Abstract
The type III secretion system of P. aeruginosa, responsible for acute infection, is composed of over twenty proteins that facilitate cytotoxin injection directly into host cells. Integral to this process is production and secretion of PcrV. Administration of a recently developed, anti-PcrV immunoglobulin, either as a therapeutic or prophylactic has previously demonstrated efficacy against laboratory strains of P. aeruginosa in a murine model. To determine if this therapy is universally applicable to a variety of P. aeruginosa clinical isolates, genetic heterogeneity of pcrV was analyzed among strains collected from three geographically distinct regions; United States, France and Japan. Sequence analysis of PcrV demonstrated limited variation among the clinical isolates examined. Strains were grouped according to the presence of non-synonymous single nucleotide polymorphisms. Representative isolates from each mutant group were examined for the ability of anti-PcrV to bind the protein secreted by these strains. The protective effect of anti-PcrV IgG against each strain was determined using an epithelial cell line cytotoxicity assay. The majority of strains tested demonstrated reduced cytotoxicity in the presence of anti-PcrV IgG. This study provides insights into the natural sequence variability of PcrV and an initial indication of the amino acid residues that appear to be conserved across strains. It also demonstrates the protective effect of anti-PcrV immunotherapy against a multitude of P. aeruginosa strains from diverse global regions with a variety of mutations in PcrV.
Keywords: Pseudomonas aeruginosa, type III secretion, anti-PcrV IgG, Single Nucleotide Polymorphism, Cytotoxicity
Introduction
Pseudomonas aeruginosa is an opportunistic gram-negative pathogen capable of eliciting acute or chronic infection of immunocompromised, burn and cystic fibrosis patients. P. aeruginosa is one of the leading causes of nosocomial infections, with a mortality rate as high as 60% among ICU patients (2-4). Although the introduction of antibiotics with anti-pseudomonal activity demonstrated initial promise, this versatile bacteria’s ability to develop resistance to multiple antimicrobials has become a significant obstacle to treating infections associated with P. aeruginosa. Given the great infectious threat to hospitalized patients, alternative means are necessary to treat and prevent acute P. aeruginosa infections.
Extensive research since the 1960’s on mechanisms of P. aeruginosa pathogenesis led to the discovery of a wide variety of virulence-associated factors. Several of these factors were used as candidates for immunotherapy or immunoprophylaxis to combat P. aeruginosa infections. Antibodies against antigenic molecules such as multivalent lipopolysaccharides (LPS), exotoxin A, flagella, mucoid exopolysacharide, and outer membrane protein had shown potential benefits against P. aeruginosa both in vitro and in vivo during initial investigations (14, 24). However, these vaccines never gained clinical acceptance due to a wide array of problems (24).
Although no P. aeruginosa vaccine has yet made its way into clinical practice, the relatively recent identification of another virulence factor, the type III secretion system (TTSS), associated with acute infection by this species (17, 14), has led to development of a novel immunotherapy effective against laboratory strains of this bacterial species (28). The TTSS is a mechanism widely exploited by gram-negative pathogens to intoxicate eukaryotic cells and permits successful multiplication of the pathogen in the host (16). This system, which involves direct injection of cytotoxins through a needle complex into eukaryotic epithelia, is necessary for P. aeruginosa virulence in acute lung injury and burn wound infection mouse models (17, 14). PcrV, a homologue of the Yersinia V-antigen LcrV, has been recognized as an important structural translocation component of the P. aeruginosa TTSS. Secretion of PcrV facilitates delivery of cytotoxins [ExoU, ExoS, ExoT and ExoY; (27)] that interfere with host cell signal transduction and a variety of cellular processes, resulting in cell death or significant alterations in the host immune response.
The protective antigenic characteristics of LcrV were reported more than 40 years ago (3, 4). Similarly, our previous studies have demonstrated that active immunization with PcrV mediates protection against lethal P. aeruginosa infection in several animal models (28). Further, a murine monoclonal anti-PcrV antibody, Mab166, was developed and its protective effects on acute lung injury demonstrated when it was co-instilled with bacterial inoculum or passively transferred to infected animals (12). The administration of either Mab166 or Fab of Mab166 exhibited comparable therapeutic effects to rabbit polyclonal anti-PcrV IgG in vivo (11, 30). The success of both active and passive PcrV immunization in animal models led to the current development of PcrV and anti-PcrV IgG as a potential human vaccine and therapeutic agent respectively, against P. aeruginosa infection.
Amino acid sequence divergence in the protective antigenic region of LcrV of Yersinia enterocolitica O8 exists (22). Therefore, serum raised against one form of Yersinia does not cross-protect the host against Yersinia enterocolitica O8 infection (22, 29, 31). In the case of PcrV, the blocking epitope for Mab166 was previously confirmed to lie between amino acid 144 - 257 of PcrV (12). For anti-PcrV immunotherapy to be universally effective against all clinical strains of P. aeruginosa, amino acid sequence of the antigenic region should be highly conserved or, if amino acid variations exist they should not compromise anti-PcrV IgG binding capacity. Our current study was constructed in an attempt to avoid some of the pitfalls of the LcrV immunotherapy, and to serve as a proof of principle for anti-PcrV applicability against a range of clinical isolates. We investigated polymorphisms in the pcrV coding sequence of P. aeruginosa strains isolated from various patient samples (blood, urine and airway samples) in three geographically distinct regions and determined resulting amino acid alterations. Isolates were grouped based on amino acid sequence and the ability of anti-PcrV IgG to bind to the PcrV variants was assessed. In addition, cytotoxicity of each strain in the presence and absence of anti-PcrV IgG in vitro was determined. This study provides insight into the sequence variability of PcrV and demonstrates efficacy of anti-PcrV IgG administration against a range of clinical isolates. It also provides a platform for targeted amino acid investigations into the molecular function of this crucial Type III secretion protein.
Materials and Methods
Bacterial strains used in this study
DNA and amino acid sequences of the pcrV gene were analyzed from a total of 90 P. aeruginosa isolates, collected from blood, urine and respiratory sources in three globally distinct regions, United States (28 isolates) France (32 isolates) Japan (30 isolates). Each isolate was collected from an individual patient.
RAPD PCR Analysis of P. aeruginosa isolates
As the US cohort had previously been shown to be non-clonal by RAPD PCR analysis (1) P. aeruginosa isolates from the French and Japanese cohorts were analyzed for clonality. Strains were cultured in 200 μl of LB medium in 96-well plates overnight at 37°C without shaking. Cultures were diluted 1:10 or 1:100 in sterile water. Reactions consisted of either RAPD primer 272 or 208 (5′-AGCGGGCCAA-3′ or 5′-ACGGCCGA CC-3′ respectively; 25 μM final concentration in 5 μl), 5μl diluted culture, 25μl of 2x Taq PCR Mastermix (Qiagen, CA) and 15μl sterile water. Reactions were performed in a GeneAmp thermocycler (Applied Biosystems, CA) as follows: 95°C, 5 min, followed by 45 cycles of 95°C, 1 min, 32°C, 1 min 72°C 1 min. PCR products were visualized on a 1.5% agarose gel stained with ethidium bromide and viewed under UV light. Strains were considered clonal if the entire banding pattern was identical.
Determination of gene heterogeneity
Bacterial genomic DNA was purified from each isolate using a genomic DNA purification kit (Clontech, CA). The coding region of pcrV is 884 bp in length, the entire gene was amplified using a specific primer set designed just outside the coding region: [PVOT1-5 (nt -29 upstream of pcrV) 5′-TGCGTGGCTTGTTGATCTGA-3′; PVOT1-3 (nt +904 downstream of pcrV) 5′-TGCTGGTCGGTGTCGGAA-3′). Reactions contained 15 pmol of each primer, 1 μl of genomic DNA, 25 μl 2 x iProof HF mastermix (Biorad, CA; to assure faithful amplification of PvrV gene sequence) and 14 μl H2O. PCR reactions were carried out as follows: initial denaturation at 94° C for 5 min; 36 cycles of 94° C for 30 s, 52° C for 30 s, 72° C for 1 min; and a final extension step at 72° C for 7 min. Amplified pcrV from each isolate was cloned into the plasmid vector pCR-Blunt II-TOPO (Invitrogen, CA) and sequenced in both directions by the University of California San Francisco Biomolecular Resource Center. The PcrV sequence of PAO1 (previously published in the Pseudomonas Genome Project) was homologous to that of our laboratory PAO1 PcrV sequence and was therefore used as the reference strain for all sequence comparisons.
Analysis of pcrV sequence
Sequence translation was performed using the nucleic acid to amino acid translation program at: http://www.biochem.ucl.ac.uk/cgi-bin/mcdonald/cgina2aa.pl. Nucleic acid and translated amino acid sequences were analyzed using ClustalX freeware available for download at: http://www.itc.virginia.edu/achs/molbio/servers/free_software.html.
Multiplex PCR Analyses of TTSS genotype
P. aeruginosa clinical isolates were cultured in 200 μl Luria broth (LB) in 96-well plates overnight at 37°C without shaking. Cultures were diluted 1:10 or 1:100 in sterile water prior to addition into the PCR mixture. Multiplex PCR reactions consisted of 5 μl diluted culture, 3.75 pmol of each primer, (exoUF, 5′-CCGTTGTGGTGCCGTTGAAG-3′, exoUR 5′-CCAGATGTTCACCGACTCGC; exoSF, 5′-GCGAGGTCAGCAGAGTAT-3′, exoSR 5′-TTCGGCGTCACTG TGGATGC-3′; exoYF, 5′-CGGATTCTA TGGCAGGGAGG-3′, ExoYR 5′-GCCCTTGATGCACTCGACCA-3′; exoTF, 5′-AATCGCCGTCCAACTGCATGCG-3′, exoTR 5′-TGTTCGCCGAGGTACTGCTC-3′) 12.5 μl of Taq PCR Mastermix (Qiagen, CA) and 5.5 μl of sterile H2O to give a final volume of 25 μl. Reactions were performed in a GeneAmp thermocycler (Applied Biosystems, CA) as follows: 94°C, 2 min, followed by 36 cycles of 94°C, 30 sec, 58°C, 30 sec, 68°C 1 min and a final extension step of 68°C for 7 min. PCR products were visualized on a 3% agarose gel, stained with ethidium bromide and viewed under UV light.
Immunoblot analysis of PcrV, ExoS and ExoU production
Representative isolates from each of the mutant groups were cultured under TTSS-inducing conditions in MIN-S medium (21). Cultures were incubated with shaking overnight at 37 °C prior to dilution to OD600 of 0.1 in fresh MIN-S medium and cultured for a further 5 h. Bacterial cells were harvested by centrifugation and the supernatant removed. Cell-free supernatant from each sample was concentrated using Centricon tubes (MWCO 10 KDa; Millipore, MA). The cell pellet was divided in half and either lysed using SDS lysis buffer and boiling or resuspended in 50 mM potassium phosphate buffer (pH 8.0) and sonicated three times for 30 sec. Concentration of protein in all preparations was determined by the Biorad Dc protein quantification kit (Biorad, CA). Standardized total protein concentrations (10 μg) were loaded onto polyacrylamide gels and run under either native or denaturing conditions. Polyacrylamide gels were transferred to PVDF membrane and immunoblotted with anti-PcrV, anti-ExoS or anti-ExoU as previously described (23).
Quantification of pcrV expression by RT-PCR
Bacterial strains were cultured as described above for immunoblot analysis. Total RNA was extracted using Trizol (Invitrogen, CA) according to manufacturer’s instructions. RNA was treated with DNaseI (RNase-free; Promega, WI) and 40 cycles of PCR were carried out to confirm absence of DNA contamination. Total RNA (1 μg) was reversed transcribed using Multiscribe (Applied Biosystems, CA). Q-PCR reaction mixtures contained 2 μl of cDNA, 5 μM each of pcrV primers (pcrVF 5′-GACAAGGTCAACGAGAAG-3′; pcrVR 5′-TGAGAATGTCGCGCAGGA-3′), 12.5 μl SYBR green master mix (Applied Biosystems, CA) and 5.5 μl of H2O to give a final volume of 25 μl. The PCR reactions were performed on Stratagene Max3000P (Stratagene, CA) as follows, a 10 min denaturation step at 95 °C, followed by 40 cycles of 30 s at 95 °C, 1 min at 58 °C and 30 s at 72 °C. 2−ΔΔCT was determined for each representative strain using rpoD as internal control.
Protective effect of anti-PcrV IgG against P. aeruginosa cytotoxicity
Two hundred μl of human immortalized BEAS-2B lung epithelial cells were seeded at a density of 2.5 × 105 ml−1 in Dulbeccos Modified Eagle Medium supplemented with 10% BCS in 96-well cultures and incubated overnight to reach confluency. Wells were divided into test and control for each strain. Test wells received 10 μg of anti-PcrV IgG in PBS, control wells received control IgG (10 μg in PBS). Plates were incubated with antibodies for 1 h prior to addition of bacterial inoculum. Each of the representative clinical isolates (from NSS groups outlined in Table 2) was cultured overnight in LB medium. Cells were pelleted, resuspended and washed three times in sterile PSB, diluted to an OD600 of 0.1 and regrown in LB medium for a further 1.5 h. Following this, cultures were harvested, washed with Lactated Ringers and finally resuspended in 100 μl Ringers:PBS solution (2:1 ratio by volume). Each well received 5 × 106 bacteria. Cytotoxicity was assayed 2 h post infection by lactate dehydrogenase release using the Cyto Tox96® kit (Promega, CA) according to the manufacturers instructions.
Table 2.
NSS’s present within the amino acid sequence of PcrV
| Substitution | Total number of strains with NSS |
Frequency in each cohort |
||
|---|---|---|---|---|
| Japanese | US | French | ||
| L6F | 30 | 7 | 14 | 9 |
| A9G | 16 | 5 | 4 | 7 |
| A9S | 2 | 2 | 0 | 0 |
| A19D | 1 | 0 | 1 | 0 |
| S21P | 32 | 8 | 10 | 14 |
| E30K | 2 | 2 | 0 | 0 |
| Q83H | 1 | 1 | 0 | 0 |
| H107Y | 2 | 1 | 0 | 1 |
| Y192Sa | 2 | 0 | 0 | 2 |
| S196Na | 1 | 0 | 1 | 0 |
| L211Va | 1 | 0 | 1 | 0 |
| L224Fa | 1 | 1 | 0 | 0 |
| S225Ra | 31 | 10 | 13 | 8 |
| S225Ka | 9 | 0 | 5 | 4 |
| S225Ga | 2 | 0 | 2 | 0 |
NSS’s within the mAB166 epitope of PcrV
Results
All isolates were tested for clonality by RAPD PCR. The US cohort had previously been confirmed non-clonal (1), the French and Japanese cohorts exhibited two strains (15 and 16) in each cohort that appeared clonal (Fig. 1A and B). To further test these strains a second RAPD PCR using an alternative primer (RAPD208) was carried out. This reaction demonstrated differential banding pattern in each of the strains (Fig. 1C). Therefore isolates in all three cohorts from geographically distinct regions were considered non-clonal.
Fig. 1.
Clonal analysis of the French (A) and Japanese (B) cohorts of P. aeruginosa clinical isolates by RAPD PCR. C. Confirmation of non-clonality of strains 15 and 16 from the French (F) and 15 and 16 from the Japanese (J) cohort by an alternative RAPD primer (RAPD 208). Results are representative of two independent reactions per isolate.
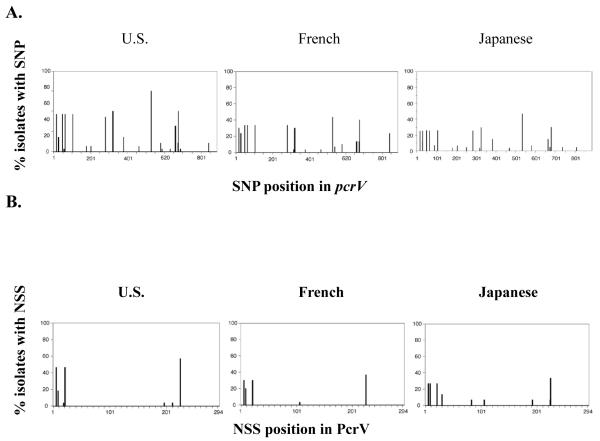
All strains encoded pcrV and no insertion or deletion mutations were detected in the coding region in any of the clinical isolates. Single nucleotide polymorphisms (SNP’s) were distributed relatively evenly throughout the pcrV gene across all three cohorts. Of the 90 isolates surveyed, a total of 33 SNP’s were identified in pcrV. The majority (63%) of these nucleotide sequence changes resulted in synonymous polymorphisms (i.e., the nucleotide change did not alter the amino acid sequence). However, approximately a third (37%) of the nucleotide changes were non-synonymous SNP’s (NSS’s; i.e. the nucleotide change resulted in an amino acid sequence alteration). The position and frequency of pcrV SNP’s and amino acid variations resulting from NSS’s are shown for each cohort in Figure 2A and B. Variations in pcrV and resulting changes in amino acid sequence (compared to the reference strain PAO1) are summarized in Table 1. Location of amino acid substitutions on the PcrV protein are illustrated in Figure 2C.
Fig. 2.
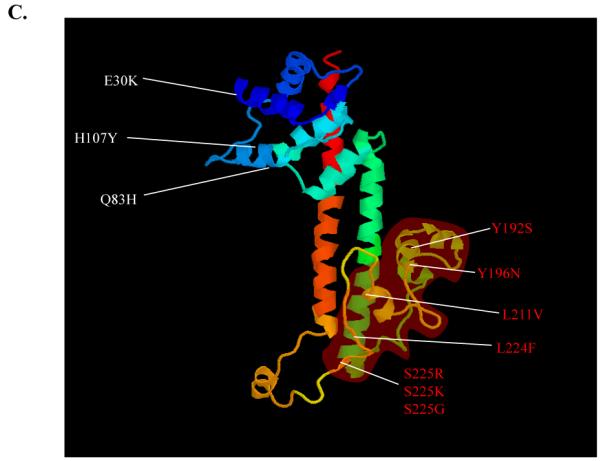
A. SNP and B. NSS analysis of clinical isolates in the US, French and Japanese cohorts. C. Model of PcrV protein from the Swissprot repository (http://swissmodel.expasy.org), indicating the positions of NSS’s. The Mab 166 epitope (amino acids 144-257) and mutations within this region are highlighted in red. The molecular model does not include the first 24 amino acids of the protein, NSS’s in this region are omitted.
Table 1.
Summary of gene sequence heterogeneity between various clinical isolate groups
| Isolate group | n | Total number of SNP’sa | Total number of NSS’sb |
|---|---|---|---|
| U.S.A | 28 | 21 | 7 |
| French | 32 | 17 | 5 |
| Japan | 30 | 21 | 9 |
|
| |||
| Combined | 90 | 59 | 12 |
SNP: single nucleotide polymorphisms
NSS: nonsynonymous single nucleotide polymorphisms.
The total number of NSS’s in all three cohorts combined was 14 (Table 2). Four of these NSS’s (L6F, 19G, S21P and S225R) were present in all three cohorts. The Japanese cohort exhibited the highest number of NSS’s (9 detected). Interestingly, although the US cohort exhibited a high frequency of SNP occurrence (21 SNP’s), only 7 NSS resulted from the nucleotide changes in this cohort. The French cohort displayed highest PcrV sequence conservation with 17 SNP’s resulting in only 5 NSS’s. The frequency of amino acid substitution was calculated at 1 substitution per 33, 42 or 59 amino acids for the Japanese, United States and French cohorts respectively indicating that PcrV is largely conserved across all geographic regions surveyed, particularly in the French cohort. Although SNP’s had been evenly distributed throughout the coding region, NSS’s appeared to cluster around the N-terminus and in the region between residues 192-225. To further examine aspects of secretion and anti-PcrV IgG protection, strains were grouped according to the NSS’s they encoded. Fifteen mutant groups resulted (Table 3), including group 1 which contained the reference strain, PAO1 and acted as control.
Table 3.
Groups representative of all NSS’s and combinations of NSS’s found in isolates from all three cohorts
| Group number | NSS | Number of strains in group |
|---|---|---|
| 1 | Reference strain (PAO1) | 45 |
| 2 | S225G | 2 |
| 3 | S196N, L211V | 1 |
| 4 | S225R | 2 |
| 5 | H107Y | 2 |
| 6 | L224F | 1 |
| 7 | Q84H, S225R | 1 |
| 8 | L6F, A9G, S21P, S225R | 11 |
| 9 | L6F, S21P, S225R | 6 |
| 10 | L6F, A9G, S21P, S225K | 4 |
| 11 | L6F, S21P, Y192S, S225K | 2 |
| 12 | L6F, S21P, A19D, S225R | 1 |
| 13 | A9G, S21P, S225R, | 1 |
| 14 | L6F, A9G, S21P, E30K, S225R | 2 |
| 15 | L6F, A9S, S21P, S225R | 2 |
In our previous study (1), we demonstrated that the epitope recognized by the blocking monoclonal antibody, Mab166, is located between amino acids 144 and 257 of the PcrV protein. Of the 14 NSS’s detected, 5 were located at this epitope (Fig. 2C). Four of these occurred at low frequency (Table 2). However, residue 225 a serine residue in PAO1 exhibited the highest frequency of substitution in all three cohorts. This residue was substituted in 42 strains (47% of total isolates). Amino acid substitutions at this site varied, 31 strains (34% of total cohort) possessed a S225R substitution, 9 strains (10%) had a S226K substitution and two strains (2%) possessed a S226G substitution.
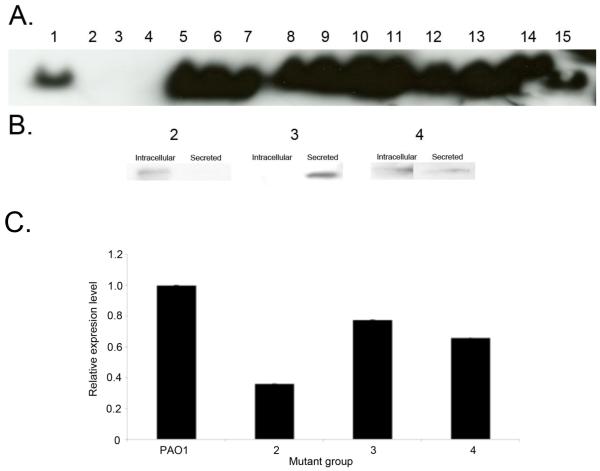
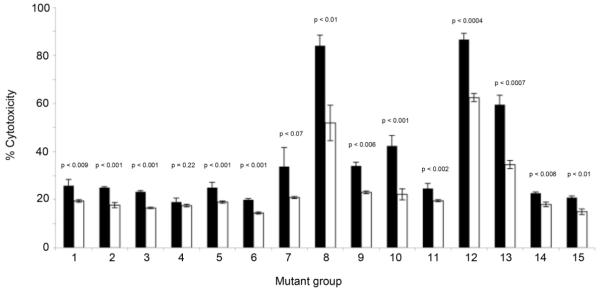
While it is important to consider the effect of amino acid substitutions at the protective epitope, substitutions outside this region could potentially disrupt PcrV protein conformation and affect secretion and/or binding of anti-PcrV IgG. Both of these aspects have the potential to affect efficacy of anti-PcrV immunotherapy. To examine the ability of various PcrV protein variants to bind anti-PcrV IgG, immunoblot analyses was performed. Native immunoblot analysis demonstrated that the majority of representative strains produced PcrV that could be detected in the secreted fraction (Fig. 3A), suggesting that these sequence variants readily bind anti-PcrV IgG. Notable exceptions included representative strains from groups 2, 3 and 4. To rule out a lack of binding due to reduced secretion, denaturing imunoblot analysis was performed on both intracellular or secreted total protein fractions of these strains. This demonstrated detctable PcrV in either the intracellular (mutant group 2) or extracellular (mutant groups 3 and 4) fraction, inferring binding to the anti-PcrV antibody (Fig. 3B). To confirm that these strains produced lower concentrations of PcrV, quantitative-PCR (Q-PCR) of pcrV mRNA was carried out for these strains and compared to PAO1. Representative strains from mutant groups 2, 3 and 4 exhibited significantly lower concentrations of pcrV mRNA compared to PAO1 (p < 0.0006; Fig. 3C). This suggests that the low level of signal produced by immunoblot analysis of these strains is likely due reduced pcrV transcription and consequent diminished PcrV production by these strains. To assess the ability of anti-PcrV immunoglobulin to block the TTSS system of all mutant groups, cytotoxicity of each of the strains in the presence of anti-PcrV IgG or control IgG was determined. Addition of 5 μg ml−1 anti-PcrV IgG to confluent BEAS-2B lung epithelial cell line prior to the addition of representative P. aeruginosa strains from each mutant group resulted in reduction of cytotoxicity towards mammalian cells for all but one mutant group (group 4; S255R substitution; Fig. 4). The most striking protection was observed with very cytotoxic strains from mutant groups 8, 12 and 13 (which also encode the S225R substitution in combination with a variety of other substitutions). In these cases treatment with anti-PcrV IgG resulted a marked reduction in cytotoxicity (mean decrease is 36% ± 4% SEM) of these isolates compared to treatment with a control IgG.
Fig. 3.
A. Native immunoblot analysis of representative strains of the PcrV NSS groups. B. Denaturing immunoblot analysis of intracellular and secreted PcrV fractions using increased total protein concentrations (30 μg) from representative strains of groups 2, 3 and 4. Western analysis results are representative of at least two independent experiments. C. Comparative Q-PCR analysis of relative pcrV expression [compared to pcrV expression of PAO1 (group 1)] by representative strains from mutant groups 1-4. Results are representative of three independent experiments carried out in triplicate.
Fig. 4.
Protective effect of anti-PcrV IgG (white bars) compared to control IgG (black bars) added prior to infection with representative strains from mutant groups 1-15. All but two strains (Groups 4 and 7) demonstrate a significant reduction in cytotoxicity in the presence of anti-PcrV IgG regardless of mutations present (p values provided above respective bars). Results are representative of at least two independent experiments carried out in triplicate.
Discussion
The 884 bp pcrV gene of P. aeruginosa encodes a protein of the P. aeruginosa Type III secretion system. Secretion of this protein is required for efficient intoxication of mammalian tissue by Type III cytotoxins. An anti-PcrV immunoglobulin has demonstrated protection against acute infection by laboratory strains of P. aeruginosa in several model systems (12, 20, 30) and is currently in clinical trials. PcrV is a V-antigen homolog of the LcrV protein of Yersinia pestis [causative agent of bubonic plague (9, 10)], and shares 57% amino acid sequence similarity with this protein. Initially, vaccination with truncates of LcrV represented a highly effective immunotherapy against both bubonic and pneumonic plague in animal models (13). Subsequently, however it was reported that antiserum against one strain of Yersinia spp (Y. enterocolitica serotype O8) did not confer immunity against other strains of Yersinia in vaccinated individuals (22). Further studies identified the existence of a hypervariable region between amino acids 225 and 232 of Yersinia LcrV (22). In this region, LcrV of Y. enterocolitica serotype O8 has a unique amino acid sequence encoding 9 extra amino acids (22). This hypervariable region represents the main sequence variability between LcrV proteins of Y. pestis strains, and is the primary cause for LcrV antiserum’s lack of cross-protective ability against different strains of Yersinia spp. To avoid the pitfalls of the Y. pestis anti-LcrV vaccine, we undertook a study of multiple non-clonal clinical isolates from various geographic regions to determine genetic variability of pcrV and examine if resulting alterations in the PcrV amino acid sequence could potentially reduce the efficacy of anti-PcrV binding and protection against P. aeruginosa type III-mediated virulence.
No deletion or insertion mutations were detected in any of the clinical isolates from any of the geographic regions surveyed which is in accordance with a previous pcrV gene analysis study on a small group of clinical isolates from patients in the United States (1). SNP analysis demonstrated 59 polymorphisms in pcrV across all three cohorts. The majority of these SNP’s did not alter the amino acid sequence of the protein; substitutions were detected at only 12 amino acid positions in the entire collection of isolates. This implies that PcrV is relatively conserved across geographically distinct regions.
Although no crystal structure exists for PcrV, molecular modeling based on sequence similarity to the corresponding regions of LcrV predicts that PcrV contains α-helical regions in its N- and C-termini (amino acids 109-156 and 227-294 respectively). The internal region containing the Mab166 epitope (amino acids 144-257) and its downstream carboxyl-terminal are the most antigenic regions of PcrV, while the N-terminal half (amino acids 1-138) is not required for optimal protection (12). This is supported by the observation that representative strains from several mutant groups exhibited diminished cytotoxicity in the presence of the anti-PcrV IgG despite the presence of a variety of mutations at their respective N-termini. The Mab166 epitope and C-terminal tail regions are thought to be important in preserving three dimensional structure that may be necessary to induce a protective immune response against P. aeruginosa infection. Mutations detected in the Mab166 epitope did not appear to affect the antigenic properties of PcrV, since multiple groups encoding individual and combinations of amino acid alterations in the antigenic region still exhibited PcrV binding and reduced cytotoxicity in the presence of this immunoglobulin. The importance of the C-terminal region in maintenance of structural integrity of the protein is underscored by the conservation of amino acid sequence in this region across all three cohorts (no amino acid substitutions were detected beyond residue 225).
The effect of amino acid residue changes on anti-PcrV IgG binding was assessed. Immunoblot analysis demonstrated that all strains produced PcrV although some groups (2, 3 and 4) produced very small quantities. Q-PCR analysis confirmed that mRNA levels of pcrV were significantly lower in representative strains from these groups when compared to PAO1, suggesting that reduced production of PcrV in these strains arose from an upstream transcriptional regulatory effect. This suggests that diminished immunoblot signal observed for these strains was due not to reduced anti-PcrV IgG binding but to a lack of PcrV protein. This is supported by in vitro cytotoxicity assays, these three strains were amongst the lest cytotoxic, presumably because they do not express or secrete sufficient concentrations of PcrV to facilitate appreciable intoxication of mammalian cells. However, addition of anti-PcrV IgG did reduce what little cytotoxicity there was, suggesting that the anti-PcrV IgG did bind and maintain efficacy against these strains.
Control of cytotoxin secretion of Y. pestis is thought to be due to LcrV (PcrV homolog) binding LcrG (PcrG homolog), formation of this heterodimer blocks cytotoxin secretion from this species (8, 18). PcrV rapidly associates (and dissociates) with PcrG in a 1:1 ratio (19), however the relevance of the PcrV:PcrG interaction to cytotoxin secretion is not yet known. Few of the molecules that interact with PcrV are known and it is believed that PcrV may play additional or alternative roles in P. aeruginosa compared to that of LcrV in Yersinia pestis. Further investigations to determine the contribution of individual and combinations of mutations identified in this study that naturally arise in a PcrV population of clinical isolates are essential to elucidating the basis for such interactions.
The SNP and NSS analyses presented here, strongly suggest that pcrV of P. aeruginosa is a relatively well-conserved gene regardless of the geographic source of clinical isolates. PcrV of the strains examined, unlike LcrV of Y. enterocolitica serotype O8, did not possess a hypervariable region that impacted immunoglobulin binding. The individual and combinations of polymorphisms that were detected in various forms of PcrV did not ameliorate the protective effect of anti-PcrV IgG against the majority of strains. These data suggest that the current PcrV antibody, which was originally designed based on P. aeruginosa PAO1 as the reference amino acid sequence represents an excellent immunotherapeutic option against the majority of P. aeruginosa strains worldwide.
ACKNOWLEDGEMENTS
This research was supported by National Institutes of Health (NIH) PH50HL74005 and HL69809 to J. P. W-K. NIH HL067600 & American Lung Association RG004N to T. S. and a UCSF Academic Senate Independent Investigator grant to S.V.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
BIBLIOGRAPHY
- 1.Allmond LR, Ajayi T, Moriyama K, Wiener-Kronish JP, Sawa T. V-antigen genotype and phenotype analyses of clinical isolates of Pseudomonas aeruginosa. J. Clin. Microbiol. 2004;42:3857–3860. doi: 10.1128/JCM.42.8.3857-3860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almirall J, Mesalles E, Klamburg J, Parra O, Agudo A. Prognostic factors of pneumonia requiring admission to the intensive care unit. Chest. 1995;107:511–516. doi: 10.1378/chest.107.2.511. [DOI] [PubMed] [Google Scholar]
- 3.Burrows TW. An antigen determining virulence in Pasteurella pestis. Nature. 1956;177:426–427. doi: 10.1038/177426b0. [DOI] [PubMed] [Google Scholar]
- 4.Burrows TW, Bacon GA. The basis of virulence in Pasteurella pestis: an antigen determining virulence. Br. J. Exp. Pathol. 1956;37:481–493. [PMC free article] [PubMed] [Google Scholar]
- 5.Craven DE, Steger KA. Nosocomial pneumonia in mechanically ventilated adult patients: epidemology and prevention in 1996. Semin Respir Infect. 1996;11:32–53. [PubMed] [Google Scholar]
- 6.Crouch Brewer S, Wunderink RG, Jones CB, Leeper KV., Jr. Ventilator-associated pneumonia due to Pseudomonas aeruginosa. Chest. 1996;109:1019–1029. doi: 10.1378/chest.109.4.1019. [DOI] [PubMed] [Google Scholar]
- 7.Dacheux D, Attree I, Toussaint B. Expression of ExsA in trans confers type III secretion system-dependent cytotoxicity on noncytotoxic Pseudomonas aeruginosa cystic fibrosis isolates. Infect. Immun. 2001;69:538–542. doi: 10.1128/IAI.69.1.538-542.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeBord KL, Lee VT, Schneewind O. Roles of LcrG and LcrV during type III targeting of effector Yops by Yersinia enterocolitica. J. Bacteriol. 2001;183:4588–4598. doi: 10.1128/JB.183.15.4588-4598.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duncan CJ, Scott S. What acused the black deadth? Postgrad. Med. J. 2005;81:315–320. doi: 10.1136/pgmj.2004.024075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elvin SJ, Eyles JE, Howard KA, Ravichandran E, Somavarappu S, Alpar HO, Williamson ED. Protection against bubonic and pneumonic plague with a single dose microencapsulated sub-unit vaccine. Vaccine. 2005;15:4433–4439. doi: 10.1016/j.vaccine.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 11.Faure K, Fujimoto J, Shimabukuro DW, Ajayi T, Shime N, Moriyama K, Spack EG, Wiener-Kronish JP, Sawa T. Effects of monoclonal anti-PcrV antibody on Pseudomonas aeruginosa-induced acute lung injury in a rat model. J. Immune Based Ther. Vaccines. 2003;1:2. doi: 10.1186/1476-8518-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank DW, Vallis A, Wiener-Kronish JP, Roy-Burman A, Spack EG, Mullaney BP, Megdoud M, Marks JD, Fritz R, Sawa T. Generation and characterization of a protective monoclonal antibody to Pseudomonas aeruginosa PcrV. J. Infect. Dis. 2002;186:64–73. doi: 10.1086/341069. [DOI] [PubMed] [Google Scholar]
- 13.Hill J, Leary SE, Griffin KF, Williamson ED, Titball RW. Regions of Yersinia pestis V antigen that contribute to protection against plague identified by passive and active immunization. Infect. Immun. 1997;65:4476–4482. doi: 10.1128/iai.65.11.4476-4482.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holder IA. Pseudomonas immunotherapy: a historical overview. Vaccine. 2004;22:831–839. doi: 10.1016/j.vaccine.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 15.Holder IA, Neely AN, Frank DW. PcrV immunization enhances survival of burned Pseudomonas aeruginosa-infected mice. Infect. Immun. 2001;69:5908–5910. doi: 10.1128/IAI.69.9.5908-5910.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hueck CJ. Type III protein secretion systems in bacterial pathogens of plants and animals. Microbiol. Mol. Biol. Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurahashi K, Kajikawa O, Sawa T, Ohara M, Gropper MA, Frank DW, Martin TR, Wiener-Kronish JP. Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J. Clin. Invest. 1999;104:743–750. doi: 10.1172/JCI7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matson JS, Nilles ML. LcrG-LcrV interaction is required for control of Yops secretion in Yersinia pestis. J Bacteriol. 2001;183(17):5082–5091. doi: 10.1128/JB.183.17.5082-5091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nanao M, Ricard-Blum S, Di Guilmi AM, Lemaire D, Lascoux D, Chabert J, Attree I, Dessen A. Type III secretion proteins PcrV and PcrG from Pseudomonas aeruginosa form a 1:1 complex through high affinity interactions. BMC Microbiol. 2003;3:21. doi: 10.1186/1471-2180-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neely AN, Holder IA, Wiener-Kronish JP, Sawa T. Passive anti-PcrV treatment protects burned mice against Pseudomonas aeruginosa challenge. Burns. 2005;31:153–158. doi: 10.1016/j.burns.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Nicas TI, Iglewski BH. Isolation and characterization of transposon-induced mutants of Pseudomonas aeruginosa deficient in production of exoenzyme S. Infect. Immun. 1984;45:470–474. doi: 10.1128/iai.45.2.470-474.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roggenkamp A, Geiger AM, Leitritz L, Kessler A, Heesemann J. Passive immunity to infection with Yersinia spp. mediated by anti-recombinant V antigen is dependent on polymorphism of V antigen. Infect. Immun. 1997;65:446–451. doi: 10.1128/iai.65.2.446-451.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy-Burman A, Savel RH, Racine S, Swanson BL, Revadigar NS, Fujimoto J, Sawa T, Frank DW, Wiener-Kronish JP. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 2001;183:1767–1774. doi: 10.1086/320737. [DOI] [PubMed] [Google Scholar]
- 24.Rumbaugh KP, Sawa T, Wiener-Kronish JP. New perspectives on prevention and management of Pseudomonas aeruginosa infections. In: Hauser AR, editor. In Severe infections caused by Pseudomonas aeruginosa. Kluwer Academic Publishers; Boston: 2003. a. J. R. [Google Scholar]
- 25.Sato H, Feix JB, Hillard CJ, Frank DW. Characterization of phospholipase activity of the Pseudomonas aeruginosa type III cytotoxin, ExoU. J. Bacteriol. 2005;187:1192–1195. doi: 10.1128/JB.187.3.1192-1195.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato H, Frank DW. ExoU is a potent intracellular phospholipase. Mol. Microbiol. 2004;53:1279–1290. doi: 10.1111/j.1365-2958.2004.04194.x. [DOI] [PubMed] [Google Scholar]
- 27.Sawa T, Wiener-Kronish JP. A therapeutic strategy against the shared virulence mechanism utilized by both Yersinia pestis and Pseudomonas aeruginosa. Anesthesiol. Clin. North America. 2004;22:591–606. doi: 10.1016/j.atc.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Sawa T, Yahr TL, Ohara M, Kurahashi K, Gropper MA, Wiener-Kronish JP, Frank DW. Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nat. Med. 1999;5:392–398. doi: 10.1038/7391. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt A, Schaffelhofer S, Muller K, Rollinghoff M, Beuscher HU. Analysis of the Yersinia enterocolitica 0:8 V antigen for cross protectivity. Microb. Pathog. 1999;26:221–233. doi: 10.1006/mpat.1998.0268. [DOI] [PubMed] [Google Scholar]
- 30.Shime N, Sawa T, Fujimoto J, Faure K, Allmond LR, Karaca T, Swanson BL, Spack EG, Wiener-Kronish JP. Therapeutic administration of anti-PcrV F(ab’)(2) in sepsis associated with Pseudomonas aeruginosa. J. Immunol. 2001;167:5880–5886. doi: 10.4049/jimmunol.167.10.5880. [DOI] [PubMed] [Google Scholar]
- 31.Une T, Brubaker RR. Roles of V antigen in promoting virulence and immunity in yersiniae. J. Immunol. 1984;133:2226–2230. [PubMed] [Google Scholar]