterns on SDS-PAGE Coomassie-stained gels (data not shown). To evaluate if the lngA-deletion mutants were unable to express the longus major subunit LngA, longus expression was evaluated by immunoblot. Strains were grown on TB to optimize conditions of longus expression. As shown in Figure 1, panel E, wild-type E9034 ETEC strain showed a 22-kDa protein band corresponding to the predicted mature product of LngA and consistent with previous report (13). Mutants E9034A ΔlngA::cat and E9034A ΔlngA were unable to express LngA. Both mutants complemented with the recombinant lngA gene carried by plasmid pAC-lngA (E9034A ΔlngA::cat(pAC-lngA) and E9034A ΔlngA(pAC-lngA) recovered the expression the mature LngA protein (Figure 1, panel E). Prepilin protein was also detected in lane 6 loaded with E9034A ΔlngA(pAC-lngA) suggesting the prepilin peptidase may not process prepilin when it is expressed in excess amounts. Negative controls E. coli DH5α and E. coli HB101 strains did not express LngA. Positive controls E. coli DH5α(pAC-lngA) strain showed the 28-kDa LngA prepilin protein band since these strains lack the prepilin peptidase gene or other biosynthetic longus genes as the E9034A wild type strain.
2.3. The lngA gene is essential for expression of the Longus type IV pilus
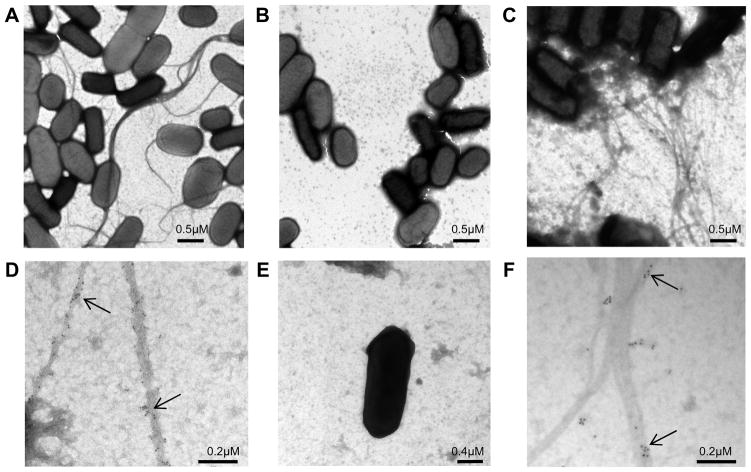
To determine if the lngA gene is not only required for expression of the LngA subunit but also to express the longus pilus, wild-type and mutant strains were examined with TEM. Wild-type ETEC E9034A strain grown on TB for optimal conditions of longus expression showed long bundle-forming pili characteristic of Longus (Figure 3, panel A), while the E9034A ΔlngA isogenic mutant strain showed no evidence of pilus-like structures (Figure 3, panel B). This result confirms that the lngA gene is the major subunit gene of longus and it is therefore essential for longus pilus expression. To corroborate that the lngA gene is the LngA major subunit essential for longus expression, E9034A ΔlngA isogenic mutant complemented in trans with the recombinant lngA, carried by the pAC-lngA plasmid, recovered expression of longus (Figure 3, panel C).
Figure 3. Transmission electron microscopy (TEM) of ETEC strains expressing longus.
Panel A. Wild-type E9034A ETEC strain grown under optimal conditions for longus expression showing typical longus filaments. Panel B. Isogenic E9034AΔlngA mutant strain lacking pili. Panel C. E9034AΔlngA (pAClngA) complemented mutant strain showing Longus-like structures. Panel D. Immunogold labeling TEM of E9034A ETEC wild-type strain. Panel E. Immunogold labeling of E9034AΔlngA mutant strain. Panel F. Immunogold labeling of E9034AΔlngA (pAClngA) complemented mutant strain. Arrows indicate gold particles specifically attached to type IV pili. Scale bar size indicated in μM.
To confirm that the type IV pili observed on the wild-type and complemented mutant E9034A ΔlngA(pAC-lngA) strains were longus, immunogold-labeling TEM was carried out using specific anti-LngA antibody. As observed in Figure 3, panel D, wild-type E9034A strain had gold particles specifically attached to the long pili structures. Similar gold particles were observed attaching to the E9034A ΔlngA(pAC-lngA)-complemented mutant strain (Figure 3, panel F) and absent in E9034AΔlngA mutant (Figure 3, panel E). Immunogold-labeling TEM of the same strains did not revealed gold particle binding to longus pili when anti-type I pili was used, indicating that the anti-longus antibody gold particle binding is specific for longus (data not shown).
2.4. Longus mediates ETEC self-aggregation
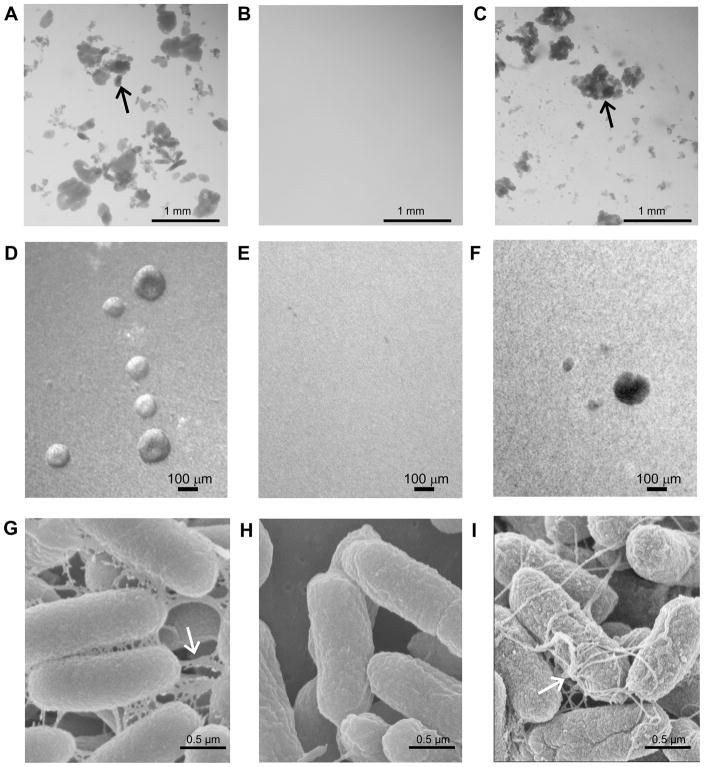
The bundle-forming pilus (BFP) of EPEC mediates self-aggregation formation during adherence to epithelial cells. This phenotype is also designated localized adherence, as bacteria self-aggregate at well defined areas on the cell surface. During ETEC growth in liquid media using optimal conditions of longus expression, the wild-type E9034A strain self-aggregated, while the E9034A ΔlngA isogenic mutant did not (Figure 4, Panel A and B). The complemented mutant E9034A ΔlngA(pAC-lngA) strain grown at similar conditions was also able to self-aggregate (Figure 4, Panel C). This phenotype, which shares similarity with the BFP, is designated self-aggregation formation phenotype, and it is not observed under growth conditions that do not support longus expression (data not shown). Light microscopy of bacterial suspension indicate that before self-aggregate are visible to the naked eye, self-aggregates start to form after 3h or incubation as spheres of bacterial aggregates. These spheric aggregates were observed with wild type and complemented mutant strains (Figure 4, panels D and F) and absent with the mutant strain (Figure 4, panel E). To examine the longus-mediated interactions within the self-aggregates SEM was performed at the high magnification. SEM images illustrate that the self-aggregation phenotype in longus-expressing ETEC is mediated by multiple pili-pili and pili-bacterial interactions. Longus interactions were only observed when the wild type E9034A and complemented mutant strains were grown under optimal conditions of longus expression (Figure 4, panels G and I). These interactions are not observed with the longus mutant strain grown in TB (Figure 4, Panel H) or with the wild-type grown in Luria broth (data not shown), indicating that self-aggregation is dependent on longus expression.
Figure 4. Longus-mediated microcolony formation phenotype.
Panels A, D and G: E9034A wild-type ETEC strain; Panels B, E and H: E9034AΔlngA; Panel C, F and I: E9034AΔlngA(pAC-lngA) complemented mutant. Arrow indicates bacterial self-aggregates. Panels A, B, and C are pictures obtained from bacterial suspensions. Scale bar sizes indicated in mm. Panels D, E, and F are pictures obtained by inverted light microscope. Panels G, H, and I are scanning electron microscopy pictures. Arrow indicates pilin-bacterial interactions. Scale bars sizes for Panels D to I indicated in μM. All strains were grown in TB.
2.5. Longus-expressing ETEC exposed to antimicrobial agents has improved survival than the longus-mutant
Longus mediated ETEC self-aggregates may result in the formation of two bacterial populations, one exposed population that surrounds the self-aggregate and another internal population that maybe protected from the external environment. To evaluate if bacteria within self-aggregates are protected against stress factors a qualitative survival assay was performed. ETEC strains preincubated under optimal conditions of longus expression, for self-aggregates to form, were exposed to increasing concentrations of antibacterials above the MIC and incubated overnight. As shown in table 3, there were surviving E9034A wild type ETEC at a 10 fold higher concentration of antibiotics, compared with the E9034AΔlngA mutant, indicating that longus has a protective effect on bacteria exposed to antibiotics. No difference in survival was noticed at any concentration of lysozyme tested.
Table 3.
Effect of self-aggregation on survival of longus-expressing ETEC after exposure to antimicrobials.
| Antibacterials | Concentration | E9034A | E9034A:lngA |
|---|---|---|---|
| Chloramphenicol | 15 μg/ml | + | + |
| 150 μg/ml | + | − | |
| Gentamicin | 25 μg/ml | + | + |
| 250 μg/ml | + | − | |
| Kanamycin | 1 mg/ml | + | + |
| 10 mg/ml | + | − | |
| Tetracycline | 250 μg/ml | + | + |
| 2.5 mg/ml | + | − | |
| Lysozyme | 0.5 mg/ml | + | + |
| 5 mg/ml | + | + |
Bacteria preincubated for 4h in TB were exposed to increasing concentrations of antimicrobials and incubated at 37°C overnight. Bacterial survival evaluated by plating overnight suspensions on LA plates. (+) indicates positive growth and (−) indicates no growth.
2.6. ETEC survival in the presence of antibacterials agents is dependent on longus expression
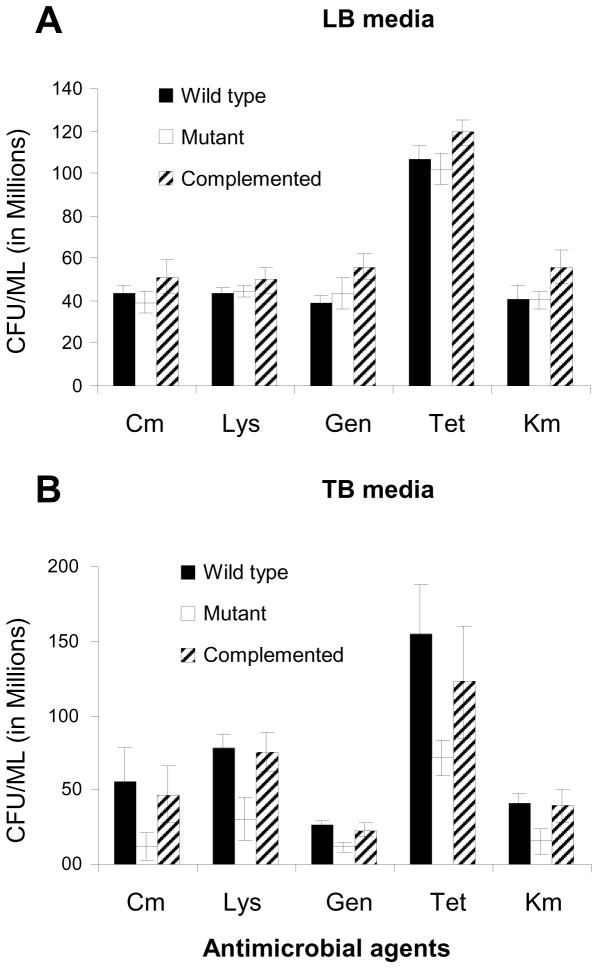
To evaluate if longus-expression if associated with protection against antimicrobials, including lysozyme, wild type, mutant and complemented mutant strains were preincubated for 4h before being exposed to a set concentration of antibacterials above the MIC. To determine quantitative differences in survival, exposed bacterial suspensions were diluted and plated for determination of CFU/ml. As shown in Figure 5 there was no difference in bacterial survival between wild type, mutant, or complemented mutant grown on LB, media not associated with optimal longus expression. Difference in survival was detected between mutant and wild type and complemented mutant grown on TB, media that induced optimal expression of longus. The assay also detected improved survival of wild type and complemented mutant in the presence of lysozyme, compared with the mutant, indicating that while lysozyme may be bacteriostatic difference in survival are observed. No effect was observed when using human sera, or lactoferrin (data not shown).
Figure 5. Effect of longus expression on ETEC survival after exposure to antimicrobials.
Panel A. Effect of antimicrobials on bacterial suspensions preincubated in LB. After overnight incubations all suspensions were processed to determine CFU/ml. Panel B. Effect of antimicrobials on bacterial suspensions preincubated in TB. Antimicrobial concentrations used: Chloramphenicol (Cm) at 24 μg/ml, Gentamicin 96 μg/ml, Tetracycline 24 μg/ml, Kanamycin 192 μg/ml, and lysozyme 768 μg/ml. Black bars: E9034A: wild type ETEC strain; white bars: E9034AΔlngA mutant; and striped bars: E9034A:lngA(pBADlngA) complemented mutant. Error bars correspond to standard deviations from experiments done in triplicate.
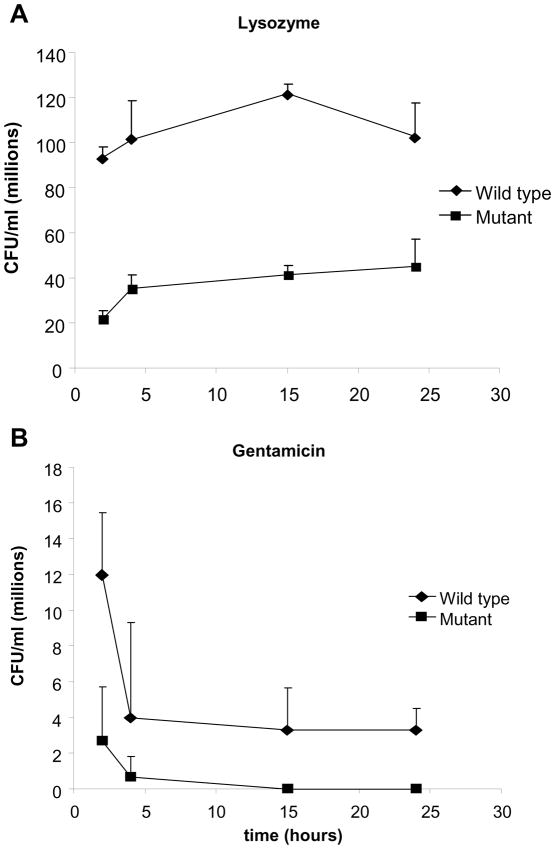
To evaluate the kinetics of protection against antibacterials wild type and mutant strains were exposed to antimicrobials for different periods of time. Bacterial survival was evaluated quantitatively by determination of CFU/ml. As shown in Figure 6, both gentamycin and lysozyme resulted in decreased mutant survival compared with the wild type. Similar effect was noticed with the remaining antibiotics (data not shown). This data indicates that longus-expressing ETEC have improved survival when exposed to antimicrobial agents.
Figure 6. Kinetics of survival after longus-expressing ETEC exposure to antimicrobials.
Panel A. Kinetics of survival after exposure of preincubated wild type and mutant to lysozyme (350 μg/ml). Panel B. Kinetics of survival after exposure of preincubated wild type and longus mutant strains to Gentamicin (48 μg/ml). Black diamons: E9034A: wild type ETEC strain; black square: E9034A:lngA ETEC mutant strain. Error bars correspond to standard deviations from experiments done in triplicate.
3. DISCUSSION
ETEC diarrhea is a leading cause of morbidity and mortality in the developing world and one of the first E. coli pathotypes to be identified. Among the 22 ETEC CSA factors described limited information is available on the role of longus (CSA21) on adherence, bacteria-bacteria interaction, or colonization. In this study we characterize the lngA gene encoding the major subunit LngA, and show that it is responsible for longus pilus expression in ETEC. Prediction sequencing analysis suggest that the longus pilin assembles as a two-domains protein. The N-terminus hydrophobic conserved domain is directly implicated in the alpha helix pilin polymerization assembly. The C-terminus variable domain facing the exterior of the alpha helix is likely responsible for the interactions with other pili, and surrounding bacteria, which may explaining the formation of bundles, and bacterial aggregates, respectively. TCP type IVb pili assembles as a left-handed three-start alpha helix structure (5), PAK pilus and the N. gonorrheae type IVa pili, instead, assembles as a unique alpha helix structure (7). While protein sequence analysis of LngA predicted the secondary and terciary structure to be highly similar to TcpA, more studies may be necessary to define the full length filamentus structure of longus. New studies may also confirm that longus may assemble as other type IVb pili in a three alpha helix polymers to form the native pilus.
By comparing wild type E9034A and lngA-mutant strain we showed that longus mediates bacterial self-aggregation in similar fashion to the BFP-mediated microcolony formation of EPEC (9, 18, 31) or pili-mediate self-aggregation of Neisseria gonorrhoeae (23). BFP-mediated pili bacterial and cell bacterial interactions are mediated by the lectin-like nature of the BFP major subunit (16, 17, 32) with specificity to N-acetyllactosamine. Longus-mediated self-aggregation is dependent on pilus-pilus and bacterial-pilus interactions based on SEM images showing pili nets interconnecting bacteria. While the exact nature of this inter-pili interaction is unknown preliminary data indicates that fucose molecules may participate in these interactions (our unpublished results), as increasing concentration of this sugar inhibit longus-mediated self-aggregation. Based on the predicted protein LngA model, the variable C-terminus domain located at the exposed region of the alpha helix structure are the regions most likely to mediate lectin-like pili-pili and bacteria-pili interactions, and ultimately responsible for the bacterial aggregates.
The ability of longus to mediate ETEC self-aggregates or microcolonies may result in two distinct bacterial populations, one surrounding and another filling the bacterial aggregate. The surface exposed population may be vulnerable to the surrounding environment, which may include gut-associated bactericidal stress factors. Longus-mediated self-aggregates may protect internal bacteria against the host-adverse environment in a similar manner as bacterial biofilms (4). Bacteria inside the self-aggregate, similar to bacteria inside the biofilm, may avoid contact with lumen intestinal bactericidal agents such as lactoferrin, lysozyme, secretory antibodies, and even antibiotics known to be detrimental for bacterial colonization. We report that the longus expressing wild type ETEC were protected from several antibacterial agents suggesting that longus mediated self-aggregation have a protective effect against stress factors. To our knowledge this is a novel property attribute to longus type IV pili that may be explored in other type IVa and IVb pili. More studies will be necessary to elucidate if the mechanism of protection is similar to the one occurring in biofilms and whether other bacterial and host factors are involved. Furthermore, studies are underway to evaluate if the longus-mediated aggregates also participate in adherence to epithelial cells and in intestinal colonization.
We speculate that longus-mediated self-aggregation may participate in ETEC colonization. Longus may bind to specific receptors on the intestinal cell surface or on the mucus layer and aggregate bacteria in microcolonies at specific sites resulting in increased colonization and virulence potential. EPEC microcolony formation on the surface of epithelial cells (26) was proven essential in virulence as a BFP mutant was less virulent than the wild-type EPEC in clinical trials (2). So far, transformed laboratory tissue culture epithelial cells (HeLa, Hep-2, CaCo-2 cells) do not seem to have membrane receptors for longus-expressing ETEC. Preliminary data suggest that mucus produced by epithelial cells rather than cell membrane-bound receptors may provide a substrate for longus-specific ETEC adherence (our unpublished results).
The initial characterization of the lngA mutation in the ETEC longus cluster has facilitated the identification of longus-associated phenotypes, and a better understanding of the role of longus in self-aggregation and ETEC protection against stress host factors. Further studies are underway to evaluate the role of longus in intestinal cell adherence, intestinal colonization, ETEC pathogenesis, and the possible role of longus regulators in longus expression and function as well as in global regulation of ETEC virulence.
4. MATERIALS AND METHODS
4.1. Bacterial strains and plasmids
The bacterial strains and plasmids used in this study are listed in Table 1. Longus-expressing ETEC E9034A wild-type strain is clinical isolate originally obtained from an individual with diarrhea in the Caribbean (20). Bacterial growth on solid or liquid media consisting of Luria agar, Triptone Soy Agar-Blood 5% (TSA-B), Luria broth (LB) or Terrific broth (TB) (25) used incubation temperatures of 30, 37, or 42°C dep ending on the assay. TB or TSA-B, were used at 37°C for optimal Longus express ion. The growth media was supplemented with chloramphenicol 25μg/mL, ampicillin 100 μg/mL, isopropyl β-D-1 galactopyranoside (IPTG) 40μg/mL, or L-arabinose 0.08% as needed.
Table 1.
Strains and plasmids used in this study.
| Strains/Plasmids | Features | SOURCE |
|---|---|---|
| E coli DH5α | Laboratory E. coli strain | (13) |
| E coli HB101 | Laboratory E. coli strain | (13) |
| E coli DH5α(pBluescript SK) | Control E. coli with plasmid vector. ampr | (13) |
| ETEC E9034A | Wild type ETEC strain, Longus pilus | (20) |
| E9034A(pKD46) | pKD46 plasmid electroporated into E9034A. ampr | This study |
| E9034A ΔlngA::FRT-cat (pKD46) | lngA:FRT-cat insertion mutant. catr and ampr, | This study |
| E9034A ΔlngA::FRT-cat | Plasmid pKD46 cured from E9034AΔlngA::cat (pKD46). catr | This study |
| E9034A ΔlngA::FRT-cat (pCP20) | pCP20 electroporated into E9034A ΔlngA::FRTcat. ampr and catr | This study |
| E9034A ΔlngA | cat cassette removed and pCP20 cured. amps and cats | This study |
| pCR2.1-ΔlngA::cat | ΔlngA::cat PCR product cloned into pCR2.1 | This study |
| pOG140 | pBluescript SKII carrying lngA; ampr | (13) |
| pBAD18 | Expression vector with AraC promoter; ampr | (15) |
| pAC-lngA | lngA gene cloned into pBAD18 under AraC promoter regulation; ampr | This study |
| pBAD-lngA::FRT-cat | ΔlngA::FRT-cat cloned into pBAD18; ampr and cmr | This study |
ampr: Ampicillin resistant. catr:chloramphenicol resistant.
4.2. Molecular biology techniques
Standard techniques of molecular biology including transformation and electroporation of bacterial strains with plasmid DNA, cloning, restriction analysis, and DNA sequencing was carry out as described before (25). Protein modeling prediction was performed by using the Swissprotein web-based software at www.expasy.org/spdbv/ (version 4.0.1) (1, 27).
The Lambda Red recombination system (6) was used to construct a site-directed mutagenesis of the lngA gene, described in the Results section. The gene disruption consisted of a marker exchange of the wild-type lngA for a truncated lngA with a chloramphenicol acetyl transferase (cat) cassette in the middle, and flanked by a 65bp sequence repeat designated FLP recombination target (FRT). FLP, a Saccharomyces cerevisiae recombinase protein with specificity for the FRT repeats (3), allows the excision DNA fragment located between repeats. The FLP-mediated excision of the cat cassette from the truncated lngA resulted in the lngA isogenic mutant of E9034A.
Fragment DNA amplification for construction of recombination DNAs, or for DNA analysis used polymerase chain reaction (PCR) using oligonucleotide primers obtained from Integrated DNA Technologies (IDT, Corallville, IA) (Table 2). PCR reaction used 1 μL (50 ng) of template, 40 pmols of each primer mix with 0.5mM MgCl2 and 2X Platinum Blue Mix Taq polymerase (Invitrogen, Carlbad, CA) at a final concentration 1X, in 100 μL final reaction volume. The PCR program cycle consisted of an initial denaturation step at 94 °C for 5 min, followed by 35 cycles of denaturation for 30 s at 94 °C, annealing for 30 s at 58°C and extension for 45 s at 72°C. The amplified product was analyzed by agarose electrophoresis. DNA amplicons were purified by Qiaquick gel extraction kit (QIAGEN, Valencia, CA) for further experiments including cloning or recombination experiments.
Table 2.
Oligonucleotides used in this study for PCR DNA amplification.
| Oligonucleotides* | Sequence | Targets | SOURCE |
|---|---|---|---|
| LngA-1F | 5′-ATGCTATCCGTGTATAACCG-3′ | lngA gene | This study |
| LngA-708R | 5′-ACGGCTACCTAAAGTAATTG-3′ | lngA gene | This study |
| C1 | 5′-ATATTTGCCCATGGTGAAAA-3′ | cat cassette | (6) |
| pKD3/lngA-F | 5′-CAACAGGAGGAATACTTATGCTATCCGTGTATAACCGGACACAGGTGTAGGCTGGAGCTGCTTC-3′ | cat, FRT, lngA | This study |
| pKD3/lngA-R | 5′-CGGCTACCTAAAGTAATTGAGTTTACCTGAGCAGTACAGGTACTATGGGAATTAGCCATGGTCC-3′ | cat, FRT, lngA | This study |
Oligonucleotides were obtained from IDT (Coralville, IA)
4.3. Construction of an insertionally inactivated lngA gene
The construction of an lngA mutant carrying the acetyl-transferase (cat) gene cassette, conferring resistance to chloramphenicol, was carried out by PCR. Tailing amplification of the cat cassette from the pKD3 plasmid template using forward (pKD3/lngA-F) and reverse (pKD3/lngA-R) primers is described in Table 2. Primers containing the 44 nucleotides upstream or downstream lngA and the 22 nucleotides upstream or downstream the FRT-flanked cat cassette of pKD3 plasmid, were used. The resulting 1,120-bp amplified PCR product designated ΔlngA::FRT-cat was cloned into the pCR2.1 vector to generate the pCR2.1-ΔlngA::FRT-cat plasmid.
4.4. Immunobloting
The immunoblots were carried out as previously described (12). Briefly, 1×108 colony-forming units (CFU) were harvested from liquid or solid media, washed in phosphate-buffered saline (PBS) pH 7.4, resuspended in 80 μL of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sampling buffer and boiled for 5 min. Then10 μL of crude extract were used for SDS-PAGE in 12% polyacrylamide gels. Proteins were transferred in a Trans-Blot Semi-Dry system (Bio-Rad, Hercules, CA) to nitrocellulose membranes (Millipore, Bradford, MA). LngA was probed overnight with a murine monoclonal anti-LngA antibody (1:3000) (13) in PBS pH7.4 plus 5% dry milk. Goat anti-mouse/horseradish peroxidase conjugate (Jackson ImmunoResearch, West Grove, PA) diluted 1:10,000 was used as a secondary antibody. Signal detection from blots was carried out by chemioluminescence using the ECL Plus western blotting system (GE healthcare, Piscataway, NJ).
4.5. Electron Microscopy
Transmission electron microscopy (TEM) experiments were performed for visualization of type IV pilus. Bacteria grown on solid media were resuspended carefully in PBS pH 7.4 and stained with phosphotungstic acid on Formvar carbon coated copper grids 200 mesh (Ted Pella, Redding, CA) before examination under TEM. Immunogold labeling of longus pilus was carried out as described before (9). Bacteria samples were fixed with 2.5% glutaraldehyde and rinsed with PBS. Samples were incubated in primary antibody (mouse monoclonal anti-lngA) for 1 h followed by rinsing with PBS. The secondary antibody, consisting of 10nm Gold particles conjugated to goat anti-mouse antibody, were added to sample, incubated for 2 h, and fixed with 2.5% glutaraldehyde for 2 min, and rinsed with PBS. The final step consisted of 2.5% uranyl acetate negative staining for 1 min before visualization with TEM.
For scanning electron microscopy (SEM) experiments, bacteria were grown in liquid media (TB for optimal longus expression) to log phase before being fixed with 2.5% glutaraldehyde for 60 min. Bacteria were then transferred onto poly-L-lysine-treated silicon wafers (Ted Pella, Inc., Redding, CA) and incubated at 37°C for 30 min. Bacteria were fixed again with 2.5% glutaraldehyde for 30 min, rinsed three times with 0.1 M Na cacodylate buffer, and negatively stained with 1% Osmium tetroxide for 1 h followed by three 15-min rinses. Samples were dehydrated in a graded series of ethanol (25%, 50%, 75%, 95%, 100%) and hexamethyldisilazane (Polysciences Inc, Warrington, PA). Finally, bacteria samples were sputter-coated before examination with SEM.
4.6. Self-aggregation assay
Bacteria self-aggregation phenotype consisting of bacterial clumping during growth in liquid media was analyzed using flat-bottom 24-well tissue culture plates (BD Biosciences, San Jose, CA). For this purpose a 1:100 dilution of an over night culture was incubated at 37 °C in TB or LB for 4h. Visualization of bacterial micro-colonies was noticed by naked eye. Recording of self-aggregates was done directly on bacterial suspension by a digital camara, or by an inverted light microscope IX81 with DP70 camera (Olympus, Center Valley, PA).
4.7. Bacterial survival in the presence of antimicrobial agents
ETEC strains able or unable of forming self-aggregates were tested for the ability to survive antibacterial factors. 100 μl of TB or LB media in 96 well plates were inoculated with overnight cultures of ETEC bacterial cultures and incubated at 37°C for 4 hours. This period was sufficient for longus-expressing ETEC strains to form self-aggregates. After 4h, stress factors were added to wells containing bacterial suspensions and incubation continued at 37°C for se veral hours. Different concentrations of stress factors were tested, and for different time periods. Concentrations used were equal or above the minimal inhibitory concentration (MIC). Quantification of surviving bacteria was performed by plating bacterial suspensions from two-fold serial dilutions on LA plates and counting CFU after overnight incubation at 37C. Qualitative evaluation of survival was done by plating suspensions on LA plate and visualizing bacterial growth.
The MIC for each antimicrobial was determined by a dilution method using either LB or TB media. In brief, 2μl of overnight bacterial culture were used to inoculate 200μl of LB or TB broth in 96-well plates and two-fold serial dilutions of stress factors added before incubation at 37C. The MIC for each one of the antimicrobials on ETECs grown on LB or LB was as follows: Chloramphenicol (ACROS, Morris Plains, NJ), 8 μg/ml for LB and 32 μg/ml for TB; Kanamycin (Fisher, Pittsburg, PA), 64 μg/ml for LB and 128 μg/ml for TB; Tetracycline (Rpi, Prospect, IL), 8 μg/ml for LB and 32 μg/ml for TB; Gentamicin (Cellgro, Herdon, VA), 32 μg/ml for LB and 64 μg/ml for TB; and Lysozyme (Sigma, St Lois, MO), 256 μg/ml for LB and 512 μg/ml for TB. The MIC was defined as the lowest concentration of antimicrobial where not visibly turbidity was detected after 16 h of incubation at 37°C. Concentrations of antimicrobial used in survival experiments varied from 1X to 100X MICs. All experiments were done several times and in triplicate.
Figure 2. Deletion mutation of the lngA gene.
Panel A. Diagram of the cat cassette insertion into the lngA gene by homologous recombination. Diagram of the wild-type lngA gene and a recombinant lngA:cat DNA fragment that by homologous recombination event result in the lngA:cat insertion mutant. Primers pKD3-lngA-F/pKD3-lngA-R to construct lngA::FRT-cat exchange marker and primers lngA-F/lngA-708R to test for lngA sequences are shown. Panel B. Detection of lngA:cat DNA recombination by PCR amplification. i) DNA PCR amplification for detection of lngA DNA using primer set lngA-F/lngA708-R. Lane 1: E. coli DH5α; Lane 2: DH5α pKD46; Lane 3: DH5α pKD3; Lane 4: E9034A; Lane 5: E9034A(pKD46); Lane 6: DH5α pAClngA:FRT-cat; Lane 7: E9034AlngA:FRT-:cat. Arrows indicate DNA fragments of 1.1 and 0.7 kb. ii) DNA PCR amplification for detection of cat insertion using primer set C1-lngAR PCR. Lanes 1 to 7: same as panel B-I above. Arrow indicates a 0.84-Kb DNA fragment. Panel C. Diagram the isogenic lngA deletion mutation construction, showing the Flp-mediated cat cassette excision by homologous recombination at the FRT sites. The resulting lngA deletion mutant contains an FRT site in the middle of lngA. Primer c1/lngA-708R for recognition of cat sequences and primers lngA-F/lngA708R for detection of lngA sequences are shown. Panel D. Detection of lngA deletion mutation by DNA PCR amplification. i) PCR amplification for detection of lngA DNA using primer set lngA-F-lngA708R. Lane 1: DH5α; Lane 2: DH5α pCP20; Lane 3: E9034A; Lane 4: E9034AlngA::FRT-cat; Lane 5: E9034AlngA::FRT-cat(pCP20); Lane 6: E9034AΔlngA. Arrows indicate 1.1, 0.7, and 0.19-Kb DNA fragments. ii) PCR amplification for detection of cat DNA using primers C1-lngA708-R. Lanes 1 to 6: same as panel D-i above. Arrow indicates a 0.84-Kb DNA fragment. M represents molecular weight markers (Bioline, Tauton, MA). Panel E. Expression of LngA. The expression of LngA was determined by immunoblot in whole cells lysates using a anti-Longus monoclonal antibody. Lane 1, E coli DH5α(pAClngA); Lane2, E coli DH5α; Lane 3, E coli HB101; Lane 4, E9034A; Lane 5, E9034AΔlngA; Lane 6, E9034AΔlngA(pAC-lngA); Lane 7, E9034AΔlngA::FRT-cat; Lane 8, E9034AΔlngA::FRT-cat (pAC-lngA). Arrows indicate the 28 kDa LngA prepilin and the 22 kDa mature LngA pilin.
Acknowledgments
This work was supported in part by the Children’s Miracle Network grant 1892, 2007 University of Iowa Children’s Hospital, the NIAID-NIH clinician scientist mentored research award (K08) No. KAI079410A, and the Robert Wood Johnson Foundation award through the Harold Amos Faculty Development Program to O.G.G.-D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 2.Bieber D, Ramer SW, Wu CY, Murray WJ, Tobe T, Fernandez R, et al. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–8. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 3.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 4.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–22. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 5.Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, et al. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell. 2003;11:1139–50. doi: 10.1016/s1097-2765(03)00170-9. [DOI] [PubMed] [Google Scholar]
- 6.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–5. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Folkhard W, Marvin DA, Watts TH, Paranchych W. Structure of polar pili from Pseudomonas aeruginosa strains K and O. J Mol Biol. 1981;149:79–93. doi: 10.1016/0022-2836(81)90261-8. [DOI] [PubMed] [Google Scholar]
- 8.Giron JA, Gomez-Duarte OG, Jarvis KG, Kaper JB. Longus pilus of enterotoxigenic Escherichia coli and its relatedness to other type-4 pili--a minireview. Gene. 1997;192:39–43. doi: 10.1016/s0378-1119(97)00039-5. [DOI] [PubMed] [Google Scholar]
- 9.Giron JA, Ho AS, Schoolnik GK. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science. 1991;254:710–3. doi: 10.1126/science.1683004. [DOI] [PubMed] [Google Scholar]
- 10.Giron JA, Levine MM, Kaper JB. Longus: a long pilus ultrastructure produced by human enterotoxigenic Escherichia coli. Mol Microbiol. 1994;12:71–82. doi: 10.1111/j.1365-2958.1994.tb00996.x. [DOI] [PubMed] [Google Scholar]
- 11.Giron JA, Viboud GI, Sperandio V, Gomez-Duarte OG, Maneval DR, Albert MJ, et al. Prevalence and association of the longus pilus structural gene (lngA) with colonization factor antigens, enterotoxin types, and serotypes of enterotoxigenic Escherichia coli. Infect Immun. 1995;63:4195–8. doi: 10.1128/iai.63.10.4195-4198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez-Duarte OG, Chattopadhyay S, Weissman SJ, Giron JA, Kaper JB, Sokurenko EV. Genetic diversity of the gene cluster encoding longus, a type IV pilus of enterotoxigenic Escherichia coli. J Bacteriol. 2007;189:9145–9. doi: 10.1128/JB.00722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez-Duarte OG, Ruiz-Tagle A, Gomez DC, Viboud GI, Jarvis KG, Kaper JB, et al. Identification of lngA, the structural gene of longus type IV pilus of enterotoxigenic Escherichia coli. Microbiology. 1999;145 ( Pt 7):1809–16. doi: 10.1099/13500872-145-7-1809. [DOI] [PubMed] [Google Scholar]
- 14.Gutierrez-Cazarez Z, Qadri F, Albert MJ, Giron JA. Identification of enterotoxigenic Escherichia coli harboring longus type IV pilus gene by DNA amplification. J Clin Microbiol. 2000;38:1767–71. doi: 10.1128/jcm.38.5.1767-1771.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–30. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyland RM, Griener TP, Mulvey GL, Kitov PI, Srivastava OP, Marcato P, et al. Basis for N-acetyllactosamine-mediated inhibition of enteropathogenic Escherichia coli localized adherence. J Med Microbiol. 2006;55:669–75. doi: 10.1099/jmm.0.46344-0. [DOI] [PubMed] [Google Scholar]
- 17.Hyland RM, Sun J, Griener TP, Mulvey GL, Klassen JS, Donnenberg MS, et al. The bundlin pilin protein of enteropathogenic Escherichia coli is an N-acetyllactosamine-specific lectin. Cell Microbiol. 2008;10:177–87. doi: 10.1111/j.1462-5822.2007.01028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knutton S, Shaw RK, Anantha RP, Donnenberg MS, Zorgani AA. The type IV bundle-forming pilus of enteropathogenic Escherichia coli undergoes dramatic alterations in structure associated with bacterial adherence, aggregation and dispersal. Mol Microbiol. 1999;33:499–509. doi: 10.1046/j.1365-2958.1999.01495.x. [DOI] [PubMed] [Google Scholar]
- 19.Kosek M, Bern C, Guerrant RL. The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull World Health Organ. 2003;81:197–204. [PMC free article] [PubMed] [Google Scholar]
- 20.Levine MM, Ristaino P, Marley G, Smyth C, Knutton S, Boedeker E, et al. Coli surface antigens 1 and 3 of colonization factor antigen II-positive enterotoxigenic Escherichia coli: morphology, purification, and immune responses in humans. Infect Immun. 1984;44:409–20. doi: 10.1128/iai.44.2.409-420.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mundy R, Pickard D, Wilson RK, Simmons CP, Dougan G, Frankel G. Identification of a novel type IV pilus gene cluster required for gastrointestinal colonization of Citrobacter rodentium. Mol Microbiol. 2003;48:795–809. doi: 10.1046/j.1365-2958.2003.03470.x. [DOI] [PubMed] [Google Scholar]
- 22.Nishimura LS, Giron JA, Nunes SL, Guth BE. Prevalence of enterotoxigenic Escherichia coli strains harboring the longus pilus gene in Brazil. J Clin Microbiol. 2002;40:2606–8. doi: 10.1128/JCM.40.7.2606-2608.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park HS, Wolfgang M, van Putten JP, Dorward D, Hayes SF, Koomey M. Structural alterations in a type IV pilus subunit protein result in concurrent defects in multicellular behaviour and adherence to host tissue. Mol Microbiol. 2001;42:293–307. doi: 10.1046/j.1365-2958.2001.02629.x. [DOI] [PubMed] [Google Scholar]
- 24.Pichel MG, Binsztein N, Qadri F, Giron JA. Type IV longus pilus of enterotoxigenic Escherichia coli: occurrence and association with toxin types and colonization factors among strains isolated in Argentina. J Clin Microbiol. 2002;40:694–7. doi: 10.1128/JCM.40.2.694-697.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Russell DW. Molecular Cloning a Laboratory Manual. 3. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 26.Scaletsky IC, Silva ML, Trabulsi LR. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect Immun. 1984;45:534–6. doi: 10.1128/iai.45.2.534-536.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–5. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strom MS, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–96. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 29.Taniguchi T, Akeda Y, Haba A, Yasuda Y, Yamamoto K, Honda T, et al. Gene cluster for assembly of pilus colonization factor antigen III of enterotoxigenic Escherichia coli. Infect Immun. 2001;69:5864–73. doi: 10.1128/IAI.69.9.5864-5873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taniguchi T, Fujino Y, Yamamoto K, Miwatani T, Honda T. Sequencing of the gene encoding the major pilin of pilus colonization factor antigen III (CFA/III) of human enterotoxigenic Escherichia coli and evidence that CFA/III is related to type IV pili. Infect Immun. 1995;63:724–8. doi: 10.1128/iai.63.2.724-728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tobe T, Sasakawa C. Role of bundle-forming pilus of enteropathogenic Escherichia coli in host cell adherence and in microcolony development. Cell Microbiol. 2001;3:579–85. doi: 10.1046/j.1462-5822.2001.00136.x. [DOI] [PubMed] [Google Scholar]
- 32.Vanmaele RP, Armstrong GD. Effect of carbon source on localized adherence of enteropathogenic Escherichia coli. Infect Immun. 1997;65:1408–13. doi: 10.1128/iai.65.4.1408-1413.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]