Abstract
Abnormal trophoblast invasion is associated with the most common and most severe complications of human pregnancy. The biology of invasion, as well as the etiology of abnormal invasion remains poorly understood. The aim of this study was to characterize the transcriptome of the HTR-8/SVneo human cytotrophoblast cell line which displays well characterized invasive and non-invasive behavior, and to correlate the activity of the transcriptome with nuclear matrix attachment and cell phenotype. Comparison of the invasive to non-invasive HTR transcriptomes was unremarkable. In contrast, comparison of the MARs on chromosomes 14–18 revealed an increased number of MARs associated with the invasive phenotype. These attachment areas were more likely to be associated with silent rather than actively transcribed genes. This study supports that view that that nuclear matrix attachment may play an important role in cytotrophoblast invasion by ensuring specific silencing that facilitates invasion.
Keywords: epigenetics, trophoblast, invasion, nuclear matrix, gene expression
Introduction
Placental development and invasion is vital for the successful continuation of pregnancy. Inadequate trophoblast invasion has been associated with abnormal pregnancy outcomes, including preeclampsia and intrauterine growth restriction. The initial events of placental invasion reflect a carefully choreographed sequence of cell differentiation events mediated through the production of various cell signaling molecular pathways [1]. The cytotrophoblast invades beyond the syncytiotrophoblast and forms columns from which emanate the EVT lineage [1]. The EVT is highly invasive and migrates through the decidua basalis [3] to the superficial third of the myometrium [1]. The invasion of the EVT terminates at the distal branches of the maternal spiral arteries. This process eventually remodels the maternal spiral arteries that are converted into flaccid channels which supply the placenta and thus the fetus with oxygenated maternal blood [1, 2]. Failure of the extravillous trophoblasts to invade the maternal spiral arteries has been associated with preeclampsia and growth restriction as well as other complications during pregnancy [3].
The eukaryotic genome contains two forms of stored information: the DNA sequence (genome) and the epigenetic information (epigenome) that influences gene expression without changing the DNA sequence itself [4]. Epigenetic marks are integral to differential developmental fates [5] that can be maintained and stably transmitted during mitosis [5, 6]. Within the nucleus this information is indexed by the nuclear matrix, an amorphous structure comprised of DNA, RNA and more than 500 proteins [7]. Interactions of the nuclear matrix can be visualized as the base of DNA loops that extend beyond the chromosome territory, appearing in association with the induction of transcription [8, 9]. When transcription is repressed, genes are often repositioned into or near nuclear lamina associated heterochromatic regions [10]. At interphase, chromosomes are specifically partitioned into territories within the nucleus [11, 12], with late-replicating and gene poor regions located at the nuclear periphery, and gene rich regions located more centrally [13, 14]. MARs can provide a boundary function and/or facilitate long range interactions of activation or repression [15–17] markedly effecting the phenotype. This is exemplified by the nuclear matrix protein SATB-1, an essential component that determines the developmental fate of T-lymphocytes [18] through its interaction with AT-rich rich-regions of the genome. The roles of many of the components of the nuclear matrix have been revealed in the pathology of various diseases typified by laminopathies. These present as a wide range of human diseases including dilated cardiomyopathy, limb girdle muscular dystrophy 1B (LGMD1B), Charcot-Marie-Tooth type 2B1(CMT2B1), Hutchinson-Gilford progeria syndrome (HGPS), and atypical early-onset Werner syndrome [19].
The role of epigenetics in early placental invasion is just beginning to be examined. Using the BeWo and JEG3 choriocarcinoma cell lines as models of invasion, Novakovic et al have shown that during invasion the active chromatin state as measured by tumor suppressor and proto-oncogene transcripts is correlated with methylation status [20, 21]. Suppression of trophoblast methylation specifically increased the levels of E-cadherin and plakoglobulin mRNAs in conjunction with impaired trophoblast migration and wound-healing [21]. This likely reflects its release from methylation induced silencing. Recently, Kimura et al, have suggested that chromatin looping may be required to facilitate the expression of placental specific regions of the human growth hormone locus [22]. This is likely mediated by nuclear matrix attachment. However, the role of MARs and chromatin looping in trophoblast differentiation or invasion in normal or pathological pregnancies remains to be elucidated.
To fill this void we have characterized the chromosomal-wide matrix attachment binding using an invasive human cytotrophoblast model, the immortalized cell line HTR-8/SVneo [41]. These first trimester cytotrophoblast cells proliferate essentially maintaining an undifferentiated state until grown on Matrigel basement membrane, which induces their extravillous differentiation to an invasive phenotype [36]. Like primary cultures of first trimester cytotrophoblast cells [23, 24], HTR-8/SVneo cells switch their expression of integrins from ITGA6 to ITGA1 and begin to produce HLA-G protein during culture on Matrigel [36], providing useful protein markers of extravillous differentiation. Accordingly, we have used the HTR-8/SVneo cytotrophoblast model to investigate changes in matrix attachment during extravillous differentiation. A significantly higher density of matrix attachment was observed in the invasive cytotrophoblast cells. This is consistent with the tenet that genomic plasticity is likely reduced during differentiation thereby directing cell fate [23, 24].
Materials and Methods
Cell Culture Conditions
HTR-8/SVneo cells, maintained at passage 38–45, were cultured in mass at the Cell Culture Facility of the Cleveland Clinic (Cleveland OH, USA) in 1:1 Ham’s F12: DMEM medium containing 10% fetal bovine serum and antibiotics (penicillin and streptomycin). Cells were grown to confluence at 37°C in a humidified 5%CO2/95% air incubator. Prior to assessing invasion, the adherent HTR-8/SVneo cells were harvested following treatment with trypsin/EDTA then transferred to roller bottles coated with either 10 µg/ml fibronectin (diluted with sterile PBS), or Matrigel ™, diluted 1:10 with sterile PBS. The cells were then rinsed free of serum then cultured on Matrigel ™, to induce an invasive phenotype as described aboveor fibronectin (to allow for cell attachment to the bottle, resulting in cell proliferation in a monolayer without differentiation) for 24 hours in serum free 1:1 Ham’s F12: DMEM medium supplemented with 5mg/ml BSA. Subsequent to fibronectin or Matrigel ™culture the cells were harvested with trypsin/EDTA. Western blot analysis for human leukocyte antigen-G (HLA-G) and α1 integrin (ITGA1) was used to confirm the phenotype as extravillous when cultured on Matrigel™ or non-invasive when cultured on fibronectin.
Spectral Karyotyping
HTR-8/SVneo cells at passage 38 were cultured for 1 day at 37°C in 1:1 Ham's F12: DMEM containing 10% fetal bovine serum. Mitotic cells were harvested and then treated with colcemid for two hours. Chromosomal slides were prepared essentially as described [25, 26]. After pepsin treatment and fixation with formaldehyde followed by dehydration, the chromosomal slides were denatured then hybridized to the denatured painting probes (SkyPaint, Applied Spectral Imaging: Vista, CA) for at least 48 hours at 37°C. The chromosomes were stained with DAPI and mounted with antifade subsequent to washing and hybridization detection [25, 26]. Twenty mitotic figures were randomly selected for SKY image analysis providing that high-quality hybridization signals with minimal overlapping chromosomes for the mitotic spread were observed. The degree of chromosomal abnormality was not a selection criterion. Chromosomes were analyzed by the color and size using software developed by Applied Spectral Imaging (Vista, CA).
Expression Analysis
The expression profiles of the HTR-8/SVneo cells grown on Matrigel ™ or fibronectin were determined and compared using the Illumina Sentrix Human-8 v2 Expression BeadChip arrays [27]. Total RNA was isolated using the RNeasy kit (Qiagen Inc., Valencia, CA, USA). The resulting RNA was then amplified and labeled by in vitro transcription using the Illumina RNA amplification system (Ambion, Austin, TX, USA). A 750 ng aliquot of the in vitro transcribed probe was used for hybridization. The array analyses was carried out in duplicate for HTR-8/SVneo cells grown on either Matrigel ™ or fibronectin, for a total of four independent isolations. The data was analyzed using Illumina Bead Studio. The average signal for each reporter across bead replicates was cubic spline normalized to determine a standardized expression value. Expressed genes were identified by signal values higher than the internal spike-in controls for expression (Smin > 3000). The biological replicates exhibited a correlation coefficient (r) of µ0.99 for both the HTR-F and the HTR-M replicates.
Nuclear Matrix and Loop Extractions
To determine the optimal time to prepare nuclear matrices from HTR-8/SVneo cells, nuclei from cells grown on either Matrigel ™ or fibronectin were exposed to 2 M NaCl for varying amounts of time essentially as described [28]. Both HTR-F and HTR-M, matrix and loop DNA were isolated after optimal extraction as described [29] [30]. Briefly, the halo structures were gently washed with REact® 3 restriction buffer (Invitrogen, Carlsbad, CA, USA) for 20 minutes at room temperature then centrifuged at 1000 × g at 4°C. This was repeated a total of three times to remove any residual non-nuclear matrix proteins. After the third wash, the DNA loops were separated from the matrix bound DNA by restriction digestion with 400 U of EcoRI (Invitrogen) at 37°C for 3 hours. Subsequent to restriction digestion, the DNA was centrifuged at 16,000 × g for five minutes at 4°C to pellet the nuclear matrix bound portion of the DNA. The supernatant, containing the loop-associated DNA, was then transferred to a separate tube. The nuclear matrix bound DNA was washed 2 additional times with React ® 3 (Invitrogen) buffer to minimize loop contamination of the matrix-bound DNA. The remaining residual proteins were removed from both the loop and matrix associated DNA by overnight digestion with 50 µg/mL proteinase K buffered with 50 mM Tris-HCl buffer, pH 8.0, with 50 mM NaCl, 25 mM EDTA and 0.5% SDS. DNA from each fraction was then purified using the Quantum-Prep Matrix kit (BioRad, Hercules, CA, USA), and then resuspended in nuclease-free deionized water and stored at −20°C.
Verification of fractionation and aCGH hybridization
The fractionation of loop and matrix associated DNA was confirmed by real time PCR as previously described [17, 27, 31]. Regions that were previously described as loop or matrix associated including the human growth hormone locus (unpublished data) were initially amplified in triplicate to verify fractionation. Purified DNA from each nuclear matrix and loop associated fraction was then analyzed using array 7 of the NimbleGen Systems CGAR0150-WHG8 CGH isothermal oligonucleotide 8 array system (NimbleGen Systems Inc., Madison, WI, USA). Median probe spacing is 713 bp interrogating chromosomes 14–18. This includes the human growth hormone locus that is known to be under epigenetic control in human placenta [22]. Two biological replicates were completed for the cells grown under either culture condition. All hybridizations were performed by NimbleGen (Reykjavik, Iceland). The loop and matrix array signals exhibited a correlation coefficient of 0.976 for the cells grown on fibronectin and 0.966 for the cells grown on Matrigel ™.
Identification of MARs by aCGH
To assess similarity between replicates, CGH data was initially evaluated using SignalMap (v. 1.9) (Roche NimbleGen Inc., Madison, WI, USA). Methods previously detailed [27, 30] were used to evaluate the dataset and to identify a parsimonious sets of MARs found in both cell types and unique to either cell type. Briefly, the sites of significance were identified if the putative MAR fulfilled three criteria. First, the probe must exhibit a log2 signal ratio in the lower 2.5% of the ranked signal on both replicates. Second, two additional probes with concordant signal within a 3 kb region on each side of the site were required. Third, the average signal across the restriction fragment that was interrogated was required to be concordant. Restriction fragments meeting these requirements but with inconsistent signal across the length of these fragments were excluded. Regions meeting these criteria in both replicates were then selected for further analysis. The significance of associations between MAR fragments and other genomic features was assessed relative to randomly permuted datasets in which the MAR coordinates had been arbitrarily assigned to a chromosome.
Validation of MARs identified by aCGH
Twenty-six regions spread across all chromosomes were randomly selected (see Supplemental Table 1) to confirm the aCGH results by real-time PCR as previously described [16, 17, 27, 31]. The regions selected for confirmation by real-time PCR included both genic and intergenic regions and were targeted to both loop and matrix associated regions. All real-time PCR reactions were performed in triplicate, starting with 5 ng of loop or matrix associated DNA. Initial template was calculated by the KLab PCR algorithm and ratios were compared to aCGH results [31]. Concordance between array data and PCR validation was observed.
Western Blot Analysis
Lysates were prepared in SDS sample buffer from a portion of the cytotrophoblast cells after culture on fibronectin or Matrigel. Subsequent analysis by western blotting was as previously described [33]. Each lane contained 30 µg of total protein and was labeled with antibodies against beta-actin (rabbit polyclonal; Cell Signaling Technology, Danvers, MA), HLA-G (mouse monoclonal, clone G233; Exbio, Prague, Czech Republic) or ITGA1 (mouse monoclonal, clone 5E8D9; Millipore, Billerica, MA).
Data Analysis
Correlations were calculated using RegionMiner (Genomatix Software GmbH) and an in-house suite of bioinformatics tools. These included MARs as a function of phenotype, genes expressed as well as silent genes. Gene expression (expressed/repressed) and phenotype (invasive/proliferative) were treated as binary variables. A signal threshold of 3000 (the level of the internal spike-in control) was used as the lower limit to assign a gene to an expressed state when comparing MARs as a function of expressed vs. repressed genes. A 2-fold difference in expression was the lower limit for defining a significant difference in expression when comparing phenotypes. SPSS (SPSS Inc., Chicago, IL, USA) was employed to evaluate statistical significance. The combinations of expression, location, and phenotype were compared with chi-square followed by Yates correction for continuity to calculate p-values for any association.
Results and Discussion
Dysregulated trophoblast invasion is associated with many adverse clinical outcomes. However, what governs early first trimester trophoblast differentiation and invasion remains unclear. Trophoblast invasion presents a carefully choreographed series of cell differentiation and maturational steps. Studies on multiple cell lines have sought to clarify the signaling pathways important to trophoblast invasion [1, 19, 21, 32–40]. Messenger RNA profiling with microarrays has also been carried out in syncytiotrophoblast cultures derived from term and preterm placentas [32], as well as from growth restricted fetuses. Transcription factors have been implicated as one group of regulators of trophoblast invasion [39]. Orchestrated, by the epigenetic state of the nucleus, this symphony of processes is guided by the nuclear matrix. Interestingly, this structure has yet to be characterized during trophoblast differentiation and development. To address this matter we examined the role that this nuclear organizer assumes during trophoblast invasion.
SKY
There are significant technical barriers to using first trimester trophoblast cells obtained from ongoing pregnancies that can be overcome by using a cell culture system. For example, tissues from pregnancies undergoing spontaneous abortions would not be acceptable as they could not be presumed to reflect normal pregnancies. The use of tissue from pregnancies undergoing therapeutic or voluntary termination of pregnancy is also not well suited since the outcome of the pregnancy would never be known. The tissue that would be obtained from CVS of ongoing pregnancies is likely to be heterogeneous and until delivery the outcome of the pregnancy would never be known.
Several trophoblast cell lines are available for use including HTR-8/SVneo, SW-71, JEG-3, and BeWo. The use of JEG-3 or BeWo cell line was precluded since they are choriocarcinoma derived cell lines and genomic stability would be a concern. In comparison, the HTR-8/SVneo cell line is widely available and possibly one of the most successfully applied as a model of trophoblast invasion. Accordingly the HTR-8/SVneo cell line was considered further. The HTR-8/SVneo cell line was created by transfecting first-trimester villous explants with the SV40 virus large T-antigen. This created a homogenous, immortalized but non-tumorigenic first trimester human cytotrophoblast cell line [36, 38, 41]. The environment can be altered such that the cells display an invasive phenotype. When the HTR-8/SVneo cells are induced to invade, they behave in a similar manner as primary cultured extravillous invasive cytotrophoblasts, displaying integrin switching, accumulation of HLA-G and can invade Matrigel ™ [33].
To ensure that we selected a comparatively stable genomic model, 10 mitotic figures for the HTR-8/SVneo cell line was assessed by spectral karyotyping. The average chromosome number for the HTR-8/SVneo cell line was 60 and varied from 51–95. Twenty-two specific clonal chromosome aberrations (CCAs) were identified by SKY analysis as illustrated in Figure 1. Within the HTR-8/SVneo cell line, 30% of the cells examined displayed at least one non-clonal chromosomal aberration (NCCA), including t(4;5;8;5), t(1:1), t(11;6;8;2), and t(12;18).
Figure 1. SKY Analysis of HTR-8/Svneo Cells.
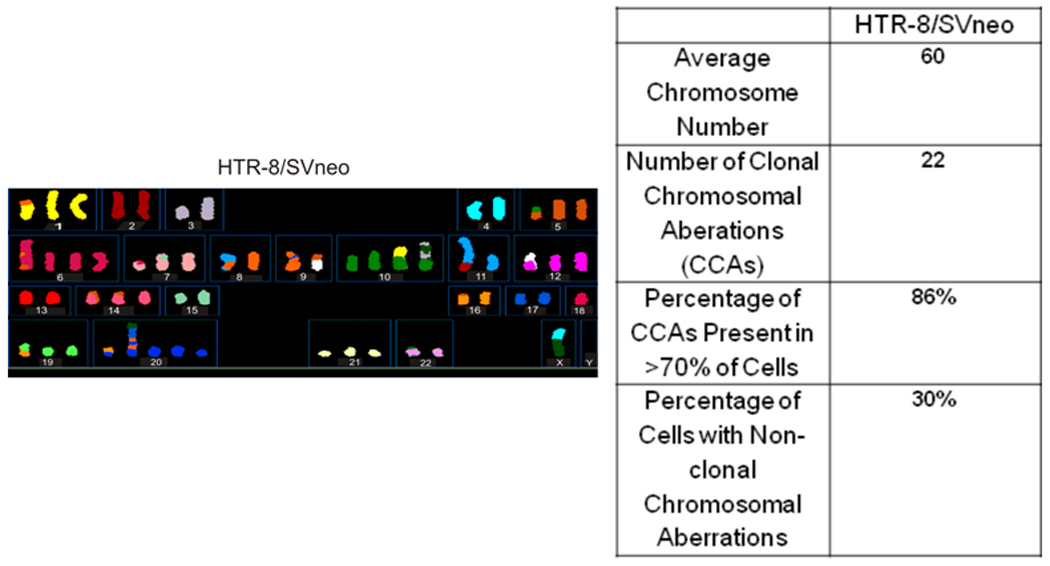
SKY analysis was performed to assess suitability of the cells for this study. The HTR-8/SVneo cells SV40 virus large T-antigen transformed chromosomal profile was determined by SKY (left panel). The right panel details the specific clonal chromosomal aberrations identified in 20% of the HTR cells.
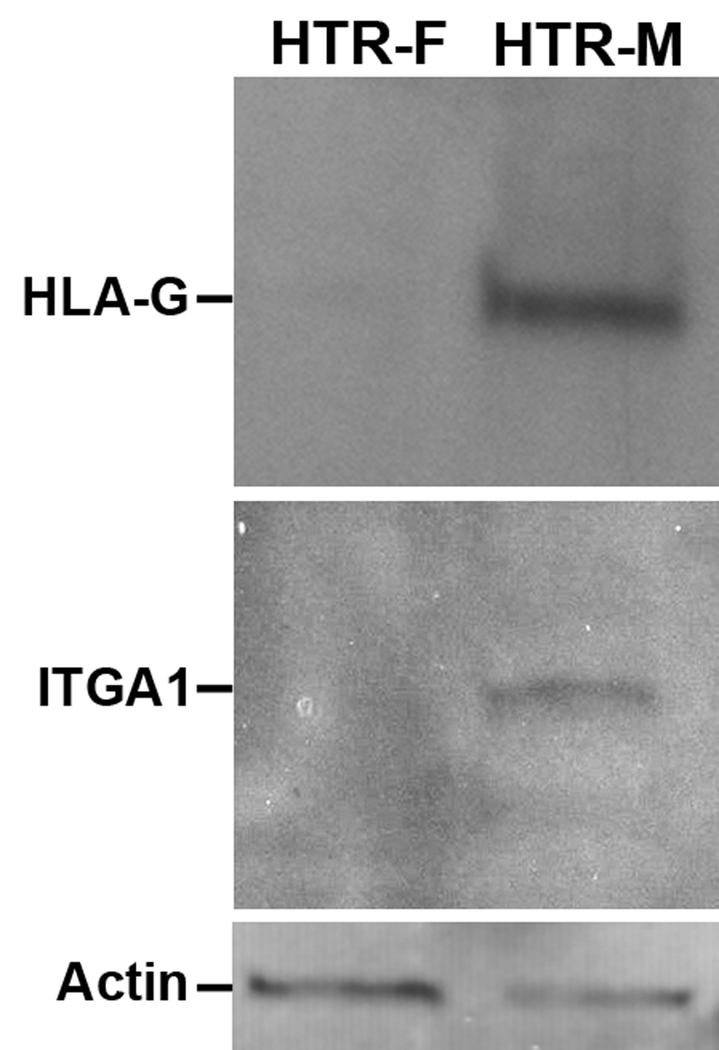
The vast majority of the chromosomal changes in the HTR-8/SVneo cells were clonal, and most of the genomic rearrangements were stable. Western blot analysis shown in Figure 2, subsequently confirmed that HLA-G and ITGA1, markers of invasion were appropriately upregulated upon culture with Matrigel (Figure 2). Accordingly this cell line was deemed an appropriate model system to examine the long-range chromatin effects during invasion.
Figure 2. Western Blot Analysis confirming invasive phenotype.
The phenotype of the cells grown under the different culture conditions was assessed by western blotting. The HTRF cells grown on fibronectin do not show expression of HLA-G or ITGA1, in comparison to the HTRM cells grown on Matrigel, although both express actin. The induction of HLA-G and ITGA1 expression is reflective of an invasive condition.
Expression Analysis
Comparison of the transcriptomes, as measured on the Illumina Sentrix Human-8 v2 Expression BeadChip arrays, from the HTR- and HTR-M cells revealed no significant differences between invasive (HTR-M) and non-invasive (HTR-F) cells. Somewhat conservative measures of expression were used since we considered a minimum signal of 3000 units to indicate expression above a general background level of approximately 80 units. This was in addition to requiringed at least a 2-fold change in signal of 3000 units to assign differential expression. The markers of phenotypic changes in these cells, HLA-G, and the integrins ITGA1, ITGA5 and ITGA6 were examined. The cell types showed HLA-G mRNA near the minimal level, without any significant difference between cell phenotypes. The alpha integrin mRNA, represented on this array by a single probe, was not detected. This is in an interesting contrast to the western blot results, which confirmed that the characteristic markers of invasive phenotypes, HLA-G and ITGA1, were appropriately upregulated when cultured with Matrigel (Figure 2). These findings suggest that the accumulation of HLA-G and ITGA1 are due to post-transcriptional events, which has been shown for HLA-G in primary cultures of cytotrophoblast cells [42]. Together, this data supports the view that the marked phenotypic differences [36] between the invasive and non-invasive HTR-8/SVneo cells reflect post-transcriptional regulatory events.
The Preparation of the Nuclear Matrix for analysis
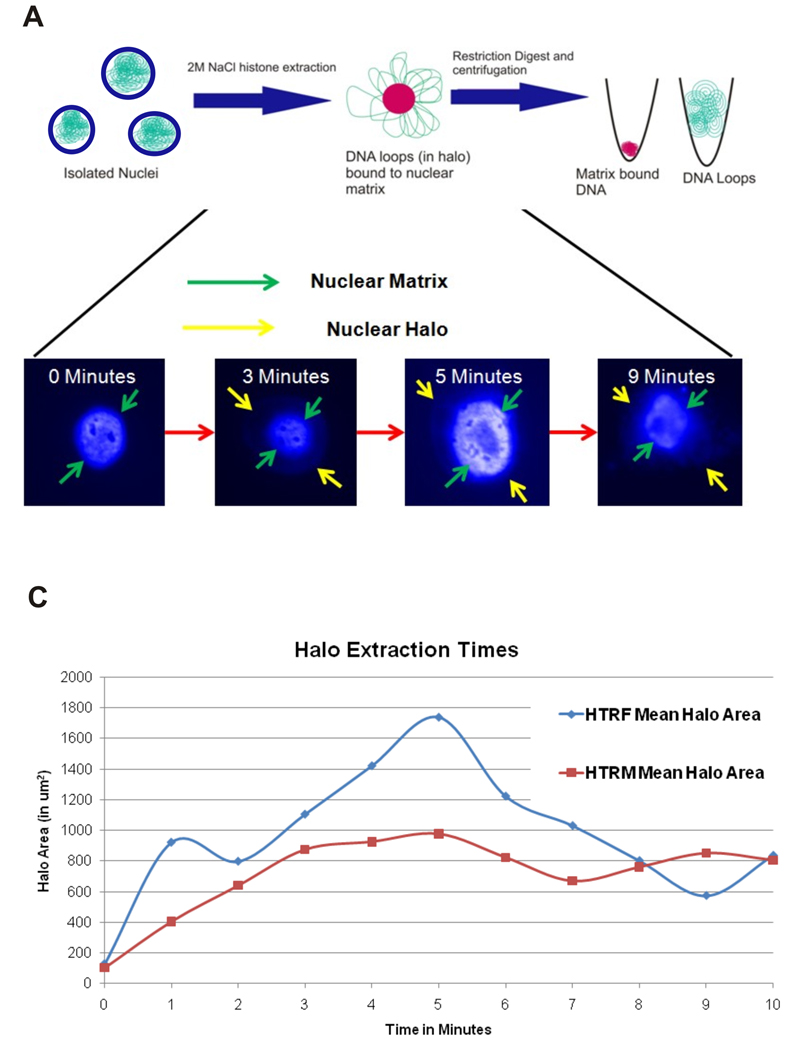
The protocol to ensure that the chromatin loops were efficiently separated from the MARs is outlined in Figure 3. It is essential to determine the optimal time for extraction for each cell-type. After the nuclei were prepared, the loops were extracted by exposure to 2 M NaCl for varying amounts of time. The area of the NM was subtracted from the total area of the structure to determine the halo area. The time point corresponding to the maximal halo area was defined as the optimal extraction time (Figure 3). Although the optimal extraction time for both the HTR-F cells and the HTR-M cells was 5 minutes, the maximal halo area of the HTR-F (1738 µm2) was significantly larger than that of the HTR-M (977 µm2). This observation suggests that loop size in the HTR-M cells is significantly smaller than that in the HTR-F cells.
Figure 3. Optimizing Nuclear Matrix Extraction.
A: Nuclei were isolated and then treated with 2M NaCl to determine the time required to induce maximum loop-sized halos. Pseudocolor images of DAPI stained nuclei during extraction are shown. Similar images were obtained from HTRF cells. The bright center area corresponds to DNA bound to the nuclear matrix. With increasing time of extraction, the nuclear halo grows until 5 minutes, and by 9 minutes appears to collapse onto itself. The maximally extracted loop and matrix fractions (5 minutes in both the HTR-F (invasive) and HTR-M (noninvasive) cells were then separated by restriction digestion and fractions subsequently hybridized to NimbleGen oligonucleotide CGH arrays.
B: The time course of extraction for the HTR-F (noninvasive) and HTR-M (invasive) trophoblast cells. The measurements shown on the x-axis are the halo area, and the y-axis shows the corresponding time. The maximal and optimal extraction time was found to be 5 minutes in each cell type. Interestingly, the HTR-M cells had significantly smaller halos (Independent T-test, p<0.05) than the HTR-F cells. This corresponds to their smaller loop size as shown by the aCGH results.
Characterization of Nuclear Matrix Attachment Sites
To evaluate differences in nuclear organization, the regions of nuclear matrix attachment for both the HTRM and HTRF cells along chromosomes 14–18 were identified by aCGH (Supplemental Tables 1 and 2). Real-time PCR validation was undertaken with various randomly located primers. As shown in Supplemental Table 3, greater than 80% of primers in both the HTR-F and the HTR-M validated, with some exhibiting enhanced values.
Loop size was inferred by MAR-MAR spacing and appeared similar each showing a similar broad distribution. The majority range from as small as 8 kb to as large as 260 kb. The mean loop size (or MAR spacing) in the non-invasive HTRF cytotrophoblasts was 110 kb with a median of 33 kb while the mean loop size in the invasive HTRM cytotrophoblasts was 89 kb with a median of 28 kb (Mann-Whitney-U p<0.001). A total of 3594 MARs were identified in the invasive HTRM cells and 2951 MARs in the non-invasive HTF cells. HTRF nuclear matrix shared 1945 MARs with the HTRM while the HTRM shared 1901 MARs with the HTRF cells. In comparison, the HTR-M cells (1703) had significantly more MARs that were unique to that cell type than the HTR-F cells (1006; Fisher’s Exact p<0.0001).
The location of each MAR in comparison to expressed and silent genes was then assessed. As above, no significant differences between invasive (HTR-M) and non-invasive (HTR-F) cells were revealed. MAR-gene associations and position in the HTR-F and HTR-M cells were remarkably similar. However, when assessed as a function of MARs, their association with silent genes was apparent (Table 1; OR=7.2 with a 95% CI: 6.3–8.4).
Table 1. Nuclear matrix attachment is associated with silent genes.
The distribution of nuclear matrix attachment regions unique and common to both cell types outside of and within genes is shown. The HTR-F cells had significantly fewer unique (or total, data not shown) MARs than the HTR-M cells. A–B: The distribution of HTR-F and HTR-M unique MARs is shown. Most MARs are intergenic, and when MARs are located within a gene, the genes they are located within are not likely to be expressed (OR 0.040, 95% CI: 0.026–0.061; Fisher’s Exact p <0.001). The number of HTR-F MARs across the chromosomes are shown. C–D: The pattern of intragenic MAR association with gene silencing is evident. HTR-F and HTR-M have the same relative distribution of MARs in silenced vs. expressed genes for both the unique (OR=0.94, 95% CI: 0.64–1.39) and Mars found in both cell types (OR=1.00, 95% CI: 0.71–1.40) MARs.
| A: HTR-F Unique MARs | ||||
|---|---|---|---|---|
| Chromosome | Unique MARs | Unique MARs in Genes (% of Unique MARs) |
Unique MARs in Expressed Genes (% of MARs in Genes) |
Unique MARs in Silent Genes (% of MARs in Genes) |
| 14 | 268 | 82 (31%) | 19 (23% | 63 (77%) |
| 15 | 191 | 62 (32%) | 12 (19%) | 51 (82%) |
| 16 | 194 | 49 (25%) | 6 (12%) | 43 (88%) |
| 17 | 166 | 52 (31%) | 8 (15%) | 45 (87%) |
| 18 | 187 | 55 (29%) | 5 (9%) | 50 (91%) |
| Total | 1006 | 300(29%) | 50(17%) | 252(84%)* |
| B: HTR-M Unique MARs | ||||
|---|---|---|---|---|
| Chromosome | Unique MARs | Unique MARs in Genes (% of MARs) |
Unique MARs in Expressed Genes (% of MARs in Genes) |
Unique MARs in Silent Genes (% of MARs in Genes) |
| 14 | 488 | 144(30%) | 32 (22%) | 113 (78%) |
| 15 | 309 | 98 (32%) | 21 (21%) | 78 (80%) |
| 16 | 351 | 73 (21%) | 11 (15%) | 63 (89%) |
| 17 | 229 | 68 (30%) | 9 (13%) | 59 (87%) |
| 18 | 316 | 86 (27%) | 9 (10%) | 77 (90%) |
| Total | 1703 | 469 (28%) | 82 (17%) | 390 (83%) |
| C: HTR-M Common MARs | ||||
|---|---|---|---|---|
| Chromosome | Common MARs |
MARs in Genes (% of Common MARs) |
Mars in Expressed Genes (% of MARs in Genes) |
MARs in Silent Genes (% of MARs in Genes) |
| 14 | 536 | 174 (32%) | 35 (20%) | 141 (81%) |
| 15 | 462 | 118 (26%) | 23 (19%) | 98 (83%) |
| 16 | 372 | 75 (20%) | 7 (9%) | 69 (92%) |
| 17 | 328 | 90 (27%) | 7(8%) | 83 (92%) |
| 18 | 203 | 47 (23%) | 6 (13%) | 41 (87%) |
| Total | 1901 | 504 (27%) | 78 (15%) | 432 (86%) |
| D: HTR-F Common MARs | ||||
|---|---|---|---|---|
| Chromosome | Common MARs |
MARs in Genes |
Mars in Expressed Genes |
MARs in Silent Genes |
| 14 | 548 | 169 (31%) | 34 (20%) | 137 (81%) |
| 15 | 477 | 122 (26%) | 23 (19%) | 102 (84%) |
| 16 | 382 | 75 (20%) | 7 (9%) | 69 (92%) |
| 17 | 333 | 89 (27%) | 7 (8%) | 82 (92%) |
| 18 | 205 | 44 (21%) | 6 (14%) | 38 (86%) |
| Total | 1945 | 499 (26%) | 77 (15%) | 428 (86%) |
Conclusions
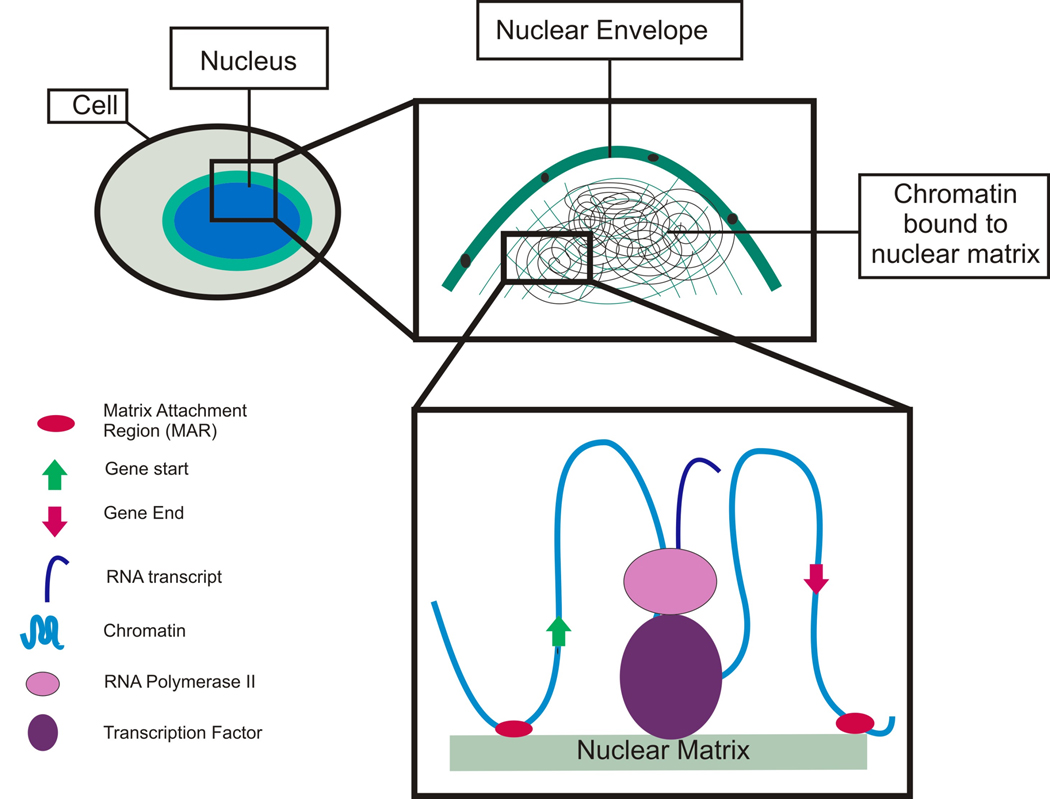
Trophoblast invasion is an important mediator of pregnancy. Although multiple studies have helped to shed some light on the biology of placental invasion, an integrated understanding of the mechanisms that produce an invasive phenotype in first trimester trophoblast cells remains elusive. The nuclear matrix is a candidate global regulator of the processes that control cellular differentiation and therefore likely plays a role in the determination of trophoblast phenotype. This is the first study to survey nuclear matrix attachment sites in trophoblast cells, and the first to correlate sites of attachment with an invasive phenotype. Through these experiments, we have shown that an increased density of nuclear matrix attachment sites (or smaller loop size) is associated with differentiation towards an invasive phenotype in HTR-8/SVneo cells. MAR density in the HTR-8/SVneo cells grown under conditions promoting an invasive phenotype is increased relative to the MAR density in conditions promoting a proliferative phenotype. MARs appear to play a role in the biology of invasion and perhaps along with a function, provide a structural framework for organizing these nuclear events, (Figure 4).
Figure 4. Nuclear Matrix Model.
Nuclear matrix attachment regions (MARs) may serve a variety of functional roles, including regulation of gene expression or silencing as well as coordination of DNA replication. MARs appear to be clustered near the 5’ and 3’ ends of genes which may serve to position the genes for expression, regulatory factor binding, or for post-transcriptional regulation. When MARs are located within genes they appear to be associated with gene silencing. Perhaps these MARs are associated with a broad category of functional elements that affect cell phenotype in different ways.
Previous studies have shown that the nuclear matrix plays a role in insulating genic domains, positioning transacting factors for both expression as well as replication [6, 11, 14, 15, 17, 18, 27, 43–45] (Figure 4). Others have shown that epigenetic events, such as DNA methylation and histone modification, are important regulators for invasion associated factors [21], exerting their influence by varying the level of transcripts. While many epigenetic regulatory mechanisms (such as histone modification and DNA methylation) affect the overall quantity of transcription, the transcriptomes of the invasive and non-invasive cytotrophoblast cells were similar. We identified a large number of nuclear matrix attachment sites within genes, which were eight times more likely to be associated with silent rather than expressed genes. This suggests that the NM is acting to affect phenotype by restricting subsequent choice as it nudges the cells along their developmental path. It may well be that the invasive cytotrophoblast phenotype is more determined by suppression of particular genes rather than activation. Consistent with our observations, one would expect that as observed post-translational regulation during invasion and differentiation provides a mechanism to fine tune the response. Given that cell-specific differentiation events have been previously associated with the nuclear matrix [18, 27], this likely represents another example of the NM playing a vital role in differentiation and invasion. Ultimately understanding the mechanisms of production of an invasive phenotype in first trimester trophoblast cells may give us a keen insight in to the biology, diagnosis and treatment of major disorders during pregnancy.
The process of trophoblast invasion seems to have a parallel in the example of malignant invasion. In contrast to trophoblast invasion, the development of cancer represents an escape from normal regulation, whereas trophoblast invasion represents a highly ordered sequence of events. However, investigations into genomic structure and invasion in cancer reveal that certain protein constituents of the nuclear matrix (e.g., p53 and SATB1) can serve as tumor suppressor genes or proto-oncogenes, where the respective loss of function or activation is associated with increasingly malignant behavior in tumor cells. For example SATB1 association with breast cancer and p53 with most types of cancer [46].
Instructively, mutations effecting the expression of p53 are associated with Li Fraumini syndrome, an autosomal dominant hereditary cancer predisoposition syndrome. P53 normally functions as a cell-cycle checkpoint protein and when p53 expression is decreased or absent, patients develop a wide variety of cancers at young ages (under age 45). In fact, loss-of-function mutations in p53 are present in more than fifty percent of human malignant tumors [47]. Comparison of p53 expressing and p53 null tumors has revealed that the loss of this nuclear matrix binding protein allows a cell to escape from the normal G1 arrest of the cell cycle during which DNA damage is repaired[47]. This loss of binding to the nuclear matrix may disrupt the normal local looping patterns and expose origins of replication or result in the loss of nuclear matrix associated gene silencing [30]. In turn, this would likely result in the aberrant expression of proteins that allow progression through the cell cycle. Thus, Li Fraumeni syndrome serves as an example in which loss of appropriate nuclear matrix binding is part of a cascade leading to uncontrolled invasion, in contrast to placental invasion, which is a very controlled event.
An example of increased nuclear matrix binding associated with increasing invasive capacity is demonstrated by breast cancer. Increased production of the nuclear matrix protein SATB1 is highly correlated with an increased invasive capacity and malignant behavior in breast cancer [48]. This offers evidence that increased nuclear matrix binding may be associated with an increased invasive capacity in human cancers, which is consistent with our observation in the HTR cell line. It is possible that disruptions in the regulation of proteins that increase binding to the nuclear matrix may ultimately be found to be associated with placenta accreta, a disorder of increased placental invasion. Disruptions of nuclear matrix binding may be involved in the molecular pathogenesis of disorders involving inadequate invasion, such as fetal growth restriction and preeclampsia.
Supplementary Material
The chromosomal locations for HTR-F MARs.
The chromosomal locations for HTR-M MARs.
PCR confirmation of fractionation with various randomly located primers. The column at the right shows the ratio of loop: matrix template as determined by real-time PCR. The mean loop to matrix ratio from the aCGH chips was: 0.999, with a standard deviation of 0.3. In order to be conservative, 1 standard deviation difference between the expected and actual signal ratio confirms fractionation. Indeterminate validation was defined as more than 1 but less than 2 standard deviation differences. When evaluating the PCR validation reactions, several unexpected instances of extreme loop and matrix partitioning were revealed. Greater than 80% of primers in both the HTR-F and the HTR-M validated. The * indicate that the sample was indeterminate or did not validate by PCR.
Acknowledgements
The authors would like to thank the Wayne State University department of Obstetrics and Gynecology for funding this research through a fellow-in training grant to KJD. Additionally we would like to thank Brian Kilburn, Claudia Lalancette, Graham Johnson, and Robert Goodrich for technical support for this project as well as Ed Sendler for lending his analytical expertise to this project. This research was supported in part by the Intramural Research Program of the NICHD, NIH. The authors would also like to thank Dr. Charles Graham of Queens University, Kingston, Ontario, for providing the HTR-8/SVneo cell line.
List of Abbreviations
- aCGH
array Comparative Genomic Hybridization
- CVS
Chorionic Villous Sampling
- EVT
Extravillous Cytotrophoblasts
- HTR
Human Trophoblast Cells Obtained From HTR-8/Svneo Cell Line
- HTR-M
HTR-8/Svneo Cells Grown On Matrigel (with invasive phenotype)
- HTR-F
HTR-8/Svneo Cells Grown On Fibronectin (with proliferative phenotype)
- MAR
Matrix Attachment Region
- NM
Nuclear Matrix
- SKY
Spectral Karyotyping
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors do not have any potential or actual personal, political, or financial interests in the material, information, or techniques described in this paper.
The array data reported in this publication are available at GEO as GSE17501
References
- 1.Lyall F. Mechanisms regulating cytotrophoblast invasion in normal pregnancy and pre-eclampsia. Aust N Z J Obstet Gynaecol. 2006;46(4):266–273. doi: 10.1111/j.1479-828X.2006.00589.x. [DOI] [PubMed] [Google Scholar]
- 2.Lyall F, et al. Human Trophoblast Invasion and Spiral Artery Transformation: The role of PECAM-1 in Normal Pregnancy, Preeclampsia, and Fetal Growth Restriction. American Journal of Pathology. 2001;158(5):1713–1721. doi: 10.1016/S0002-9440(10)64127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball E, et al. Late sporadic miscarriage is associated with abnormalities in spiral artery transformation and trophoblast invasion. Journal of Pathology. 2006;208:535–542. doi: 10.1002/path.1927. [DOI] [PubMed] [Google Scholar]
- 4.Lewin B. Genes IX. Sudbury, MA: Jones and Bartlett Publishers; 2008. [Google Scholar]
- 5.Keenen B, Serna ILDL. Chromatin Remodeling in Embryonic Stem Cells: Regulating the Balance Between Pluripotency and Differentiation. J . Cell Physiol. 2009;219:1–7. doi: 10.1002/jcp.21654. [DOI] [PubMed] [Google Scholar]
- 6.Verschure PJ. Chromosome organization and gene control: it is difficult to see the picture when you are inside the frame. J Cell Biochem. 2006;99(1):23–34. doi: 10.1002/jcb.20957. [DOI] [PubMed] [Google Scholar]
- 7.Barboro P, et al. Proteomic analyis of the nuclear matrix in the early stages of rat liver carcinogenesis: Identification of differentially expressed and MAR-binding proteins. Experimental Cell Research. 2009;15(2):226–239. doi: 10.1016/j.yexcr.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 8.Iarovia OV, et al. Induction of transcription within chromosomal DNA loops flanked by MAR elements causes an association of loop DNA with the nuclear matrix. Nucleic Acids Research. 2005;33(13):4157–4163. doi: 10.1093/nar/gki733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heng HHQ, et al. Chromatin loops are selectively anchored using scaffold/matrix-attachment regions. Journal of Cell Science. 2004;117(7):999–1008. doi: 10.1242/jcs.00976. [DOI] [PubMed] [Google Scholar]
- 10.Reddy KL, et al. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;52:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 11.Bode J, et al. From DNA structure to gene expression: mediators of nuclear compartmentalization and dynamics. Chromosome Res. 2003;11(5):435–445. doi: 10.1023/a:1024918525818. [DOI] [PubMed] [Google Scholar]
- 12.Mekhail K, et al. Role for perinuclear chromosome tethering in maintenence of genome stability. Nature. 2008;456(7222):667–670. doi: 10.1038/nature07460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Courbet S, et al. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature. 2008;455:557–560. doi: 10.1038/nature07233. [DOI] [PubMed] [Google Scholar]
- 14.Kumaran RI, Thakar R, Spector DL. Chromatin Dynamics and Gene Positioning. Cell. 2008;132:929–934. doi: 10.1016/j.cell.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martins RP, Ostermeier GC, Krawetz SA. Nuclear Matrix Interactions at the Human protamine Domain: A Working Model of Potentiation. The Journal of Biological Chemistry. 2004;279(50):51862–51868. doi: 10.1074/jbc.M409415200. [DOI] [PubMed] [Google Scholar]
- 16.Platts AE, Quayle AK, Krawetz SA. In-silico prediction and observations of nuclear matrix attachment. Cell Mol Biol Lett. 2006;11(2):191–213. doi: 10.2478/s11658-006-0016-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ostermeier GC, et al. Nuclear matrix association of the human beta-globin locus utilizing a novel approach to quantitative real-time PCR. Nucleic Acids Research. 2003;31:3257–3266. doi: 10.1093/nar/gkg424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ottaviani D, et al. Anchoring the Genome. Genome Biology. 2008;9(1):201.1–201.6. doi: 10.1186/gb-2008-9-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malhas A, et al. Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression. The Journal of Cell Biology. 2007;176(5):593–603. doi: 10.1083/jcb.200607054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novakovic B, et al. Specific tumour-associated methylation in normal human term placenta and first-trimester cytotrophoblasts. Mol Hum Reprod. 2008;14(9):547–554. doi: 10.1093/molehr/gan046. [DOI] [PubMed] [Google Scholar]
- 21.Rahnama F, et al. Epigenetic Regulation of Human Trophoblastic Cell Migration and Invasion. Endocrinology. 2006;147:5275–5283. doi: 10.1210/en.2006-0288. [DOI] [PubMed] [Google Scholar]
- 22.Kimura A, Liebhaber SA, Cooke NE. Epigenetic Modifications at the Human Growth Hormone Locus Predict Distinct Roles for Histone Acetylation and Methylation in Placental Gene Activation. Molecular Endocrinology. 2004;18(4):1018–1032. doi: 10.1210/me.2003-0468. [DOI] [PubMed] [Google Scholar]
- 23.Damsky CH, et al. Integrin switching regulates normal trophoblast invasion. Development. 1994;(120):3657–3666. doi: 10.1242/dev.120.12.3657. [DOI] [PubMed] [Google Scholar]
- 24.McMaster MT, et al. Human placental HLA-G expression is restricted to differentiated cytotrophoblasts. J. Immunology. 1995;154:3771–3778. [PubMed] [Google Scholar]
- 25.Heng HHQ, Tsui L-C. Modes of DAPI banding and simultaneous in situ hybrdization. Chromosoma. 1993;102:325–332. doi: 10.1007/BF00661275. [DOI] [PubMed] [Google Scholar]
- 26.Ye CJ, et al. Combined multicolor-FISH and immunostaining. Cytogenetic and Genome Research. 2006;114:227–234. doi: 10.1159/000094205. [DOI] [PubMed] [Google Scholar]
- 27.Linnemann AK, Platts AE, Krawetz SA. Differential nuclear scaffold/matrix attachment marks expressed genes. Human Molecular Genetics. 2009;18(4):645–654. doi: 10.1093/hmg/ddn394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krawetz SA, et al. In silico and wet-bench identification of nuclear matrix attachment regions. Methods in Molecular Medicine. 2005;108:439–458. doi: 10.1385/1-59259-850-1:439. [DOI] [PubMed] [Google Scholar]
- 29.Krawetz SA, et al. In silico and wet-bench identification of nuclear matrix attachment regions. In: Fennel JP, Baker AH, editors. Methods in Molecular Medicine: Hypertension: Methods and Protocols. New Jersey: Humana Press; 2004. pp. 443–463. [DOI] [PubMed] [Google Scholar]
- 30.Linnemann AK, Krawetz SA. Silencing by nuclear matrix attachment distinguishes cell-type specificity: association with increased proliferation capacity. Nucleic Acids Research. 2009;37(9):2779–2788. doi: 10.1093/nar/gkp135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Platts AE, et al. Real-time PCR quantification using a variable reaction efficiency model. Annals of Biochemistry. 2008;380:315–322. doi: 10.1016/j.ab.2008.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aronow BJ, Richardson BD, Handwerger S. Microarray analysis of trophoblast differentiation: gene expression reprogramming in key gene function categories. Physiol Genomics. 2001;6(2):105–116. doi: 10.1152/physiolgenomics.2001.6.2.105. [DOI] [PubMed] [Google Scholar]
- 33.Burrows TD, King A, Loke YW. Trophoblast migration during human placental implantation. Hum Reprod Update. 1996;2(4):307–321. doi: 10.1093/humupd/2.4.307. [DOI] [PubMed] [Google Scholar]
- 34.Cohen M, Meisser A, Bischof P. Metalloproteinases and human placental invasiveness. Placenta. 2006;27(8):783–793. doi: 10.1016/j.placenta.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Handwerger S, Aronow B. Dynamic changes in gene expression during human trophoblast differentiation. Recent Prog Horm Res. 2003;58:263–281. doi: 10.1210/rp.58.1.263. [DOI] [PubMed] [Google Scholar]
- 36.Kilburn BA, et al. Extracellular Matrix Composition and Hypoxia Regulate the Expression of HLA-G and Integrins in a Human Trophoblast Cell Line. Biology of Reproduction. 2000;62:739–744. doi: 10.1095/biolreprod62.3.739. [DOI] [PubMed] [Google Scholar]
- 37.Knofler M, et al. Regulation of Trophoblast Invasion-- A Workshop Report. Placenta. 2008;28(22):S26–S28. doi: 10.1016/j.placenta.2007.11.008. Supplement A: Trophoblast Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leach RE, et al. Heparin-binding EGF-like growth factor regulates human extravillous cytotrophoblast development during conversion to the invasive phenotype. Dev Biol. 2004;266(2):223–237. doi: 10.1016/j.ydbio.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 39.Loregger T, Pollheimer J, Knofler M. Regulatory transcription factors controlling function and differentiation of human trophoblast--a review. Placenta. 2003;24 Suppl A:S104–S110. doi: 10.1053/plac.2002.0929. [DOI] [PubMed] [Google Scholar]
- 40.Reister F, et al. Altered protease expression by periarterial trophoblast cells in severe early-onset preeclampsia with IUGR. J Perinat Med. 2006;34(4):272–279. doi: 10.1515/JPM.2006.052. [DOI] [PubMed] [Google Scholar]
- 41.Graham CH, et al. Establishment and Characterization of First Trimester Human Trophoblast Cells with Extended Lifespan. Exp Cell Res. 1993;206:204–211. doi: 10.1006/excr.1993.1139. [DOI] [PubMed] [Google Scholar]
- 42.McMaster M, et al. Human Placental HLA-G expression is restricted to differentiated cytotrophoblasts. Journal of Immunology. 1995;154(8):3771–3778. [PubMed] [Google Scholar]
- 43.Bode J, et al. Transcriptional augmentation: modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements) Crit. Rev. EukaryotGene Expr. 2000;10:73–90. [PubMed] [Google Scholar]
- 44.Dobreva G, et al. SATB2 Is a Multifunctional Determinant of Craniofacial Patterning and Osteoblast Differentiation. Cell. 2006;125:971–986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 45.Ellies DL, Krumlauf R. Bone Formation: The Nuclear Matrix Reloaded. Cell. 2006;125:840–842. doi: 10.1016/j.cell.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 46.Linnemann AK, Krawetz SA. Maintenance of a functional higher order chromatin structure: The role of the nuclear matrix in normal and disease states. Gene Therapy and Molecular Biology. 2009;13 [PMC free article] [PubMed] [Google Scholar]
- 47.Gu J, et al. Mechanism of Functional Inactivation of a Li-Fraumeni Syndrome p53 That Has a Mutation Outside of the DNA-binding Domain. Cancer Research. 2001;6:1741–1746. [PubMed] [Google Scholar]
- 48.Han H, et al. SATB1 reprogrammes gene expression to promote breast tumor growth and metastasis. Nature. 2008;452(7184):187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The chromosomal locations for HTR-F MARs.
The chromosomal locations for HTR-M MARs.
PCR confirmation of fractionation with various randomly located primers. The column at the right shows the ratio of loop: matrix template as determined by real-time PCR. The mean loop to matrix ratio from the aCGH chips was: 0.999, with a standard deviation of 0.3. In order to be conservative, 1 standard deviation difference between the expected and actual signal ratio confirms fractionation. Indeterminate validation was defined as more than 1 but less than 2 standard deviation differences. When evaluating the PCR validation reactions, several unexpected instances of extreme loop and matrix partitioning were revealed. Greater than 80% of primers in both the HTR-F and the HTR-M validated. The * indicate that the sample was indeterminate or did not validate by PCR.