Abstract
Genuine La proteins contain two RNA binding motifs, a La motif (LAM) followed by a RNA recognition motif (RRM), arranged in a unique way to bind RNA. These proteins interact with an extensive variety of cellular RNAs and exhibit activities in two broad categories: i) to promote the metabolism of nascent pol III transcripts, including precursor-tRNAs, by binding to their common, UUU-3’OH containing ends, and ii) to modulate the translation of certain mRNAs involving an unknown binding mechanism. Characterization of several La-RNA crystal structures as well as biochemical studies reveal insight into their unique two-motif domain architecture and how the LAM recognizes UUU-3’OH while the RRM binds other parts of a pre-tRNA. Recent studies of members of distinct families of conserved La-related proteins (LARPs) indicate that some of these harbor activity related to genuine La proteins, suggesting that their UUU-3’OH binding mode has been appropriated for the assembly and regulation of a specific snRNP (e.g., 7SK snRNA assembly by hLARP7/PIP7S). Analyses of other LARP family members (i.e., hLARP4, hLARP6) suggest more diverged RNA binding modes and specialization for cytoplasmic mRNA-related functions. Thus it appears that while genuine La proteins exhibit broad general involvement in both snRNA-related and mRNA-related functions, different LARP families may have evolved specialized activities in either snRNA or mRNA related functions. In this review, we summarize recent progress that has led to greater understanding of the structure and function of La proteins and their roles in tRNA processing and RNP assembly dynamics, as well as progress on the different LARPs.
La exhibits general and broad activity in both snRNA and mRNA-related functions
La proteins are factors found in nearly all eukaryotes examined [1] and have important functions in RNA metabolism. Human La (hLa, also called Sjogren’s Syndrome Antigen B, or SS-B) was first identified as an autoantigen in patients suffering from autoimmune disorders such as systemic lupus erythematosus, Sjogren’s syndrome and neonatal lupus [2, 3]. Although it remains unclear why La is targeted as an autoantigen in some persons, its phosphorylation status, subcellular localization and susceptibility to proteases in the context of apoptosis are believed to contribute to its antigenicity in the susceptible population [4–7]. La is an essential protein in all examined metazoans, including Drosophila, mice, as well as in Trypanosoma and Arabidopsis [8–11], but can be deleted in budding and fission yeast [12, 13], in which much of the work describing fundamental La function has been done. Clues to its cellular functions came from early experiments indicating that La preferentially associates with RNAs ending in stretches of uridylate residues, as are found at the ends of all nascent RNA polymerase III (pol III) transcripts due to their d(T)n transcription termination signals [14–16].
For the largest class of pol III transcripts, the precursor tRNAs, the uridylate-containing 3’ ends are removed during processing, and consistent with the rapid turnover of most La ribonucleoproteins (RNPs) [17], La occupancy is not observed on the mature RNAs. Pol III transcripts bound by La include pre-tRNAs, pre-5S rRNA, pre-U6 snRNA, the nascent forms of 7SL SRP RNA, 7SK RNA, Y RNAs, Alu, B1, B2 and some other cellular RNAs [13, 18–26] as well as the pol III transcribed viral RNAs EBER I & II and adenoviral VA RNAs [27–29]. Its presence in a human pol III holoenzyme, as well as chromatin immunoprecipitation with pol III transcribed genes in yeast and human, suggests that La is involved in pol III activity and is positioned to be the first factor to bind nascent pol III transcripts [30–32]. In addition to Pol III transcripts, La has also been found to influence the metabolism of short-lived intermediates of the Pol II transcribed U1, U2, U3, U4 and U5 snRNAs, in yeast [33–35], and associate with a U1 precursor in human cells [36]. Close examination revealed that these intermediates also end in UU-3’OH as a result of post-transcriptional processing [37–39]. For RNA targets terminating in UU-3’OH, the simplest and most fundamental role of La proteins is to bind and promote their maturation, in large part by protecting them from 3’ exonucleolytic degradation or excessive trimming. As will be discussed in upcoming sections, more complex functions have also been described for human and yeast La proteins, both for nascent snRNAs, and for other RNAs.
While function in pre-tRNA processing has been well documented and described mechanistically, La has also been strongly implicated in the translation and/or metabolism of many UUU-3’OH-lacking cellular and viral mRNAs, apparently involving a broad range of activities [40–58]. While understanding the roles of La-mediated activities in mRNA metabolism is an important subject, the diversity of data suggests that it may function differently for different mRNAs, and a review of each would be beyond the scope of this review. However, we believe it is worthwhile to note that the mRNAs whose translation is modulated by La protein contain atypical translational initiation contexts, including viral and cellular mRNAs with internal ribosome entry sites (IRESs) [59], 5’ terminal oligopyrimidine tracts (5’TOPs) [51, 60, 61] or upstream open reading frames (uORFs) [50]. While many positive effects of La on translation have been reported, its effects can also be negative, and in some cases the outcome is dependent on the conditions of the experiment and the presence of poly-A binding protein (PABP) [51, 62–64]. In general, most mRNAs for which La positively effects translation have lengthy and complex 5’ UTRs (see refs above), whereas studies of specific mRNAs whose outcomes revealed a negative effect on translation were on mRNAs with short presumably nonstructured 5’ UTRs such as the very short 5' UTR containing 5'TOP mRNAs [51, 62–64]. In this regard we note evidence that suggests that the nonphosphorylated (cytoplasmic) isoform of the human La protein can recognize the 5’ GpppN cap (and related structures) present on cellular mRNAs [65]. This leads us to speculate as to how both the negative and positive influences on translation may be effected by this cryptic cap-binding activity of La. Binding to the caps of 5' TOP mRNAs by nonphospho-La may inhibit cap-mediated translation initiation. The same activity on complex, e.g., IRES mRNAs can conceivably shift the balance toward internal initiation. This would not necessarily exclude a more direct role for La at the IRES element (HCV mRNA) itself or just upstream of the AUG, as for MDM2 mRNA [50].
La domain architecture: intricate juxtaposition of two RNA binding motifs
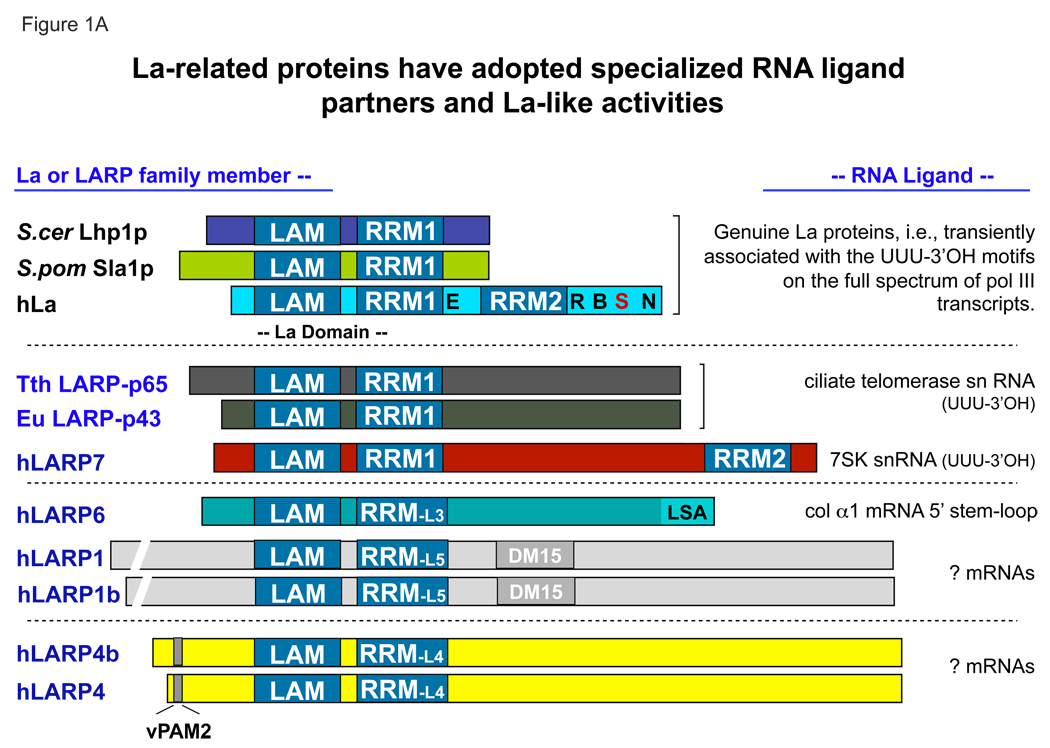
La proteins have a conserved architecture, with an N-terminal La domain consisting of a La motif (LAM; amino acids 6–100, hLa numbering [66]) and an RNA recognition motif (RRM1; 106–195) separated by a short linker (Fig. 1A). Since the original description of the La motif in genuine La proteins, it has also been found in a number of other La-related proteins (LARPs) that segregate into distinct families conserved throughout evolution. Four LARP families have been discerned in addition to La, LARPs 1, 4, 6 and 7. Although earlier gene annotation also noted LARPs 2 and 5, further comparative analyses argue that these should be referred to as LARPs 1b and 4b respectively since they are highly sequence related to their main families [1].
Figure 1.
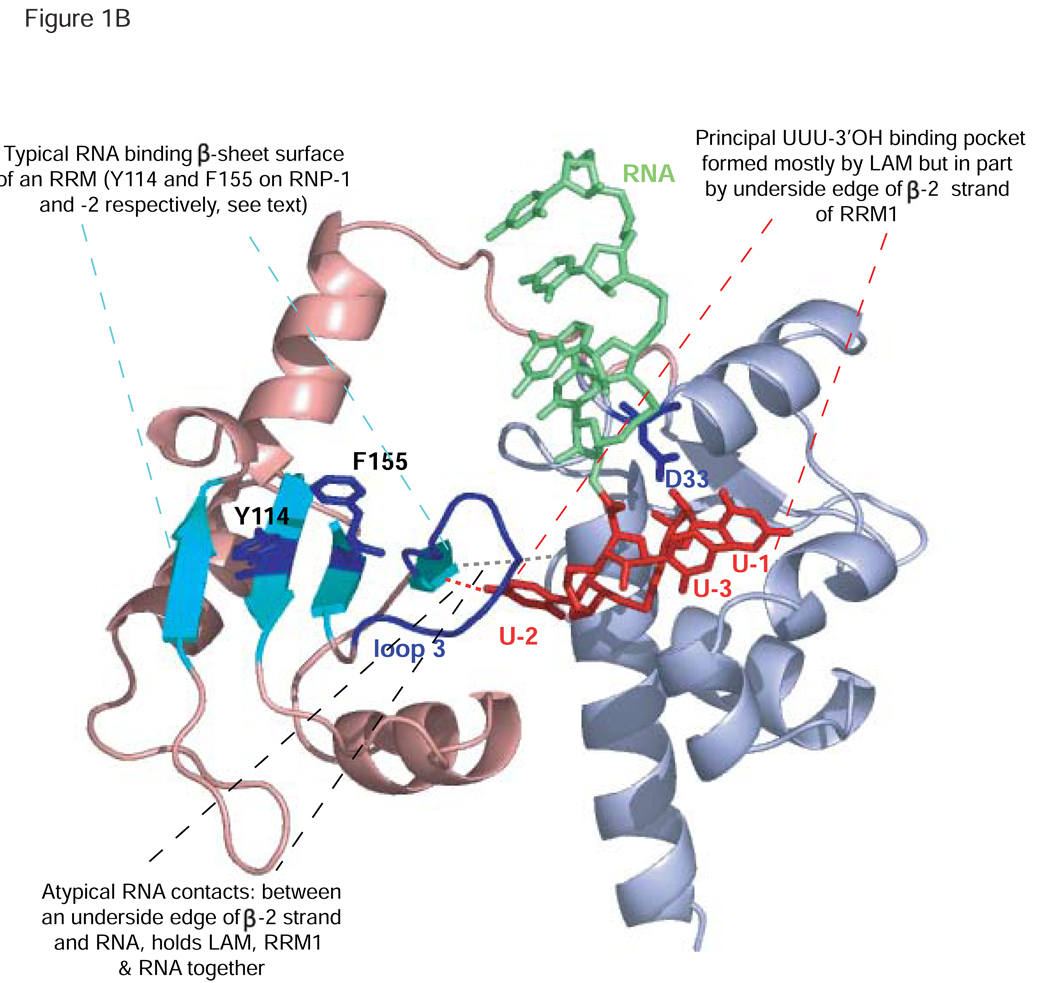
A) Schematic alignment indicating the conserved LAM-RRM domain architecture of the genuine La proteins of three species (top, S. cerevisiae Lhp1p, S. pombe Sla1p, and human hLa) followed by LARPs, arranged in order of their sequence conservation in their LAMs (see B below). Numbered RRM-L3, RRM-L4, RRM-L5 motifs reflect variations of these from the canonical RRM and their conservation of these across LARP families [1]. Note that while the RRM of LARP4 is routinely predicted as such by the NCBI BLAST server, the RRMs of LARPs 1 and 6 were not (unpublished observations) but were found using special methods [1]. Three characterized members of the LARP7 family are shown, Tth: Tetrahymena thermophila LARP-p65; Eu: Eupliotes aediculatus LARP-p43, and hLARP7, also known as PIP7S [21, 143, 150]. The LARPs 6, 1, & 4 families are represented by their human members, along with coexisting subfamily members hLARPs 1b and 4b (see text). Features indicated in the hLa schematic and described in the text include a nuclear export element in RRM1 (E; [112]), a nuclear retention element (R; [110]), a short basic motif (B; [73]), the CKII phosphorylation site S366 (S; [160]) and a nuclear import sequence (N; [108]). B) The RRM1 of La provides two surfaces for pre-tRNA binding: i) at the interface between the RRM and the LAM [76], responsible for UUU-3’OH dependent interaction (RNA corresponding to U-3 to U-1 shown in red), and ii) RRM1 loop-3 (shown in blue), which is important for UUU-3’OH independent binding to pre-tRNAs and possibly other La targets [87]. The D33 side chain (in the LAM) which makes bidentate contacts to the 2' and 3' OH groups of the terminal U (U-1) of the RNA, is shown in blue. RRM1 β-sheet surface residues Y114 and F155, whose side chains are indicated in blue, also function in pre-tRNA processing [80]. Also depicted are two hydrogen bonds that connect the LAM and an underside edge of RRM1, in part through La Y23, and in part through the U-2 base of the RNA.
As will be discussed later in the review, it is hypothesized that the LARP families have evolved distinct and conserved functions compared to genuine La proteins, despite their retention of the La motif critical for classical La function, as described below. Structural studies of the LAM and RRM from human La revealed that the LAM harbors a winged-helix type fold [66–68], common in DNA binding transcription factors and some RNA-binding proteins [69–71], while RRM1 adopts a classic β1α1β2β3α2β4 topology similar to that found in other members of the abundant class of RRM proteins [72]. Since earlier studies established that both the LAM and RRM1 were required for recognition of UUU-3’OH by La [73, 74], it was expected that their typical nucleic acid interaction surfaces would be involved in La-RNA target interactions [75]. On the contrary, a co-crystal structure containing the La domain from human La (hLa) and a short RNA ending in UUU-3’OH revealed that this mode of binding surprisingly occurs through contacts other than to the expected surfaces (described in more detail in [76] and below), but nonetheless confirming the requirement for both motifs for RNA binding while leaving the function of the expected nucleic acid interaction surfaces unclear. This mode of UUU-3’OH recognition was confirmed and expanded upon by additional cocrystal structures [76, 77].
Although many RNA binding proteins utilize more than one RNA binding motif for target RNA binding, the way that La does this is more complex than usual. For example, while poly-A binding protein and sex lethal each use the typical RNA binding surfaces of adjacent RRMs to contact a contiguous stretch of RNA, La uses two spatially separate regions of its RRM1 to form parts of two RNA binding surfaces that contact different regions of an RNA. Thus, for La, a single RRM1 contributes to two separate RNA binding surfaces, one of which is also formed in part by the juxtaposed LAM (Fig. 1B). As will be discussed below, arrangement of the LAM and RRM1 in hLa supports formation of the two RNA binding surfaces, mediated in part by contacts to the RNA segment that bridges the RRM1 and LAM, and in part by two additional direct contacts between the LAM and the RRM1.
La promotes tRNA processing via two distinct mechanisms, simple 3' end binding and a more complex activity
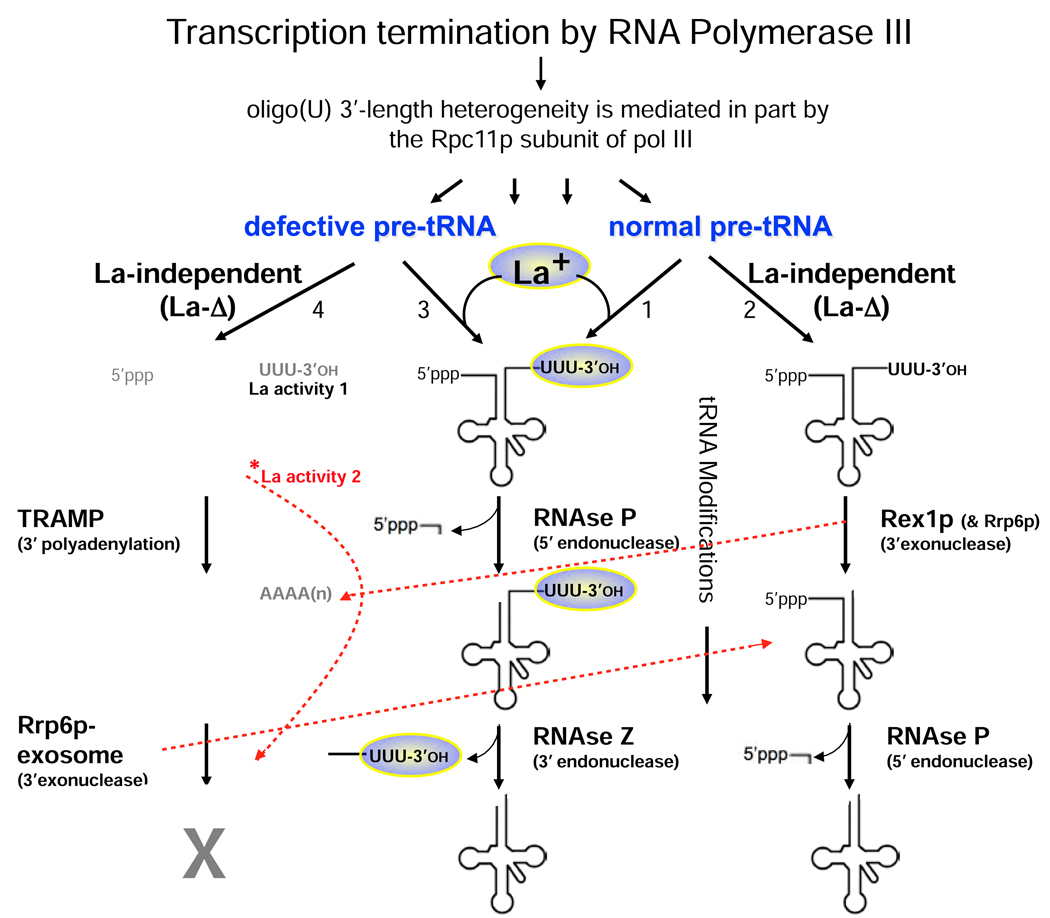
The best-characterized substrates for La binding are pre-tRNAs, and La binding to these and other pol III transcripts is dependent of the length of the 3' terminal oligo(U) tract. Human La protein binds best to RNAs that contain 3 or more terminal Us [14–16], whereas a direct comparison indicates that S. pombe La clearly requires 4 or more terminal Us for efficient binding [78]. As the S. pombe pol III requires at least 5T residues as a minimal terminator whereas human pol III requires only 4 [79], these co-variations suggest that binding of nascent pol III RNAs by La protein is functionally linked to the pol III termination system. Moreover, transcription termination by pol III on a particular gene produces a heterogeneous set of transcripts that differ in the length of their 3' oligo-U tracts. Typically, the most abundant RNA species end with one less rU residue than the minimal number of dT residues required for termination, and as alluded to above, it is this nascent RNA species that can be efficiently engaged by La protein. However a substantial fraction of the nascent pol III transcripts from the same gene ends with fewer U residues than required for efficient La binding [15, 78]. By following the phenotype of a functional suppressor-tRNA in S. pombe, Huang et al. observed that this 3' end heterogeneity leads nascent transcripts that differ in their La binding to two different posttranscriptional tRNA maturation pathways [78]. Moreover, the extent to which the pol III subunit, Rpc11p, executes its 3' exonuclease activity during the transcription termination process can be a determinant of terminal oligo-U length and therefore the degree to which the nascent transcripts are engaged by La [78] (Fig. 2). One consequence of this heterogeneity is that the La-associated pre-tRNAs and the La-independent pre-tRNAs are differentially sensitive to the 3'-5' exosome-TRAMP nuclear surveillance system [80, 81] (see below).
Figure 2. Scheme depicting the role of the 3' end binding activity of La protein in different pathways of precursor-tRNA processing in yeast.
La is nonessential in yeast but its absence affects the processing of most precursor-tRNAs [12, 13]. La protein is depicted as a colored oval. The degree to which pre-tRNAs require La for maturation depends on their structural integrity. Four pathways are depicted labeled 1–4 under the diagonal arrows leading into the pathways. Most pre-tRNAs probably go through pathway 1. Some pre-tRNAs do not functionally engage La even when present [88], and these would presumably use pathway 2, a La-independent pathway. Pathway 4 reflects the nuclear surveillance pathway for pre-tRNAs [94]. Defective pre-tRNAs are structurally impaired due to mutations that disrupt basepairs in the anticodon stem or variable arm, or lack of appropriate modifications (see text). A curved dashed red line depicts a separate decay pathway due to absence of La activity 2 which is required for the maturation of structurally impaired pre-tRNAs that are degraded even in the absence of Rrp6p [80] (see text); *La activity 2 is mediated by the conical RNA β-surface (and loop-3) of RRM1. Additional red dashed lines depict connectivity and crosstalk between the pathways (see [81, 90]).
Studies in budding and fission yeast indicate that in the absence of La most pre-tRNAs follow a variant tRNA processing pathway (Fig. 2). During pre-tRNA processing, La binds and protects the 3’ ends of pre-tRNAs from at least two exonucleases, the 3’ exonuclease Rex1p (pathway 1 in Fig. 2) and for defective pre-tRNAs, the nuclear exosome-Rrp6 (pathway 4 in Fig. 2) [80, 81]. After processing of the 5’ leader by RNAse P, the 3' end-protected trailer is processed by the endonuclease RNAse Z (pathways 1 & 3 in Fig. 2) precisely after the discrimator base, the first unpaired nucleotide following the acceptor stem [82–86], leading to dissociation of La from the tRNA and addition of CCA to the new 3' end [87]. In the absence of La, and for those pre-tRNAs that do not engage La even when it is present [88] (pathway 2 in Fig. 2), the order of pre-tRNA processing may be altered such that exonucleolytic trimming of 3’ trailers precedes 5’ leader cleavage by RNAse P [13, 18, 89].
Yet, precisely how La and other factors contribute to the 3’ end metabolism of all pre-tRNAs in the cell is not fully clear. For example, as noted above although La is required for normal 3’ endonucleolytic processing of many pre-tRNAs in S. cerevisiae [18], some pre-tRNAs do not use the La-dependent pathway even when it is present [88]. Rex1p [81, 90] is a 3' riboexonuclease that functionally processes pre-tRNAs in the absence of La and presumably also those pre-tRNAs that undergo 3' exonucleolytic processing even when La is present. For those pre-tRNAs that do use it, La appears to promote endonucleolytic processing by blocking access of the 3’ ends to the processing exonucleases, i.e., Rex1p [81, 90]. Accordingly, since La is nonessential in yeast, and in its absence, pre-tRNAs gain access to Rex1p for productive processing, it might have been expected that in cells lacking La, the 3’ endonuclease, RNAse Z, would be nonessential. Intriguingly, this is not the case in fission yeast, S. pombe [91]. It remains to be determined if this is also true in S. cerevisiae and also if it reflects the possibility that RNAse Z may be required for an essential yeast function other than pre-tRNA processing..
It has been shown that La deletion is synthetically lethal with yeast mutations that disrupt the secondary structure of essential tRNAs [18, 92]. This leads to susceptibility of the structurally-impaired pre-tRNA to degradation, in this case, by a second class of 3’ exonuclease distinct from Rex1p, the surveillance activities of the TRAMP polyadenylation complex and the nuclear exosome which degrades aberrant RNAs [93, 94]. Furthermore, ectopic La can complement the deletion of tRNA modification enzymes that catalyze addition of modifications that presumably stabilize tRNA structure [93, 95, 96]. The absence of proper modification can sensitize some pre-tRNAs to degradation by the surveillance exosome, which can be averted by the 3’ end binding activity of La [93]. In the case of pre-tRNAiMet, failure to be methylated on A58 by the tRNA methyltransferase TRM6-TRM61, leads to nuclear degradation of the hypomodified precursor tRNA; remarkably however, overexpression of La can avert this degradation, permitting the hypomethylated but otherwise mature tRNAiMet to persist and be exported to the cytoplasm where it apparently functions in translation [93]. This latter observation suggests that a critical function of methylation of A58 of tRNAiMet is to avoid decay of the pre-tRNAiMet in the nucleus.
Some functions of La in pre-tRNA processing reflect a La activity beyond simple 3’end protection [80]. Several lines of evidence indicate that La exhibits a complex function related to RNA structure/folding. In addition to the capacity of La to complement the loss of tRNA modifications thought to stabilize tRNA structure, La mutants have been described which harbor wild-type 3’ end binding and protection activity but are still incapable of rescuing the maturation of certain structurally impaired pre-tRNAs, even when Rrp6, a 3' exonuclease component of the nuclear surveillance exosome, is deleted [80]. These results are consistent with functions for La in pre-tRNA processing that are more complex than simple UUU-3’OH binding and protection from the 3’ surveillance exonucleases [80]. Indeed, the 3' end binding activity and the second, more complex activity which allows La to function in the rescue of structurally-impaired pre-tRNAs, have been localized to the two different RNA binding surfaces of the LAM and RRM1 of the human and S. pombe La proteins [80, 87].
Attempts to reconcile simple vs. complex functions in RNA metabolism have become a critical theme in the study of the mechanisms that control La function: simple functions related to UUU-3’OH mediated RNA binding and 3’ end protection, vs. complex La functions that involve enhancing the folding and/or stability of various RNA targets via poorly understood binding mode(s) and mechanisms. Although this has begun to be addressed in the context of pre-tRNA processing [80, 87], understanding precisely how the coordinated use of the separate RNA binding surfaces of the LAM and RRM1 contribute to other activities such as in mRNA-related functions, during RNA chaperone activity, and for La binding to its more stably associated pol III transcripts such as cellular Y RNAs, as well as viral encoded EBER and VA1 RNAs remains a challenge.
As described in detail below, recent crystallographic and biochemical studies have advanced understanding of the variations of La-RNA binding modes and associated functions significantly. Another advance related to these outstanding issues involves the increasingly studied LARPs [21, 97–105]. Just as genuine La proteins combine various mechanisms of RNA binding to provide complex functions, it would appear that an emerging theme in LARP function might also involve variations on similar modes to provide distinct functions on their own sets of RNA targets. The diversity of function of La proteins and LARPs, and their expected shared and divergent manners of RNA recognition, will be the topic of the remainder of this review article.
Controlling the activity of the La domain by C-terminal regions
The C-terminal domain of La proteins is more divergent, and increases significantly in size from yeast to higher organisms (Fig. 1A). In higher eukaryotes, a second RRM of unknown function (RRM2 or RRMc) is often found following RRM1, although whether this RRM in human La binds RNA is questionable since it has chemical features unusual for a RRM and its β-sheet surface is obscured by an α-helix that constitutes a nuclear retention element [106].
La shuttles between the nucleus and cytoplasm [107], and the C-terminal domain contains several determinants important for La intracellular trafficking, including a nuclear localization signal (NLS) found at the end of all La proteins examined except S. cerevisiae in which it is located more centrally in the protein [108, 109], and a nuclear retention element [110, 111] which masks a conserved nuclear export element in RRM1 [112] (Fig. 1A). Disruption of these trafficking elements causes mislocalization of La and malfunction in tRNA maturation, and in some cases leads to disorder of some of the pre-tRNA processing steps [110, 112].
In addition to direct effects on tRNA processing, the intracellular trafficking of La is an important determinant of the types of RNA substrates it encounters for binding [60]. This is noteworthy since several conditions have been described including viral infection that lead to a redistribution of La to the cytoplasm where it is presumably used for viral replication [55] (more extensively reviewed in [113]).
Another motif in the hLa CTD has been implicated in RNA binding. In higher organisms, a highly charged short-basic motif (SBM) of about forty amino acids that includes a Walker A like motif immediately following the NRE appears to be a RNA binding element that increases the overall affinity and stability of La-RNA interactions whose binding activity can be inhibited by phosphorylation at ser-366. The unphosphorylated SBM inhibits 5’-processing of pre-tRNAs by RNAse P presumably by stabilizing and/or reorganizing pre-tRNA binding to La [73, 89, 114]. This inhibition of 5' processing is relieved by phosphorylation of S366 by protein kinase CK2 [51], a pathway that is faithfully executed upon hLa expression in yeast [89].
Interestingly, phospho-specific antibodies have localized S366-phosphorylated hLa to the nucleoplasm, the locale of the immature pol III transcripts that comprise the primary binding targets of La, while non-phosphorylated hLa is concentrated in the nucleolus and also accumulates in the cytoplasm where it is found associated with certain transcripts including the 5' terminal oligopyrimidine (5'TOP) mRNAs [60, 115]. Thus, the highest affinity ligands for La are UUU-3'OH bearing pre-tRNAs (Kd 2–10 nM) and other snRNAs which are most abundant in the nucleus, and their processing is facilitated by phospho-La. Although the higher-affinity RNA binding isoform, nonphospho-La resides in the cytoplasm (and nucleolus), there are far fewer UUU-3'OH bearing RNAs in this compartment and nonphospho-La uses its higher affinity binding mode to associate with its next best high affinity ligands, the 5' terminal oligopyrimidine (5'TOP) mRNAs [60]. Accordingly, the relative concentrations of hLa S366 isoforms in different subcellular compartments, in conjunction with the relative concentrations of specific competing RNA ligands in these compartments, would appear to determine the differential association of nonphospho-La and phospho-La with their respective classes of different RNAs [60]. While phosphorylation of S366 has been described only for hLa, other compensatory modifications could confer comparable discriminatory functions in other species [116].
Several other phosphorylation sites have also been mapped to La [117] and one of these, T302, promotes nuclear export and polysome association of mouse La in response to the mitogen activated kinase, Akt [54]. Retrograde transport of La in sensory neuron axons is controlled by sumoylation of residue K41 [118].
Yeast La proteins are also phosphorylated at multiple sites including a site for the master control kinase, Sch9, dependent on TAP42 and in response to rapamycin, although no connection to RNA metabolism has yet been established [119, 120]. Since no single phosphorylation site appears to have been conserved across widely diverged species (unpublished observation), it seems that different organisms use La phosphorylation to different degrees and this may coincide with species-specific recruitment of different La kinases, as observed for mouse and human La [116].
The importance of C-terminal determinants in RNA target discrimination has also been revealed by a study of the LARP7 family member hLARP7/PIP7S ([21] described in detail in a section below). While La and hLARP7/PIP7S have highly homologous N-terminal domains, especially in the residues implicated in UUU-3’OH recognition (later section), hLARP7/PIP7S effectively discriminates against pre-tRNAs to preferentially bind its natural RNA target, the UUU-3’OH containing pol III transcript, 7SK snRNA [21]. This is reflected in the lack of pre-tRNAs as compared to 7SK snRNA bound to PIPS7 in human cells as monitored by immunoprecipitation [21] (R.Y. & R.M., unpublished). We note that differences in the precise subcellular localization of the La and LARP7 proteins within the nucleus may contribute to targeting their different ligands toward this discrimination although this remains to be determined. Nonetheless, while full-length hLARP7/PIP7S is incapable of functioning in a pre-tRNA-mediated suppression assay in S. pombe, as compared to wild-type S. pombe or human La which are highly active, deletion of the C-terminus of PIP7S, however, activates it for tRNA–mediated suppression. This reflects significant RNA 3' end protection activity by the hLARP7 La domain, and is consistent with its C-terminal region playing an important function in preventing PIP7S from interacting with pre-tRNAs [21]. Given the divergence of C-terminal domains between genuine La proteins and the LARPs, it is likely that this region in these factors play important roles in RNA target discrimination.
Apparent conservation of RNA binding modes by La proteins and LARPs
The recent publication of multiple high-resolution La-domain/UUU-3’OH co-crystal structures has greatly enhanced our understanding of this mode of RNA recognition [76, 77]. Although the structure of a LARP-RNA interaction has yet to be solved, it is tempting to speculate that the conserved juxtaposition of the LAM and RRM, as well as high conservation by the LARPs of residues important in UUU-3’OH recognition by La, may reflect similarities in RNA binding modes, perhaps most especially for LARP7 and the p65- and p43 LARPs since these show the highest homology to the La proteins [1], and appear to exhibit UUU-3’OH recognition.
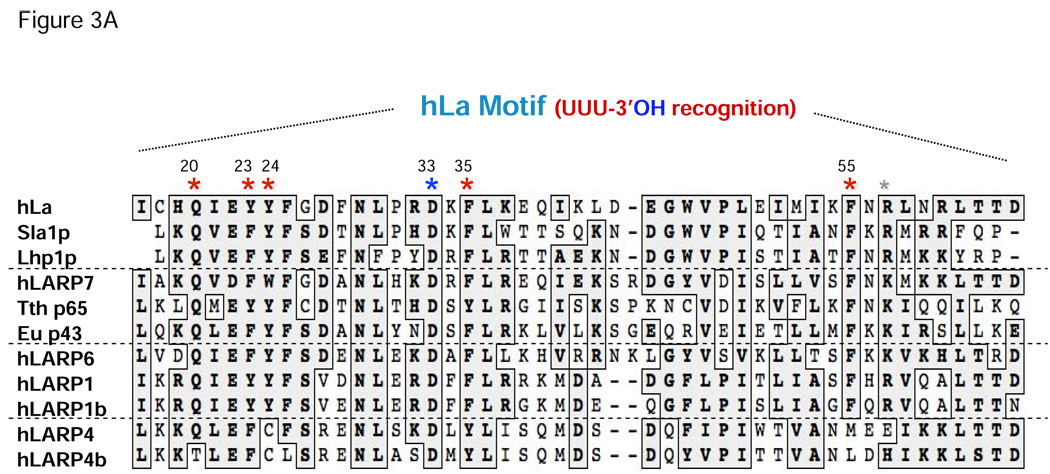
The greatest degree of homology between these various LARP classes of proteins occurs in the LAM, reminiscent of the high degree of homology in this motif in genuine La proteins across species (e.g., Fig. 3A). The RRM found adjacent to the LAM in all La proteins is also present in a high proportion of LARPs. RRMs in many proteins contain aromatic residues in their RNP-1 and RNP-2 motifs (short sequence tracts on the β-3 and β-1 strands respectively of the RRM β-sheet surface) that stack on RNA target bases during canonical RNA binding [72, 121]. As alluded to above, although the canonical RRM β-sheet surface is not used by La for UUU-3'OH binding, it is used for an additional independent mode of RNA binding [87]. For hLa, the LAM and RRM1 bind different regions of a pre-tRNA [87]. The same may be true for LARPs, although it is also plausible that the LAM and RRM of LARPs may be arranged to form a contiguous RNA binding site. However, it is entirely conceivable that the paucity of intermotif contacts between the LAM and RRM1 of La protein, and the apparent reliance of the orientation of these motifs on the UUU-3’OH RNA itself, may allow arrangements of the LAM and RRM1 that are significantly different from that observed in the hLa-UUU-3’OH structures. Moreover, the observation that the RNA ligand mediates much of the LAM-RRM1 interface in the hLa structures suggests that different RNA targets may program different orientations of the LAM and RRM even for La or a LARP.
Figure 3.
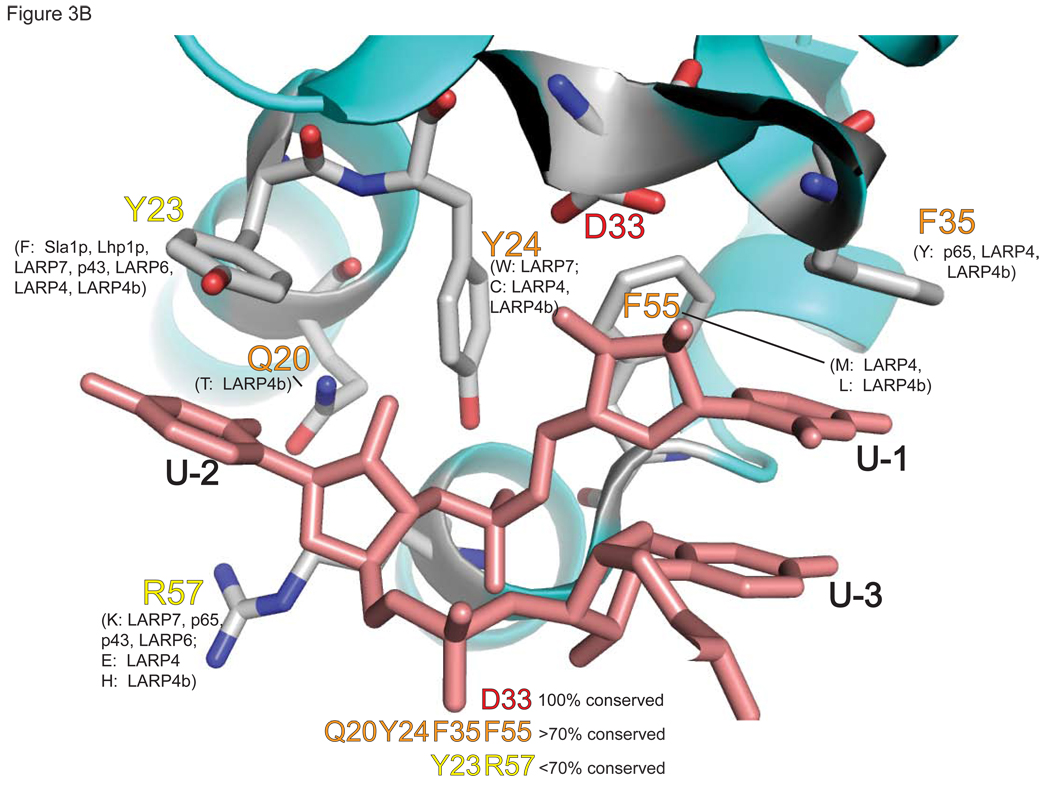
A) CLUSTAL-W alignment of the LAM sequences of various La proteins and LARPs. The residues involved in UUU-OH recognition by hLa [76, 77] are indicated by asterisks above and numbering according to hLa, below (see text). B) High resolution structure derived from [77], to depict conservation of residues involved in the UUU-3’OH recognition RNA binding pocket. Crystallographic assignment of residues shown to contact the terminal UUU-3’OH by hLa are indicated (hLa structure and numbering [77]), along with the percentage of identity conservation for the noted La proteins (Sla1p, Lhp1p) and LARP family members (hLARP1, hLARP1b, hLARP4, hLARP4b, hLARP6, hLARP7, p43 and p65).
Notable is the conservation of RRM1 RNP-1/2 motifs among genuine La proteins and some LARPs despite the lack of RNP-RNA contacts in the La-UUU-3’OH co-crystals. In any case, evidence suggests that the RRM may be used to different degrees by different LARPs since point-mutation of hLa F155 (RNP-1) decreases hLa binding to the respiratory syncytial virus leader RNA [55] but not pre-tRNA [80], and mutation of LARP7 Y127 (RNP-2) has a significant effect on LARP7 binding to 7SK RNA (see below,[21]). While some LARPs contain RRMs of sufficient homology to be recognized computationally as such, others lack the aromatic residues in the RRM RNP-1 and/or RNP-2 [72, 121] even though they are predicted to adopt the β1α1β2β3α2β4 RRM fold [1]. It might therefore be expected that La proteins and LARPs might utilize residues other than, or in addition to, those in the RNP-1/2 motifs for RRM-mediated RNA recognition.
Recent work [87] suggests that during pre-tRNA processing, the loop connecting the β2 and β3 strands of hLa RRM1 (loop-3) (Fig. 1B) makes UUU-3’OH independent contacts to the main body of tRNA using an RNA binding mode distinct from the typically observed RNP-mediated RRM mechanism, in that for La binding to pre-tRNA, loop-3 appears to confer substantially more affinity than the aromatic residues in RNP-1 and −2. This is not unprecedented, as many RRMs depend on the loops between their β-strands and α-helices for RNA recognition, sometimes exclusively [122]. This alternative binding mode leaves open the possibility that the RRM-like motifs in the more divergent LARPs might still engage RNA more similarly to La than might otherwise be anticipated. The loop-3 dependent mechanism has been hypothesized to play an important function in tRNA precursor-product discrimination by La before and after endonucleolytic pre-tRNA 3’ trailer processing: specifically, La preferentially binds intact pre-tRNAs using tRNA contacts to RRM1 and UUU-3’OH contacts to the LAM, compared to single site lower affinity binding to the tRNA and trailer after 3’ endonucleolytic processing. The resulting lower affinity for the products of pre-tRNA 3’ processing would allow La to preferentially dissociate from these and recycle onto new tRNA precursors [87]. Consistent with this RRM1 binding mode playing important functions in La activities that are more complex than simple 3’ trailer binding and protection, residues in loop-3 are also important for a characterized RNA chaperone activity of hLa [87, 123].
Sequence conservation of the RRM1 loop-3 is very high within the genuine La protein family and within individual LARP families, yet is the most highly divergent portion of RRM1 across LARP families and between these and La proteins [1, 87]. RRM1 loop-3 is therefore an attractive candidate for a determinant of RNA specificity and functional divergence across La and LARP families, in addition to that conferred by the LAM (see later section). An appealing model for the importance of a UUU-3’OH independent binding mode in LARP function relates to the interaction of hLARP7/PIP7S to its 7SK snRNA target. La binds stably to pre-tRNA as a result of combined use of two separate binding surfaces. Contrary to the dynamic turnover of pre-tRNA-La RNPs that occurs rapidly during tRNA processing, the hLARP7/PIP7S-7SK snRNA interaction, which would appear to be more stable, may be so because 7SK snRNA undergoes little if any RNA 3' oligo(U) end cleavage/modification as part of its posttranscriptional processing. It is tempting to postulate that the LARP7–7SK snRNA interaction is analogous to the stable interaction of La with the pre-tRNA prior to 3' trailer cleavage; because 7SK RNA does not undergo 3' processing, the UUU-3’OH dependent and UUU-3’OH independent binding modes would be expected to retain their additive effect resulting in a long-lived hLARP7/PIP7S-7SK snRNA interaction. However, as noted above, the C-terminal domain of hLARP7/PIP7S also contributes to 7SK binding. Nevertheless, the remodeling of the 7SK snRNP that is associated with physiological responses to regulate P-TEFb activity [21, 103, 104] likely uses additional mechanisms distinct from those observed for La-pre-tRNA RNPs.
Binding of the LARP7 family members p43 and p65 to their UUU-3’OH-containing telomerase RNAs, described in more detail in a later section, may employ a similar additive use of their LAMs and RRM1s, in addition to their C-terminal regions ([124]).
Conservation of LAM amino acid side chains involved in UUU-3’OH contacts by genuine La proteins and LARPs
During UUU-3’OH recognition by La, most contacts between the terminal uridylates and the La domain are made to highly conserved residues on the LAM (Fig. 3A, B). Of particular interest are Q20 (hLa numbering), which makes uridylate-specific contacts to the penultimate base (U-2) of the RNA; F35 and F55, which stack on the base and sugar of the last uridine (U-1), respectively; Y23, which in addition to making the only direct hydrogen bond protein-protein contact between the LAM and RRM1 (to N139), stacks on U-2; and D33, which by making bidentate contacts to the 2’-OH and 3’-OH groups of the terminal sugar residue, allows La to discriminate between DNA ends and RNA ends, and to discriminate between 3’OH-containing transcripts and 3’ phosphate-containing RNAs which result from cleavage by some ribonucleases. The only UUU-3'OH contact to RRM1 is an uridylate specific contact between the O4 of U-2 and the peptide backbone of I140. As diagrammed, the conservation of residues implicated in RNA binding varies across representative LARPs from different families, with the LARP7 family (including Tth p65 and Eae p43) having the greatest homology and LARP4 and 4b having the least (Fig. 3A). With the high conservation of these residues by La and the LARP7 family members hLARP7/PIP7S, p65 and p43, it would seem likely that LARP7 members engage their pol III synthesized targets (telomerase RNA is a pol III transcript in ciliates) via the same UUU-3’OH mediated mechanism.
One unanticipated result of the assignment of residues important for La binding to UUU-3’OH was the surprising lack of apparent RNA sequence specificity [76]. These studies revealed an exact sequence requirement only for the penultimate (U-2) uridylate of the UUU’3-OH motif, with mutations in the RNA at the terminal (U-1) or other 3’ uridylates accompanied by only moderate drops in affinity. Conversely, mutation of individual La residues revealed by the structure as important for RNA binding resulted in significant to catastrophic decreases in affinity [76]. The data are consistent with these La residues fulfilling important functions in RNA binding without necessitating a strict requirement for RNA sequence identity. Indeed a high degree of the amino acid side chains in the UUU-3’OH binding pocket provide aromatic groups that stack with the RNA. Consistent with this high degree of apparent flexibility, more recent structures that capture interactions between the La domain and other UUU-3’OH containing ligands have revealed a significant degree of plasticity in binding [77]. These structures compared the nature of La-UUU-3’OH binding to several small RNA targets varying in length, composition and the number of terminal uridylates. It was found that the orientations of the RNAs ending in three versus four uridylates were qualitatively distinct: even though the identity of amino acids and functional groups involved in the uracil specific contacts were maintained, the identity of which uracil (i.e., U-4 vs. U-3) took part in these contacts varied. These studies also showed divergence in the path taken by the 5’ ends of the RNAs upstream of the conserved uridylate contacts, especially as compared to the original crystal structure [76] (for direct comparison see Supplementary Fig. 8 in [87].
These data raise the possibility that the potential to use a variety of RNA-binding mechanisms may be an inherent feature of the presumed dynamic nature of the LAM-RRM1 arrangement to accommodate various RNA targets. Consistent with this, the high-resolution co-crystal structures reveal a surprising paucity of contacts restricting the spatial orientation of the RRM1 with respect to the bound RNA and the LAM. As noted above, these consist of a hydrogen bond between the bound U-2 RNA and RRM1 residue I140, a salt bridge between LAM residue R57 and RRM1 residue D125, and a hydrogen bond between the hydroxyl of Y23 of the LAM and RRM1 residue N139. Strikingly, even this Y23-N139 intermotif protein-protein contact is poorly conserved in genuine La proteins (Fig. 3A), as Y23 is replaced by phenylalanine in yeast La, a residue which could still stack on U-2 of the bound RNA, but with the loss of the tyrosine-specific hydroxyl functional group would lose its ability to contact RRM1 by this mechanism. It is intriguing that this replacement is associated with a requirement by Sla1p for more terminal uridylates than human La for optimal binding [78]. This raises the possibility that the additional U residue recognized by Sla1p [78] mediates an additional LAM-RRM1 interface interaction, i.e., similar to the role of U-2, in the absence of Y23.
Consistent with a lack of spatial restriction between the LAM and RRM1, NMR studies have found that these motifs, although tethered to each other by the linker, tumble independently of one another in the absence of RNA [77]. Taken together, the information from several structural studies are consistent with common themes that include binding site plasticity as a means to accommodate a variety of RNA ligands.
The modality of La-RNA binding as illustrated by differences in the existing crystal structures may provide insight into the expected divergence of RNA target recognition by the LARPs. The significant but non-absolute conservation of residues important for UUU-3’OH recognition in some of the LARPs might mean these same residues also play important functions in RNA binding, without necessitating a concomitant conservation of RNA target identity. At one end of this proposed spectrum is the highly homologous LARP7 family which also binds to RNAs terminating in UUU-3’OH, such as hLARP7/PIP7S binding to the 7SK snRNA and p43 and p65 binding the ciliate pol III transcribed telomerase RNAs. As discussed previously, hLARP7 also depends on specificity determinants in its C-terminal sequences to bind 7SK and prevent non-productive interactions with RNAs normally bound by genuine La. At the other end of the spectrum is LARPs 4 (& 4b) which have significantly less conservation in the LAM, and exhibit RNA sequence binding characteristics distinct from hLa (R. M. and S. Gaidamakov, unpublished observations), and one of these, LARP4, appears to function in mRNA metabolism (R.Y and R.M, submitted). These observations are consistent with a model in which the various LARP families have evolved specialized roles involving the UUU-3’OH dependent or UUU-3’OH independent functions of La proteins. It will be interesting to evaluate how the variations in amino acids important in La-UUU-3’OH recognition correlate with target discrimination by the LARPs. Determination of which binding modes provide substrate specificity across LARP families should prove to be of primary importance for this emerging field.
LARPs: Appropriating an RNA binding mode for divergent functions
While the function of La in the metabolism of nascent pol III transcripts has been extensively studied, the study of LARPs is still in its infancy. LARPs can be broadly defined as factors containing a high degree of homology with genuine La proteins in the LAM-RRM arrangement of their La domain, but have taken on independent functions from those associated with La. A recent phylogenetic analysis of LAM containing proteins reveals that these are found in all eukaryotes, with the exception of some protists [1]. A subset of these includes factors that contain, like genuine La proteins, an RRM or RRM-like motif (RRM-L) immediately following the LAM (i.e., containing a La domain), and these factors segregate into conserved families across evolution. One significant facet of this analysis was the discovery that La proteins and LARPs are totally absent from the archaea, suggesting these proteins evolved early after the archaeal/eukaryal radiation. While investigating the function of these factors is still in the initial stages, an emerging theme appears to be the apparent appropriation of La-like mechanisms for RNA binding interactions. As noted above this is most apparent for the LARP7 family members.
A previously underappreciated element of LARP conservation is the predominant co-evolution of an RRM or RRM-L immediately following the LAM. Early study of LARP function focused on the budding yeast proteins Sro9p and Slr1p, which unlike genuine La contain a LAM but no adjacent RRM [98–100]. The more recent phylogenetic analysis of LARPs indicates that this RRM-less architecture of LAM-containing proteins is the exception rather than the rule; out of 134 examined LAM containing sequences, 113 contain an RRM (or RRM-L) immediately following the LAM, similar to what is always found for genuine La proteins [1]. With respect to RRM1 conservation, according to Bousquet-Antonelli and Deragon, only LARP7 contains a canonical RRM1 as is found in genuine La proteins, while the other LARP families contain variations of RRM-L [1]. It is noteworthy that the degree of divergence from the canonical RRM varies for the different LARPs. For example, we found that while the RRM of LARP4 was routinely predicted as such by the NCBI BLAST server, the RRMs of LARPs 1 and 6 were not (unpublished observations) but were found using special methods [1]. While the preponderance of RRM or RRM-L motifs in LARPs is high, their sequence conservation varies such that they segregate LARPs into distinct groups, previously noted by a numbered classification of RRM-L motifs (Figure 1) [1]. Sequence alignment of these indicate high similarity within families but significant divergence across them, suggesting that these LARP families likely have distinct functions that are broadly conserved across species, some of which have been investigated experimentally in a variety of model organisms. To date, the best-characterized family is LARP7, which includes the human protein hLARP7/PIP7S and its homologs in numerous species [125] as well as the telomerase subunits p43 and p65, of two ciliate species.
Characterization of the human LARP7 family member
Human PIP7S, also called hLARP7 and HDCMA18, is an RNA binding protein with high homology to human La in its La domain but with a divergent C-terminal region. First identified in a comprehensive screen of genes showing microsatellite instability in human gastric tumors, LARP7/HDCMA18 was the second most susceptible gene to acquire a frameshift mutation in a short homopolymeric dN tract that leads to stop codons truncating RRM2 [126]. An independent analysis of factors associated with various transcription machineries identified hLARP7/PIP7S as a P-TEFb interacting partner, although no function for hLARP7/PIP7S was apparent at that time [127]. P-TEFb, comprised of the Cdk9 kinase and cyclin T1, is recruited to sites of active transcription and phosphorylates the CTD of RNA polymerase II, relieving a block to transcriptional elongation [128]. Additional analyses revealed that hLARP7/PIP7S specifically binds to and enhances the stability of the 7SK snRNA [21, 103, 104], which along with the HEXIM1 & 2 proteins and the 5' methylphosphate capping enzyme, MeCPE, forms a complex (the 7SK snRNP) that sequesters P-TEFb and inhibits both its activity and the capping activity of the MeCPE [129–132]. siRNA-mediated knockdown of hLARP7/PIP7S resulted in decreased levels of 7SK RNA and the 7SK snRNP, resulting in an increase in P-TEFb dependent transcription with consequent biological effects [21, 103, 104].
Consistent with a role as a tumor-suppressor, hLARP7/PIP7S knockdown was also shown to increase P-TEFb occupancy at two tumor-related genes and was sufficient to cause the transformation of a cell line used to study carcinogenesis [21]. In retrospect the tumor suppressor activity of hLARP7/PIP7S is consistent with the LARP7 Drosophila ortholog multisex-combs (mxc) encoding a tumor-suppressor [133] although it wasn’t noted to be homologous to La protein. Recently, a Drosophila homolog of 7SK RNA was identified [134], and HEXIM, 7SK and LARP7 homologs have also been identified in many metazoa [125]; this presumably reflects the evolutionary conservation of a P-TEFb or like activity that is controlled by 7SK RNA and LARP7. This leads to a very interesting question: what advantages are afforded through the regulation of P-TEFb activity by the regulated assembly and dissassembly of a RNP (see below), as opposed to the more commonly observed and typically simpler protein-mediated regulation?
A kinetic coupling model of transcription suggests that the elongation rate of pol II can determine the efficiency with which some splice acceptor or donor sites are recognized during cotranscriptional splicing. Moreover, recruitment of splicing factors to the CTD of pol II can affect alternative splicing. Consistent with this, hLARP7-mediated alterations of P-TEFb activity induces gene-specific alternative splicing [135].
The La domains of hLARP7/PIP7S and human La are highly homologous, and hLARP7/PIP7S shares with La all the residues previously implicated in specific UUU-3’OH recognition [76, 77] (Fig. 3A, B). As 7SK RNA is a pol III transcript ending in UUU-3’OH, it seems likely that hLARP7/PIP7S would use a binding mode similar to La for the engagement of the 7SK 3’ end. In support of this, replacement of the LAM (where the majority of UUU-3’OH specific contacts are made) from hLARP7/PIP7S with that of La results in only a modest decrease in 7SK RNA binding or 7SK snRNP formation in vivo [21]. Consistent with this, other regions provide RNA substrate specificity and these differ for La and hLARP7/PIP7S. First, the Y127D mutation in hLARP7/PIP7S, located in the RRM1 of the La domain, significantly reduces the affinity for 7SK RNA, while (a less drastic) mutation of the equivalent residue in human La (Y114A) has little effect on pre-tRNA binding [21, 80]. More significantly, and consistent with a 7SK specificity determinant residing in the RRM of the La domain of hLARP7/PIP7S, replacing the hLARP7/PIP7S RRM1 with the hLa RRM1 led to significant decreases in 7SK RNA binding and 7SK snRNP formation in vivo [21]. While the loop-3 of the RRM1 of hLa provides significant affinity for pre-tRNA, it remains unclear if this region of hLARP7/PIP7S is important for 7SK RNA binding. Because of the basic nature of loop-3 of hLa RRM1 it is tempting to speculate that the hLa RRM1 exhibits general affinity for RNA, perhaps recognizing RNA backbone or other features of structure with little requirement for sequence specificity, consistent with its ability to engage a wide variety of different pre-tRNA sequences as well as other RNAs, whereas the RRM1 of hLARP7/PIP7S is likely to have adopted a sequence-specific interaction with 7SK RNA.
As alluded to above, the function of the C-terminus of hLARP7/PIP7S in RNA recognition appears to be complex, since deletion of the hLARP7/PIP7S C-terminal region (or transformation-associated frameshift mutations producing premature stop codons upstream of RRM2) results in significant defects in 7SK RNA binding and 7SK snRNP formation in vivo [21], while other studies suggest more modest defects by the N-terminal La domain in in vitro 7SK RNA binding assays [104]. Furthermore, the La domain of hLARP7/PIP7S, but not the full-length protein, is capable of partially complementing La 3’ end protection function in pre-tRNA maturation, as monitored using a red-white screen for tRNA-mediated suppression in S. pombe [21]. In sum, the data suggest that the La domain of both La and hLARP7/PIP7S provide specific and high-affinity binding to UUU-3’OH, but the LARP7 C-terminus enhances binding to 7SK RNA while limiting binding to UUU-3'OH containing RNAs other than 7SK RNA. An attractive model is that the decreases in 7SK RNA levels observed in the context of C-terminally truncated hLARP7/PIP7S in vivo could be due to increased hLARP7/PIP7S occupancy by other UUU-3’OH containing RNAs which can now better compete for binding. The C-terminal region of hLARP7/PIP7S has also been shown to be important for interaction with CDK9 in the absence of 7SK RNA [104].
La-7SK snRNA-hLARP7 dynamics
It had been known that a small fraction (2–5%) of 7SK snRNA was associated with La protein, consistent with its transcription by pol III and the idea that La is only transiently associated with nascent pol III RNAs that then go on to become part of more mature RNPs [20, 136]. Older as well as recent work from several laboratories now provide the beginnings of a model for the transfer of 7SK snRNA from La during the dynamic assembly of the 7SK snRNP. La can bind to the UUU-3'OH of nascent 7SK RNA. The short basic motif (SBM) of human La that includes a Walker A like motif independently recognizes the 5' pppG end of nascent pol III transcripts [114]. However, a small number of pol III transcripts, including 7SK snRNA, U6 snRNA and B2 SINE RNAs, are modified on their 5' ends by the 5’ methyl capping enzyme, MeCPE to produce the monomethyl phosphate cap, CH3pppG [137]. The 5' monomethyl cap of 7SK RNA decreases the affinity for La protein [65] and has also been shown to stabilize the 7SK RNA in an in vivo system [138]. Moreover, MeCPE appears to remain associated with 7SK snRNA[139]. Thus it would appear that hLARP7/PIP7S and the MeCPE may displace La from nascent 7SK RNA to form a RNP in which the RNA 5' and 3' ends are stabilized against exonucleolytic attack by MeCPE and LARP7/PIP7S respectively [132]. Although the kinetic components of the 7SK snRNP assembly pathway remain to be determined, it would be useful to determine if LARP7/PIP7S plays a role in the 7SK RNP assembly and/or disassembly dynamics that contribute to regulating P-TEFb activity in a manner similar to what has been described for LARP-p65 during hierarchical assembly of the telomerase RNP [140].
Other LARP7 family members function in telomerase RNPs
Other well studied LARPs that segregate phylogenetically into the LARP7 family [1] include the ciliate homologs p43 (Eupliotes aediculatus) and p65 (Tetrahymena thermophila), which for p65 has been shown to assist in the correct folding of the telomerase RNA and hierarchal assembly of the RNP [140–144]. Unlike in yeast and vertebrates, where telomerase RNA is transcribed by pol II, in ciliates telomerase RNA is synthesized by pol III and retains its terminal uridylates in the telomerase RNP [145–148]. In the absence of p43 or p65, binding of the telomerase reverse transcriptase (TERT) to telomerase RNA and the associated telomerase activity is compromised. While this suggests that these LARPs may undertake a molecular chaperone function for the telomerase RNA similar to that which has been hypothesized for La proteins [75, 87, 92, 123, 149], the role that p65 plays in the assembly is different from that provided by general chaperone activity [144]. Affinity, competition and footprinting experiments indicate that while p43 and p65 do have higher affinity for telomerase substrates ending in UUU-3’OH, this is not the exclusive or even principal binding determinant [141, 150, 151]. Other telomerase RNA binding determinants, in particular the stem-loop preceding the terminal uridylates, have been mapped [124, 141, 151], and similar to human hLARP7/PIP7S-7SK RNA, domains in the C-terminal region have importance in the mechanism of telomerase RNA binding. Indeed, it appears the C-terminal region of p65 is critical for any interaction with the telomerase RNA [124, 151]. This significant involvement of the C-terminal region of LARP7 family members may have special functional relevance to the nature of their RNA target interactions: human LARP7 binding to 7SK RNA and p43/p65 binding to telomerase RNA constitute stable interactions, in contrast to La, where the N-terminal La domain is thought to be largely sufficient for its more transient interactions with immature pol III transcripts that must undergo extensive processing and RNP remodeling that includes dissociation from La [73, 87, 149, 152]. However, with regard to the possibility of a molecular handoff of nascent pol III transcribed telomerase RNA during ciliate telomerase RNP assembly, we note that no genuine La protein sequence has been found in the as yet incomplete Euplotes database [1]. Moreover, while four LAM-containing sequences have been found in Tetrahymena, none of these grouped with genuine La proteins [1].
The LARP6 family
LARP6 family members are characterized by having a more central, RRM-L3 containing La domain and a newly described motif of unknown function at the extreme C-terminus called the LSA motif [1]. The first studied member of the LARP6 family is the moth Manduca sexta homolog Acheron. Originally identified in a screen for factors induced during apoptosis of intersegmental muscles during M. sexta development [102], Acheron shares homology with the human protein hLARP6 [1]. Mammalian LARP6 has been implicated in cellular differentiation of myotubes and has been shown to interact with the developmental transcription factor CASK-C, a novel CASK/Lin-2 family member [56, 153]. Thus, studies of the LARP6 members of moth, fish and mammals indicate that LARP6 interacts with transcription factors, and functions upstream of the transcription factor MyoD, to control muscle development [56, 102, 153]. As a transcription factor-associated protein, LARP6 may function in the control of mRNA synthesis, and this could conceivably be related to the way LARP7 controls P-TEFb activity, although no small RNA target (i.e., akin to 7SK snRNA) has been identified for LARP6.
More recently, human LARP6 has been shown to specifically bind with high affinity (Kd 1.4 nM) to a very highly conserved stem-loop motif in the 5’UTR of α1 collagen mRNAs and is hypothesized to regulate their localized translation, suggesting a function for LARP6 in mRNA metabolism [154] distinct from that proposed for Acheron. Additional data showed that hLARP6 was found associated with collagen related mRNAs that share a similar stem-loop motif [154]. Overexpression of hLARP6 led to redistribution of collagen mRNAs off of polyribosomes [154], similar to 5’TOP mRNAs upon overexpression of hLa [51]. Knockdown of hLARP6 also led to decreased collagen synthesis suggesting that hLARP6 levels should be precisely controlled to regulate its effects on translation [154].
Like LARP7, LARP6 has conserved all of the amino acid side chains involved in UUU-3'OH recognition by genuine La proteins (Fig 3A & B). Indeed, mutating a single U to A in the single stranded bulge of the α1 collagen mRNA stem-loop abolished hLARP6 binding [154]. However, it is not yet clear if this uracil-dependent hLARP6 binding to the α1 collagen mRNA stem-loop is mediated by the LAM binding pocket. It would be important to determine this because if so it suggests that the LAM RNA binding pocket of LARPs can bind RNAs that do not end in 3’OH. This is a significant issue because the LARPs contain the hLa equivalent of D33 which recognizes the 3’OH and discriminates against 3’ phosphate. By some estimates, it is thought that in its 3'OH binding mode, the LAM binding pocket of hLa can not accommodate a 3' phosphate. Although hLa binding to 5' TOP mRNAs suggests that some rearrangement may occur to accommodate a 3' phosphate, this is speculative.
The LARP1 family
The LARP1 family [1] is more generally divergent from genuine La proteins than the LARP6 and 7 families, although this is not reflected in Fig. 3A. LARP1 members lack a typical RRM after the LAM. Instead, many LARP1 family members are predicted to contain an RRM-L5 that adopts the RRM fold but lacks the RNP-1 and RNP-2 consensus sequences, though some members of this family may have no recognizable RRM adjacent to the LAM [1]. Furthermore, members of this family may or may not harbor a C-terminal DM15 motif whose function is unknown. As indicated in Fig. 3A, the amino acids in the LAM that are used by genuine La proteins for UUU-3'OH binding are conserved in hLARP1.
Both LARP1 and LARP1b are related sequences throughout their length [1]. The hLARP1 and hLARP1b amino acid sequences are 59% identical and 73% similar. Yet, each appears to have been inherited as separate lineages from an ancient genome since both are found in a wide variety of organisms.
The first characterized member of the LARP1 family was the D. melanogaster protein dlarp, a Hox target shown to be important in spermatogenesis and later found to bind PABP [155–157]. Another characterized member is LARP1 from C. elegans. Like human La, the La domain of LARP1 has the ability to bind poly (U) and poly (G) [105]. Surprisingly, the C-terminal region of the protein (which includes the DM15 motif) was also reported to have this ability in isolation from the La domain. LARP1 null worms are viable, but display defects in oogenesis. Deletion of C. elegans LARP1 resulted in an increase in the steady state levels of certain mRNAs. Although it was not determined whether the increase in mRNA levels in the larp-1 deletion mutant was due to alterations in transcription or decay, LARP1 protein appeared to colocalize with P bodies which are sites of mRNA degradation, and this led to the suggestion that LARP1 functions to lower mRNA levels by promoting mRNA decay [105]. However, the loci observed were not distinguished from other loci such as stress granules which are linked to distinct effects on mRNA metabolism. These results are somewhat reminiscent of observations made for the distantly related S. cerevisiae LARPs Sro9p and Slr1p, which bind mRNAs on translating ribosomes, and data that localize Sro9p to punctate bodies in the cytoplasm [100]. Curiously, overexpression of Sro9p led to increases in mRNA levels that were in part due to augmented transcription by pol II and in part to increased mRNA stability [158]. Consistent with a possible link to the LARP1 family, Sro9p and Slr1p also display a domain architecture found in certain LARP1 family members, specifically, the presence of a more C-terminally located LAM without an adjacent RRM [1]. While the present data point to a role for the LARP1 family in mRNA metabolism, questions about the nature of this function and possible mechanisms for target RNA binding and recognition are still unclear.
The LARP4 family
The LARP4 family is notably distinguished from La proteins and other LARP families by the lack of conservation of 2 or 3 of the key side chains in the LAM that are used by La proteins to recognize UUU-3'OH (Fig. 3A). In humans (and many other organisms) two LARP4 family members are found (hLARP4 and hLARP4b, previously called hLARP5) [1], which share 37% identity and 53% similarity throughout their amino acid sequence. Accordingly, the gene for each appears to have been inherited as separate lineages from an ancient genome since both are found in a wide variety of organisms, sometimes on different chromosomes.
Unlike other LARPs, LARP4 and LARP4b diverge in several of the amino acids critical for canonical UUU-3’OH recognition, suggesting that they may have distinct functions and RNA targets. Their divergence from genuine La and each other is most notable at position 24 equivalent to hLa proteins where C replaces the invariant aromatic (usually F), as well as at position 55 where M (LARP4) or L (LARP4b) replace the invariant F, and position 57 where E (LARP4) or H (LARP4b) replace either K or R (Fig. 3A). Moreover, LARP4b bears T in place of Q20, striking because Q20 is one of the few side chains in hLa that makes specific contacts to the key U-2 base of UUU-3'OH. These observations indicate that the LARP4 family is the most diverged from genuine La proteins and other LARPs in the RNA binding pockets of their LAMs.
We are aware of only one functional study of a LARP4 family member, human LARP4 (R.Y. and R.M., submitted), which suggests that hLARP4 is a cytoplasmic, polyribosome-associated protein that interacts with poly-A binding protein (PABP). LARP4 overexpression stabilizes the mRNAs whose levels it affects as determined by actinomycin D treatment followed by half-life analyses (R.Y. and R.M., submitted). Thus, LARP4 appears to act as a positive factor to promote mRNA stability, in contrast to what is suspected for LARP1, i.e., that it promotes mRNA decay. However, notable similarities for LARP4 and LARP1 exist. Drosophila LARP1 was also found to be associated with PABP in vivo, also in a manner resistant to RNAse [157]. The Yang et al. study shows that LARP4 uses a highly conserved PAM2 motif [159] to interact with PABP as do several other PABP-associating proteins that are involved in translation and/or posttranscriptional mRNA metabolism. The presence of a highly conserved PAM2 in LARP4 and 4b family members suggests that these are constituents of a larger system of PABP-interacting proteins that compete for the PAM2 interaction site on PABP. We could not identify a candidate PAM2 sequence in LARP1 (unpublished observation), and the mechanism by which LARP1 associates with PABP remains to be determined.
Closing remarks
In summary, most LARPs have conserved the LAM-RRM arrangement and exhibit varying degrees of conservation of the amino acid side chains in the LAM that are involved in UUU-3'OH binding by genuine La proteins. However, as noted for human La, these amino acid side chains make surprisingly few contacts that are U-specific, and this appears to account for the ability of human La to recognize sequences other than U at the −1 and −3 position with minimal loss of affinity. On the basis of these observations together with the observed plasticity of the human La UUU-3’OH binding pocket, there would appear to be great potential for plasticity of the LAM binding pockets of LARPs, including the ability to bind sequences other than UUU-3'OH. Nonetheless, based on the degree to which these amino acids have been conserved by LARP7 family members which have been well documented to recognize UUU-3’OH on their RNA ligands, it is tempting to speculate that sequence specificity of the UUU-3’OH binding pockets of LARPs may be dictated by their conservation of these amino acid side chains. A single U to A mutation in the highly conserved collagen mRNA 5’ UTR abolishes binding by hLARP6 in vitro suggesting that U recognition is maintained by the LAM of LARP6, which has also conserved all of these key amino acids, in a manner similar to hLa although mutagenesis of hLARP6 residues will be needed to confirm this. A corollary to this view is that some LARPs, including those with the lowest conservation of these amino acids, such as LARPs 4 and 4b may be expected to recognize RNA sequence other than UUU-3'OH.
Moreover, the phylogenetic analysis presented in Fig. 3A, which reveals lack of conservation of Y at position 23 of genuine La proteins, the side chain that makes one of only two protein-protein interactions between the LAM and RRM1 and thereby contributes to fixation of the LAM and RRM orientations relative to each other, suggests additional plasticity in how the LAM and RRM1 may be arranged to work together during RNA binding. For hLa protein, the independent nature of the LAM and RRM1 binding sites is illustrated by their differential sensitivity to magnesium ions [87]. The relative contributions of the LAM and RRM1 to RNA binding may vary for the different LARPs. For example, for La protein, the LAM is used for sequence-specific binding whereas the RRM1 appears to be an ancillary binding site that can accommodate different RNAs. It is suspected that for some LARPs the RRM1 may dominate over the LAM in sequence specificity and/or affinity. Future biochemical and structural studies are expected to contribute largely to our understanding of these interesting proteins.
Finally, a theme from emerging studies seems to be that some LARPs are involved in RNA stability, as is the case for genuine La proteins which protect their ligands against 3’ end-mediated decay. Thus, LARP7 would appear to protect 7SK snRNA from 3’ end digestion whereas LARPs −1, −4 and −5 may be involved in mRNA stability or other aspects of mRNA metabolism although many details and confirmatory studies will be necessary to shed more light on this.
ACKNOWLEDGEMENTS
We are grateful to Sasi Conte, Stephen Curry, Cécile Bousquet-Antonelli and Ying Huang for critical comments and discussion, and to Qiang Zhou and Cécile Bousquet-Antonelli for sharing data prior to publication. This work was supported by the National Science and Engineering Council of Canada (M.B.) and the Intramural Research Program of the NICHD, NIH (R.M and R.Y.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Bousquet-Antonelli C, Deragon J. A comprehensive analysis of the La-motif protein superfamily. RNA. 2009 doi: 10.1261/rna.1478709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattioli M, Reichlin M. Heterogeneity of RNA protein antigens reactive with sera of patients with systemic lupus erythematosus. Description of a cytoplasmic nonribosomal antigen. Arthritis Rheum. 1974;17:421–429. doi: 10.1002/art.1780170413. [DOI] [PubMed] [Google Scholar]
- 3.Reichlin M. Current perspectives on serological reactions in SLE patients. Clin Exp Immunol. 1981;44:1–10. [PMC free article] [PubMed] [Google Scholar]
- 4.Casciola-Rosen L, Andrade F, Ulanet D, Wong W, Rosen A. Cleavage by granzyme B is strongly predictive of autoantigen status: implications for initiation of autoimmunity. J Exp Med. 1999;190:815–826. doi: 10.1084/jem.190.6.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romero V, Fellows E, Jenne DE, Andrade F. Cleavage of La protein by granzyme H induces cytoplasmic translocation and interferes with La-mediated HCV-IRES translational activity. Cell Death Differ. 2009;16:340–348. doi: 10.1038/cdd.2008.165. [DOI] [PubMed] [Google Scholar]
- 6.Rosen A, Casciola-Rosen L. Clearing the way to mechanisms of autoimmunity. Nat Med. 2001;7:664–665. doi: 10.1038/89034. [DOI] [PubMed] [Google Scholar]
- 7.Terzoglou AG, Routsias JG, Avrameas S, Moutsopoulos HM, Tzioufas AG. Preferential recognition of the phosphorylated major linear B-cell epitope of La/SSB 349–368 aa by anti-La/SSB autoantibodies from patients with systemic autoimmune diseases. Clin Exp Immunol. 2006;144:432–439. doi: 10.1111/j.1365-2249.2006.03088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park JM, Kohn MJ, Bruinsma MW, Vech C, Intine RV, Fuhrmann S, Grinberg A, Mukherjee I, Love PE, Ko MS, DePamphilis ML, Maraia RJ. The multifunctional RNA-binding protein La is required for mouse development and for the establishment of embryonic stem cells. Mol Cell Biol. 2006;26:1445–1451. doi: 10.1128/MCB.26.4.1445-1451.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai C, Tolias PP. Genetic analysis of a La homolog in Drosophila melanogaster. Nucleic Acids Res. 2000;28:1078–1084. doi: 10.1093/nar/28.5.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arhin GK, Shen S, Perez IF, Tschudi C, Ullu E. Downregulation of the essential Trypanosoma brucei La protein affects accumulation of elongator methionyl-tRNA. Mol Biochem Parasitol. 2005;144:104–108. doi: 10.1016/j.molbiopara.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Fleurdepine S, Deragon JM, Devic M, Guilleminot J, Bousquet-Antonelli C. A bona fide La protein is required for embryogenesis in Arabidopsis thaliana. Nucleic Acids Res. 2007;35:3306–3321. doi: 10.1093/nar/gkm200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoo CJ, Wolin SL. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: a yeast homolog of the La autoantigen is dispensable for growth. Mol. Cell Biol. 1994;14:5412–5424. doi: 10.1128/mcb.14.8.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Horn DJ, Yoo CJ, Xue D, Shi H, Wolin SL. The La protein in Schizosaccharomyces pombe: a conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA. 1997;3:1434–1443. [PMC free article] [PubMed] [Google Scholar]
- 14.Stefano JE. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3' termini of RNA polymerase III transcripts. Cell. 1984;36:145–154. doi: 10.1016/0092-8674(84)90083-7. [DOI] [PubMed] [Google Scholar]
- 15.Mathews MB, Francoeur AM. La antigen recognizes and binds to the 3'-oligouridylate tail of a small RNA. Mol. Cell. Biol. 1984;4:1134–1140. doi: 10.1128/mcb.4.6.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy R, Henning D, Tan E, Busch H. Identification of a La protein binding site in a RNA polymerase III transcript (4.5 I RNA) J. Biol. Chem. 1983;258:8352–8356. [PubMed] [Google Scholar]
- 17.Hendrick JP, Wolin SL, Rinke J, Lerner MR, Steitz JA. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol. Cell. Biol. 1981;1:1138–1149. doi: 10.1128/mcb.1.12.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoo CJ, Wolin SL. The yeast La protein is required for the 3' endonucleolytic cleavage that matures tRNA precursors. Cell. 1997;89:393–402. doi: 10.1016/s0092-8674(00)80220-2. [DOI] [PubMed] [Google Scholar]
- 19.Rinke J, Steitz JA. Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La lupus antibodies. Cell. 1982;29:149–159. doi: 10.1016/0092-8674(82)90099-x. [DOI] [PubMed] [Google Scholar]
- 20.Chambers JC, Kurilla MG, Keene JD. Association between the 7 S RNA and the lupus La protein varies among cell types. J Biol Chem. 1983;258:11438–11441. [PubMed] [Google Scholar]
- 21.He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, Luo K, Zhou Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell. 2008;29:588–599. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boire G, Craft J. Human Ro ribonucleoprotein particles: characterization of native structure and stable association with the La polypeptide. J Clin Invest. 1990;85:1182–1190. doi: 10.1172/JCI114551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen CKJ, Maniatis T. The organization, structure, and in vitro transcription of Alu family RNA polymerase III transcription units in the human α-like globin gene cluster: precipitation of in vitro transcripts by lupus anti-La antibodies. J. Mol. Applied Genet. 1982;1:343–360. [PubMed] [Google Scholar]
- 24.Chang DY, Hsu K, Maraia RJ. Monomeric and dimeric Alu RNAs induced by adenovirus assemble into SRP9/14-containing RNPs in HeLa cells. Nucl. Acids Res. 1996;24:4165–4170. doi: 10.1093/nar/24.21.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maraia R, Zasloff M, Plotz P, Adeniyi-Jones S. Pathway of B1-Alu expression in microinjected oocytes: Xenopus laevis proteins associated with nuclear precursor and processed cytoplasmic RNAs. Mol. Cell Biol. 1988;8:4433–4440. doi: 10.1128/mcb.8.10.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maraia RJ, Kenan DJ, Keene JD. Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol. Cell. Biol. 1994;14:2147–2158. doi: 10.1128/mcb.14.3.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosa MD, Gottlieb E, Lerner MR, Steitz JA. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol. Cell. Biol. 1981;1:785–796. doi: 10.1128/mcb.1.9.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinmann R, Brendler TG, Raskas HJ, Roeder RG. Low molecular weight viral RNAs transcribed by RNA polymerase III during adenovirus 2 infection. Cell. 1976;7:557–566. doi: 10.1016/0092-8674(76)90206-3. [DOI] [PubMed] [Google Scholar]
- 29.Lerner MR, Andrews NC, Miller G, Steitz JA. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A. 1981;78:805–809. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fairley JA, Kantidakis T, Kenneth NS, Intine RV, Maraia RJ, White RJ. Human La is found at RNA polymerase III-transcribed genes in vivo. Proc Natl Acad Sci U S A. 2005;102:18350–18355. doi: 10.1073/pnas.0506415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.French SL, Osheim YN, Schneider DA, Sikes ML, Fernandez CF, Copela LA, Misra VA, Nomura M, Wolin SL, Beyer AL. Visual analysis of the yeast 5S rRNA gene transcriptome: regulation and role of La protein. Mol Cell Biol. 2008;28:4576–4587. doi: 10.1128/MCB.00127-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Luo T, Roeder RG. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev. 1997;11:2371–2382. doi: 10.1101/gad.11.18.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xue D, Rubinson DA, Pannone BK, Yoo CJ, Wolin SL. U snRNP assembly in yeast involves the La protein. EMBO J. 2000;19:1650–1660. doi: 10.1093/emboj/19.7.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kufel J, Allmang C, Chanfreau G, Petfalski E, Lafontaine DL, Tollervey D. Precursors to the U3 Small Nucleolar RNA Lack Small Nucleolar RNP Proteins but Are Stabilized by La Binding. Mol Cell Biol. 2000;20:5415–5424. doi: 10.1128/mcb.20.15.5415-5424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kufel J, Allmang C, Verdone L, Beggs J, Tollervey D. A complex pathway for 3' processing of the yeast U3 snoRNA. Nucleic Acids Res. 2003;31:6788–6797. doi: 10.1093/nar/gkg904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madore SJ, Wieben ED, Pederson T. Eukaryotic small ribonucleoproteins. Anti-La human autoantibodies react with U1 RNA-protein complexes. J Biol Chem. 1984;259:1929–1933. [PubMed] [Google Scholar]
- 37.Chanfreau G, Elela SA, Ares M, Jr, Guthrie C. Alternative 3'-end processing of U5 snRNA by RNase III. Genes Dev. 1997;11:2741–2751. doi: 10.1101/gad.11.20.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seipelt RL, Zheng B, Asuru A, Rymond BC. U1 snRNA is cleaved by RNase III and processed through an Sm site-dependent pathway. Nucleic Acids Res. 1999;27:587–595. doi: 10.1093/nar/27.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. Embo J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meerovitch K, Svitkin YV, Lee HS, Lejbkowicz F, Kenan DJ, Chan EK, Agol VI, Keene JD, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svitkin YV, Pause A, Sonenberg N. La autoantigen alleviates translational repression by the 5' leader sequence of the human immunodeficiency virus type 1 mRNA. J. Virol. 1994;68:7001–7007. doi: 10.1128/jvi.68.11.7001-7007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pellizzoni L, Cardinali B, Lin-Marq N, Mercanti D, Pierandrei-Amaldi P. A Xenopus laevis homologue of the La autoantigen binds the pyrimidine tract of the 5' UTR of ribosomal protein mRNAs in vitro: implication of a protein factor in complex formation. J Mol Biol. 1996;259:904–915. doi: 10.1006/jmbi.1996.0368. [DOI] [PubMed] [Google Scholar]
- 43.McLaren RS, Caruccio N, Ross J. Human La Protein: a Stabilizer of Histone mRNA. Mol. Cell Biol. 1997;17:3028–3036. doi: 10.1128/mcb.17.6.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ali N, Pruijn GJ, Kenan DJ, Keene JD, Siddiqui A. Human La antigen is required for the hepatitis C virus internal ribosome entry site (IRES)-mediated translation. J Biol Chem. 2000;275:27531–27540. doi: 10.1074/jbc.M001487200. [DOI] [PubMed] [Google Scholar]
- 45.Kim YK, Back SH, Rho J, Lee SH, Jang SK. La autoantigen enhances translation of BiP mRNA. Nucleic Acids Res. 2001;29:5009–5016. doi: 10.1093/nar/29.24.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pudi R, Abhiman S, Srinivasan N, Das S. Hepatitis C virus internal ribosome entry site-mediated translation is stimulated by specific interaction of independent regions of human La autoantigen. J Biol Chem. 2003;278:12231–12240. doi: 10.1074/jbc.M210287200. [DOI] [PubMed] [Google Scholar]
- 47.Pudi R, Srinivasan P, Das S. La protein binding at the GCAC site near the initiator AUG facilitates the ribosomal assembly on the hepatitis C virus RNA to influence internal ribosome entry site-mediated translation. J Biol Chem. 2004;279:29879–29888. doi: 10.1074/jbc.M403417200. [DOI] [PubMed] [Google Scholar]
- 48.Costa-Mattioli M, Svitkin Y, Sonenberg N. La Autoantigen Is Necessary for Optimal Function of the Poliovirus and Hepatitis C Virus Internal Ribosome Entry Site In Vivo and In Vitro. Mol. Cell. Biol. 2004;24:6861–6870. doi: 10.1128/MCB.24.15.6861-6870.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heise T, Guidotti LG, Chisari FV. Characterization of Nuclear RNases That Cleave Hepatitis B Virus RNA near the La Protein Binding Site. J Virol. 2001;75:6874–6883. doi: 10.1128/JVI.75.15.6874-6883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trotta R, Vignudelli T, Pecorari L, Intine RV, Guerzoni C, Santilli G, Candini O, Byrom MW, Goldoni S, Ford LP, Caligiuri MA, Maraia R, Perrotti D, Calabretta B. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell. 2003;I3:145–160. doi: 10.1016/s1535-6108(03)00020-5. [DOI] [PubMed] [Google Scholar]
- 51.Schwartz E, Intine RV, Maraia RJ. CK2 is responsible for phosphorylation of human La protein serine-366 and can modulate 5'TOP mRNA metabolism. Mol Cell Biol. 2004;24:9580–9591. doi: 10.1128/MCB.24.21.9580-9591.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inada M, Guthrie C. Identification of Lhp1p–associated RNAs by microarray analysis in Saccharomyces cerevisiae reveals association with coding and noncoding RNAs. Proc Natl Acad Sci U S A. 2004;101:434–439. doi: 10.1073/pnas.0307425100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ni Q, Chen Z, Yao HP, Yang ZG, Liu KZ, Wu LL. Inhibition of human La protein by RNA interference downregulates hepatitis B virus mRNA in 2.2.15 cells. World J Gastroenterol. 2004;10:2050–2054. doi: 10.3748/wjg.v10.i14.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brenet F, Socci ND, Sonenberg N, Holland EC. Akt phosphorylation of La regulates specific mRNA translation in glial progenitors. Oncogene. 2009;28:128–139. doi: 10.1038/onc.2008.376. [DOI] [PubMed] [Google Scholar]
- 55.Bitko V, Musiyenko A, Bayfield MA, Maraia RJ, Barik S. Cellular La protein shields nonsegmented negative-strand RNA viral leader RNA from RIG-I and enhances virus growth by diverse mechanisms. J Virol. 2008;82:7977–7987. doi: 10.1128/JVI.02762-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weng H, Kim C, Valavanis C, Wang Z, Schwartz LM. Acheron an novel LA antigen family member, binds to CASK and forms a complex with Id transcription factors. Cell Mol Biol Lett. 2009;14:273–287. doi: 10.2478/s11658-008-0046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhattacharyya S, Das S. An apical GAGA loop within 5' UTR of the coxsackievirus B3 RNA maintains structural organization of the IRES element required for efficient ribosome entry. RNA Biol. 2006;3:60–68. doi: 10.4161/rna.3.2.2990. [DOI] [PubMed] [Google Scholar]
- 58.Mondal T, Ray U, Manna AK, Gupta R, Roy S, Das S. Structural determinant of human La protein critical for internal initiation of translation of hepatitis C virus RNA. J Virol. 2008;82:11927–11938. doi: 10.1128/JVI.00924-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 60.Intine RV, Tenenbaum SA, Sakulich AS, Keene JD, Maraia RJ. Differential phosphorylation and subcellular localization of La RNPs associated with precursor tRNAs and translation-related mRNAs. Molecular Cell. 2003;12:1301–1307. doi: 10.1016/s1097-2765(03)00429-5. [DOI] [PubMed] [Google Scholar]
- 61.Cardinali B, Carissimi C, Gravina P, Pierandrei-Amaldi P. La protein is associated with terminal oligopyrimidine mRNAs in actively translating polysomes. J Biol Chem. 2003;278:35145–35151. doi: 10.1074/jbc.M300722200. [DOI] [PubMed] [Google Scholar]
- 62.Svitkin YV, Evdokimova VM, Brasey A, Pestova TV, Fantus D, Yanagiya A, Imataka H, Skabkin MA, Ovchinnikov LP, Merrick WC, Sonenberg N. General RNA-binding proteins have a function in poly(A)-binding protein-dependent translation. Embo J. 2009;28:58–68. doi: 10.1038/emboj.2008.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Svitkin YV, Ovchinnikov LP, Dreyfuss G, Sonenberg N. General RNA binding proteins render translation cap dependent. EMBO J. 1996;15:7147–7155. [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu J, Hayakawa A, Kakegawa T, Kaspar RL. Binding of the La autoantigen to the 5' untranslated region of a chimeric human translation elongation factor 1A reporter mRNA inhibits translation in vitro. Biochim Biophys Acta. 2001;1521:19–29. doi: 10.1016/s0167-4781(01)00277-9. [DOI] [PubMed] [Google Scholar]
- 65.Bhattacharya R, Perumal K, Sinha K, Maraia R, Reddy R. Methyl-phosphate cap structure in small RNAs reduces the affinity of RNAs to La protein. Gene Expr. 2002;10:243–253. doi: 10.3727/000000002783992398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alfano C, Sanfelice D, Babon J, Kelly G, Jacks A, Curry S, Conte MR. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat Struct Mol Biol. 2004;7–14:7–12. doi: 10.1038/nsmb747. [DOI] [PubMed] [Google Scholar]
- 67.Kenan DJ, Keene JD. La gets its wings (News and Views) Nat Struct & Mol Biol 11. 2004;11:303–305. doi: 10.1038/nsmb0404-303. [DOI] [PubMed] [Google Scholar]
- 68.Dong G, Chakshusmathi G, Wolin SL, Reinisch KM. Structure of the La motif: a winged helix domain mediates RNA binding via a conserved aromatic patch. EMBO J. 2004;23:1000–1007. doi: 10.1038/sj.emboj.7600115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 70.Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 71.Yoshizawa S, Rasubala L, Ose T, Kohda D, Fourmy D, Maenaka K. Structural basis for mRNA recognition by elongation factor SelB. Nat Struct Mol Biol. 2005;12:198–203. doi: 10.1038/nsmb890. [DOI] [PubMed] [Google Scholar]
- 72.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. Febs J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 73.Goodier JL, Fan H, Maraia RJ. A carboxy-terminal basic region controls RNA polymerase III transcription factor activity of human La protein. Mol Cell Biol. 1997;17:5823–5832. doi: 10.1128/mcb.17.10.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Horke S, Reumann K, Schulze C, Grosse F, Heise T. The La motif and the RNA recognition motifs of human La autoantigen contribute individually to RNA recognition and subcellular localization. J Biol Chem. 2004;279:50302–50309. doi: 10.1074/jbc.M407504200. [DOI] [PubMed] [Google Scholar]
- 75.Maraia RJ, Bayfield MA. The La protein-RNA complex surfaces. Mol Cell. 2006;21:149–152. doi: 10.1016/j.molcel.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 76.Teplova M, Yuan YR, Phan AT, Malinina L, Ilin S, Teplov A, Patel DJ. Structural basis for recognition and sequestration of UUU(OH) 3' temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol Cell. 2006;21:75–85. doi: 10.1016/j.molcel.2005.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kotik-Kogan O, Valentine ER, Sanfelice D, Conte MR, Curry S. Structural analysis reveals conformational plasticity in the recognition of RNA 3' ends by the human La protein. Structure. 2008;16:852–862. doi: 10.1016/j.str.2008.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang Y, Intine RV, Mozlin A, Hasson S, Maraia RJ. Mutations in the RNA Polymerase III Subunit Rpc11p That Decrease RNA 3' Cleavage Activity Increase 3'-Terminal Oligo(U) Length and La-Dependent tRNA Processing. Mol Cell Biol. 2005;25:621–636. doi: 10.1128/MCB.25.2.621-636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hamada M, Sakulich AL, Koduru SB, Maraia R. Transcription termination by RNA polymerase III in fission yeast: A genetic and biochemical model system. J Biol Chem. 2000;275:29076–29081. doi: 10.1074/jbc.M003980200. [DOI] [PubMed] [Google Scholar]
- 80.Huang Y, Bayfield MA, Intine RV, Maraia RJ. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat Struct Mol Biol. 2006;13:611–618. doi: 10.1038/nsmb1110. [DOI] [PubMed] [Google Scholar]
- 81.Copela LA, Fernandez CF, Sherrer RL, Wolin SL. Competition between the Rex1 exonuclease and the La protein affects both Trf4p–mediated RNA quality control and pre-tRNA maturation. Rna. 2008;14:1214–1227. doi: 10.1261/rna.1050408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hagenbuchle O, Larson D, Hall GI, Sprague KU. The primary transcription product of a silkworm alanine tRNA gene: identification of in vitro sites of initiation, termination and processing. Cell. 1979;18:1217–1229. doi: 10.1016/0092-8674(79)90234-4. [DOI] [PubMed] [Google Scholar]
- 83.Schiffer S, Rosch S, Marchfelder A. Assigning a function to a conserved group of proteins: the tRNA 3'-processing enzymes. Embo J. 2002;21:2769–2777. doi: 10.1093/emboj/21.11.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nashimoto M, Nashimoto C, Tamura M, Kaspar RL, Ochi K. The inhibitory effect of the autoantigen La on in vitro 3' processing of mammalian precursor tRNAs. J Mol Biol. 2001;312:975–984. doi: 10.1006/jmbi.2001.5026. [DOI] [PubMed] [Google Scholar]
- 85.Dubrovsky EB, Dubrovskaya VA, Levinger L, Schiffer S, Marchfelder A. Drosophila RNase Z processes mitochondrial and nuclear pre-tRNA 3' ends in vivo. Nucleic Acids Res. 2004;32:255–262. doi: 10.1093/nar/gkh182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.de la Sierra-Gallay IL, Pellegrini O, Condon C. Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z. Nature. 2005;433:657–661. doi: 10.1038/nature03284. [DOI] [PubMed] [Google Scholar]
- 87.Bayfield MA, Maraia RJ. Precursor-product discrimination by La protein during tRNA metabolism. Nat Struct Mol Biol. 2009;16:430–437. doi: 10.1038/nsmb.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kufel J, Tollervey D. 3'-processing of yeast tRNATrp precedes 5'-processing. RNA. 2003;9:202–208. doi: 10.1261/rna.2145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Intine RVA, Sakulich AL, Koduru SB, Huang Y, Pierstorrf E, Goodier JL, Phan L, Maraia RJ. Transfer RNA maturation is controlled by phosphorylation of the human La antigen on serine 366. Mol Cell. 2000;6:339–348. doi: 10.1016/s1097-2765(00)00034-4. [DOI] [PubMed] [Google Scholar]
- 90.Ozanick SG, Wang X, Costanzo M, Brost RL, Boone C, Anderson JT. Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:298–308. doi: 10.1093/nar/gkn925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao W, Su W, Yuan S, Huang Y. Functional conservation of tRNase ZL among Saccharomyces cerevisiae, Schizosaccharomyces pombe and humans. Biochem J. 2009;422:483–492. doi: 10.1042/BJ20090743. [DOI] [PubMed] [Google Scholar]
- 92.Chakshusmathi G, Kim SD, Rubinson DA, Wolin SL. A La protein requirement for efficient pre-tRNA folding. EMBO J. 2003;22:6562–6572. doi: 10.1093/emboj/cdg625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, Hinnebusch AG. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Anderson JT, Wang X. Nuclear RNA surveillance: no sign of substrates tailing off. Crit Rev Biochem Mol Biol. 2009;44:16–24. doi: 10.1080/10409230802640218. [DOI] [PubMed] [Google Scholar]
- 95.Copela LA, Chakshusmathi G, Sherrer RL, Wolin SL. The La protein functions redundantly with tRNA modification enzymes to ensure tRNA structural stability. Rna. 2006;12:644–654. doi: 10.1261/rna.2307206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Johansson MJ, Bystrom AS. Dual function of the tRNA(m(5)U54)methyltransferase in tRNA maturation. RNA. 2002;8:324–335. doi: 10.1017/s1355838202027851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu W, Farrell RA, Stillman DJ, Winge DR. Identification of SLF1 as a new copper homeostasis gene involved in copper sulfide mineralization in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2464–2472. doi: 10.1128/mcb.16.5.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tsukada M, Gallwitz D. Isolation and characterization of SYS genes from yeast, multicopy suppressors of the functional loss of the transport GTPase Ypt6p. J Cell Sci. 1996;109((Pt 10)):2471–2481. doi: 10.1242/jcs.109.10.2471. [DOI] [PubMed] [Google Scholar]
- 99.Kagami M, Toh-e A, Matsui Y. SRO9 a multicopy suppressor of the bud growth defect in the Saccharomyces cerevisiae rho3-deficient cells, shows strong genetic interactions with tropomyosin genes, suggesting its role in organization of the actin cytoskeleton. Genetics. 1997;147:1003–1016. doi: 10.1093/genetics/147.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sobel SG, Wolin SL. Two yeast La motif-containing proteins are RNA-binding proteins that associate with polyribosomes. Mol Biol Cell. 1999;10:3849–3862. doi: 10.1091/mbc.10.11.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lan C, Lee HC, Tang S, Zhang L. A novel mode of chaperone action: heme activation of Hap1 by enhanced association of Hsp90 with the repressed Hsp70-Hap1 complex. J Biol Chem. 2004;279:27607–27612. doi: 10.1074/jbc.M402777200. [DOI] [PubMed] [Google Scholar]
- 102.Valavanis C, Wang Z, Sun D, Vaine M, Schwartz LM. Acheron, a novel member of the Lupus Antigen family, is induced during the programmed cell death of skeletal muscles in the moth Manduca sexta. Gene. 2007;393:101–109. doi: 10.1016/j.gene.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, Coulombe B, Price DH. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Markert A, Grimm M, Martinez J, Wiesner J, Meyerhans A, Meyuhas O, Sickmann A, Fischer U. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008;9:569–575. doi: 10.1038/embor.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nykamp K, Lee MH, Kimble J. C. elegans La-related protein, LARP-1, localizes to germline P bodies and attenuates Ras-MAPK signaling during oogenesis. Rna. 2008;14:1378–1389. doi: 10.1261/rna.1066008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jacks A, Babon J, Kelly G, Manolaridis I, Cary PD, Curry S, Conte MR. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure (Camb) 2003;11:833–843. doi: 10.1016/s0969-2126(03)00121-7. [DOI] [PubMed] [Google Scholar]
- 107.Fok V, Friend K, Steitz JA. Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. J Cell Biol. 2006;173:319–325. doi: 10.1083/jcb.200601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Simons FH, Broers FJ, Van Venrooij WJ, Pruijn GJ. Characterization of cis-acting signals for nuclear import and retention of the La (SS-B) autoantigen. Exp Cell Res. 1996;224:224–236. doi: 10.1006/excr.1996.0132. [DOI] [PubMed] [Google Scholar]
- 109.Rosenblum JS, Pemberton LF, Bonifaci N, Blobel G. Nuclear Import and the Evolution of a Multifunctional RNA-binding Protein. J Cell Biol. 1998;143:887–899. doi: 10.1083/jcb.143.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Intine RV, Dundr M, Misteli T, Maraia RJ. Aberrant nuclear trafficking of La protein leads to disordered processing of associated precursor tRNAs. Mol Cell. 2002;9:1113–1123. doi: 10.1016/s1097-2765(02)00533-6. [DOI] [PubMed] [Google Scholar]
- 111.Brenet F, Dussault N, Borch J, Ferracci G, Delfino C, Roepstorff P, Miquelis R, Ouafik L. Mammalian peptidylglycine alpha-amidating monooxygenase mRNA expression can be modulated by the La autoantigen. Mol Cell Biol. 2005;25:7505–7521. doi: 10.1128/MCB.25.17.7505-7521.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bayfield MA, Kaiser TE, Intine RV, Maraia RJ. Conservation of a masked nuclear export activity of La proteins and its effects on tRNA maturation. Mol Cell Biol. 2007;27:3303–3312. doi: 10.1128/MCB.00026-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maraia RJ, Intine RV. La protein and its associated small nuclear and nucleolar precursor RNAs [review] Gene Expression. 2002;10:41–57. [PMC free article] [PubMed] [Google Scholar]
- 114.Fan H, Goodier JL, Chamberlain J, Engelke DR, Maraia RJ. 5' Processing of tRNA precursors can be modulated by the human La antigen phosphoprotein. Mol. Cell Biol. 1998;18:3201–3211. doi: 10.1128/mcb.18.6.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Intine RV, Dundr M, Vassilev A, Schwartz E, Zhao Y, Depamphilis ML, Maraia RJ. Nonphosphorylated human La antigen interacts with nucleolin at nucleolar sites involved in rRNA biogenesis. Mol Cell Biol. 2004;24:10894–10904. doi: 10.1128/MCB.24.24.10894-10904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Park JM, Intine RV, Maraia RJ. Mouse and human La proteins differ in kinase substrate activity and activation mechanism for tRNA processing. Gene Expr. 2007;14:71–81. doi: 10.3727/105221607783417619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Broekhuis CH, Neubauer G, van der Heijden A, Mann M, Proud CG, van Venrooij WJ, Pruijn GJ. Detailed analysis of the phosphorylation of human La (SS-B) autoantigen. (De)phosphorylation does not affect subcellular distribution. Biochemistry. 2000;39:3023–3033. doi: 10.1021/bi992308c. [DOI] [PubMed] [Google Scholar]
- 118.van Niekerk EA, Willis DE, Chang JH, Reumann K, Heise T, Twiss JL. Sumoylation in axons triggers retrograde transport of the RNA-binding protein La. Proc Natl Acad Sci U S A. 2007;104:12913–12918. doi: 10.1073/pnas.0611562104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Long KS, Cedervall T, Walch-Solimena C, Noe DA, Huddleston MJ, Annan RS, Wolin SL. Phosphorylation of the Saccharomyces cerevisiae La protein does not appear to be required for its functions in tRNA maturation and nascent RNA stabilization. RNA. 2001;7:1589–1602. [PMC free article] [PubMed] [Google Scholar]
- 120.Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 122.Dominguez C, Allain FH. NMR structure of the three quasi RNA recognition motifs (qRRMs) of human hnRNP F and interaction studies with Bcl-x G-tract RNA: a novel mode of RNA recognition. Nucleic Acids Res. 2006;34:3634–3645. doi: 10.1093/nar/gkl488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Belisova A, Semrad K, Mayer O, Kocian G, Waigmann E, Schroeder R, Steiner G. RNA chaperone activity of protein components of human Ro RNPs. Rna. 2005;11:1084–1094. doi: 10.1261/rna.7263905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.O'Connor CM, Collins K. A novel RNA binding domain in tetrahymena telomerase p65 initiates hierarchical assembly of telomerase holoenzyme. Mol Cell Biol. 2006;26:2029–2036. doi: 10.1128/MCB.26.6.2029-2036.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marz M, Donath A, Verstraete N, Nguyen VT, Stadler PF, Bensaude O. Evolution of 7SK RNA and its Protein Partners in Metazoa. Mol Biol Evol. 2009 doi: 10.1093/molbev/msp198. [DOI] [PubMed] [Google Scholar]
- 126.Mori Y, Sato F, Selaru FM, Olaru A, Perry K, Kimos MC, Tamura G, Matsubara N, Wang S, Xu Y, Yin J, Zou TT, Leggett B, Young J, Nukiwa T, Stine OC, Abraham JM, Shibata D, Meltzer SJ. Instabilotyping reveals unique mutational spectra in microsatellite-unstable gastric cancers. Cancer Res. 2002;62:3641–3645. [PubMed] [Google Scholar]
- 127.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, Chabot B, Poirier GG, Hughes TR, Blanchette M, Price DH, Coulombe B. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 129.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 130.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 131.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 132.Xue Y, Yang Z, Chen R, Zhou Q. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp977. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Remillieux-Leschelle N, Santamaria P, Randsholt NB. Regulation of larval hematopoiesis in Drosophila melanogaster: a role for the multi sex combs gene. Genetics. 2002;162:1259–1274. doi: 10.1093/genetics/162.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gruber AR, Kilgus C, Mosig A, Hofacker IL, Hennig W, Stadler PF. Arthropod 7SK RNA. Mol Biol Evol. 2008;25:1923–1930. doi: 10.1093/molbev/msn140. [DOI] [PubMed] [Google Scholar]
- 135.Barboric M, Lenasi T, Chen H, Johansen EB, Guo S, Peterlin BM. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc Natl Acad Sci U S A. 2009;106:7798–7803. doi: 10.1073/pnas.0903188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wassarman DA, Steitz JA. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol. Cell. Biol. 1991;11:3432–3445. doi: 10.1128/mcb.11.7.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Singh R, Gupta S, Reddy R. Capping of mammalian U6 small nuclear RNA in vitro is directed by a conserved stem-loop and AUAUAC sequence: conversion of a noncapped RNA into a capped RNA. Mol Cell Biol. 1990;10:939–946. doi: 10.1128/mcb.10.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shumyatsky G, Wright D, Reddy R. Methylphosphate cap structure increases the stability of 7SK, B2 and U6 small RNAs in Xenopus oocytes. Nucleic Acids Res. 1993;21:4756–4761. doi: 10.1093/nar/21.20.4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xue Y, Yang Z, Chen R, Zhou Q. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Stone MD, Mihalusova M, O'Connor C M, Prathapam R, Collins K, Zhuang X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–461. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Aigner S, Postberg J, Lipps HJ, Cech TR. The Euplotes La motif protein p43 has properties of a telomerase-specific subunit. Biochemistry. 2003;42:5736–5747. doi: 10.1021/bi034121y. [DOI] [PubMed] [Google Scholar]
- 142.Aigner S, Cech TR. The Euplotes telomerase subunit p43 stimulates enzymatic activity and processivity in vitro. RNA. 2004;10:1108–1118. doi: 10.1261/rna.7400704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Witkin KL, Collins K. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 2004;18:1107–1118. doi: 10.1101/gad.1201704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Teixeira MT, Gilson E. La sets the tone for telomerase assembly. Nat Struct Mol Biol. 2007;14:261–262. doi: 10.1038/nsmb0407-261. [DOI] [PubMed] [Google Scholar]
- 145.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 146.Shippen-Lentz D, Blackburn EH. Functional evidence for an RNA template in telomerase. Science. 1990;247:546–552. doi: 10.1126/science.1689074. [DOI] [PubMed] [Google Scholar]
- 147.Lingner J, Hendrick LL, Cech TR. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- 148.McCormick-Graham M, Romero DP. Ciliate telomerase RNA structural features. Nucleic Acids Res. 1995;23:1091–1097. doi: 10.1093/nar/23.7.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pannone B, Xue D, Wolin SL. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;17:7442–7453. doi: 10.1093/emboj/17.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Aigner S, Lingner J, Goodrich KJ, Grosshans CA, Shevchenko A, Mann M, Cech TR. Euplotes telomerase contains an La motif protein produced by apparent translational frameshifting. EMBO J. 2000;19:6230–6239. doi: 10.1093/emboj/19.22.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Prathapam R, Witkin KL, O'Connor CM, Collins K. A telomerase holoenzyme protein enhances telomerase RNA assembly with telomerase reverse transcriptase. Nat Struct Mol Biol. 2005;12:252–257. doi: 10.1038/nsmb900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ohndorf UM, Steegborn C, Knijff R, Sondermann P. Contributions of the individual domains in human La protein to its RNA 3'-end binding activity. J Biol Chem. 2001;276:27188–27196. doi: 10.1074/jbc.M102891200. [DOI] [PubMed] [Google Scholar]
- 153.Wang Z, Glenn H, Brown C, Valavanis C, Liu JX, Seth A, Thomas JE, Karlstrom RO, Schwartz LM. Regulation of muscle differentiation and survival by Acheron. Mech Dev. 2009 doi: 10.1016/j.mod.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 154.Cai L, Fritz D, Stefanovic L, Stefanovic B. Binding of LARP6 to the Conserved 5' Stem-Loop Regulates Translation of mRNAs Encoding Type I Collagen. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chauvet S, Maurel-Zaffran C, Miassod R, Jullien N, Pradel J, Aragnol D. dlarp, a new candidate Hox target in Drosophila whose orthologue in mouse is expressed at sites of epithelium/mesenchymal interactions. Dev Dyn. 2000;218:401–413. doi: 10.1002/1097-0177(200007)218:3<401::AID-DVDY1009>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 156.Ichihara K, Shimizu H, Taguchi O, Yamaguchi M, Inoue YH. A Drosophila orthologue of larp protein family is required for multiple processes in male meiosis. Cell Struct Funct. 2007;32:89–100. doi: 10.1247/csf.07027. [DOI] [PubMed] [Google Scholar]
- 157.Blagden SP, Gatt MK, Archambault V, Lada K, Ichihara K, Lilley KS, Inoue YH, Glover DM. Drosophila Larp associates with poly(A)-binding protein and is required for male fertility and syncytial embryo development. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 158.Tan Q, Li X, Sadhale PP, Miyao T, Woychik NA. Multiple mechanisms of suppression circumvent transcription defects in an RNA polymerase mutant. Mol Cell Biol. 2000;20:8124–8133. doi: 10.1128/mcb.20.21.8124-8133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Albrecht M, Lengauer T. Survey on the PABC recognition motif PAM2. Biochem Biophys Res Commun. 2004;316:129–138. doi: 10.1016/j.bbrc.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 160.Fan H, Sakulich AL, Goodier JL, Zhang X, Qin J, Maraia RJ. Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell. 1997;88:707–715. doi: 10.1016/s0092-8674(00)81913-3. [DOI] [PubMed] [Google Scholar]